Abstract
Chromatin remodelers are ATP-dependent enzymes that reorganize nucleosomes within all eukaryotic genomes. Here we report a complex of the Chd1 remodeler bound to a nucleosome in a nucleotide-free state, determined by cryo-electron microscopy (cryo-EM) to 2.3 Å resolution. The remodeler stimulates the nucleosome to absorb an additional nucleotide on each strand at two different locations: on the tracking-strand within the ATPase binding site and on the guide-strand one helical turn from the ATPase motor. Remarkably, the additional nucleotide on the tracking strand is associated with a local transformation toward an A-form geometry, explaining how sequential ratcheting of each DNA strand occurs. The structure also reveals a histone-binding motif, called ChEx, which can block opposing remodelers on the nucleosome and may allow Chd1 to participate in histone reorganization during transcription.
Introduction
Chromatin remodelers are ATP-driven motors that determine the placement and composition of nucleosomes throughout the genome. Proper reorganization of nucleosomes is critical for fundamental cellular processes such as transcription, DNA replication, and DNA repair, making chromatin remodeling essential for normal cell growth and development 1-4. Chd1 is intimately tied to transcriptional activation, frequently localized to transcribed genes and found to interact with several transcriptional elongation factors 5-9. Within gene bodies, Chd1 and ISWI-type remodelers help maintain proper nucleosome packaging during passage of RNA polymerases, which is essential for preventing inappropriate transcriptional initiation at cryptic start sites 10. Chd1 and ISWI remodelers can shift nucleosomes back and forth and can generate regularly spaced arrays by using regulatory domains to control the direction of nucleosome sliding 11-14. Given their inherent two-fold symmetry, nucleosomes can simultaneously accommodate two remodelers 15-18. Nucleosomes can only be shifted unidirectionally by each remodeler, with back-and-forth sliding resulting from alternating action of opposing remodelers 14. How remodelers on opposite sides of the nucleosome may compete or cooperate with each other is presently unclear.
Chd1 is named for three characteristic domains: a pair of chromodomains, a helicase-like ATPase motor, and a DNA-binding domain 19. Using its ATPase motor, Chd1 repositions nucleosomes by binding and translocating DNA past the histone core at an internal site called superhelix location 2 (SHL2)16,20. Chd1 was shown to alter DNA twist at SHL2 in a nucleotide-dependent fashion, which was proposed to underlie a twist defect mechanism of translocating nucleosomal DNA by single base pairs steps 21,22. The twist defect mechanism has been bolstered by cryo-EM structures of Chd1 and other remodelers in different nucleotide states, where nucleosomal DNA was found to be distorted with the ATPase motor in ADP-bound and nucleotide-free states 23-26. Structures of Chd1 bound to the nucleosome have so far been reported with the ATP-mimic ADP•BeF3−, where nucleosomal DNA at SHL2 maintains the same twist as the nucleosome alone 17,27. Here we describe a high resolution Chd1-nucleosome complex in a nucleotide-free state, which shows that changes in DNA twist and initial translocation arise from local distortion toward an A-form geometry. The structure also reveals a previously unanticipated histone-binding motif, which we call ChEx (Chd1 Exit-side binding), that can antagonize competing remodelers on the opposite side of the nucleosome.
Results
We resolved a 1:1 complex of yeast Chd1 bound to a nucleosome by cryo-electron microscopy (cryo-EM) to 2.3 Å resolution (Fig. 1a, Table 1, Supplementary Video 1). Mononucleosomes with 40 bp flanking DNA on each side (40N40) were made using a variant of the Widom 601 sequence and cross-linked to Chd1 using GraFix 28. Starting with ~10.9 million particles from 6907 micrographs, the Chd1-nucleosome complex was reconstructed through in-depth in silico classification and refinement using 450,433 particles. After further processing, a complex consisting of the histone core, wrapped nucleosomal DNA, Chd1 chromodomains and Chd1 ATPase motor was obtained at 2.3 Å resolution (0.143 FSC), with a calculated local resolution of the remodeler from 2.2 to 2.8 Å (Extended Data Fig. 1 and 2, Supplementary Video 2). In addition, using focused classification, we were able to obtain a 2.7 Å Chd1-nucleosome complex where the Chd1 DNA-binding domain was visible, a 2.6 Å structure of the nucleosome alone, and a 2.9 Å structure of the nucleosome with only an N-terminal portion of Chd1, called ChEx (Fig. 1b,c and Extended Data Fig. 3 and 4). We first describe the DNA conformation and interactions observed in the high-resolution structure, then discuss other subsets. Finally, we focus on the N-terminal ChEx segment of Chd1 that binds to the histone core on the exit side of the nucleosome.
Figure 1. Structure of the Chd1 remodeler bound to a nucleosome in the nucleotide-free state.
a, Three views of a cryo-EM reconstruction of the Chd1-nucleosome complex at 2.3 Å resolution. b, Three additional cryo-EM reconstructions from the same dataset: a subclassification focused on the Chd1 DNA-binding domain at 2.7 Å resolution, and two nucleosome-focused subclassifications with and without the ChEx segment at 2.9 Å and 2.6 Å resolution, respectively. c, A composite model of Chd1 bound to the unwrapped nucleosome (PDB code: 7TN2), built using high-resolution and DNA-binding domain subclassification maps. See also Extended Data Fig. 1-4 and Supplementary Video 1.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| #1 Chd1-nucleosome (EMD-25479) (PDB 7TN2) |
#2 Chd1-nucleosome with stronger DBD (EMD-25480) (PDB 7TN2) |
#3 Nucleosome only (EMDB-25481) (PDB 7SWY) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 81,000 | 81,000 | 81,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 49.9 | 49.9 | 49.9 |
| Defocus range (μm) | −1.0 to −2.2 | −1.0 to −2.2 | −1.0 to −2.2 |
| Pixel size (Å) | 0.972 | 0.972 | 0.972 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 10,900,948 | 10,900,948 | 10,900,948 |
| Final particle images (no.) | 450,533 | 75,867 | 823,685 |
| Map resolution (Å) | 2.3 | 2.7 | 2.6 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 3.8 – 2.2 | 4.5 – 2.4 | 5.0 – 2.5 |
| Refinement | |||
| Initial model used (PDB code) | 3LZ0, 3MWY | 3LZ0, 3MWY | 3LZ0 |
| Model resolution (Å) | 2.58 | 3.08 | 2.6 |
| FSC threshold | 0.5 | 0.5 | 0.143 |
| Model resolution range (Å) | 5.0 – 2.58 | 5.0 – 3.08 | 5.5 – 2.6 |
| Map sharpening B factor (Å2) | −30 | −15 | −30 |
| Model composition | |||
| Non-hydrogen atoms | 19507 | 19507 | 11786 |
| Protein residues | 1654 | 1654 | 751 |
| Nucleotides | 320 | 320 | 286 |
| B factors (Å2) | |||
| Protein | 24.78/177.50/67.23 | 24.78/177.50/67.23 | 24.20/115.53/36.99 |
| Nucleotide | 52.97/847.08/169.71 | 52.97/847.08/169.71 | 20.00/469.56/168.90 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.008 | 0.006 |
| Bond angles (°) | 0.646 | 0.646 | 0.664 |
| Validation | |||
| MolProbity score | 1.63 | 1.63 | 1.28 |
| Clashscore | 4.42 | 4.42 | 3.77 |
| Poor retainers (%) | 0.00 | 0.00 | 0.00 |
| Ramachandran plot | |||
| Favored (%) | 93.86 | 93.86 | 97.41 |
| Allowed (%) | 6.14 | 6.14 | 2.59 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 |
Although the Chd1-nucleosome complex was prepared in the presence of ATPγS, the GraFix procedure was carried out without nucleotide. Notably, using the variant Widom sequence with an insertion of an A•T bp at one of the two SHL2 sites 21, nucleosomes bind more strongly to Chd1 in the nucleotide-free state than with ATPγS (Extended Data Fig. 5). In the structure, the nucleotide-binding site is empty, and the conformation of the ATPase motor matches the open state reported for other remodelers in ADP-bound and nucleotide-free states (Supplementary Video 3). We therefore consider this structure to represent Chd1 bound to the nucleosome in the nucleotide-free state.
Chd1 induces an A-form-like DNA geometry at SHL2
Like all superfamily 1 (SF1) and SF2-type ATPases, chromatin remodelers have a bi-lobed ATPase motor that, when bound to ATP, can achieve a tightly closed state, and when ADP-bound or nucleotide-free, favors a more open conformation 22,29. Compared to Chd1-nucleosome complexes bound to ADP•BeF3−, where the ATPase motor is in a closed state 17,27, the open state ATPase has a distinct placement of lobe 2. Although it maintains a similar location on the guide strand, ATPase lobe 2 appears to have shifted along the tracking strand by 1 nucleotide (Fig. 2a). This shift of lobe 2 relative to the closed state is consistent with the inchworm mechanism of translocation proposed for SF1 and SF2 ATPases 29-31. However, distinct from translocases that only bind to single-stranded nucleic acid substrates, remodeler ATPases translocate with respect to duplex DNA.
Figure 2. DNA distortions induced by the nucleotide-free state of the Chd1 ATPase motor.
a, Comparison of Chd1 in the closed, ADP•BeF3−-bound state (5O9G; light gray) and the open, nucleotide-free state (7TN2; colored). These structures were aligned using the histone core. b, Views comparing nucleosomal DNA in the presence (7TN2; orange, yellow) and absence (7SWY; gray) of the ATPase motor. Orange and gray rectangles highlight the orientations of base pairs in the ATPase-bound and ATPase-free structures, respectively. See also Supplementary Video 4. c, A comparison of nucleosomal DNA bound to the nucleotide-free and ADP•BeF3−-bound structures of Chd1, aligned on ATPase lobe 2.
The engagement of the open state ATPase at SHL2 is coupled with a distortion of nucleosomal DNA (Fig. 2b, Supplementary Video 4). At the ATPase binding site, the altered structure of DNA accommodates an additional nucleotide on the tracking strand but not the complementary guide strand. This DNA distortion is similar to those observed in other ADP-bound and nucleotide-free remodeler-nucleosome complexes (Supplementary Video 3) 23-25. Thus, the local changes in duplex DNA observed here likely represent a common geometric transformation induced by the open state of remodeler ATPases.
