Abstract
Background
Extramedullary manifestations (EM) are rare in acute myeloid leukemia (AML) and their impact on clinical outcomes is controversially discussed.
Methods
We retrospectively analyzed a large multi-center cohort of 1583 newly diagnosed AML patients, of whom 225 (14.21%) had EM.
Results
AML patients with EM presented with significantly higher counts of white blood cells (p < 0.0001), peripheral blood blasts (p < 0.0001), bone marrow blasts (p = 0.019), and LDH (p < 0.0001). Regarding molecular genetics, EM AML was associated with mutations of NPM1 (OR: 1.66, p < 0.001), FLT3-ITD (OR: 1.72, p < 0.001) and PTPN11 (OR: 2.46, p < 0.001). With regard to clinical outcomes, EM AML patients were less likely to achieve complete remissions (OR: 0.62, p = 0.004), and had a higher early death rate (OR: 2.23, p = 0.003). Multivariable analysis revealed EM as an independent risk factor for reduced overall survival (hazard ratio [HR]: 1.43, p < 0.001), however, for patients who received allogeneic hematopoietic cell transplantation (HCT) survival did not differ. For patients bearing EM AML, multivariable analysis unveiled mutated TP53 and IKZF1 as independent risk factors for reduced event-free (HR: 4.45, p < 0.001, and HR: 2.05, p = 0.044, respectively) and overall survival (HR: 2.48, p = 0.026, and HR: 2.63, p = 0.008, respectively).
Conclusion
Our analysis represents one of the largest cohorts of EM AML and establishes key molecular markers linked to EM, providing new evidence that EM is associated with adverse risk in AML and may warrant allogeneic HCT in eligible patients with EM.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-022-01267-7.
Keywords: Acute myeloid leukemia, Extramedullary, Leukemia cutis, Chloroma, Myeloid sarcoma
Background
Extramedullary manifestation (EM) of infiltrating clonal blast populations in a variety of organs and tissues in acute myeloid leukemia (AML) is defined as a distinct entity in the 2016 WHO classification of myeloid neoplasms, myeloid sarcoma [1], (or synonymously granulocytic sarcoma [2] or chloroma [3]) and can either present concurrently with bone marrow and/or peripheral blood involvement, or isolated, and in some cases even antecedent to bone marrow involvement or relapse [4]. While manifestations can be found in a wide variety of organs, most frequent locations include lesions in the connective tissues, intestinal organs and the skin, where EM AML is often referred to as leukemia cutis [4, 5].
Previous reports have estimated EM to be present in 2–9% of AML cases [6–8]. However, the recent PET-AML trial has reported a frequency of 17% in newly diagnosed AML patients [9]. Nevertheless, genetic events and molecular mechanisms that lead to the formation of EM in AML and the impact on clinical outcomes are not well understood and previous studies are commonly confined to small samples or case series, often with controversial results [4, 10, 11].
We here present a large multi-center cohort of newly diagnosed and intensively treated AML patients to compare cytogenetic and molecular profiles as well as clinical presentations and outcomes between AML presenting with concurrent EM and non-EM AML.
Methods
Patient cohort
We analyzed 1583 adult patients with newly diagnosed AML from previous multi-center clinical trials (AML96 [12], AML2003 [13], AML60+ [14], and SORAML [15]) and the multi-center German Study Alliance Leukemia (SAL) AML registry (NCT03188874). Eligibility was determined by age ≥ 18 years, diagnosis of AML according to WHO criteria [1], curative treatment approach with intensive induction therapy and available biomaterial at initial diagnosis. AML without antecedent history of malignancy or radio-/chemotherapy was defined as de novo, while AML not fulfilling these criteria or with prior history of myeloid neoplasms was defined as secondary AML (sAML) and AML with prior radio- and/or chemotherapy for the treatment of non-myeloid malignancies was defined as therapy-associated AML (tAML). Early death (ED30) was defined as death from any cause within 30 days after initial diagnosis. EM AML status was determined at baseline by clinical examination for the entire cohort. Additional histopathologic confirmation of EM in the respective organ was available in 38 cases. The investigation was carried out under the auspices of the SAL registry and received approval of the Institutional Review Board of the Technical University Dresden (EK 98032010). Written informed consent was obtained from all participants according to the Declaration of Helsinki.
Molecular and cytogenetic profiling
All studies were performed on pre-treatment bone marrow aspirates or peripheral blood. Cytogenetic profiling was done using standard techniques for chromosome banding and fluorescence in situ hybridization (FISH). Molecular profiling was done using high resolution fragment analysis for FLT3-ITD [16], NPM1 [17] or CEBPA [18]. For additional alterations, characterization was done using the TruSight Myeloid assay (Illumina, San Diego, CA, USA) as described in detail previously [19, 20]. Briefly, the panel targets 54 genes that are frequently mutated in myeloid neoplasms (Additional file 1: Table S1). Genomic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) and quantified using the NanoDrop (ThermoFisher, Waltham, MA, USA) spectrophotometer. For each reaction, 50 ng of genomic DNA was used. Samples were sequenced paired-end on a NextSeq (150 bp PE) or MiSeq (300 bp PE) NGS-instrument (Illumina, San Diego, CA, USA). A 5% variant allele frequency (VAF) mutation calling cut-off was used.
Statistical analysis
Statistical analysis was performed and visualizations were created using STATA BE 16.0 (Stata Corp, College Station, TX, USA) and R 4.1.2 (R Foundation, Vienna, Austria). Categorical variables were compared using the two-sided Fisher’s exact test. Normality was evaluated using the Shapiro–Wilk test. If the assumption of normality was met, continuous variables between two groups were analyzed using the two-sided unpaired t-test. If the assumption of normality was violated, continuous variables between two groups were analyzed using the Wilcoxon rank sum test. Univariable analysis to test for the effect of different mutations on EM AML status as well as EM AML’s impact on ED30 and CR was carried out using logistic regression. For survival analysis including the evaluation of event-free survival (EFS), relapse-free survival (RFS) and overall survival (OS), the Kaplan–Meier method and the log-rank test were used. For univariable and multivariable analysis of prognostic markers, Cox-proportional hazard models were used. For both odds ratios (OR) and hazard ratios (HR), 95%-confidence intervals (95%-CI) are reported. Statistical significance was determined using a significance level α of 0.05.
