Abstract
The monitoring of profile concentrations of chemical markers in saliva samples can be used to diagnose COVID-19 patients, and differentiate them from healthy individuals. Here, this purpose is achieved by designing a paper-based colorimetric sensor with an origami structure, containing general receptors such as pH-sensitive organic dyes, Lewis donors or acceptors, functionalized nanoparticles, and ion metal complexes. The color changes taking place in the receptors in the presence of chemical markers are visually observed and recorded with a digital instrument. Different types and amounts of the chemical markers provide the sensor with a unique response for patients (60 samples) or healthy (55 samples) individuals. These two categories can be discriminated with 84.3% accuracy. This study evidences that the saliva composition of cured and healthy participants is different from each other with accuracy of 85.7%. Moreover, viral load values obtained from the rRT-PCR method can be estimated by the designed sensor. Besides COVID-19, it may possible to simultaneously identify smokers and people with kidney disease and diabetes using the specified electronic tongue. Due to its high efficiency, the prepared paper device can be employed as a rapid detection kit to detect COVID-19.
Keywords: COVID-19, Colorimetric detection, Metabolomics, Paper-based device, Sensor array, Saliva
Graphical abstract
1. Introduction
As the most available fluid in the human body, saliva has been used to diagnose various diseases such as autoimmune, cardiovascular, diabetic, cancerous and renal diseases as well as genetic disorders, and viral, bacterial and fungal infections [1,2]. Since the saliva collection is a simple, cost-effective, and non-invasive procedure with the ability to detect virus RNA in the oropharyngeal cavity, it has attracted the attention of researchers for the detection of COVID-19 disease [[3], [4], [5], [6], [7], [8]]. Conventionally, the analysis of viral genetic materials is performed by the rRT-PCR gold standard method [3]. Although this method provides highly valuable information, false negative responses might be obtained in some cases due to PCR limiting factors or sequence changes during the experiment [9].
The screening of chemical markers in body fluids is a simple and effective way for the disease diagnosis [10]. In fact, chemical markers are small-sized organic compounds, cations and anions, whose concentrations change by the body's immune mechanism against a natural or pathological biological process [11]. The markers of various tissues are transferred from the bloodstream to the saliva sample through intracellular or paracellular diffusion [12]. These chemicals are characterized by spectroscopy, nuclear magnetic resonance and chromatography methods, allowing for the evaluation of the concentration of one or more specific materials, and for the discrimination between two different saliva compositions [13]. In the latter case, the results obtained by the liquid chromatography mass spectrometry have indicated that the chemicals such as C25H36N6O6, dl-phenylalanine, C7H10N6O2, 2-linoleoyl-sn-glycero-3-phosphoethanolamine, C39H71N2O16P3, 1-hexadecanoylpyrrolidine, taurine, C47H84N9O13P3, and C22H36N8O9 are different in the saliva samples of healthy individuals and COVID-19 patients [14].
The instrumental methods provide a fingerprint response for each sample, giving rise to detailed information on the type and concentration of its constituent compounds. However, they are considered non-user-friendly assays due to the device complexity, high price, and need of certain laboratory conditions, skilled technicians and long-term experiments [15].
Sensory devices with one or more sensing components, monitor chemical markers by chemical or biological receptors, and interact with a particular analyte or a wide range of compounds, leading to electrical and optical changes [16]. Despite the high sensitivity and selectivity of bioreceptors, the preparation, storage conditions and fabrication of sensors based on these materials are not cost-effective [17]. Of course, it is possible to achieve unique responses using multiple sensors known as electronic tongues (E-tongues) [18].
The E-tongue configuration comprises an array of chemical reagents with different structural properties involved in the proton and electron transfer processes or electrostatic and hydrophobic interactions [19]. In the case of the colorimetric-based E-tongue, the interactions taking place in solution or on a paper substrate, are associated with the sensor discoloration [20,21]. Having the ability to fabricate a sensor with miniature dimensions, the capillary property in transferring the sample, high flexibility, portability, low consume of the sample, and simple usage are among the advantages that have resulted in developing various paper-based sensors for detection of chemical markers of food, environmental specimens and biological fluids [22]. In this regard, the paper-based E-tongue has been applied to detect antibiotics [23], antioxidants [24], biogenic amines [25], and the metabolites of tuberculosis [26] and sepsis in serum and urine samples [27]. However, no reports have so far been published on the fabrication of a salivary sensor array for detection of COVID-19 disease.
Due to their different viscosities, saliva samples may not move at the same flow rate on the paper substrate with two-dimensional structures, including microfluidic, lateral flow, and distance-based formats. The devices based on origami structures allow the species to penetrate vertically into the paper texturethrough overlapping the injection and detection areas [28]. In turn, this makes the time performance of the experiment shorter, while also needing the low volume of samples. By miniaturizing the sensor dimensions, it is also possible to reduce the effect of viscosity [28].
In our research group, we have tried to develop sensing tools based on array structure to track the difference between the metabolite profiles of patients caused by COVID-19 disease and healthy people in the various body fluids such as saliva, serum and urine and in the exhaled breath. As the first report, this study proposes a miniature E-tongue with an origami structure to diagnose COVID-19 disease by observing chemical markers in saliva samples. Chemical receptors are selected from four categories; dye-based proton donors and acceptors, material-based electron donors and acceptors, metal ion complexes, and nanoparticles. In this way, the feasibility of the sensor for the detection of small amounts of the chemical markers is enhanced, creating different patterns between patients and healthy individuals. The diversity of sensor components raises the possibility of discerning the chemical markers associated with the other comorbidities. Since the sensor components have different sensitivities to saliva compounds, a reasonable correlation is expected to be found between the responses of one or more sensing components and the disease severity.