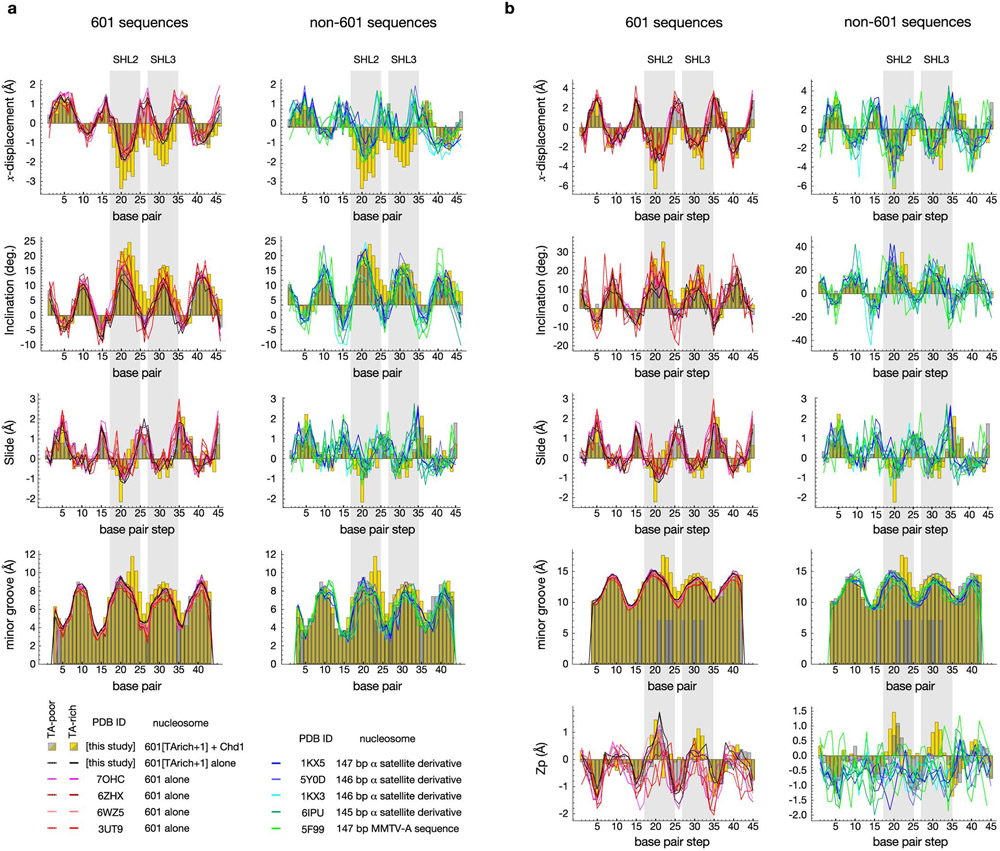
Locally, the DNA distortion displays several features that resemble an A-form-like geometry. At SHL2, the duplex appears underwound, with the bases shifting away from the central axis of DNA, and the base pairs are inclined relative to the helical axis. These DNA characteristics are reflected in the helical parameters x-displacement and inclination, which for naked B-form DNA are normally ~0 Å and 2 ± 9°, respectively 32-34. As defined by CURVES+ 35, the distortion at SHL2 displays a peak x-displacement of approximately -3 Å and an inclination of 24°, which fall in the range of A-form (Extended Data Fig. 6). Two other parameters that discriminate between A-form and B-form are Slide, which is the displacement of a base pair along its long axis with respect to the neighboring base pair, and Zp, the mean phosphate position along z between two adjacent base pairs 32,36-38. For the remodeler-induced distortion, the Slide parameter shows a sharp deviation from isolated nucleosomes in a single base pair step, 20-21 bp from the nucleosome dyad, that is consistent with A-form. Zp, in contrast, is borderline A-form at base step 21, and has similar values in isolated 601 nucleosomes (Extended Data Fig. 6).
Bound to the nucleotide-free ATPase motor, the DNA at SHL2 shows a distinct geometry from that bound in the ADP•BeF3− state. Although the resolution of the available ADP•BeF3− states is limited, when aligned relative to ATPase lobe 2, both DNA structures show a similar placement of backbone phosphates, despite the different register of the tracking strands (Fig. 2c). This similar placement reflects the phosphate-specific and base-insensitive binding of lobe 2. In this manner, ATPase lobe 2 ensures that the DNA distortion is accompanied by a full one-nucleotide shift of the tracking strand, since a partial shift would not satisfy all phosphate-binding sites.
DNA distortion outside the remodeler binding site
By altering the geometry of the duplex, the ATPase motor enforces asymmetry on downstream DNA. Although the A-form-like geometry is highly localized, the increase in angle of inclination and more negative x-displacement, initiated at the site of the SHL2 distortion, continues outside of the ATPase binding site (Fig. 2b, Extended Data Fig. 6). Unlike previously reported nucleotide-free and ADP-bound structures 23,24, the nucleosomal DNA in our structure regains its canonical geometry by SHL3.5 due to an additional nucleotide absorbed in the guide strand at SHL3 (Fig. 2b, Supplementary Figure 1, Supplementary Video 4). With a slight unwinding of the duplex, SHL3 demarcates both the extent of the distortion initiated at SHL2 and the front edge of the DNA segment that has ratcheted 1 bp further onto the nucleosome. Although the distortion at SHL3 was induced by the remodeler (Supplementary Figure 2), it was also likely facilitated by the insertion of an A•T base pair 22 bp from the dyad, which disrupts DNA phasing on the TA-rich side. Combined with the additional nucleotide on the tracking strand at SHL2, the additional nucleotide on the guide strand at SHL3 allowed the TA-step at SHL3.5 and TG-step at SHL4.5 to recover the optimal placement of the canonical 601 sequence. Despite this sequence bias, which likely explains the higher affinity of nucleotide-free Chd1 (Extended Data Fig. 5), the distortion at SHL3 was attained without direct contacts from the ATPase motor, which suggests that absorption of an additional nucleotide in the guide strand may be a natural response to remodeler-induced DNA strain on the nucleosome.
Nucleosomes have been shown to accommodate single bp differences at SHL2 and SHL5 (ref. 39). At SHL3, the sugar-phosphate backbone of the guide strand resembles how SHL2 and SHL5 absorb DNA, with the phosphate backbone smoothly arcing away from its previous position (Extended Data Fig. 7). In unbound nucleosomes, SHL2 and SHL5 accommodate a full bp within the same helical turn of the duplex in a staggered manner, with the two sites of nucleotide absorption occurring ~5 bp away from each other 22. Here, the guide strand is prevented from accommodating an additional nucleotide at SHL2 due to the grip of the ATPase motor in the open state, requiring that each strand absorb an additional nucleotide on distinct helical turns of DNA.
A high-resolution view of remodeler–nucleosome interactions
The nucleotide-free structure reveals an asymmetry in the distribution of ATPase contacts with DNA, with lobe 2 showing more extensive interactions than lobe 1 on the tracking strand (Fig. 3). Like other SF2 enzymes 29,40, lobe 2 of Chd1 coordinates phosphates −19 to −21 of the tracking strand using backbone and side chain contacts from helicase motifs IV and V. In addition, lobe 2 hydrogen bonds to phosphates −19 and −20 using a remodeler-specific motif called Block D, and coordinates the sugar-phosphate backbone around phosphate −22 using side chains from a remodeler-specific insertion called the gating helix (Fig. 3c). In contrast, ATPase lobe 1 only makes direct hydrogen bonds with two phosphates, −16 and −17, through K497 and Y494 (Fig. 3b). Interestingly, compared to other SF2 ATPases, lobe 1 of Chd1 appears 2-3 Å farther away from the tracking strand, which prevents DNA from directly hydrogen bonding with the conserved T491 of motif Ic and the backbone of motif Ia.
Figure 3. Interactions between the Chd1 ATPase motor and DNA at SHL2.
a, Schematic representation of key interactions. Helicase motifs and remodeler-specific blocks are based on refs. 61 and 62. b-e, Interactions between ATPase lobe 1 (purple) and lobe 2 (blue) with the DNA tracking strand (orange) and guide strand (yellow). Hydrogen bonds 3.2 Å or shorter are shown with green, dashed lines, and those between 3.2-3.5 Å with yellow dots. See also Supplementary Video 2.
As expected for the open state, ATPase lobe 1 and 2 are separated by a ~6 Å gap between motifs 1a and V, with each domain having its own distinct footprint on the tracking strand. On the guide strand, however, both lobes contact one another where they coordinate phosphates 23 and 24 (Fig. 3d). Two highly conserved residues at the interface are K836 (brace helix, lobe 2) and N547 (motif III, lobe 1). K836 forms a triad of hydrogen bonds that connects three distinct regions: the side chain of N547 (lobe 1), the backbone carbonyl of W793 (lobe 2), and phosphate 24 on the guide strand. N547 hydrogen bonds to E551 and is within 3.5 Å of phosphate 23. In addition to these contacts, ATPase lobe 1 binds to the guide strand through hydrogen bonds to phosphates 21 and 23 and van der Waals contacts with phosphate 22 using a conserved motif (Block G) first identified in Rad54 (ref. 41; Fig. 3e). ATPase lobe 2 makes additional contacts with the guide strand through the aromatic ring of W793 (Block L), which packs against the sugar-phosphate backbone on the 3’ side of phosphate 23, and side chains of both R772 (motif V) and N794 (Block L; Fig. 3d).