Results
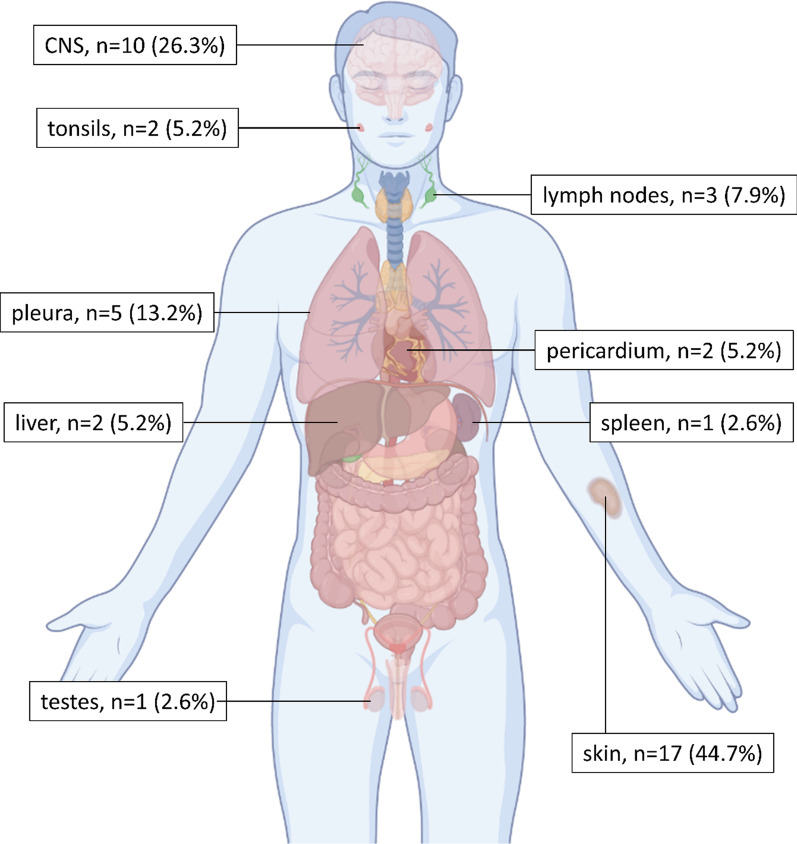
EM AML was found in 225 of 1583 patients (14.2%), only two of whom presented with isolated extramedullary tissue infiltration, i.e., myeloid sarcoma [1], while the majority had concurrent bone marrow involvement. In 38 cases, additional biomaterial of the affected organ was available for histopathologic evaluation. Most frequently affected sites were skin (44.7%), central nervous system (26.3%), and pleura (13.2%). Figure 1 displays the distribution of histopathologically confirmed EM.
Fig. 1.
Distribution of histopathologically confirmed extramedullary manifestations. For 38 out of 225 patients, biomaterial of the affected site was available for histopathological confirmation of extramedullary manifestations (EM). Most frequently, EM was found in the skin, central nervous system (CNS) and pleura. Three patients had two affected EM sites and one patient had three EM sites
Regarding baseline patient characteristics (Table 1) we did not find differences in occurrence of EM AML between male or female patients (p = 0.773), de novo, sAML or tAML (p = 0.6587), or regarding age (p = 0.765). Patients with EM AML compared to patients without EM AML had significantly higher white blood cell (WBC) count (p < 0.0001), higher LDH (p < 0.0001), peripheral blood blast (PBB) counts (p < 0.0001) and bone marrow blast (BMB) counts (p = 0.019) while hemoglobin levels and platelet counts did not differ. Every increase of WBC by 5*109/l led to an increase in the odds of EM AML of 0.005 (Additional file 1: Fig. S1A) and every one-percent increase in BMB (Additional file 1: Fig. S1B) and PBB (Additional file 1: Fig. S1C) counts led to an increase in the odds of EM AML of 0.01 for both. For LDH, every increase by 50 U/l led to an increase in the odds of EM AML by 0.002 (Additional file 1: Fig. S1D).
Table 1.
Baseline patient characteristics
| Parameter | Non-EM AML | EM AML | p |
|---|---|---|---|
| n/N (%) | 1358/1583 (85.79) | 225/1583 (14.21) | |
| Age (years), median (IQR) | 53 (42–60) | 53 (42–61) | 0.7652 |
| Sex, n (%) | 0.7730 | ||
| Female | 656 (48.31) | 106 (47.11) | |
| Male | 702 (51.69) | 119 (52.89) | |
| Disease status, n (%) | 0.6587 | ||
| De novo | 1154 (84.98) | 196 (87.11) | |
| sAML | 143 (10.53) | 21 (9.33) | |
| tAML | 45 (3.31) | 7 (3.11) | |
| ELN-Risk 2017, n (%) | 0.0817 | ||
| Favorable | 423 (31.15) | 81 (36.0) | |
| Intermediate | 412 (30.34) | 77 (34.22) | |
| Adverse | 425 (31.30) | 56 (24.89) | |
| Complex karyotype, n (%) | 0.535 | ||
| No | 1106 (81.44) | 201 (89.33) | |
| Yes | 122 (8.93) | 18 (8.0) | |
| Normal karyotype, n (%) | 0.504 | ||
| No | 535 (39.40) | 88 (39.11) | |
| Yes | 727 (53.53) | 129 (57.33) | |
| Laboratory, median (IQR) | |||
| WBC (109/l) | 14.5 (3.3–47.2) | 32.24 (11.1–87.0) | < 0.0001 |
| HB (mmol/l) | 5.9 (5.0–7.1) | 5.9 (5.1–7.0) | 0.5814 |
| PLT (109/l) | 52 (28–96.5) | 53 (28–94) | 0.8999 |
| LDH (U/l) | 429 (260–724) | 605 (462–1008) | < 0.0001 |
| PBB (%) | 33 (8–70) | 55 (19–79.5) | < 0.0001 |
| BMB (%) | 63 (43–79) | 68.5 (47.5–82.5) | 0.019 |
Bold typing indicates statistical significance (p < 0.05)
AML acute myeloid leukemia, sAML secondary AML, tAML therapy-associated AML, BMB bone marrow blasts, EM extramedullary, HB hemoglobin, IQR interquartile range, n/N number, PBB peripheral blood blasts, PLT platelet count, WBC white blood cell count.