2. Experimental section
2.1. Equipment
The printing process was performed by HP LaserJet printer 1200. The pH of solutions was observed by a Metrohm 632 pH-meter (Model 780 pH lab). To inject the chemical receptors on the respective detection zones, a micropipette (BRAND Transferpette® S, Germany) was used. The image of sensor was achieved by a canon scanner (CanoScan LiDE 220). The software including AutoCAD 2016, Image J (1.51n, National Institutes of Health, USA), MATLAB R2015 and SPSS (Version 22; Chicago, IL, USA) were applied to design the sensor structure, extract the color element values, run the classification methods and perform the other statistical analysis, respectively.
2.2. Chemicals
Bovine serum albumin (BSA), caffeic acid (CA), poly glutamic acid (PGA), sodium citrate, bromophenol red (R1), 2, 4-dinitrophenylhydrazine and phenylboronic acid (PBA), pyrocatechol violet (Py), vanadyl sulfate pentahydrate (VOSO4·5H2O) were purchased from Sigma Aldrich. Gold (III) chloride trihydrate (HAuCl4·3H2O), bromocresol purple (R2), iron (II) chloride tetrahydrate (Fecl2.4H2O), copper (II) nitrate trihydrate (Cu(NO3)2·3H2O), sulfuric acid (H2SO4), sodium borohydride (NaBH4), sodium hydroxide (NaOH), boric acid (H3BO3) and ethanol (EtOH) were achieved from Merck chemical company. The porphyrine based dyes such as [meso-tetraphenylporphyrin]Iron (III) chloride (Fe(III)TPPCl), [meso-tetraphenylporphyrin]-Tin (II) (Sn(II)TPP) and meso-tetrakis (4-hydroxyphenyl) porphyrin-manganese (III) acetate (Mn(III)T (4-OH)PP(OAc)) were obtained from Sharghi group at Shiraz University, Shiraz, Iran. The preparation processes of these materials have been found in the previous reports [[29], [30], [31]]. Whatman® Grade NO.2 filter paper was selected to create an origami paper based substrate.
Except the pyrocatechol violet that dissolved in the deionized water, the solutions of the other dyes and the porphyrins were prepared in EtOH. To provide the DWES solution, a container poured by 2, 4-dinitrophenylhydrazine (0.4 g), EtOH (10.0 mL), deionized water (3.0 mL) and H2SO4 (2.0 mL) were stirred for 10 min. The mixture was filtered and the result solution was used for further studies [32].
2.3. Chemical receptors
The nanoparticle based receptors were provided by synthesis of gold nanoparticles (AuNPs) coated by some capping agents consist of BSA, CA and PGA. The methods for preparation of these NPs were described in supplementary materials. Each NPs solution was subjected to a freeze-dryer and the result powder was crushed fine in a mortar. The desired NPs solution was obtained by re-dispersing a specified amount of the powder in the deionized water.
The dye based receptors was created by mixing R1 and R2 dye solutions and an additive such as DWES or PBA (2.0 mol L−1) solutions. The dye concentration and the volume ratio of mixture components were optimized. The porphyrin solutions with the optimal concentration were used as sensing element without any modifications.
To have the metal ion complexes, the mixture of Py (50.0 μL) and a metal ion solution including V (IV), Fe (II) and Cu (II) was added to 100.0 μL of 0.1 mol L−1 borate buffer (pH = 9.0) [33]. The same and optimized concentration of dye and metal ion was used to prepare this mixture.
2.4. Participants
The participants of this study were men (having an age range of 21–80 years), and divided into three groups: patients (60 samples), healthy individuals (55 samples), and cured people (15 samples). The patients were examined by a pulmonologist, and their disease was confirmed using chest imaging and rRT-PCR results. It should be noted that the patients who had taken medication were excluded from the study. After two months, the ones undergoing the treatment process (cured samples) were recalled for the followings. All the participants were interviewed by a physician, and their medical documents were observed. They became aware about the experiment process, and filled an informed consent form. An overview of the demographic information is provided in Table S1. The sampling procedure and other analyses were performed in Baqiyatallah Hospital (Tehran, Iran) between the years 2020 and 2021.
2.5. Sensor structure
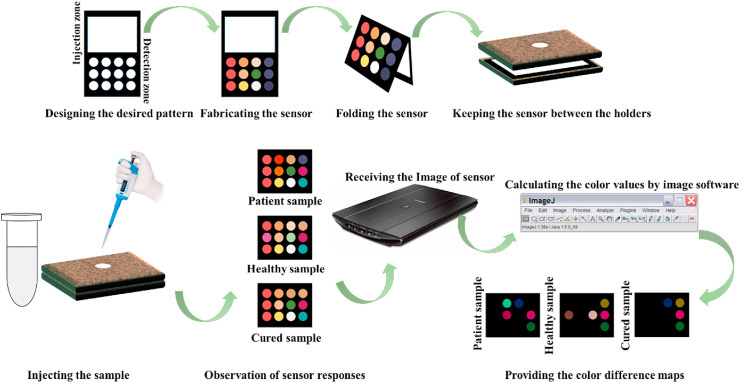
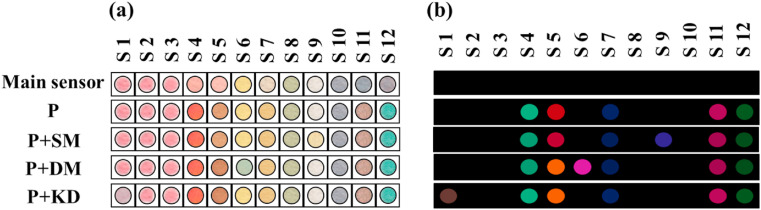
To design the device with an origami structure, a rectangular pattern with dimensions of 1.5 cm × 1 cm was drawn in AutoCAD software. The pattern consisted of injection (white rectangle) and detection (12 small circles) parts (see Fig. 1 a). The design was implemented on a paper substrate using a printer. The black regions became hydrophobic by heating the paper at 200 °C for 45 min [28]. The small circles were filled with the relevant chemical receptors (0.12 μL). The fabricated sensor and the location of each receptor are shown in Fig. 1b and c,respectively.