Focused subclassifications of DNA-binding domain and bridge
The Chd1 ATPase motor is bound at SHL2 and stimulates unwrapping of DNA on the opposite nucleosome gyre, referred to as exit DNA. In previous complexes where Chd1 was bound to the ATP mimic ADP•BeF3−, unwrapped exit DNA was bound by the Chd1 DNA-binding domain 17,27. In our dataset, the DNA-binding domain and unwrapped DNA had much weaker density than the chromo- and ATPase domains. Through focused classification, we found a subset from a minority (~9.4%) of particles (76,867 of 817,698), where the DNA-binding domain is positioned on exit DNA as in previous structures (Fig. 1b, c, Extended Data Figs. 3 & 4). Although this structure has an overall resolution of 2.7 Å, the local resolution of the DNA-binding domain and exit DNA is lower, between 3.2 - 4.8 Å. In addition, density from other subsets suggested different orientations of the DNA-binding domain on exit DNA (Extended Data Fig. 8). For ~43% (351,371 of 817,698 particles), the DNA-binding domain was not visible. Thus, the DNA-binding domain appears to dynamically associate with exit DNA in the nucleotide-free state.
In other remodeler-nucleosome complexes, the ATPase motor makes a primary contact at SHL2, and simultaneously, a secondary contact with DNA at SHL6 on the opposite gyre of the nucleosome 18,24,25,42-44. Disruption of opposite gyre contacts was reported to diminish nucleosome sliding activity for Snf2 and ISWI remodelers 42,44. Due to unwrapping of exit DNA, the Chd1 ATPase motor contacts opposite gyre DNA differently, with a loop (S. cerevisiae residues 475-481) that packs into the DNA major groove (Extended Data Fig. 9). To determine the importance of this interaction for nucleosome sliding activity, we tested variants with four simultaneous substitutions (K478D/G479A/K480D/K481A, called Chd1[DADA]) and a replacement of residues 475-481 with an Ala-Gly linker (called Chd1[Δ475-481]). Both mutants showed a two-fold reduction in sliding activity (Extended Data Fig. 9). Thus, while this region impacts activity and may convey the unwrapped nature of exit DNA, it does not appear to be essential for nucleosome repositioning.
Bound to the opposite DNA gyre, the DNA-binding domain contacts the chromodomains through a conserved interface. In previous work, we showed that disruption of this interface stimulated ATPase activity approximately two-fold, suggesting that this interface may be inhibitory 16. Disruption of four different patches at this interface yielded at most only a modest (less than two-fold) increase in nucleosome sliding rate (Extended Data Fig. 9). Thus, this interface does not appear to strongly impact sliding of end-positioned nucleosomes, and further work will be needed to clarify the role that this conserved interface plays in remodeler action.
The Chd1 DNA-binding domain regulates the direction of nucleosome sliding through an autoinhibitory element called the bridge 14. The bridge, which lies between the ATPase motor and DNA-binding domain in the primary sequence, has been visualized as an extended segment that can stably bind to an inactive conformation of the ATPase 45. In our electron density maps, we observed poorly defined density against the ATPase in the vicinity where the bridge is bound in the Chd1-alone structure. Using focused refinement, we attempted to obtain a stable class with a clear and well-defined bridge density, but this approach revealed high heterogeneity of the region (Extended Data Fig. 8). We suspect that density in this region corresponds to the bridge adopting multiple conformations, which would be poised to inhibit the ATPase motor. We have proposed that the bridge stimulates remodeler dissociation from the nucleosome by blocking full ATPase engagement with DNA 14. Here, however, the ATPase motor is fully engaged with nucleosomal DNA at SHL2, which may make it incompatible with stable bridge binding.
The Chd1 ChEx segment binds to the exit-side acidic patch
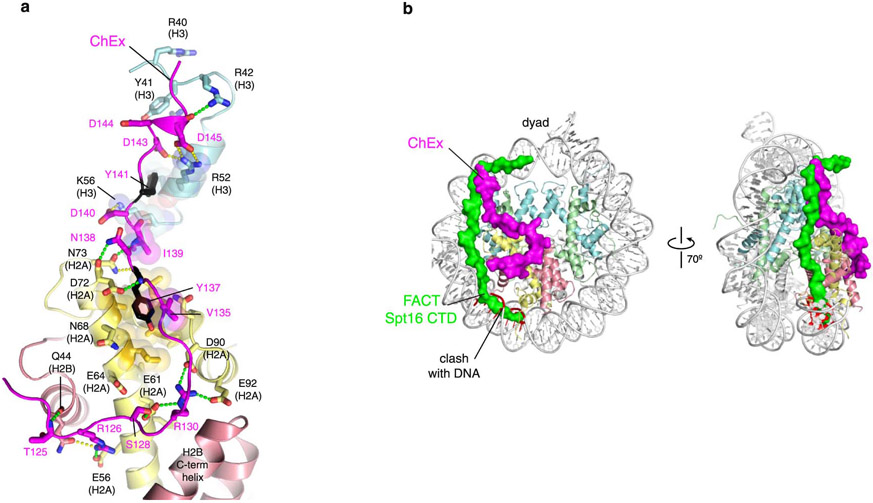
The Chd1-nucleosome complexes also revealed a CHD-family motif bound to the histone core. We identified electron density contacting the αN helix of histone H3 and extending to the acidic patch of the histone H2A/H2B dimer (Fig. 4a). Based on the shape and chemical environment, this density appeared to match the sequence of an N-terminal segment preceding the Chd1 chromodomains (Fig. 4b). To confirm the identity of this segment, we performed cross-linking mass spectrometry using disuccinimidyl dibutyric urea (DSBU) and 1,1'-carbonyldiimidazole (CDI). The N-terminal segment of Chd1 was highly reactive, and all cross-links from this region of Chd1 to the histone core were consistent with the cryo-EM model (Figure 4c, d, Supplementary Figure 3, Supplementary Dataset 1). Relative to the ATPase motor, this 28 amino acid segment binds the acidic patch on the exit-side H2A/H2B dimer (Fig. 1a). We therefore refer to this segment as ChEx, for Chd1 Exit-side binding.
Figure 4. The Chd1 ChEx segment binds the histone core.
a, Overview of the ChEx electron density (magenta) on the histone core (H2A, yellow; H2B, salmon; H3, blue-gray; H4, green). b, Sequence and model of the Chd1 ChEx segment. c, Connectogram of cross-links between Chd1 and histone proteins using cross-linkers CDI (teal), DSBU (purple), or both CDI and DSBU (black). See also Supplementary Figure 3. d, Analysis of ChEx cross-links using Jwalk 63. Residues 116-120, not visible in the structure, were modeled by PyRosetta. Black paths show minimal SASDs. H2A is yellow and H2B is salmon. e, Native gel nucleosome binding assays, using 120 nM Chd1, 30 nM 40-601-40 nucleosome, 1 mM AMP-PNP, and salmon sperm DNA as competitor (0.0625, 0.125, 0.25, 0.5, 1, 2, 4 mg ml −1). These gels are representative of eight independent experiments. f, Quantification of nucleosome sliding reactions using 100 nM Chd1 and 40 nM 80-601-0 nucleosomes. Shown are data from multiple independent experiments for Chd1[wt] (n=8) and Chd1[ΔChEx] (n=9), with fits to averaged data. g, Nucleosome sliding rates of Chd1, possessing (black circles) or lacking ChEx (open circles), plotted as a function of Chd1 concentration (nM).
The ChEx segment helps stabilize the remodeler on its nucleosome substrate, since its deletion allows Chd1 to be more easily competed off of nucleosomes (Fig. 4e). Deletion of the ChEx segment also reduced sliding activity (Fig. 4f), though with saturating amounts of Chd1, the difference in maximal sliding rates of Chd1[wt] and Chd1[ΔChEx] was less than 2-fold (Fig. 4g). The slower sliding of Chd1[ΔChEx] matches the reduced sliding activity that was observed for wild-type Chd1 on nucleosomes with a mutated exit-side acidic patch 46, corresponding to where our model shows ChEx binding to the nucleosome. Given the modest decrease in activity, ChEx appears to facilitate but not be essential for nucleosome sliding.
The H2A acidic patch is perhaps the most commonly recognized epitope of the nucleosome 47,48. The ChEx segment presents two arginine residues to the H2A acidic patch: Chd1(R126) is bound to H2A(E56), and Chd1(R130) – which is analogous to “arginine anchor” residues found in other acidic patch binding elements 48 – is bound to the acidic triad consisting of H2A residues E61, D90, and E92, (Fig. 5a). In addition to these arginines, S128 hydrogen bonds to the side chain of H2A(E61). The ChEx segment also contains a highly conserved Tyr residue (Y137) that packs in a hydrophobic pocket on H2A formed by L65, A69, L85 and A86 and makes a backbone hydrogen bond with H2A(D72) (Extended Data Fig. 10). This region is followed by acidic residues (143-DDDLLE-148) that are electrostatically matched by basic histone residues on the αN helix of H3. The αN helix contacts DNA in fully wrapped nucleosomes, but it becomes exposed when exit-side DNA is unwrapped by Chd1. Notably, other CHD-family members show homology to the ChEx region of yeast Chd1 (Fig. 5b). Recent work suggested that this region of CHD7 interacts with the acidic patch of the nucleosome 49, strengthening the idea of a common CHD-family motif for binding the histone surface.
Figure 5. The ChEx segment competes with H2A/H2B acidic patch binding elements.
a, Superposition of the LANA-bound nucleosome (1ZLA, gray) with the Chd1-bound nucleosome, focused on the region of overlap at the acidic patch. The backbone trace of Chd1-ChEx (magenta) and LANA (gray) are shown as tubes, with key residues highlighted as sticks. Acidic side chains of histone H2A in the Chd1-nucleosome complex are shown as spheres. b, Sequence alignment of the ChEx region of CHD family members. c, The Chd1 ChEx segment stimulates displacement LANA from the nucleosome. A Dabcyl-labeled LANA peptide was titrated in the presence of 30-601-30/Cy3-H2B nucleosomes (gray axis). After quenching the bulk of the fluorescence signal (273 nM LANA), Chd1 variants were titrated in the presence of AMP-PNP. Greater fluorescence recovery suggests effective competition with Dabcyl-LANA. Error bars show the standard deviations from mean values, obtained from n independent experiments: Chd1[wt] (n=4); Chd1[ΔChEx] (n=5); Chd1[R126A/R130A] (n=4); Chd1[Y137A] (n=4); no Chd1, LANA only (n=3).