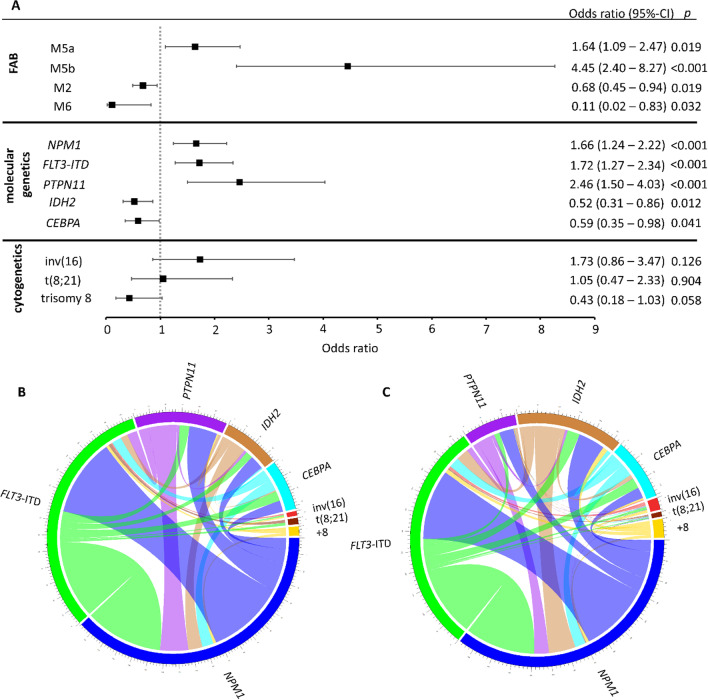
Regarding cytomorphologic subtypes according to the French-American-British (FAB) classification [21], we found significantly increased odds for the presence of EM AML for FAB-M5a (OR: 1.64 [95%-CI: 1.09–2.47], p = 0.019) and FAB-M5b (OR: 4.45 [95%-CI: 2.40–8.27], p < 0.001) while decreased odds were found for FAB-M2 (OR: 0.68 [95%-CI: 0.49–0.94], p = 0.019) and FAB-M6 (OR: 0.11 [95%-CI: 0.02–0.83], p = 0.032). As for molecular genetics, significantly increased odds of EM AML were found for PTPN11 (OR: 2.46 [95%-CI: 1.50–4.03], p < 0.001), NPM1 (OR: 1.66 [95%-CI:1.24–2.22], p < 0.001), and FLT3-ITD (OR: 1.72 [95%-CI: 1.27–2.34], p < 0.001) with an increase in FLT3-ITD ratio leading to a corresponding increase in the odds of EM AML (Additional file 1: Fig. S1E). However, mutations in IDH2 (OR: 0.52 [95%-CI: 0.31–0.86], p = 0.012) and CEBPA (OR: 0.59 [95%-CI: 0.35–0.98], p = 0.041) were associated with a decrease in the odds of EM AML. No significant associations with EM AML were found for inv [16] (OR: 1.73 [95%-CI: 0.86–3.47], p = 0.126), t(8;21) (OR: 1.05 [95%-CI: 0.47–2.33], p = 0.904), trisomy 8 (OR: 0.43 [95%-CI: 0.18–1.03], p = 0.058), or other common cytogenetic aberrations. Odds ratios and confidence intervals for these parameters are summarized in Fig. 2.
Fig. 2.
Parameters associated with extramedullary manifestations in AML. Logistic regression was used to obtain univariable odds ratios for presence or absence of extramedullary manifestations (EM) in AML patients (A). We found the presence of EM to be significantly associated with cytomorphologic phenotypes according to the French-American-British (FAB) classification. FAB-M5a and -M5b were associated with increased odds while -M2 and -M6 were associated with decreased odds. As for molecular genetics, mutations of NPM1, FLT3-ITD and PTPN11 were associated with EM while IDH2 and CEBPA were less likely to be associated with EM. Previous reports have suggested an association of inv [16], t(8;21) and trisomy 8 with EM, however in our analysis we did not find a statistically significant association. Molecular and cytogenetic interconnections of patients with (B) or without (C) EM AML are displayed
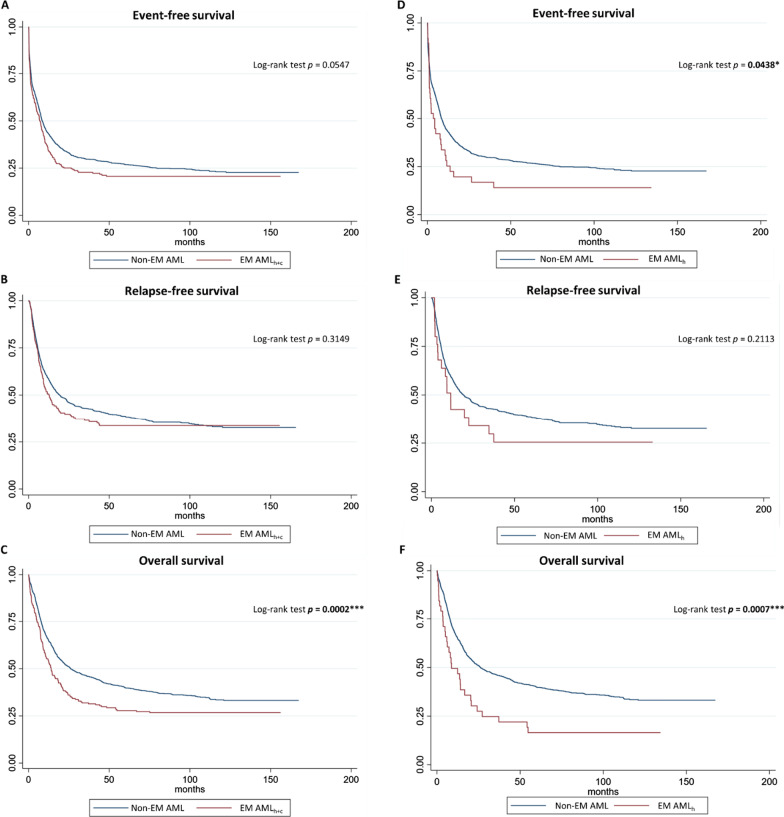
With respect to clinical outcomes, the odds of achieving CR with intensive induction therapy were significantly decreased for patients with EM AML (OR: 0.62 [95%-CI: 0.45–0.86], p = 0.004) while the odds of early death within 30 days after initial diagnosis were significantly increased (OR: 2.23 [95%-CI: 1.31–3.78], p = 0.003). However, relapse rates did not differ between patients with or without EM AML (OR: 1.01 [95%-CI: 0.76–1.35], p = 0.947). With regard to survival (Table 2), patients with EM AML compared to patients without EM did not differ significantly regarding median event-free survival (7.1 months [95%-CI: 4.8–9.1] vs. 8.4 months [95%-CI: 7.7–9.6], HR: 1.17 [95%-CI: 1.00–1.37], Cox regression p = 0.056, Fig. 3A) and median relapse-free survival (12.6 months [95%-CI: 9.3–18.4] vs. 19.3 