Fig. 1.
(a) The designing pattern and (b) the image of proposed sensor array. (c) The list of the receptors used for the fabrication of sensor.
2.6. Practical process
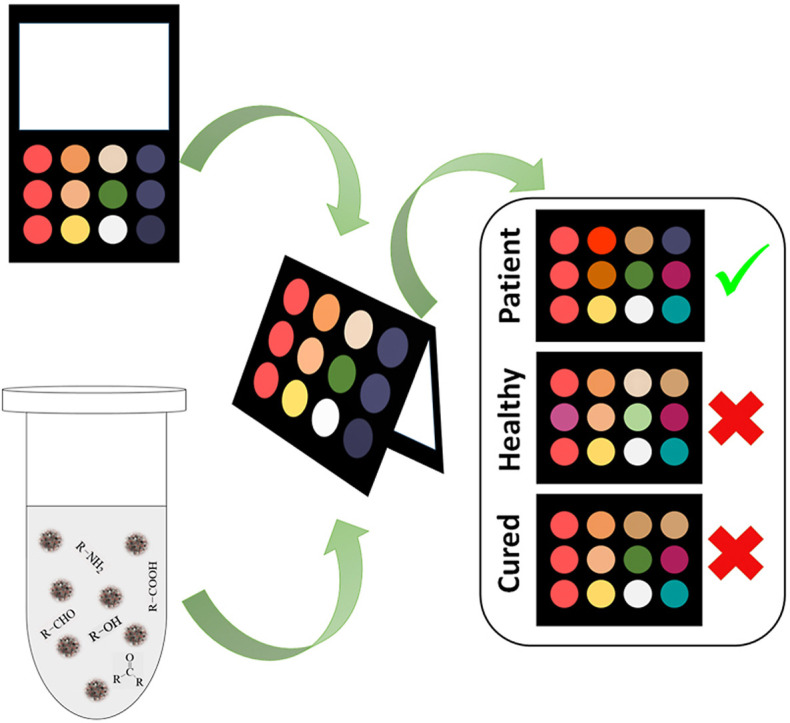
The candidates were asked to rinse their mouths twice with water to remove physical particles from the saliva samples. After 15 min, 500.0 μL of saliva of each person was collected in a sterile Falcon tube, and stored in an ice pack before use. Oral impurities were removed by centrifugation of the saliva sample at 10,000 rpm and 4 °C for 10 min. In the origami sensor, the injection and detection parts were connected to each other by folding the paper. To provide appropriate alignment, the paper was placed between two planar holders. The sensor was exposed to 40.0 μL of a clear supernatant sample. The chemical markers in saliva simultaneously interacted with all the sensing components. The efficient interactions caused the receptors to change color. In this respect, a scanner was used to monitor the discoloration. Image analysis software was employed to evaluate the color of receptors. Three numerical values corresponding to red, green and blue color components were calculated for each receptor. Since the array had 12 components, a vector containing 36 elements was generated for each experiment. The schematic diagram for fabrication and application of salivary E-tongue were shown in Scheme 1 .
Scheme 1.
The schematic diagram for fabrication and application of salivary E-tongue.
2.7. Statistical analysis
The image analysis data were aligned in three individual matrices, including patient-healthy, patient-cured and healthy-cured classes, with sizes of (115 × 36), (75 × 36) and (70 × 36), respectively. The evaluation of the sensor discrimination ability was performed by principle component analysis discriminate analysis (PCA-DA). This classification analysis was validated by calculating statistical parameters such as the sensitivity, specificity, accuracy, and error rate.
In the quantitative analysis, the total response of each saliva sample was calculated by the following equation:
where is the ith numerical value of the data vector obtained for each sample and n is equal to 36. On the other hand, this euation was used to obtain the total response of each sensing component as follows:
where, , and are the color component values for the ith chemical receptor in the data vector.
The effect of some parameters such as age, O2 saturation and disease severity on the total response of the sensor (or the sensing component) was investigated by determining the Pearson correlation coefficient in SPSS environment.
3. Results and discussion
The simplicity of paper-based E-tongues and their possibility for on-site analysis have made them applicable for diagnosis and treatment of diseases. For example, this study examined the ability of these sensors to detect COVID-19 disease. Generally, the E-tongues use non-specific receptors that are chemical compounds with specific physicochemical properties, interacting with a wide range of species in complex samples such as saliva. The chemical markers are ionic species or compounds with amino, carboxylic, aldehyde, hydroxyl, and ketone functional groups [11]. The color changes of the sensor depend on two factors: first, the interaction between the saliva components and the active sites in the receptor structure, and second, the changes in the experiment conditions (e.g., pH of media) arising from the binding of the chemical markers to the additives (DWES or PBA). The opto-E-tongue responses are shown by colorimetric patterns, being the ID cards for infected and healthy samples.
3.1. The optimization process
Since the sensor was designed to detect the disease, and discriminate the patients from the healthy individuals, the optimization process needs to be carried out in such a way that it will lead to the highest accuracy classification. In this regard, the experiments were performed on infected (5 samples) and healthy (5 samples) saliva secretions. Each sample was undergone for three times in this test. For each sample, the total response of the array was used to calculate the ratio of within-group variations and between-group variations, according to the discrimination ability function (DFA) given below [34]:
where n is the number of the studied classes, describes the total response of the array to the jth sample in the ith class, indicates the average of the total responses of the samples in the ith class, and is defined as the average response of the array for 30 measurements. Basically, the best assay discriminatory ability will be observed at the highest DAF value.