To see whether ChEx can displace other acidic patch binding elements, we developed a fluorescence-based competition assay using the well-studied LANA peptide from Kaposi’s sarcoma-associated herpesvirus 50. By addition of a Dabcyl moiety on the LANA N-terminus, acidic patch binding places the Dabcyl close enough to quench a Cy3 dye coupled to H2B(K120C) (Figure 5c). Consistent with this design, Cy3 fluorescence decreased upon titration of the Dabcyl-LANA peptide (gray circles, lower X-axis). To see whether Chd1 could compete with LANA, Dabcyl-LANA was first added to achieve ~80% quenching, and Chd1 was then titrated in (upper X-axis). When Chd1[wt] was used, Cy3 fluorescence recovered significantly, reaching ~50% of its previous intensity (black circles). However, when Chd1[ΔChEx] was used, the Cy3 fluorescence recovered only ~10% of its signal (magenta circles). To see what residues of ChEx were required for LANA displacement, we tested two variants, R126A/R130A and Y137A. With Chd1[R126A/R130A], Cy3 fluorescence recovery was equivalent to Chd1[ΔChEx] (blue circles), signifying that these two arginine residues were key for acidic patch competition by ChEx. With Chd1[Y137A], Cy3 recovery was ~40% of initial intensity (orange circles), indicating a retained ability to bind to the H2A acidic patch, yet with reduced affinity.
The acidic patch is essential for robust nucleosome sliding by INO80 and ISWI remodelers 51. Due to the two-fold symmetry of the nucleosome, each acidic patch is on the entry side relative to one SHL2 site and the exit side relative to the other SHL2. Thus, when Chd1 is bound to one SHL2 site, ChEx could potentially compete with the entry acidic patch for another remodeler at the other SHL2. The entry-side acidic patch has been shown to be required for the ISWI remodeler for robust nucleosome sliding 46,52, and we wondered whether ChEx could interfere with ISWI activity. Since both Chd1 and ISWI slide nucleosomes, we created nucleosomes with the ChEx segment fused to histone H3[ΔN] to evaluate the potential of ChEx to antagonize another remodeler. On nucleosome substrates where the H3 tail had been deleted (H3[ΔN]), both Snf2h (the ATPase subunit of human ISWI) and ACF (from Drosophila) showed faster sliding activity than on canonical nucleosomes. However, with nucleosomes where the H3 tail had been replaced by ChEx (H3[ΔN+ChEx]), both ISWI remodelers showed a 50-fold or more decrease in sliding rate (Fig. 6a, b).
Figure 6. ChEx can block other remodelers from sliding nucleosomes.
a, Nucleosome sliding reactions, with nucleosomes (40 nM) made with 0-601-80-FAM DNA and histone cores with histone H3 that was wildtype, with a deleted N-terminus (H3[ΔN]), or with a ChEx-H3 fusion (H3[ΔN+ChEx]). Remodeling reactions were carried out with Snf2h (200 nM) or ACF (100 nM) and analyzed by native PAGE. b, Quantification of nucleosome sliding reactions. Each line shows the fit to the average of four or more reactions. Averaged rates for Snf2h were 1.13±0.8 min−1 (H3[wt]), 3.4±0.6 min−1 (H3[ΔN]), and estimated to be <0.03 min−1 for H3[ΔN+ChEx]. Averaged rates for ACF were 7.7±0.4 min−1 (H3[wt]), 10.6±0.6 min−1 (H3[ΔN]), and 0.10±0.02 min−1 (H3[ΔN+ChEx]). Error bars show standard deviations. c, Model for acidic patch competition by ChEx.
These experiments demonstrate that ChEx can effectively compete for access to the acidic patch, with the potential to limit nucleosome binding and/or recognition by other factors during remodeling by Chd1 (Fig. 6c).
Discussion
Chromatin remodelers like Chd1 reposition nucleosomes by creating local distortions in nucleosomal DNA. Our structural analysis reframes the initial step of DNA translocation as an ATPase-induced alteration of DNA geometry. As described here, the nucleotide-free state of the remodeler favors an A-form-like geometry of nucleosomal DNA. Compared to B-form, a local A-form-like geometry can accommodate an additional nucleotide in one strand. By toggling DNA between two geometries, the remodeler ATPase splits the translocation of a DNA base pair into two half-reactions (Fig. 7). In the first step, the open state ATPase stimulates a one-nucleotide shift of the tracking strand. Subsequently, closure of the ATPase upon ATP binding ratchets the guide strand by one-nucleotide as DNA is forced back toward B-form. Although current structures suggest that ATP binding imposes a local B-form-like geometry, higher resolution structural information will be needed to understand how the structural deformations by a closed ATPase motor may contribute to DNA mobility. Subsequent to the initial process of selectively translocating the tracking strand, an important missing step in the cycle that awaits future work is how ATP binding and closure of the ATPase motor stimulate a shift of both tracking and guide strands past its binding site.
Figure 7. Model for DNA translocation based on changes in duplex geometry.
Red indicates where an additional nucleotide is absorbed on one strand, orange indicates DNA distortion due to an asymmetric shift in the DNA backbone. See also Supplementary Video 4.
As the open state remodeler alters DNA geometry at SHL2, the nucleosome can respond by a compensatory change at a spatially distinct site. The distortion at SHL2 inherently creates strain, which directionally propagates along the DNA duplex in the form of base pair inclination and negative x-displacement. As shown in our structure, this altered geometry can be relieved by a distortion outside of the remodeler binding site that accommodates an additional nucleotide on the guide strand. In previously solved remodeler-nucleosome complexes, a compensatory change was not visualized 23,24. Here, an additional nucleotide at SHL3 resolved the altered DNA geometry initiated at SHL2, defining the edge of the remodeler-induced distortion. We anticipate that, during this first step of nucleosome sliding, such a compensatory change may occur at multiple sites on entry DNA, governed by DNA sequence and energetics.
The ability of the nucleosome to accommodate a DNA distortion at a previously unobserved location (SHL3) has implications for propagation of twist defects throughout the nucleosome. During nucleosome sliding by ISWI and CHD-family remodelers, DNA exits the nucleosome in bursts of ~3-4 and ~6-7 bp 53,54. As shown by 3-color FRET, DNA exiting the nucleosome showed an ATP-dependent delay compared with DNA being pulled on, suggesting that the nucleosome absorbs one or more bp during sliding 55. The distortion at SHL3 indicates an ability to diffuse structural tension created elsewhere on the nucleosome, suggesting that sites other than SHL2 and SHL5 may serve as transient reservoirs for translocating DNA (twist defects) during nucleosome sliding.
Our discovery of the ChEx segment on the exit-side acidic patch suggests an unexpected means for remodeler competition. Several families of remodelers, including ISWI and INO80, rely on recognition of the H2A acidic patch for robust nucleosome sliding activity 51,56. Our results suggest that Chd1 and other CHD family members may use ChEx to block remodelers poised to shift nucleosomes in the opposite direction (Fig. 6c). Given its ability to compete with the LANA peptide, ChEx would be expected to also affect other chromatin binding factors that target the acidic patch.
Finally, the ChEx segment also implies an expanded role of Chd1 in transcription. During transcription, the nucleosome is contacted by histone chaperones, such as FACT, that aid in histone disassembly and reassembly 57,58. As shown in a recent cryo-EM complex, Chd1 can bind to the DNA gyre of the nucleosome opposite RNA polymerase II 59. As RNA polymerase II transcribes through the first gyre, this placement of Chd1 would allow ChEx to bind to the H2A/H2B dimer being unwrapped by the polymerase. The binding site of ChEx overlaps with a short stretch of the FACT Spt16 C-terminus on the α-N helix of histone H3 (ref. 60), yet both proteins also have extensive non-overlapping interactions with H2A/H2B (Extended Data Figure 10). We speculate that these interactions of ChEx, reminiscent of histone chaperones, could help stabilize H2A/H2B while it is unwrapped, and thus participate in transcription-coupled remodeling along with FACT.
Methods
Protein and nucleosome purification.
The construct for Saccharomyces cerevisiae Chd1 consisted of residues 118-1274 (Chd1[wt]) or 142-1274 (Chd1[ΔChEx]). All variant Chd1 proteins (Supplementary Table 1) were expressed in E. coli with cleavable N-terminal 6xHis tags. Chd1 constructs were purified using a Cytiva HisTrap HP (nickel) column followed by cation exchange chromatography via HiTrap SP FF (Cytiva) columns16,45. The 6xHis tag was removed with overnight digestion by Precission protease, which left a residual Gly-Pro at the N-terminus. Samples were repassed over a HisTrap HP column, and flow through was collected and concentrated. For samples used for cryo-EM and XLMS, buffer for the final size-exclusion HiLoad 16/600 Superdex 200 column (Cytiva) contained 30 mM HEPES-KOH, pH 7.6, 150 mM NaCl, and 10% glycerol.
Human Snf2h protein with a TEV protease cleavable 6xHis tag (plasmid containing Snf2h was a kind gift from Geeta Narlikar) was recombinantly expressed in E. coli and purified similarly to Chd1 46. Snf2h was purified using a HisTrap HP (Cytiva) column followed by anion exchange chromatography using a HiPrep 16/10 Q FF (Cytiva) column. During overnight dialysis (4°C) into SEC buffer (25 mM HEPES-KOH, pH 7.5, 300 mM KCl, 2 mM beta-mercaptoethanol), the 6xHis tag was removed with TEV protease. The sample was then run over a HisTrap HP column, concentrated, and further purified over a HiLoad 16/600 Superdex 200 column in SEC buffer. Pooled fractions were concentrated, supplemented with 10% glycerol and flash frozen in liquid nitrogen. Drosophila ACF protein (cat #31509) was purchased from Active Motif.