months [95%-CI: 16.3–23.7], HR: 1.11 [95%-CI: 0.90–1.36], Cox regression p = 0.315, Fig. 3B). However, patients with EM AML showed significantly decreased median overall survival (14.0 months [95%-CI: 10.6–18.4] vs. 26.2 months [95%-CI: 22.4–32.6], HR: 1.38 [95%-CI: 1.16–1.63], Cox regression p < 0.001, Fig. 3C). In a multivariable model adjusting for ELN2017 risk groups and age (Additional file 1: Table S1), EM AML remained an independent marker of reduced OS (HR: 1.43 [95%-CI: 1.21–1.70], Cox regression p < 0.001).
Table 2.
Survival times of patients with or without extramedullary manifestations in the entire patient cohort
| Survival times | EM AML | Non-EM AML | Hazard ratio | Cox regression p-value |
|---|---|---|---|---|
| Event-free survival | 7.1 [4.8–9.1] | 8.4 [7.7–9.6] | 1.17 [1.00–1.37] | 0.056 |
| Relapse-free survival | 12.6 [9.3–13.4] | 19.3 [16.3–23.7] | 1.11 [0.90–1.36] | 0.315 |
| Overall survival | 14.0 [10.6–18.4] | 26.2 [22.4–32.6] | 1.38 [1.16–1.63] | < 0.001 |
Survival times in months. Cox-proportional hazard models were used to obtain univariable hazard ratios. Brackets show 95%-confidence intervals. Statistically significant p-values are marked in bold
Fig. 3.
Impact of extramedullary manifestations on survival in acute myeloid leukemia. For the comparison between both clinically and histologically determined EM AML and non EM-AML, both event-free (A) and relapse-free survival (B) did not differ significantly while overall survival (C) was significantly reduced. When we focused only on histologically confirmed EM AML (n = 38) excluding cases for whom only clinical diagnosis of EM AML was available, we found that both event-free survival (D) as well as overall survival (F) were significantly reduced compared to AML patients without EM while relapse-free survival did not differ (E). Significance was determined at α = 0.05; * p < 0.05, ** p < 0.01, ***p < 0.001; EM AMLh+c = histologically and clinically diagnosed cases of EM AML (panel A–C); EML AMLh = only histologically confirmed cases of EM AML (panel D–F)
For 38 patients, additional biomaterial of the affected EM AML site was available. When we compared only these patients to non-EM AML patients and excluded those EM AML patients for whom only clinically diagnosed EM were available, the impact on outcome became more evident (Table 3). The odds ratio to achieve CR was significantly reduced for patients with histologically confirmed EM (OR: 0.47 [95%-CI: 0.24–0.92], p = 0.029) as well as both median EFS (3.6 months [95%-CI: 1.2–8.4], HR: 1.43 [95%-CI: 1.01–2.04], Cox regression p = 0.046, Fig. 3D) and OS (8.7 months [95%-CI: 5.1–20.3], HR: 1.84 [95%-CI: 1.29–2.64], Cox regression p < 0.001, Fig. 3F) while RFS did not differ (Fig. 3E). Again, in multivariable analysis adjusting for ELN2017 risk groups and age, EM remained an independent marker of reduced EFS (HR: 1.44 [95% CI: 1.01–2.05], Cox regression p = 0.042, Additional file 1: Table S2) as well as OS (HR: 1.67 [95% CI: 1.17–2.40], Cox regression p = 0.005, Additional file 1: Table S3).
Table 3.