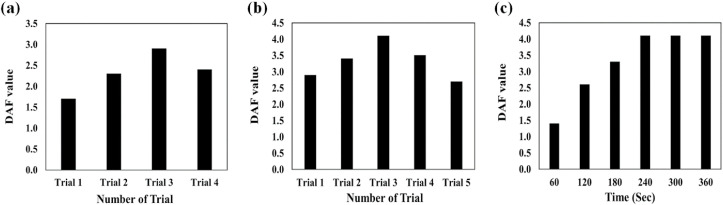
The results of DAF-based optimization experiments are presented in Fig. 2 . As inferred, the sensor response can be changed by varying the sensing component concentrations. The undesirable results are obtained for the lower concentration due to the reduction in the sufficient reactive sites for capturing the saliva species [35]. At higher concentrations, the the color changes arisen from interactions were masked by the color of receptors leading to create the unsuitable DAF values. [35]. Among the trails designed for this study (Table S2), the third one provides an appropriate concentration for the fabrication of receptor arrays (Fig. 2a).
Fig. 2.
Finding the optimization conditions: DAF results for (a) the concentration of each chemical receptors, (b) different types of mixing solution containing the organic dyes and additives and (c) the incubation of the saliva sample and chemical receptors in a period of time between 0 and 360 s. The information about each trial is represented in Table S2 and S3 in supporting information document.
The receptors can be mixed with additives in order to identify a specific class of markers. The amount of additives needs to be adjusted at an appropriate value, thus allowing for a complete interaction with the saliva chemical compositions, while also preventing the active sites of receptors from blockage. Five different trials were designed for this study (Table S3), and the highest DAF value was obtained for the mixture with a volume ratio of receptor: additive (3: 1) (Fig. 2b).
The responses of the sensor for the infected and healthy samples were compared with each other for a specific period of time. The results observed in Fig. 2c illustrate that the changes in the color of receptors become fixed after 4 min of sample injection, indicating an equilibrium between the chemical markers and sensor components. This time period was selected to collect the colorimetric data of the subsequent experiments.
3.2. Colorimetric responses
The sensor components should be capable of both detecting saliva compounds with different functional groups, and distinguishing between species with similar chemical structures. It is also cost-effective to perform the discrimination process with the lower number of receptors [36]. In addition, the use of the receptors with different reactivity in the array structure increases the sensor efficiency to differentiate between the infected and healthy samples.
Due to their various advantages such as the surface plasmon resonance, high surface-to-volume ratio and molecular recognition properties, gold nanoparticles (AuNPs) can detect the ions and chemical compounds with high sensitivity and selectivity [37]. The functional groups, size, and electrical charge of coating agents can also affect the selectivity of these receptors [37]. The organic dyes with tri-aryl methane configuration are sensitive to changes in the pH of media. They can be mixed with DWES and PBA additives. The former identifies the aldehyde and ketone species [32], whereas the latter responds to diols such as sugars [38]. The resulting interactions change the concentration of hydronium ions of the media, leading to discoloration of the organic indicators. Metalloporphyrins participate in the electron donor or acceptor reactions, so that their tendency to different analytes depends on the hardness and chemical properties of the central metal and the substituents of porphyrin cage [39,40]. The last group of receptors consists of metal ion complexes in which a metal ion is chelated with a dye. If the metal ion tends to bind the ligand less than the chemical marker, the ligand will be released, thereby forming a metal-analyte complex. In some cases, the analyte can simultaneously interact with the metal ion and functional groups in the ligand structure, creating a ternary complex [41,42].
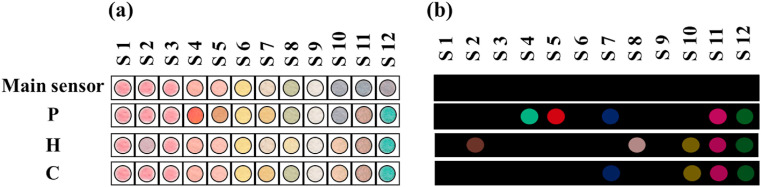
Each column of the proposed sensor array represents unique receptor classes, including NPs, pH sensors, porphyrins, and metal ion complexes. By exposing the origami-based sensor to saliva sample markers, one can observe the color changes after 4 min. The visual observations and the colorimetric pattern obtained from the image analysis are depicted in Fig. 3 .
Fig. 3.
(a) The visual observations and (b) the colorimetric pattern obtained from the image analysis for patient infected by COVID-19 (P), Healthy control (H) and Cured sample (C). The results were provided after 4 min. The sensor was fabricated at the optimum conditions shown in Fig. 2.
Among NP-based receptors, the color of CA-AuNPs changes in the presence of healthy saliva samples. Alternatively, other NPs have no tendency to interact with the metabolites of both healthy and infected saliva secretions. The metabolites of the infected samples change the color of the two organic dyes mixed with DWES. The responses of porphyrins to the different studied groups strongly depend on the central metal ion and constituents of porphyrin cage. The infected sample illuminates the sensing component containing Fe(III)TPPCl, whereas the normal saliva species interact with Mn(III)T (4-OH)PP(OAc). The V(IV)-Py complex participates in the indicator displacement reaction with the chemical markers of healthy samples. All the studied groups can affect the color of Fe(II)-Py and Cu(II)-Py complexes. Furthermore, no responses are recorded from Sn(II)TPP and bromocresol purple + PBA.
During the treatment process, the composition of the saliva of cured samples shifted from the patient to healthy profiles. The detection process was repeated for the cured people after a month. As clarified in Fig. 3, the markers of saliva samples alter the color of S7 (among S4, S5, and S7 as the specific receptors of infected samples) and S10 (from the sensory elements (S2, S8 and S10) responding to healthy samples). The results explain that the saliva metabolite profiles of the cured people are different from the compounds of the healthy samples due to the body disorders caused by the viral infections.
3.3. Evaluation of the discrimination ability
The potential of the sensor to detect COVID-19 disease was evaluated for 60 patients, and 55 healthy and 15 cured samples. The corresponding colorimetric responses are summarized in Fig. S1-S3. To determine the accuracy of the method for the correct diagnosis of each group members, the color difference data were collected in matrices with sizes of (115 × 36), (75 × 36), and (70 × 36) for patient-healthy, patient-cured, and healthy-cured classes, respectively. These data were inserted into PCA-DA algorithm for cluster statistical analysis.