All histones were recombinantly expressed Xenopus laevis proteins, which were purified and reconstituted into octamers 64. The sequences for H3[ΔN] and H3[ΔN+ChEx] are given in Supplementary Table 1. For the LANA-displacement assays, nucleosomes contained Cy3-labeled H2B(K120C) that had been labeled with Cy3-maleimide, refolded with H2A, and purified as H2A/H2B dimers. All other histone octamers were made with H3(C110A), H4, H2A, and H2B(S53C). Nucleosomal DNA contained the 145 bp Widom 601 sequence 65 and was flanked by either 40 bp of linker DNA on both sides (40N40) or zero bp on one side and 80 bp on the other (0N80 or 80N0). DNA, which was FAM-labeled on one end, was amplified by PCR, purified, and reconstituted into nucleosomes via salt gradient dialysis 66. The 601[TA-rich +1] variation contained an additional A•T located 22 bp from the dyad 21. For cryo-EM and XLMS experiments, Prep Cell (Bio-Rad) purified FAM-40-601[TA-rich+1]-40 nucleosomes were dialyzed overnight at 4°C in buffer containing 20 mM HEPES-KOH, pH 7.6, 2.5 mM KCl, and 0.1 mM EDTA. Nucleosome samples were supplemented with glycerol (10% final concentration), aliquoted and flash frozen in liquid nitrogen.
Preparation of the Chd1-nucleosome complex for CryoEM.
Prior to gradient fixation (GraFix) 28, purified S. cerevisiae Chd1 was dialyzed into Buffer A (20 mM HEPES, pH 7.55, 60 mM KCl, 2 mM MgCl2, 5% glycerol, 1.5 mM DTT). Chd1 was then mixed with purified mononucleosomes in a 2:1 molar ratio and incubated on ice with 1mM ATP-γS on ice for 20 minutes. Using Buffer B (20 mM HEPES, pH 7.55, 60 mM KCl, 2 mM MgCl2, 10% glycerol, 1.5 mM DTT) and Buffer C (20 mM HEPES, pH 7.55, 60 mM KCl, 2 mM MgCl2, 40% glycerol, 1.5 mM DTT, 0.1% glutaraldehyde), a gradient was formed with a Biocomp gradient maker (Gradient Master). The sample was then laid on top of the gradient and centrifuged in an Optima XE-90 ultracentrifuge (SW40Ti rotor, Beckman-Coulter) at 35,000 rpm (~150,000 rcf) for 16 hours at 4°C. Using a syringe, the sample was manually fractionated into separate tubes, and cross-linking reactions were quenched by adding 100 mM Tris-HCl pH 7.5 to each tube. Fractions were then analyzed using a 4% native gel stained with ethidium bromide. Using negative stain EM, the fractions containing monodisperse particles were selected, dialyzed into the final buffer (20 mM HEPES, pH 7.0, 60 mM KCl, 1.5 mM DTT, 1 mM MgCl2), and concentrated to 1.8 mg ml−1.
Cryo-EM data collection and analysis.
Cryo-EM grids of the Chd1-nucleosome complex were prepared using an established procedure 67. First, Quantifoil 2/2 grids (200 mesh, 2-um hole size) were glow-discharged for 10 s at 15 mA in an easiGlow system; 3 μl of the glutaraldehyde-immobilized complex at 4 μM concentration were applied on the grid surface, and the grids were plunge-frozen in liquid ethane maintained at liquid nitrogen temperatures, using FEI Vitrobot IV maintained at 4 °C and 100% humidity. The data was recorded on an FEI Titan Krios operated at 300 kV in Nanoprobe mode, equipped with a Gatan K3 operated in Super-resolution at a nominal magnification of 81,000x at a physical pixel size of 1.08 Å (Super-resolution 0.54 Å). The collection resulted in 6906 images, each fractionated into 40 frames, accumulating 50 e/Å2 total dose (0.0825 s/frame, 1.25 e/Å/frame).
The movie frames were motion-corrected using UCSF MotionCor2 v1.4.0 68 using full frame for global, and patched alignment (11x9) for local motion. The resulting dose-weighted images were imported into CryoSPARC 69 where their CTF was calculated using “Patch CTF estimation (multi)”. Using visual screening and the CTF calculations, the images were chosen for quality using “Manually Curate Exposures”. Initially, 100 images were processed to obtain 2D templates for particle picking: “Blob picker”, 2D classification, “Ab initio” and “Homogeneous Refinement (Legacy)”, were used to generate a 3D reconstruction, which was then used to generate 2D templates (“Create Templates”) and 10,900,948 particles were picked from the entire dataset (Extended Data Fig. 3). Using a 300,000-particle subset, five “Ab initio” 3D templates were generated and used to classify the full dataset. One class contained a clear nucleosome moiety. Using particles in this class, we used multiple “Heterogeneous Refinement” 3D classifications to improve the Chd1-nucleosome complex quality. A 2.6 Å reconstruction was obtained using 450,433 particles with the “Non-uniform Refinement (New)” procedure 70. This reconstruction revealed very well-resolved density for the nucleosome, as well as the ATPase motor, chromodomains and ChEx segment of Chd1, but a significantly weaker DNA-binding domain (DBD) density. To better characterize the DBD, Relion 71 was used to perform focused classification without alignment on the 450,433 particles using a spherical mask centered on the DBD. The best of the classes was then selected and refined in CryoSPARC to 3.05Å from 32,394 particles, showing a much-improved DBD domain. Subsequently, we decided to see if we could obtain more DBD-containing particles from a larger set of particles. Using 817,698 particles, we classified particles inside a spherical mask centered on the DBD using Relion (Extended Data Fig. 3), marked with a single asterisk), obtaining a number of different reconstructions variable in the DBD and DNA (Extended Data Fig. 8). One of these classes, containing 75,867 particles, exhibited very well-resolved DBD density, with high resolution features and was refined to 3 Å. With these well-resolved 2.6 and 3.0 Å reconstructions, further quality improvement was pursued using Relion’s Bayesian Polishing approach 72. Starting with the best-resolved subset, 450,433 particles were extracted in Relion, refined using Refine3D, optimized for beam and CTF parameters 73, and then used in training and Bayesian polishing. After a total of two rounds of Bayesian polishing, we obtained a reconstruction yielding 2.3 Å (as reported from CryoSPARC, 2.4 Å from Relion). Applying the same approach improved the DBD-containing subset to 2.7 Å.
Additionally, we analyzed a subset of the particles that did not contain Chd1 ATPase, chromodomains or DBD density, but exhibited very-well resolved nucleosome features (Supplementary Figure 3, marked with double asterisk). The nucleosome-only subset was refined to 2.6 Å. Focusing on the ChEx element, a 2.9 Å reconstruction of the nucleosome was obtained through 3D classification, with a single ChEx segment bound to the histone core. The final resolutions are reported using Fourier Shell Correlation (FSC) at 0.143 cutoff 74 following gold-standard refinement. The conversions between cryoSPARC and Relion file formats were performed using the UCSF pyem v0.5 package written by Daniel Asarnow from Yifan Cheng's laboratory (doi:10.5281/zenodo.3576630).
Model building.
The primary basis for the model was the best-resolved 2.3 Å Coulomb potential density, which allowed for unambiguous histone side chain, DNA backbone and base assignment, as well as construction of the Chd1 ChEx segment, chromodomains, and ATPase domains. In this map, the highest quality is exhibited in the core of the nucleosome, with the ATPase and chromodomains of Chd1 being clearly resolved, allowing unambiguous side chain modeling. The quality of the ChEx density allowed for the sequence-level assignment (Figure 4b). However, the DNA-binding domain and the exit-side DNA were not well resolved in this map; thus, we used the 2.7 Å reconstruction with improved DNA-binding domain density to fit and adjust them. In addition to the maps sharpened with a range of B-factors, we also used maps obtained using phenix.local_aniso_sharpen 75 and DeepEMhancer 76.
Initially, X-ray structures were rigid-body fit into cryo-EM reconstructions, with PDB 3LZ0 77 used for the nucleosome, 3MWY 45 for the Chd1 ATPase and chromodomains, and 5J70 for the Chd1 DNA-binding domain. 3MWY was divided into individual domains and fit into the 2.3 Å map, optimized locally using the UCSF Chimera 78 “Fit in map” option. 5J70 was fit into the 2.7 Å map. Coot 79 was used to connect, optimize local fits and side-chain placement, and to build previously unstructured parts of the model. The ChEx segment of Chd1 (residues 121-148) was built without prior templates, based solely on the cryo-EM reconstruction, as were some of the hitherto unresolved loops. The final composite model, containing the nucleosome, ChEx, chromodomains, ATPase motor and DNA-binding domain was refined using phenix.real_space_refine 75 with secondary structure, rotamer and Ramachandran restraints. The final checkup of the pdb was performed manually in Coot, and the model-vs-map validation was done using Molprobity 80. Figures for the model and the cryo-EM reconstructions were prepared using UCSF ChimeraX 81, Coot, and PyMOL (Version 2.0 Schrödinger, LLC). UCSF ChimeraX is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. DNA parameter analysis was performed using CURVES+35 and 3DNA 82.
Cross-linking and mass spectrometry.
For XLMS experiments, after thawing, nucleosomes were dialyzed overnight against two 1 L changes of 20 mM HEPES-KOH, pH 7.9, 2.5 mM KCl, 0.1 mM EDTA, pH 8.0. Reaction mixtures (100 μl) contained a 1:1 mixture of 40N40 601[TA-rich +1] nucleosomes and Chd1[wt], each at 4.6 μM, in 20 mM HEPES-KOH, pH 7.9, 60 mM KCl, 2 mM MgCl2, 0.2 mM EDTA, pH 8.0, and 1.25 mM ATPγS. Cross-linking was initiated by adding a 100 mM stock of either disuccinimidyl dibutyric urea (DSBU; Thermo Fisher, cat #A35459) or carbonyldiimdazole (CDI; Sigma-Aldrich cat #115533), both in anhydrous DMSO, to a final concentration of 1 mM. After a 45 min incubation at room temperature, reactions were quenched by addition of Tris-HCl pH 8.0 to a final concentration of 100 mM 83. After cross-linking, proteins were unfolded by addition of solid urea to 8 M, reduced with addition of 10 mM DTT at 37°C for 45 min, and cysteines were alkylated with 40 mM iodoacetamide (IAA) for 30 min at room temperature. Samples were diluted to 2 M urea with the addition of 20 mM sodium phosphate pH 8.0 and digested with trypsin (Pierce, cat# 90058) at a 1:50 enzyme:substrate ratio overnight at 25°C. Following the primary digest, samples were divided into equal volumes. Half of the sample was prepared for serial digestion with Glu-C (Pierce, cat# 90054) by dilution to a final concentration of 0.8 M urea with 20 mM sodium phosphate pH 8.0. Glu-C was added to these samples at a 1:50 enzyme:substrate ratio and digested overnight at 30°C.