Survival times of patients with or without histologically confirmed extramedullary manifestations
| Survival times | Hist. EM AML | Non-EM AML | Hazard ratio | Cox regression p-value |
|---|---|---|---|---|
| Event-free survival | 3.6 [1.2–8.4] | 8.4 [7.7–9.6] | 1.43 [1.01–2.04] | 0.046 |
| Relapse-free survival | 11.6 [3.9–34.6] | 19.3 [16.3–23.7] | 1.35 [0.84–2.15] | 0.213 |
| Overall survival | 8.7 [5.1–20.3] | 26.2 [22.4–32.6] | 1.84 [1.29–2.64] | < 0.001 |
In comparison to Table 2, patients with only clinical diagnosis of extramedullary manifestations (EM) were excluded and only data for patients with histologically confirmed EM are shown (n = 38). Survival times in months. Cox-proportional hazard models were used to obtain univariable hazard ratios. Brackets show 95%-confidence intervals. Statistically significant p-values are marked in bold
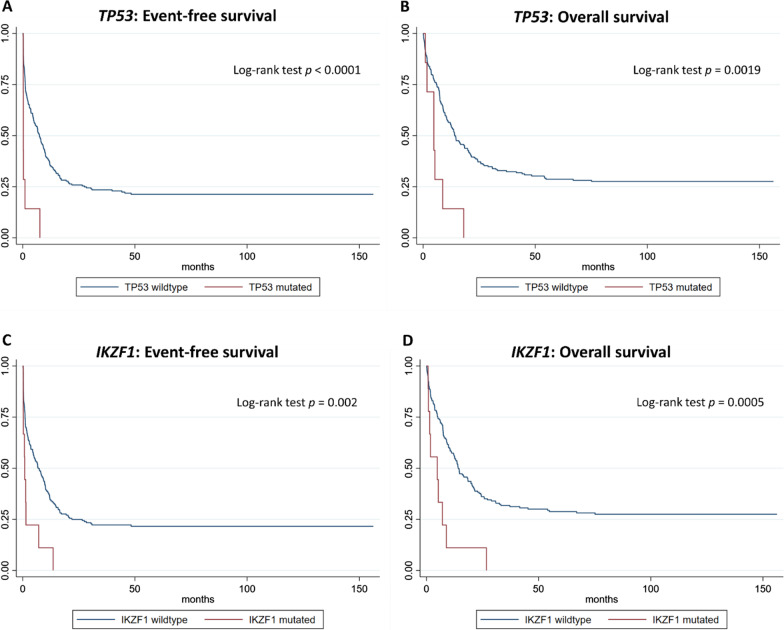
For patients with EM AML (both clinical and histological), there were no significant differences regarding survival between either patients harboring normal or complex aberrant karyotypes. With respect to the impact of molecular genetics on survival in patients with EM AML, although mutations of TP53 (7/225, 3.1%) and IKZF1 (9/225, 4.0%) were rare, both alterations were significantly associated with decreased EFS and OS. In univariable analysis, patients with EM AML and mutated TP53 compared to EM AML patients with wild-type TP53 had decreased median EFS (0.26 months [95% CI: 0.23–0.97] vs. 7.43 months [95% CI: 5.00–9.40], HR: 4.77 [95% CI: 2.20–10.35], Cox regression p < 0.001, Fig. 4A) and OS (4.80 months [95% CI: 0.99–8.74] vs. 14.10 months [95% CI: 11.21–19.86], HR: 3.16 [95% CI: 1.47–6.82], Cox regression p = 0.003, Fig. 4B). In a multivariable analysis adjusting for age and ELN2017 risk groups, TP53 remained an independent marker of reduced EFS (HR: 4.45 [95% CI: 1.94–10.20], Cox regression p < 0.001, Additional file 1: Table S4) and OS (HR: 2.48 [95% CI: 1.11–5.52], Cox regression p = 0.026 Additional file 1: Table S5). Reduced median EFS (0.85 months [95%-CI: 0.23–7.10] vs. 6.84 months [95%-CI: 4.67–9.70], HR: 2.77 [95%-CI: 1.40–5.47], Cox regression p = 0.003, Fig. 4C) and OS (4.78 months [95%-CI: 0.72–8.84] vs. 14.10 months [95%-CI:12.06–19.86], HR: 3.18 [95%-CI: 1.60–6.30], Cox regression p = 0.001, Fig. 4D) was also found for EM AML patients with mutated IKZF1 compared to wildtype EM AML patients. Again, multivariable analysis adjusting for age and ELN2017 risk groups revealed mutated IKZF1 as an independent marker of reduced EFS (HR: 2.05 [95%-CI: 1.02–4.13], Cox regression p = 0.044, Additional file 1: Table S6) and OS (HR: 2.63 [95%-CI: 1.29–5.39], Cox regression p = 0.008, Additional file 1: table S7). No significant differences in survival times of EM AML patients were found for mutations of NPM1, FLT3-ITD, PTPN11, ASXL1, RUNX1, or CEBPA.
Fig. 4.
Risk factors in acute myeloid leukemia with extramedullary manifestations. Among AML patients bearing extramedullary manifestations (EM) mutations of TP53 and IKZF1 were rare (7/225 [3.1%] and 9/225 [4.0%], respectively). However, EM AML patients with TP53 mutations showed significantly decreased event-free (A) and overall survival (B). Likewise, EM AML patients with IKZF1 mutations also showed significantly decreased event-free (C) and overall survival (D)
In our cohort, 573 (36.20%) of patients received an allogeneic hematopoietic cell transplantation, of whom 66 (11.52%) had EM AML. In a post-transplant setting, relapse rates did not differ between patients with and without EM AML (OR: 1.00 [95%-CI: 0.60–1.68], p = 0.992]. In contrast to the overall patient cohort, survival rates of patients harboring EM AML compared to those who did not present with EM did not differ (Table 4).
Table 4.