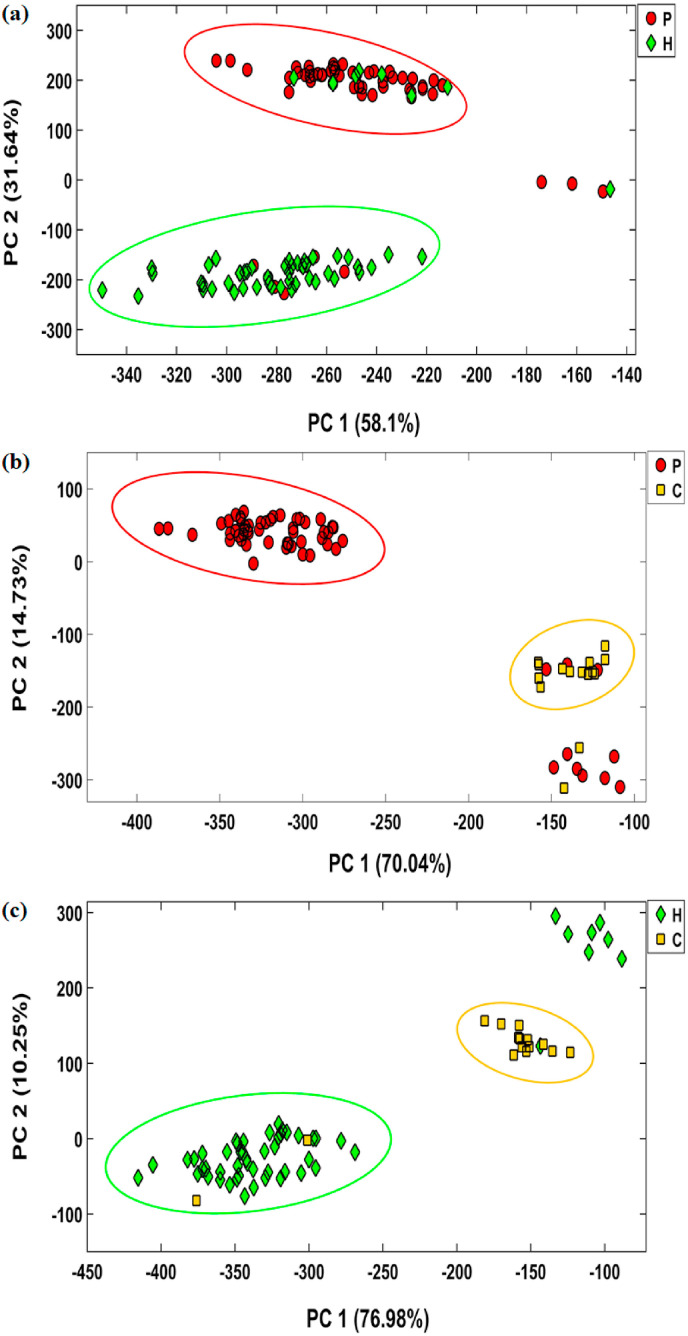
The score plots depicted in Fig. 4 show that over 84% of the total data variances are distributed in the space of the two first principal components. The diagram indicates a good distinction between the two classes of each matrix. However, the sensor misdiagnoses some members of each studied group. By considering the results of the statistical analysis, only 50 patients, 47 healthy individuals, and 13 cured samples can be classified in their corresponding classes with sensitivities of 83.3%, 85.4%, and 86.6%, respectively. Among all the participants, 20 men are incorrectly classified. The statistical parameters obtained from PCA-DA analysis are presented in Table 1 , indicating that the assay efficiency for the discrimination of the studied population is equal to 84.3% for patient-healthy, 84.0% for patient-cured, and 85.7% for healthy-cured datasets.
Fig. 4.
The results obtained by PCA-DA analysis for classification of patients, healthy controls and cured participants. The results were provided after 4 min incubation of sensor and saliva samples. The sensor was fabricated at the optimum conditions shown in Fig. 2.
Table 1.
Classification parameters obtained by PCA-LDA analysis.
| Parameters | Patient vs Healthy | Patient vs Cured | Healthy vs Cured |
|---|---|---|---|
| Sensitivity (%) | 83.3 | 83.3 | 85.4 |
| Specificity (%) | 85.4 | 86.6 | 86.6 |
| Accuracy (%) | 84.3 | 84.0 | 85.7 |
| Error rate (%) | 15.7 | 16.0 | 14.3 |
3.4. Sensing observations of comorbidities
Saliva samples can contain the chemical markers associated with other diseases. Therefore, the potential of the assay for screening the people with cardiovascular, chronic kidney, asthma, diabetes, chronic obstructive pulmonary, chronic liver and hypertension diseases was investigated. For each disease, the specific chemicals would appear in the saliva secretions. As an example, a person with kidney disease can be diagnosed by monitoring the amounts of urea, creatinine, and uric acid [43]. The concentration of compounds such as glucose, 1,3-dihydroxyacetone, lactate, citrulline, ornithine, leucine, and ethanolamine varies in the saliva composition of diabetic and healthy subjects [44]. The difference between smokers and non-smokers is achieved by determining the amounts of some species such as thiocyanate, ionizable iodine, OH-cotinine, tyramine and cadaverine [45,46].
By delving into the sensor results, it can be found that the chemical composition of saliva samples obtained from the individuals with kidney disease leads to the aggregation of BSA-AuNPs, while also changing the NP color from red to purple. Furthermore, the bromocresol purple combined with PBA turns blue in the presence of salivary metabolites of diabetic patients. The response of Sn(II)TPP receptor is only visible for smokers' samples. The visual information and difference patterns from this assay is clearly shown in Fig. 5 . Based on the saliva marker analysis, the performance of the sensor for detection of smokers, and people with diabetes and kidney disease are calculated to be 70%, and 65% and 66.6%, respectively.