Single and serial digests were acidified by the addition of TFA to a final concentration of 0.5%. Desalting was carried out using Sep-Pak C18 solid phase extraction cartridges (Waters, Milford, MA, USA). Cartridges were loaded onto a vacuum manifold and conditioned using 1 mL 80% ACN, 0.5% TFA, then equilibrated with 4X 1 mL 0.5% TFA. Samples were slowly loaded onto the cartridges under a diminished vacuum and washed with 4X 1 mL 0.5% TFA. After removing cartridges from vacuum, peptides were eluted by adding 1 mL 80% ACN, 0.5% TFA to the cartridges and centrifuging at 350xg for 2 minutes. Eluates were transferred to microfuge tubes, dried, and stored at −80 °C. Before analysis, the peptides were vigorously resuspended in 0.1% FA to a final concentration of 1 mg mL−1.
Samples were analyzed by LC-MS/MS on an UltiMate3000 UHPLC system (Thermo Fisher) coupled with a Q-Exactive HF-X Orbitrap mass spectrometer (Thermo Fisher). Approximately 1 μg of peptides were injected onto the column for each run, and each sample was run in duplicate. Column and trap cartridge temperatures were maintained at 40°C, and the flow rate was set to 300 μL/min for the duration of the run. Solvent A consisted of 0.1% FA in 2% ACN, 98% water, and solvent B consisted of 0.1% FA in ACN. Peptides were accumulated onto a trap column (Acclaim PepMap 100, C18, 75 μm x 2 cm, 3μm, 100 Å column). After washing the trap column with solvent A for 10 minutes, the trap column was switched to be in-line with the separating column, an Acclaim Pepmap RSLC, C18, 75 μm x 25 cm, 2μm, 100 Å column (Thermo Fisher). Peptides were resolved on a linear gradient from 2% B to 35% B (100 minutes), 35% to 40% B (25 minutes), 40% to 90% B (5 minutes). Residual peptides were purged from the column with a saw-tooth gradient at the end of each injection.
Data were acquired in data-dependent MS/MS mode. Each full MS scan used a mass range from 350-1500 m/z, a resolution of 120,000 at 200 m/z, an AGC target of 3e6, and a maximum injection time of 100 ms. Peptides were fragmented with stepped higher-energy collision-induced dissociation and normalized collision energies of 22%, 25%, and 28%. For each full MS scan, ten data-dependent MS/MS scans were collected using a resolution of 15,000 at 200 m/z, an AGC target of 2E5, a minimum AGC target of 8E3, a maximum injection time of 250 ms, an isolation window of 2.0 m/z, and a dynamic exclusion window of 60.0 s. Fragments with charges of <3, and >8 were excluded from analysis, and all spectra were recorded in positive ion mode.
The msConvert GUI from the ProteoWizard Toolkit was used to centroid and convert the raw data to the mzML file format 84. MeroX Version 2.0 was used for crosslink identification 85. For tryptic digests, protease sites were allowed after Arg and Lys residues, with Lys blocked by Pro as a cleavage site. For serial digests, protease sites were allowed at Arg, Asp, Glu, and Lys with Lys blocked by Pro as a cleavage site. For both single and serial digests, a maximum of three missed cleavages were allowed, the minimum peptide length was set to 3, and the maximum peptide length was set to 30. A maximum of one variable oxidation of methionine was allowed, and Cys alkylation by IAA was defined as a static modification. The proper crosslinker (CDI or DSBU) was defined for each sample, and the default MeroX settings were used for each. For the mass comparison, precursor precision was set to 5 ppm, fragment ion precision was 10 ppm, the mass recalibration on both the MS1 and MS2 level were 9.0 ppm, the mass range was 200.0-6000.0 Da, the signal-to-noise ratio was 2.0, b and y ions were selected, the minimum number of fragments per peptide was 3, the minimum charge was 2, and deisotoping mode was used. Searches were conducted in quadratic mode. The FDR cut off was set to 1%, and all scores above the FDR cutoff were accepted.
Crosslink analysis and visualization.
For cross-links where both residues were present in the model, solvent-accessible surface distances (SASD) were calculated using Jwalk v1.1 with default parameters 63, and visualized in PyMOL. DSBU with a 12.5 Å spacer has a greater allowable SASD in comparison to CDI with a spacer length of 2.6 Å. To evaluate whether cross-links to the disordered N-terminus, which immediately precedes ChEx, were consistent with the structure, five residues not visible in the cryo-EM structure were modeled using PyRosetta 86. After extending the N-terminus of the cryo-EM model using an energy-minimized peptide (GPQSTVKIPTRF), the N-terminus was relaxed again with 20 unique DSBU cross-links as atom-pair constraints, using the flat-harmonic potential with x0 = 15 Å, tol = 15 Å, σ = 1.0, following the method of Kahrahman et al. 87.
DNA competition assays.
Experiments were performed in 10 μl reactions by mixing together 30 nM 40N40-FAM nucleosome (601[canonical] or 601[TA-rich +1]) and 120 nM Chd1, in either nucleotide-free conditions or 1 mM AMP-PNP, in buffer containing 15 mM HEPES-KOH, pH 7.6, 50 mM KCl, 2.5 mM MgCl2, 0.1 mg ml−1 BSA, 1 mM DTT, and 5% sucrose. To each reaction, salmon sperm DNA (Thermo Fisher, cat #15632011) was added to a final concentration of 0, 0.0625, 0.125, 0.25, 0.5, 1.0, 2.0, 4.0 μg μl−1. After a 15 min incubation at room temperature, 2 μl of each reaction was loaded on a 4.25% native PAGE gel. After electrophoresing for 90 min (0.25x TBE, 4°C, 100V), gels were scanned on a Typhoon 5 imager (Cytiva).
Nucleosome sliding assays.
Nucleosome sliding reactions were carried out in the presence of remodeler enzyme with 40 nM FAM-80-601-0 or 0-601-80-FAM nucleosomes, using 2.5 mM ATP and 1x Slide buffer (20 mM HEPES-KOH, pH 7.6, 100 mM KCl, 5mM MgCl2, 0.1 mg ml−1 BSA, 1 mM DTT, 5% sucrose). Remodeler concentrations are as indicated in figure legends. After initiating each 30 μl reaction with addition of ATP, 1 μl aliquots were taken out at each time point and stopped by adding to separate tubes containing 7.5 μl quench buffer (20 mM Hepes-KOH, pH 7.6, 50 mM KCl, 0.1 mg ml−1 BSA, 1 mM DTT, 5% sucrose, 40 mM EDTA, 2 μg μl−1 salmon sperm DNA). For each experiment, 2.5 μl aliquots of each quenched reaction were resolved on 6% native PAGE, and scanned on a Typhoon 5 imager (Cytiva).
LANA displacement assay.
Experiments were performed in 120 μl reactions using 27.5 nM 30-601-30 nucleosomes containing H2B(K120C)-Cy3, and a Dabcyl-modified version of the LANA N-terminus, [MAPPGMRLRSGRSTGAPLTRGSC], in buffer containing 20 mM HEPES-KOH pH 7.6, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 0.1 mg ml−1 BSA, 1 mM DTT and 0.02% NP40. To monitor competition between Chd1 and LANA, the LANA titration was halted at 273 nM and Chd1 variants were titrated in. Reactions where Chd1 was added additionally contained 1 mM AMP-PNP. Cy3 emission intensity was measured on an Aviv ATF-105 spectrofluorometer at 562 nm using 540 nm excitation.
Extended Data
Extended Data Fig. 1. Cryo-EM raw data and analysis of the 2.3 Å Chd1-nucleosome complex.
a, Representative cryo-EM micrographs of Chd1-nucleosome complex. b, Selected 2D class averages generated from the particles used to reconstruct the Chd1-nucleosome complex. c, Four orthogonal views of the 2.3 Å structure. Coloring of nucleosome and Chd1 domains is according to Figure 1. d, Euler angle distribution of all the particles used in the 2.3 Å 3D-reconstruction. The distribution of particles in a specific orientation is proportional to the length of each cylinder. e, The final 2.3 Å reconstruction of the Chd1-nucleosome complex colored according to local resolution with blue and red representing the highest and lowest resolution, respectively. f, Four orthogonal views of the Chd1 ATPase and chromodomains from the 2.3 Å reconstruction, without the nucleosome density, colored according to local resolution. g, FSC curve calculated between two independent half-maps from refinements in CryoSPARC (2.3 Å, blue) and Relion (2.4 Å, purple) reported at 0.143 FSC cutoff. The Relion and CryoSPARC refinements were independent from each other. Arrows indicate the reported resolutions in Ångstroms.
Extended Data Fig. 2. Views of 2.3 Å resolution density maps of the Chd1-nucleosome complex.
The coloring of the figure follows that from Fig. 1 (guide DNA strand, yellow; tracking DNA strand, orange; ChEx, magenta; double chromodomains, light blue; ATPase lobe 1, purple; ATPase lobe 2, blue). a-f, Selected views of Chd1-ATPase interactions with DNA. g,h, Views of the DNA duplex at SHL2 and SHL3.
Extended Data Fig. 3. Overview of 3D classification and reconstruction of the Chd1-nucleosome complex dataset.
Flowchart of data processing, refinement, and classification towards the final reconstructions of Chd1-nucleosome dataset (left). Single asterisk marks the bifurcation where particles were classified according to presence of the DNA-binding domain (bottom right). Double asterisk marks the classification of the nucleosome alone or nucleosome with only ChEx (top right). Triple asterisk marks the classification for the “bridge” region connecting the brace helix and the DBD. Darker squares report significant reconstructions.