Survival times of patients with or without extramedullary manifestations who received allogeneic hematopoietic stem cell transplantation
| Survival times of patients who received HCT | EM AML | Non-EM AML | Hazard ratio | Cox regression p-value |
|---|---|---|---|---|
| Event-free survival | 9.8 [6.7–14.8] | 10.7 [9.1–13.2] | 0.98 [0.73–1.33] | 0.910 |
| Relapse-free survival | 13.0 [8.2–37.5] | 16.3 [12.7–20.0] | 0.99 [0.72–1.38] | 0.975 |
| Overall survival | 30.8 [18.2–54.2] | 72.1 [48.7–103.7] | 1.29 [0.93–1.80] | 0.130 |
Survival times for AML patients with and without extramedullary manifestations (EM) that received allogeneic hematopoietic cell transplantation (HCT, n = 573, 36.20%) are shown. Survival times in months. Cox-proportional hazard models were used to obtain univariable hazard ratios. Brackets show 95%-confidence intervals
Further, we analyzed patients harboring EM by ELN2017 subgroup with regard to the impact of HCT. For EM AML patients in the ELN2017 favorable risk group, we found no significant difference between patients who received or who did not receive HCT with respect to EFS, RFS, and OS. For EM AML patients in the ELN2017 intermediate risk group, RFS did not differ between patients who received or did not receive HCT. However, EFS (median 9.1 vs. 3.6, HR: 0.55 [95%-CI: 0.32–0.98], Cox regression p = 0.043) and OS (median 44.1 vs. 7.5 months, HR: 0.40 [95%-CI: 0.21–0.77], Cox regression p = 0.006) were significantly prolonged for ELN intermediate EM AML patients who received HCT. Finally for EM AML patients in the ELN2017 adverse risk group, again both EFS (median 7.4 vs. 1.2 months, HR: 0.48 [95%-CI: 0.27–0.85], Cox regression p = 0.013) and OS (median 21.0 vs. 7.1 months, HR: 0.31 [95%-CI: 0.17–0.58], Cox regression p < 0.001) were significantly longer for patients who received HCT compared to patients who did not receive HCT while RFS did not differ. Table 5 summarizes the impact of HCT on survival times for EM AML patients according to ELN2017 risk groups.
Table 5.
Impact of hematopoietic stem cell transplantation in for AML patients with extramedullary manifestations in ELN2017 groups
| Survival times of EM AML patients according to ELN2017 groups | HCT | no HCT | Hazard ratio | Cox regression p-value |
|---|---|---|---|---|
| ELN2017 favorable | n = 20 | n = 61 | ||
| Event-free survival | 13.5 [4.8-n.r.] | 12.6 [7.8–27.5] | 0.86 [0.45–1.65] | 0.659 |
| Relapse-free survival | 29.0 [7.5-n.r.] | 43.6 [12.6-n.r.] | 1.35 [0.66–2.78] | 0.410 |
| Overall survival | 54.0 [11.0-n.r.] | 27.2 [13.6-n.r.] | 0.81 [0.41–1.59] | 0.537 |
| ELN2017 intermediate | n = 22 | n = 55 | ||
| Event-free survival | 9.1 [3.7–44.1] | 3.6 [1.8–7.5] | 0.55 [0.32–0.98] | 0.043 |
| Relapse-free survival | 10.8 [4.6-n.r.] | 8.4 [2.1–14.4] | 0.69 [0.35–1.35] | 0.277 |
| Overall survival | 44.1 [9.1-n.r.] | 7.5 [3.6–13.1] | 0.40 [0.21–0.77] | 0.006 |
| ELN2017 adverse | n = 23 | n = 33 | ||
| Event-free survival | 7.4 [1.1–13.6] | 1.2 [0.3–3.0] | 0.48 [0.27–0.85] | 0.013 |
| Relapse-free survival | 11.6 [4.2–44.0] | 6.0 [2.3–9.8] | 0.51 [0.24–1.08] | 0.077 |
| Overall survival | 21.0 [13.6–48.4] | 7.1 [3.1–9.2] | 0.31 [0.17–0.58] | < 0.001 |
Survival times for AML patients harboring extramedullary manifestations (EM) are compared for patients receiving or not receiving hematopoietic stem cell transplantation (HCT). Survival times in months. Cox-proportional hazard models were used to obtain univariable hazard ratios. Brackets show 95%-confidence intervals. n.r. = not reached (due to small sample sizes and thus limited numbers of events within the ELN subgroups of EM AML patients who underwent HCT not all upper confidence interval limits can be estimated)
Discussion
We analyzed a large cohort of newly diagnosed AML patients, 14% of whom harbored EM based on clinical and/or histological diagnostics. Regarding clinical presentation, AML with EM was associated with significantly higher WBC, PBB, BMB and LDH at diagnosis. With the exception of two patients, all EM AML patients had concurrent bone marrow involvement. An increase in bone marrow infiltration as well as an increase in peripheral blood involvement increased the odds of EM. However, it has to be noted that our analysis likely underestimates the incidence of myeloid sarcoma, i. e. EM without infiltration of the bone marrow, as these cases were not eligible to be included in the four previous clinical trials pooled for this analysis and the cases stem from the SAL bioregistry. Previous reports have suggested a higher incidence of EM AML in FAB-M2, -M4, and -M5 [22]. In our sample, EM were significantly more frequent in patients with FAB-M5a and FAB-M5b while FAB-M2, -M6, and -4 rarely presented with EM.
For cytogenetics, previous reports in smaller cohorts and case series have linked the occurrence of EM AML with cytogenetic aberrations like t(8;21) [23, 24], trisomy 8 [25, 26] and inv [16, 27]. In our analysis, we did not find a statistically significant association between EM AML and either t(8;21) or inv [16] and for trisomy 8 there was even a trend for a lower prevalence of EM AML. Regarding molecular genetics, mutations of NPM1 and FLT3-ITD have been associated with EM AML. For NPM1, Ovcharenko et al. [28] report 13 out of 15 EM AML patients to harbor mutated NPM1 in a cohort of 89 patients with AML, Falini et al. [29] reported mutated NPM1 in 14% of 181 EM AML samples, and Döhner et al. [30] reported a correlation of mutated NPM1 and gingival hyperplasia. Ansari-Lari et al. [31] also identified FLT3-ITD in 15% of EM AML samples. Further, recent studies employing next-generation sequencing described the presence of mutations of KIT, WT1, TET1, ASXL1, SF3B1 and EZH2 [32] as well as NPM1, NRAS, and DNMT3A [33] in EM AML. In line with these previous findings, we found significantly increased odds for the presence of EM AML for mutations of NPM1 and FLT3-ITD while higher FLT3-ITD ratio was associated with higher odds of EM AML. Additionally, we found mutations of PTPN11 to be significantly associated with the presence of EM AML while the odds for EM AML were significantly decreased in IDH2- or CEBPA-mutated AML. PTPN11 has recently been described as an independent marker of poor outcome in AML [34, 35] and has been associated with EM [36]. PTPN11 encodes for the Src homology region 2 domain-containing phosphatase-2 (SHP-2) which functions as a signal enhancer for cell growth and differentiation downstream of numerous intracellular pathways including RAS/ERK/MAPK, JAK/STAT as well as PI3K/AKT and FLT3 signaling therefore playing a critical role in leukemogenesis [37–41]. However, its role in the formation of EM is insufficiently defined and further investigations are warranted to shed light on the mechanisms of how disrupted SHP2 signaling drives EM formation. While mutations of CEBPA were associated with significantly decreased odds of EM AML in our cohort, this effect was not seen individually in biallelic mutations of CEBPA, CEBPA-TAD or CEBPA-bZIP, rendering the mechanism unclear and thereby calling for more detailed investigations as subtypes of CEBPA have been reported to show differences in clinical outcomes [18].