Fig. 5.
(a) The visual observations and (b) the colorimetric difference pattern for patients infected only by COVID-19 (P) and the participants having viral infection and the other disease consist of diabetes (DM), chronic kidney (KD) and who were smoker (SM). The results were provided after 4 min. The sensor was fabricated at the optimum conditions shown in Fig. 2.
3.5. Statistical assessments
The metabolic profile of saliva samples can be affected by age and O2 saturation factors. Any changes in the type and concentration of chemical markers would vary the color of the receptor, and the total response of the sensor. Based on the data presented in Table S4, the weak Pearson correlation coefficients and the P-value> 0.05 are indicative of the lack of good and significant relationship between the sensor response and age of participants in both healthy and patient groups. In the case of the relation between the total sensor responses and O2 saturation, the corresponding Pearson coefficient is obtained to be 0.050 (P-value = 0.715) for patients and 0.055 (P-value = 0.693) for healthy individuals, indicating a weak correlation between them.
The viral load value (based on the N gene cycle threshold (CT) value) obtained from PCR analysis is one of the criteria for determining the disease severity, which can be divided into 4 groups: low, moderate, high and very high. In this experiment, the response of the sensor array (or any receptor) for each sample was compared with the respective viral load value. Fig. S4 shows a positive and pronounced linear relationship between the color changes of receptor S5 and virus load value with a Pearson correlation coefficient of 0.898 (P-value < 0.001). This study helps estimate both the visually and numerically results of PCR test.
4. Conclusions
The feasibility of a colorimetric sensor array with non-specific receptors for the diagnosis of COVID-19 disease was studied by analyzing the metabolites of saliva samples. The sensor was developed by a miniature template, and required a small amount of the analyte collected through a non-invasive process. Unlike other screening methods that report only positive or negative results, this sensor provided fingerprint colorimetric patterns for patient, healthy, and cured samples. The color changes depended only on the type and amount of metabolites, and were not influenced by other factors such as age or O2 saturation. Meanwhile, it could determine the severity of the disease by estimating the amount of viral load associated with PCR analysis. The salivary-based colorimetric electronic tongue provided a chance for simultaneously screening the non-COVID-19 diseases. The findings of our study suggested that the proposed sensor can be developed as a fast and cost-effective method for on-site detection of various diseases without the involvement of laboratory devices.
Compliance with ethical standards
The research ethics committee of Baqiyatallah University of Medical Sciences has approved the project (Approval ID: IR. BMSU.REC.1399.508).
Credit author statement
Mohammad Mahdi Bordbar: Project administration, Methodology, Investigation, Formal analysis, Writing – original draft. Hosein Samadinia: Conceptualization, Validation, Resources. Azarmidokht Sheini: Methodology, Validation. Jasem Aboonajmi: Methodology, Validation. Hashem Sharghi: Conceptualization, Validation. Pegah Hashemi: Methodology, Validation. Hosein Khoshsafar: Methodology, Validation. Mostafa Ghanei: Conceptualization, Validation, Resources. Hasan Bagheri: Conceptualization, Investigation, Supervision, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare the following competing financial interest(s): Five authors (Mohammad Mahdi Bordbar (M.M.B.), Pegah Hashemi (P.H.), Hosein Khoshsafar (H. Kh.), Mostafa Ghanei (M. Gh) and Hasan Bagheri (H. B) have filed a provisional patent application on the technology described in this manuscript. entitled “A Method For Rapid Detection Of Covid-19 Using Salivary Metabolites”. The remaining authors declare that they have no competing interests.
Acknowledgments
The authors gratefully acknowledge the support from the Research Councils of Baqiyatallah University of Medical Sciences. Also, The authors would like to thank the Clinical Research Development Unit of Baqiyatallah Hospital, for all their support and guidance during carrying out this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123537.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Pfaffe T., Cooper-White J., Beyerlein P., Kostner K., Punyadeera C. Diagnostic potential of saliva: current state and future applications. Clin. Chem. 2011;57:675–687. doi: 10.1373/clinchem.2010.153767. [DOI] [PubMed] [Google Scholar]
- 2.Malathi N., Mythili S., Vasanthi H.R. ISRN Dent; 2014. Salivary Diagnostics: A Brief Review; pp. 1–8. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandes L.L., Pacheco V.B., Borges L., Athwal H.K., de Paula Eduardo F., Bezinelli L., Correa L., Jimenez M., Dame-Teixeira N., Lombaert I.M.A., Heller D. Saliva in the diagnosis of COVID-19: a review and new research directions. J. Dent. Res. 2020;99:1435–1443. doi: 10.1177/0022034520960070. [DOI] [PubMed] [Google Scholar]