Extended Data Fig. 4. Cryo-EM analysis of nucleosome-bound Chd1 with defined DBD, nucleosome-ChEx and nucleosome-only subsets.
a, Selected 2D class averages generated from the particles used to reconstruct the Chd1-nucleosome complex at 2.7 Å with the well-defined DNA-binding domain. b, Four orthogonal views of the 2.7 Å structure containing the well-defined DNA-binding domain. Coloring of nucleosome and Chd1 domains is done according to Figure 1. c, Euler angle distribution of all the particles used in the 2.7 Å 3D reconstruction. d, The final 2.7 Å map of the Chd1-nucleosome complex colored according to local resolution; blue represents the highest resolution and red the lowest. e, Selected 2D class averages generated from the particles used to obtain the nucleosome-only reconstruction at 2.6 Å. f, Four orthogonal views of the 2.6 Å structure of the nucleosome. g, Euler angle distribution of all the particles used in the 2.6 Å 3D reconstruction. h, The final 2.6 Å nucleosome reconstruction colored according to local resolution; blue represents the highest resolution and red the lowest. i, Selected 2D class averages generated from the particles used to reconstruct the nucleosome-bound ChEx at 2.9 Å. j, Four orthogonal views of the 2.9 Å structure of ChEx bound to the nucleosome. Coloring of the nucleosome and ChEx is done according to Figure 1. k, Euler angle distribution of all the particles used in the 2.9 Å 3D reconstruction. l, The final 2.9 Å reconstruction of the nucleosome-bound ChEx colored according to local resolution; blue represents the highest resolution and red the lowest. m, n, o, FSC curves were calculated between two independently refined half-maps, before (red) and after (blue) masking, and reported at 0.143 FSC cutoff. Shown are the curves for the nucleosome-Chd1 complex with well-defined DBD (2.7 Å, m), the nucleosome alone (2.6 Å, n), and nucleosome with ChEx only (2.9 Å, o). Arrows indicate the reported resolutions in Ångstroms.
Extended Data Fig. 5. Nucleosome-Chd1 complexes in the absence of nucleotide are more resistant to competitor DNA when the 601[TA-rich +1] sequence is used.
a, 40-601-40 nucleosomes (30 nM) were preincubated with 120 nM Chd1 in the presence or absence of ATPγS for 15 min at room temperature, with or without salmon sperm DNA. Reactions were separated on 4.25% native acrylamide gels. Shown are representative gels. b, Quantification of nucleosome binding in the presence of competitor. Fits to averaged data points gave apparent Kd values of 0.43 ± 0.15 mg ml−1 and 0.11 ± 0.05 mg ml−1 for nucleotide-free Chd1[wt] for 601[TArich +1] and 601[canonical], respectively, and 0.11 ± 0.05 mg ml−1 and 0.07 ± 0.04 mg ml−1 for ATPγS-bound Chd1[wt] for 601[TArich +1] and 601[canonical], respectively. Data shown are averages of six replicates. Error bars represent standard deviations.
Extended Data Fig. 6. DNA parameters.
Shown are DNA parameters calculated with (a) CURVES+ 35 and (b) 3DNA 82, with X-axis indicating the distance from the dyad. For the Chd1-bound structure in the nucleotide-free state, yellow bars represent the Chd1-bound (TA-rich) side and gray bars represent the unbound (TA-poor) side, with dark green indicating overlap of the two sides. For the 601 sequences, solid lines show parameter values for the TA-rich sides and dotted lines show those of the TA-poor sides.
Extended Data Fig. 7. Absorption of single nucleotides on the nucleosome.
Shown are crystal and cryo-EM structures of the nucleosome, aligned based on the histone core. Each view shows the DNA minor groove facing away from the histone core. Note that in each case, the bulging strand, which remains fully base-paired, contains an additional nucleotide.
Extended Data Fig. 8. Variability in density for the Chd1 DNA-binding domain, exit DNA, and the bridge.
a, Overview of a filtered Chd1-nucleosome reconstruction where the DNA-binding domain is well-defined. b, Zoomed-in views showing different sub-classes obtained from the dataset, exhibiting variability in the exit DNA and the DNA-binding domain. c-f, Visualization of variability of the connection (“bridge”) between the brace helix and the DNA-binding domain. c, Model of the nucleosome-bound Chd1. d, Model of the nucleosome-bound Chd1 model converted into a map, filtered to 10 Å. e, Separated unmodeled density (purple) from the 2.3 Å Coulomb potential density reconstruction as shown against the map from d. f, Unmodeled densities (various colors) from subsets obtained using focused classification, shown against the map from d.
Extended Data Fig. 9. Nucleosome sliding assays of Chd1 variants.
a, Overview of the Chd1-nucleosome complex, highlighting the opposite-gyre DNA interacting loop (residues 475-481), a loop on the chromodomains that contacts the DNA-binding domain (residues 295-302), and conserved residues on the DNA-binding domain that contact the chromodomains (residues 1199-1202). b, Quantification of nucleosome sliding reactions. Sliding reactions were carried out at room temperature with 200 nM Chd1 and 40 nM FAM-80-601-0 nucleosomes. In the top graphs, data are presented as mean values ± SD, with lines showing the fit to the averaged data at each time point. Each variant was measured in multiple independent experiments: Chd1[wt] (n=11); Chd1[K478D/G479A/K480D/K481A] (n=6); Chd1[Δ475-481] (n=8); Chd1[Δ296-302] (n=7); Chd1[D1033A/E1034A/D1038A] (n=6); Chd1[R1199A/D1200A] (n=6); Chd1[D1201A/P1202A] (n=5). The bar graph shows mean sliding rates ± SD, calculated from individual fits. For comparison of rates to Chd1[wt], a two-tailed t-test yielded the following p values: Chd1[K478D/G479A/K480D/K481A] (p=2.5x10−6); Chd1[Δ475-481] (p=7.6x10−12); Chd1[Δ296-302] (p=0.29); Chd1[D1033A/E1034A/D1038A] (p=9.0x10−4); Chd1[R1199A/D1200A] (p=8.1x10−3); Chd1[D1201A/P1202A] (p=0.022).
Extended Data Fig. 10. The Chd1 ChEx segment, devoid of secondary structure, lays over the histone surface similarly to extended peptide segments of histone chaperones.
a, Interactions of ChEx region with histone core. The main chain amide of T125 hydrogen bonds to the C-terminal end of alpha helix 1 of H2B. Neighboring this region are interactions with the acidic patch: R126 ChEx hydrogen bonds with E56 of H2A, and the arginine anchor, R130, hydrogen bonds with residues in the canonical acidic patch binding pocket (E61, D90, E92). Adjacent to the acidic pocket, the conserved Y137 of ChEx packs against a hydrophobic surface of H2A, consisting of L65, A69, L85, and A86. The aromatic ring of Y137 is protected from solvent by V135 of ChEx. Several side chain/main chain hydrogen bonds are formed between H2A and ChEx around Y137: the backbone of Y137 hydrogen bonds with side chains of H2A D72 and N73; the backbone amide of I139 hydrogen bonds with the H2A N73 side chain; and the side chain of ChEx N138 hydrogen bonds with the backbone carbonyl of H2A D72. A second tyrosine (Y141) of ChEx packs against the aliphatic regions of side chains of R52 and K56 of the αN helix of H3. At the C-terminal end of ChEx, a group of acidic residues interacts with a cluster of basic residues on H3 (R42, R52, and K56). ChEx residues are labeled in magenta, and hydrogen bonds 3.2 Å or less and 3.3-3.5 Å are shown as green dashes or yellow dots, respectively. b, Comparison of nucleosome-binding footprints of the Chd1 ChEx segment with the C-terminal domain (CTD) of the FACT subunit Spt16. The FACT structure (6UPK) was aligned with the Chd1-nucleosome structure by superimposing the bound H2A-H2B dimers of each structure. With this alignment, the Spt16 CTD clashes with ChEx at the H3 binding interface and with the DNA at SHL4.5.
Supplementary Material
Supplementary Data 1 Cross-linking mass spec data for the Chd1-nucleosome complex.
Supplementary Video 1 Overview of two cryo-EM structures of the Chd1-nucleosome complex in the nucleotide-free state. The structure resolved at 2.3 Å resolution has poor density for exit DNA and the DNA-binding domain. The structure at 2.7 Å resolution resulted from focused subclassification for the DNA-binding domain, which also shows stronger density for exit DNA.
Supplementary Video 2 A tour of the interface between the Chd1 ATPase motor and nucleosomal DNA at SHL2. The electron density, shown as a mesh, is from a 2.3 Å cryo-EM map.
Supplementary Video 3 Comparison of the nucleotide-free Chd1-nucleosome structure to other remodeler-nucleosome complexes. All structures shown here were nucleotide-free or ADP-bound, with a similar “open” ATPase conformation.
Supplementary Video 4 Morphing movie illustrating the relative changes in the nucleosomal DNA upon binding of Chd1 in the nucleotide-free state. See also Figure 2.
Acknowledgements
We thank Rob Levendosky for Snf2h protein, Katie Tripp and the Center for Molecular Biophysics at Johns Hopkins for fluorometer use, and Carol Bator at the Huck Institutes of the Life Sciences Cryo-Electron Microscopy Facility for the initial cryo-EM data collection. We thank Stephen Abini-Agbomson for his help in setting up and troubleshooting the GraFix procedure, and Ulrich Baxa and Adam Wier for their support and data collection at the Frederick National Laboratory. This work was supported by NIH grants R01-GM084192 (to G.D.B.) and DP2-GM140926 (to S.D.F.). This research was also supported, in part, by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E.
Footnotes
Competing Interests
The authors declare no competing interests.
Code availability.
Scripts to analyze and visualize the structures have been deposited at github (https://github.com/gdbowman/).
Data availability.
The raw cryo-EM data has been deposited in EMPIAR (EMPIAR-10876). The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank as EMD-25479 (nucleosome-bound Chd1), EMD-25480 (nucleosome-bound Chd1 with well-defined DBD), EMD-25483 (nucleosome-ChEx) and EMD-25481 (nucleosome-only). Atomic models built using cryo-EM data have been deposited in the RCSB Protein Data Bank with PDB codes 7TN2 (nucleosome-bound Chd1) and 7SWY (nucleosome-only). The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025287. This study included analysis of previously determined nucleosome-remodeler complexes (PDB codes: 5O9G, 6IRO, 6PWF, 6IY2, 6IY3, 6FML), nucleosome-LANA complex (1ZLA), and nucleosome-only models (1KX3, 1KX5, 3UT9, 5F99, 5Y0D, 6IPU, 6WZ5, 6ZHX, 7OHC). Source data are provided with the manuscript.