With respect to clinical outcome, the impact of EM in AML is still controversial. In a retrospective analysis of 3240 AML patients, Ganzel et al. [42] found 23.7% to bear EM, however EM status was not associated with differences in survival between EM and non-EM AML. Fernandez et al. [43] found no significant associations between CR rate and disease-free survival for EM status. Accordingly, Agis et al. [44] reported no significant associations between leukemia cutis and survival in 381 AML patients and Ganzel et al. [45] did not find a difference in outcome for AML patients with CNS involvement. In contrast, in our analysis we found EM to be an independent marker of poor outcome. EM were associated with significantly decreased odds of achieving CR with standard intensive therapy while the odds of ED30 were significantly increased. Although EFS and RFS did not differ between patients with EM AML and non-EM AML when all patients including clinical and histological diagnosis of EM where investigated (Fig. 2A–C), we found OS to be significantly decreased. This finding was confirmed in multivariable analysis including age and ELN2017 risk categories. Interestingly, when we excluded patients for whom EM has been only diagnosed clinically and only included patients for whom EM has been confirmed by histology, not only was EFS also affected, but the effect on decreased OS became even more pronounced (Fig. 2D–F). For these 38 patients compared to non-EM patients (and excluding patients for whom only clinical diagnosis of EM was available), we found significantly decreased CR rates as well as significantly decreased EFS and OS both in uni- and multivariable analysis. Strikingly, by excluding patients with only clinical diagnosis of EM AML, effect sizes for decreased CR, EFS and OS became notably larger compared to our previous analysis when clinical and histological EM AML were combined suggesting that patients in the clinical EM AML group may have been false positive (possibly patients with gingival hyperplasia). Another possibility could be reduced salvageability for patients bearing EM resulting in comparable EFS but worse OS, however, this is speculative as data on response to salvage treatments except for HCT are not available. For the majority of our cohort, EM status was determined clinically at the discretion of the treating physician (which includes cases of gingival hyperplasia) and only in 38 patients biomaterial from the affected EM sites was available for histopathologic assessment. It is therefore conceivable that there were also patients with EM that have not been detected by clinical judgement alone. This discrepancy between histologically confirmed and clinically diagnosed EM could be an explanation as to why several previous reports showed discrepant rates of EM in AML and did not identify a difference in outcome for EM AML [30, 42, 43]. Therefore, the dilemma of detecting EM in AML is twofold with a considerable margin of error on both sides of the spectrum: On the one hand, in both clinical trials (including ours) the frequency of EM is likely underestimated due to insufficient screening via imaging such as PET and insufficient histological confirmation of potential EM sites which likely contributes to a substantial false negative rate. On the other hand, clinical diagnosis of EM is subjective and often performed (as in our study) at the discretion of the treating physician. This is why clinical symptoms can be misinterpreted as EM if no histological confirmation is obtained. This can potentially lead to false positives. In our analysis, clinical diagnosis of EM was followed by assessment of regression of these lesions after induction therapy according to ELN2017 recommendations for CR assessment and the label ‘CR achieved’ was only given for patients with complete regression of any clinically suspected EM lesions. However, this does not fully exclude potential false positives as misinterpreted lesions may have regressed due to other causes. Hence, it seems reasonable for future studies to focus on histologically confirmed cases of EM AML. However, due to potential clinical complications involved in taking a biopsy in AML patients with increased risk of bleeding acquiring tissue for histopathological confirmation of EM will likely not be possible in routine practice and even in the context of clinical trials it has to be evaluated on a case-by-case basis as patient safety outweighs potential knowledge gains. Given current limitations in detection mechanisms of EM via imaging or biopsy in addition to medical and ethical restrictions regarding a cost-to-benefit wager, it is questionable whether a fully representative study of the distribution of EM between sites with histological confirmation is actually obtainable given that not all suspected sites in all patients can be sampled and examined histologically due to safety reasons. The difference in outcomes could stem from the discrepancy between CR in the bone marrow while extramedullary sites may persist as can be shown by 18FDG-PET [9] and thus drive relapse. Hence, these patients constitute a special group at risk of undertreatment as they are falsely considered to having achieved CR while they actually could benefit from therapy intensification due to refractory EM that could only be detected by including more rigorous screening via imaging upon treatment response assessment. While for our cohort no data for measurable residual disease (MRD) was available, a comparative investigation of peripheral blood MRD levels for patients with or without EM in AML could provide additional insights for relapse monitoring in cases with bone marrow CR. Due to the rarity of EM AML, studies on risk factors for patients with EM are scarce. Regarding risk stratification for EM AML, Ullmann et al. reported OS to be associated with certain cytogenetic aberrations [10], while no such associations were found in our sample. Nevertheless, in univariable and multivariable analysis adjusting for age and ELN2017 risk category, we found both TP53 and IKZF1 mutations to be independently associated with reduced EFS and OS. While mutated TP53 is an established marker of adverse risk in AML in general [46], the role of IKZF1 alterations is far less well understood in AML than it is in acute lymphoblastic leukemia [47, 48]. Interestingly, IKZF1-alterations have been recently identified in patients with blastic plasmocytoid dendritic cell neoplasms, a rare hematological disease associated with cutaneous and solid organ infiltration and poor prognosis [49]. Further investigation is warranted to illuminate its role in both medullary and extramedullary disease. Consensually, patients with EM AML and high-risk cytogenetics are candidates for allogeneic HCT [50] and relapse rates as well as post-transplant survival have been reported not to differ [5, 51–53]. In line with these findings, in our analysis relapse rates as well as median EFS, RFS and OS did not differ between patients with or without EM who received allogeneic HCT. This is of special interest, as the existence of sanctuary sites and the potential risk of immune escape and EM has been described in patients with AML relapsing after allogeneic HCT [54]. Possibly, the choice of radiotherapy as conditioning therapy may be indicated in patients with a history of EM [4]. Hence, allogeneic HCT may be considered in patients with EM AML in the absence of other risk-defining cytogenetic or molecular markers.