- 4.Caixeta D.C., Oliveira S.W., Cardoso-Sousa L., Cunha T.M., Goulart L.R., Martins M.M., Marin L.M., Jardim A.C.G., Siqueira W.L., Sabino-Silva R. One-year update on salivary diagnostic of COVID-19. Front. Public Health. 2021;9:589564. doi: 10.3389/fpubh.2021.589564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalli M.A., Langmade J.S., Chen X., Fronick C.C., Sawyer C.S., Burcea L.C., Wilkinson M.N., Fulton R.S., Heinz M., Buchser W.J., Head R.D., Mitra R.D., Milbrandt J. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. 2021;67:415–424. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamazaki W., Matsumura Y., Thongchankaew-Seo U., Yamazaki Y., Nagao M. Development of a point-of-care test to detect SARS-CoV-2 from saliva which combines a simple RNA extraction method with colorimetric reverse transcription loop-mediated isothermal amplification detection. J. Clin. Virol. 2021;136:104760. doi: 10.1016/j.jcv.2021.104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaral C., Antunes W., Moe E., Duarte A.G., Lima L.M.P., Santos C., Gomes I.L., Afonso G.S., Vieira R., Teles H.S.S., Reis M.S., da Silva M.A.R., Henriques A.M., Fevereiro M., Ventura M.R., Serrano M., Pimentel C. A molecular test based on RT-LAMP for rapid, sensitive and inexpensive colorimetric detection of SARS-CoV-2 in clinical samples. Sci. Rep. 2021;11:16430. doi: 10.1038/s41598-021-95799-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeFina S.M., Wang J., Yang L., Zhou H., Adams J., Cushing W., Tuohy B., Hui P., Liu C., Pham K. SaliVISION: a rapid saliva-based COVID-19 screening and diagnostic test with high sensitivity and specifcity. Sci. Rep. 2022;12:5729. doi: 10.1038/s41598-022-09718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansson L., Hedman J. Challenging the proposed causes of the PCR plateau phase. Biomol. Detect. Quantif. 2019;17:100082. doi: 10.1016/j.bdq.2019.100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasan M.R., Suleiman M., Pérez-López A. Metabolomics in the diagnosis and prognosis of COVID-19. Front. Genet. 2021;12:721556. doi: 10.3389/fgene.2021.721556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Podzimek S., Vondrackova L., Duskova J., Janatova T., Broukal Z. Salivary markers for periodontal and general diseases. Dis. Markers. 2016:9179632. doi: 10.1155/2016/9179632. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong D.T. Salivary diagnostics. J. Calif. Dent. Assoc. 2006;34:283–285. doi: 10.3342/kjorl-hns.2011.54.11.741. [DOI] [PubMed] [Google Scholar]
- 13.Z T D., F A., R M., R K., S B., S B., A C G., T S., L D., H L., P L., E D. The human saliva metabolome. Metabolomics. 2015;11:1864–1883. [Google Scholar]
- 14.Frampas C.F., Longman K., Spick M.P., Lewis H.M., Costa C.D.S., Stewart A., Dunn-Walters D., Greener D., Evetts G.E., Skene D., Trivedi D., Pitt A.R., Hollywood K., Barran P., Bailey M.J. MedRxiv; 2021. Untargeted Saliva Metabolomics Reveals COVID-19 Severity. 2021.07.06.21260080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bordbar M.M., Hemmateenejad B., Tashkhourian J., Nami-Ana S.F. An optoelectronic tongue based on an array of gold and silver nanoparticles for analysis of natural, synthetic and biological antioxidants. Microchim. Acta. 2018;185:493. doi: 10.1007/s00604-018-3021-1. [DOI] [PubMed] [Google Scholar]
- 16.Choi J.R. Development of point-of-care biosensors for COVID-19. Front. Chem. 2020;8:517. doi: 10.3389/fchem.2020.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bordbar M.M., Nguyen T.A., Arduini F., Bagheri H. A paper-based colorimetric sensor array for discrimination and simultaneous determination of organophosphate and carbamate pesticides in tap water, apple juice, and rice. Microchim. Acta. 2020;187:621. doi: 10.1007/s00604-020-04596-x. [DOI] [PubMed] [Google Scholar]
- 18.Bordbar M.M., Nguyen T.A., Tran A.Q., Bagheri H. Optoelectronic nose based on an origami paper sensor for selective detection of pesticide aerosols. Sci. Rep. 2020;10:17302. doi: 10.1038/s41598-020-74509-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podrazka M., Báczyńska E., Kundys M., Jeleń P.S., Nery E.W. Electronic tongue-A tool for all tastes? Biosensors. 2017;8:3. doi: 10.3390/bios8010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Li S., Liu Q., Chen Z. Electronic-tongue colorimetric-sensor array for discrimination and quantitation of metal ions based on gold-nanoparticle aggregation. Anal. Chem. 2019;91:6315–6320. doi: 10.1021/acs.analchem.9b01139. [DOI] [PubMed] [Google Scholar]
- 21.Patel S., Jamunkar R., Sinha D., Monisha, Patle T.K., Kant T., Dewangan K., Shrivas K. Recent development in nanomaterials fabricated paper-based colorimetric and fluorescent sensors: a review. Trends Environ. Anal. Chem. 2021;31 doi: 10.1016/j.teac.2021.e00136. [DOI] [Google Scholar]
- 22.Fu L.M., Wang Y.N. Detection methods and applications of microfluidic paper-based analytical devices. TrAC Trends Anal. Chem. (Reference Ed.) 2018;107:196–211. doi: 10.1016/j.trac.2018.08.018. [DOI] [Google Scholar]
- 23.Taghizadeh-Behbahani M., Shamsipur M., Hemmateenejad B. Detection and discrimination of antibiotics in food samples using a microfluidic paper-based optical tongue. Talanta. 2022;241:123242. doi: 10.1016/j.talanta.2022.123242. [DOI] [PubMed] [Google Scholar]