References
- 1.Piatti P et al. Embryonic stem cell differentiation requires full length Chd1. Sci. Rep 5, 8007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman-Ayala M et al. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development 142, 118–127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basta J & Rauchman M The nucleosome remodeling and deacetylase complex in development and disease. Transl. Res 165, 36–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koh FM et al. Emergence of hematopoietic stem and progenitor cells involves a Chd1-dependent increase in total nascent transcription. Proc. Natl. Acad. Sci. U. S. A 112, E1734–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simic R et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22, 1846–1856 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krogan NJ et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol 22, 6979–6992 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner MH, Roinick KL & Arndt KM Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol 27, 6103–6115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stokes DG, Tartof KD & Perry RP CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. U. S. A 93, 7137–7142 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelley DE, Stokes DG & Perry RP CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108, 10–25 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Smolle M et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol 19, 884–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito T, Bulger M, Pazin MJ, Kobayashi R & Kadonaga JT ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145–155 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Lusser A, Urwin DL & Kadonaga JT Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol 12, 160–166 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Leonard JD & Narlikar GJ A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler. Mol. Cell 57, 850–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nodelman IM, Shen Z, Levendosky RF & Bowman GD Autoinhibitory elements of the Chd1 remodeler block initiation of twist defects by destabilizing the ATPase motor on the nucleosome. Proc. Natl. Acad. Sci. U. S. A 118, e2014498118. doi: 10.1073/pnas.2014498118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Racki LR et al. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature 462, 1016–1021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nodelman IM et al. Interdomain communication of the Chd1 chromatin remodeler across the DNA gyres of the nucleosome. Mol. Cell 65, 447–459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundaramoorthy R et al. Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. Elife 7, e35720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnung L, Ochmann M & Cramer P Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations. Elife 9, 10.7554/eLife.56178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delmas V, Stokes DG & Perry RP A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc. Natl. Acad. Sci. U. S. A 90, 2414–2418 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKnight JN, Jenkins KR, Nodelman IM, Escobar T & Bowman GD Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol. Cell. Biol 31, 4746–4759 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winger J, Nodelman IM, Levendosky RF & Bowman GD A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA. eLife 7, e34100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nodelman IM & Bowman GD Biophysics of Chromatin Remodeling. Annu. Rev. Biophys 50, 73–93 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M et al. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 567, 409–413 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Yan L, Wu H, Li X, Gao N & Chen Z Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol 26, 258–266 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Chittori S, Hong J, Bai Y & Subramaniam S Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome. Nucleic Acids Res. 47, 9400–9409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan L & Chen Z A Unifying Mechanism of DNA Translocation Underlying Chromatin Remodeling. Trends Biochem. Sci 45, 217–227 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Farnung L, Vos SM, Wigge C & Cramer P Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kastner B et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Singleton MR, Dillingham MS & Wigley DB Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem 76, 23–50 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS & Wigley DB Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Gu M & Rice CM Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. U. S. A 107, 521–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu XJ, Shakked Z & Olson WK A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol 300, 819–840 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Ng HL, Kopka ML & Dickerson RE The structure of a stable intermediate in the A B DNA helix transition. Proc. Natl. Acad. Sci. U. S. A 97, 2035–2039 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavery R & Zakrzewska K Base and base pair morphologies, helical parameters, and definitions. In Oxford Handbook of Nucleic Acid Structure (ed. Neidle S) 39–74 (Oxford University Press, 1999). [Google Scholar]
- 35.Lavery R, Moakher M, Maddocks JH, Petkeviciute D & Zakrzewska K Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 37, 5917–5929 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Hassan MA & Calladine CR Conformational Characteristics of DNA: Empirical Classifications and a Hypothesis for the Conformational Behaviour of Dinucleotide Steps. Philosophical Transactions: Mathematical, Physical and Engineering Sciences 355, 43–100 (1997). [Google Scholar]
- 37.Olson WK et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol 313, 229–237 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Marathe A, Karandur D & Bansal M Small local variations in B-form DNA lead to a large variety of global geometries which can accommodate most DNA-binding protein motifs. BMC Struct. Biol 9, 24-6807-9-24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan S & Davey CA Nucleosome structural studies. Curr. Opin. Struct. Biol 21, 128–136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fairman-Williams ME, Guenther UP & Jankowsky E SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol 20, 313–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dürr H, Korner C, Muller M, Hickmann V & Hopfner KP X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121, 363–373 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Liu X et al. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Willhoft O et al. Structure and dynamics of the yeast SWR1-nucleosome complex. Science 362, eaat7716 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Armache JP et al. Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. Elife 8, 10.7554/eLife.46057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hauk G, McKnight JN, Nodelman IM & Bowman GD The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol. Cell 39, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levendosky RF & Bowman GD Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler. Elife 8, 10.7554/eLife.45472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skrajna A et al. Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res. 48, 9415–9432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGinty RK & Tan S Principles of nucleosome recognition by chromatin factors and enzymes. Curr. Opin. Struct. Biol 71, 16–26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee E et al. A Novel N-terminal Region to Chromodomain in CHD7 is Required for the Efficient Remodeling Activity. J. Mol. Biol 433, 167114 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Barbera AJ et al. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311, 856–861 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Gamarra N, Johnson SL, Trnka MJ, Burlingame AL & Narlikar GJ The nucleosomal acidic patch relieves auto-inhibition by the ISWI remodeler SNF2h. Elife 7, 10.7554/eLife.35322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dao HT, Dul BE, Dann GP, Liszczak GP & Muir TW A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nat. Chem. Biol 16, 134–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deindl S et al. ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 152, 442–452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong Y et al. CHD4 slides nucleosomes by decoupling entry- and exit-side DNA translocation. Nat. Commun 11, 1519-020-15183-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabantsev A, Levendosky RF, Zhuang X, Bowman GD & Deindl S Direct observation of coordinated DNA movements on the nucleosome during chromatin remodelling. Nat. Commun 10, 1720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dann GP et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548, 607–611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warren C & Shechter D Fly Fishing for Histones: Catch and Release by Histone Chaperone Intrinsically Disordered Regions and Acidic Stretches. J. Mol. Biol 429, 2401–2426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou K, Liu Y & Luger K Histone chaperone FACT FAcilitates Chromatin Transcription: mechanistic and structural insights. Curr. Opin. Struct. Biol 65, 26–32 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Farnung L, Ochmann M, Engeholm M & Cramer P Structural basis of nucleosome transcription mediated by Chd1 and FACT. Nat. Struct. Mol. Biol 28, 382–387 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y et al. FACT caught in the act of manipulating the nucleosome. Nature 577, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flaus A, Martin DM, Barton GJ & Owen-Hughes T Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomä NH et al. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol 12, 350–356 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Matthew Allen Bullock J, Schwab J, Thalassinos K & Topf M The Importance of Non-accessible Crosslinks and Solvent Accessible Surface Distance in Modeling Proteins with Restraints From Crosslinking Mass Spectrometry. Mol. Cell. Proteomics 15, 2491–2500 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 64.Dyer PN et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Lowary PT & Widom J New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 66.Nodelman IM, Patel A, Levendosky RF & Bowman GD Reconstitution and Purification of Nucleosomes with Recombinant Histones and Purified DNA. Curr. Protoc. Mol. Biol 133, e130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Punjani A, Zhang H & Fleet DJ Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zivanov J, Nakane T & Scheres SHW A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanchez-Garcia R et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol 4, 874-021-02399-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vasudevan D, Chua EYD & Davey CA Crystal structures of nucleosome core particles containing the '601' strong positioning sequence. J. Mol. Biol 403, 1–10 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettersen EF et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu XJ & Olson WK 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iacobucci C et al. A cross-linking/mass spectrometry workflow based on MS-cleavable cross-linkers and the MeroX software for studying protein structures and protein-protein interactions. Nat. Protoc 13, 2864–2889 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Chambers MC et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Götze M et al. StavroX--a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom 23, 76–87 (2012). [DOI] [PubMed] [Google Scholar]
- 86.Chaudhury S, Lyskov S & Gray JJ PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kahraman A et al. Cross-link guided molecular modeling with ROSETTA. PLoS One 8, e73411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1 Cross-linking mass spec data for the Chd1-nucleosome complex.
Supplementary Video 1 Overview of two cryo-EM structures of the Chd1-nucleosome complex in the nucleotide-free state. The structure resolved at 2.3 Å resolution has poor density for exit DNA and the DNA-binding domain. The structure at 2.7 Å resolution resulted from focused subclassification for the DNA-binding domain, which also shows stronger density for exit DNA.
Supplementary Video 2 A tour of the interface between the Chd1 ATPase motor and nucleosomal DNA at SHL2. The electron density, shown as a mesh, is from a 2.3 Å cryo-EM map.
Supplementary Video 3 Comparison of the nucleotide-free Chd1-nucleosome structure to other remodeler-nucleosome complexes. All structures shown here were nucleotide-free or ADP-bound, with a similar “open” ATPase conformation.
Supplementary Video 4 Morphing movie illustrating the relative changes in the nucleosomal DNA upon binding of Chd1 in the nucleotide-free state. See also Figure 2.
Data Availability Statement
The raw cryo-EM data has been deposited in EMPIAR (EMPIAR-10876). The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank as EMD-25479 (nucleosome-bound Chd1), EMD-25480 (nucleosome-bound Chd1 with well-defined DBD), EMD-25483 (nucleosome-ChEx) and EMD-25481 (nucleosome-only). Atomic models built using cryo-EM data have been deposited in the RCSB Protein Data Bank with PDB codes 7TN2 (nucleosome-bound Chd1) and 7SWY (nucleosome-only). The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025287. This study included analysis of previously determined nucleosome-remodeler complexes (PDB codes: 5O9G, 6IRO, 6PWF, 6IY2, 6IY3, 6FML), nucleosome-LANA complex (1ZLA), and nucleosome-only models (1KX3, 1KX5, 3UT9, 5F99, 5Y0D, 6IPU, 6WZ5, 6ZHX, 7OHC). Source data are provided with the manuscript.