- 24.Park S.H., Maruniak A., Kim J., Yi G.R., Lim S.H. Disposable microfluidic sensor arrays for discrimination of antioxidants. Talanta. 2016;153:163–169. doi: 10.1016/j.talanta.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 25.Shojaeifard Z., Bordbar M.M., Aseman M.D., Nabavizadeh S.M., Hemmateenejad B. Collaboration of cyclometalated platinum complexes and metallic nanoclusters for rapid discrimination and detection of biogenic amines through a fluorometric paper-based sensor array. Sensor. Actuator. B Chem. 2021;334:129582. doi: 10.1016/j.snb.2021.129582. [DOI] [Google Scholar]
- 26.Tsai T.T., Huang C.Y., Chen C.A., Shen S.W., Wang M.C., Cheng C.M., Chen C.F. Diagnosis of tuberculosis using colorimetric gold nanoparticles on a paper-based analytical device. ACS Sens. 2017;2:1345–1354. doi: 10.1021/acssensors.7b00450. [DOI] [PubMed] [Google Scholar]
- 27.Sheini A. A point-of-care testing sensor based on fluorescent nanoclusters for rapid detection of septicemia in children. Sensor. Actuator. B Chem. 2021;328:129029. doi: 10.1016/j.snb.2020.129029. [DOI] [Google Scholar]
- 28.Sheini A., Aseman M.D., Bordbar M.M. Origami paper analytical assay based on metal complex sensor for rapid determination of blood cyanide concentration in fire survivors. Sci. Rep. 2021;11:3521. doi: 10.1038/s41598-021-83186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharghi H., Nejad A.H. Novel synthesis of meso-tetraarylporphyrins using CF3SO2Cl under aerobic oxidation. ChemInform. 2004;35:1863–1868. doi: 10.1002/chin.200424113. [DOI] [Google Scholar]
- 30.Mansuy D. Activation of alkanes : the biomimetic approach. Coord. Chem. Rev. 1993;125:129–141. doi: 10.1016/0010-8545(93)85013-T. [DOI] [Google Scholar]
- 31.Bernadou J., Meunier B., Fabiano A.S., Robert A. Redox tautomerism” in high-valent metal-oxo-aquo complexes. Origin of the oxygen atom in epoxidation reactions catalyzed by water-soluble metalloporphyrins. J. Am. Chem. Soc. 1994;116:9375–9376. doi: 10.1021/ja00099a083. [DOI] [Google Scholar]
- 32.Li J., Hou C., Huo D., Yang M., Fa H.B., Yang P. Development of a colorimetric sensor Array for the discrimination of aldehydes. Sensor. Actuator. B Chem. 2014;196:10–17. doi: 10.1016/j.snb.2014.01.054. [DOI] [Google Scholar]
- 33.Sheini A., Khajehsharifi H., Shahbazy M., Kompany-Zareh M. A chemosensor array for the colorimetric identification of some carboxylic acids in human urine samples. Sensor. Actuator. B Chem. 2017;242:288–298. doi: 10.1016/j.snb.2016.11.008. [DOI] [Google Scholar]
- 34.Brereton R.G. Data analysis for the laboratory and chemical plant. Chemometrics. 2003;8:131–132. [Google Scholar]
- 35.Mirzaei Y., Gholami A., Bordbar M.M. A distance-based paper sensor for rapid detection of blood lactate concentration using gold nanoparticles synthesized by Satureja hortensis. Sensor. Actuator. B Chem. 2021;345:130445. doi: 10.1016/j.snb.2021.130445. [DOI] [Google Scholar]
- 36.Bigdeli A., Ghasemi F., Golmohammadi H., Abbasi-Moayed S., Nejad M.A.F., Fahimi-Kashani N., Jafarinejad S., Shahrajabian M., Hormozi-Nezhad M.R. Nanoparticle-based optical sensor arrays. Nanoscale. 2017;9:16546–16563. doi: 10.1039/c7nr03311g. [DOI] [PubMed] [Google Scholar]
- 37.Bordbar M.M., Tashkhourian J., Hemmateenejad B. Structural elucidation and ultrasensitive analyses of volatile organic compounds by paper-based nano-optoelectronic noses. ACS Sens. 2019;4:1442–1451. doi: 10.1021/acssensors.9b00680. [DOI] [PubMed] [Google Scholar]
- 38.Lim S.H., Musto C.J., Park E., Zhong W., Suslick K.S. A colorimetric sensor array for detection and identification of sugars. Org. Lett. 2008;10:4405–4408. doi: 10.1021/ol801459k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suslick K., Hulkower K., Sen A., Sroka M., McNamara W. 2005. Method and Apparatus for Detecting Ammonia from Exhaled Breath. US20050171449A1. [Google Scholar]
- 40.Suslick K.S., Rakow N.A. 2002. Colorimetric Artificial Nose Having an Array of Dyes and Method for Artificial Olfaction. ww.google.com/patents/US6099288w. [Google Scholar]
- 41.Sedgwick A.C., Brewster J.T., Wu T., Feng X., Bull S.D., Qian X., Sessler J.L., James T.D., Anslyn E.V., Sun X. Indicator displacement assays (IDAs): the past, present and future. Chem. Soc. Rev. 2021;50:9–38. doi: 10.1039/c9cs00538b. [DOI] [PubMed] [Google Scholar]
- 42.Khajehsharifi H., Bordbar M.M. A highly selective chemosensor for detection and determination of cyanide by using an indicator displacement assay and PC-ANN and its logic gate behavior. Sensor. Actuator. B Chem. 2015;209:1015–1022. doi: 10.1016/j.snb.2014.10.053. [DOI] [Google Scholar]
- 43.P C., Ľ T., Š K., Ľ P., P B. Salivary markers of kidney function - potentials and limitations. Clin. Chim. Acta. 2016;453:28–37. doi: 10.1016/j.cca.2015.11.028. http://www.embase.com/search/results?subaction=viewrecord&from=export&id=L607197095 [DOI] [PubMed] [Google Scholar]
- 44.Viswanath B., Choi C.S., Lee K., Kim S. Recent trends in the development of diagnostic tools for diabetes mellitus using patient saliva. TrAC Trends Anal. Chem. (Reference Ed.) 2017;89:60–67. doi: 10.1016/j.trac.2017.01.011. [DOI] [Google Scholar]
- 45.Mueller D.C., Piller M., Niessner R., Scherer M., Scherer G. Untargeted metabolomic profiling in saliva of smokers and nonsmokers by a validated GC-TOF-MS method. J. Proteome Res. 2014;13:1602–1613. doi: 10.1021/pr401099r. [DOI] [PubMed] [Google Scholar]
- 46.Tenovuo J., Makinen K.K. Concentration of thiocyanate and ionizable iodine in saliva of smokers and nonsmokers. J. Dent. Res. 1976;55:661–663. doi: 10.1177/00220345760550042001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.