Conclusion
We analyzed a large cohort of AML patients according to the molecular and cytogenetic profiles of EM AML and its impact on survival. We found EM AML to be significantly associated with AML-M5 as well as mutations of NPM1, PTPN11 and FLT3-ITD while it was less frequent in AML with mutated IDH2 or CEBPA. For patients harboring EM AML, mutations in TP53 and IKZF1 were found to be independently associated with poor outcome. In multivariable analysis, EM represented an independent marker of reduced OS. However, survival did not differ between patients with or without EM who received HCT suggesting an important role of allogeneic transplantation in the management of EM AML.
Supplementary Information
Additional file 1: Impact of continuous variables on odds of EM manifestation and multivariable analysis for outcome.
Acknowledgements
This study was carried out under the auspices of the German Study Alliance Leukemia patient registry. We thank all associated clinicians, laboratories, nurses and especially the patients for their valuable contributions.
Abbreviations
- AML
Acute myeloid leukemia
- sAML
Secondary acute myeloid leukemia
- tAML
Therapy-associated acute myeloid leukemia
- BMB
Bone marrow blast count
- EFS
Event-free survival
- EM
Extramedullary manifestations
- FISH
Fluorescence in situ hybridization
- HCT
Hematopoietic cell transplantation
- MRD
Measurable residual disease
- PBB
Peripheral blood blast count
- RFS
Relapse-free survival
- OS
Overall survival
- WBC
White blood cell count
Author contributions
J-NE and JMM designed the study. J-NE performed the statistical analysis and wrote the draft. CT and SS performed the molecular diagnostics. All authors contributed patient samples, analyzed and interpreted the data, and provided critical scientific insights. All authors revised the manuscript and approved its final version.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The investigation was carried out under the auspices of the SAL registry and received approval of the Institutional Review Board of the Technical University Dresden (EK 98032010). Written informed consent was obtained from all participants according to the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Yilmaz AF, Saydam G, Sahin F, Baran Y. Granulocytic sarcoma: a systematic review. Am J Blood Res. 2013;3(4):265–270. [PMC free article] [PubMed] [Google Scholar]
- 3.King A. Case of chloroma. Mon J Med Sci. 1853;8(44):97. [Google Scholar]
- 4.Bakst RL, Tallman MS, Douer D, Yahalom J. How I treat extramedullary acute myeloid leukemia. Blood. 2011;118(14):3785–3793. doi: 10.1182/blood-2011-04-347229. [DOI] [PubMed] [Google Scholar]
- 5.Goyal G, Bartley AC, Patnaik MM, Litzow MR, Al-Kali A, Go RS. Clinical features and outcomes of extramedullary myeloid sarcoma in the United States: analysis using a national data set. Blood Cancer J. 2017;7(8):e592–e592. doi: 10.1038/bcj.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiernik PH, Serpick AA. Granulocytic sarcoma (chloroma) Blood. 1970;35(3):361–369. doi: 10.1182/blood.V35.3.361.361. [DOI] [PubMed] [Google Scholar]
- 7.Liu PI, Ishimaru T, McGregor DH, Okada H, Steer A. Autopsy study of granulocytic sarcoma (chloroma) in patients with myelogenous leukemia, Hiroshima-Nagasaki 1949–1969. Cancer. 1973;31(4):948–955. doi: 10.1002/1097-0142(197304)31:4<948::AID-CNCR2820310428>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 8.Avni B, Koren-Michowitz M. Myeloid sarcoma: current approach and therapeutic options. Ther Adv Hematol. 2011;2(5):309. doi: 10.1177/2040620711410774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stölzel F, Lüer T, Löck S, Parmentier S, Kuithan F, Kramer M, et al. The prevalence of extramedullary acute myeloid leukemia detected by 18FDG-PET/CT: final results from the prospective PETAML trial. Haematologica. 2020;105(6):1552. doi: 10.3324/haematol.2019.223032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullman DI, Dorn D, Jones JA, Fasciano D, Ping Z, Kanakis C, et al. Clinicopathological and molecular characteristics of extramedullary acute myeloid leukaemia. Histopathology. 2019;75(2):185–192. doi: 10.1111/his.13864. [DOI] [PubMed] [Google Scholar]
- 11.Solh M, Solomon S, Morris L, Holland K, Bashey A. Extramedullary acute myelogenous leukemia. Blood Rev. 2016;30(5):333–339. doi: 10.1016/j.blre.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Röllig C, Thiede C, Gramatzki M, Aulitzky W, Bodenstein H, Bornhäuser M, et al. A novel prognostic model in elderly patients with acute myeloid leukemia: results of 909 patients entered into the prospective AML96 trial. Blood. 2010;116(6):971–978. doi: 10.1182/blood-2010-01-267302. [DOI] [PubMed] [Google Scholar]
- 13.Schaich M, Parmentier S, Kramer M, Illmer T, Stölzel F, Röllig C, et al. High-dose cytarabine consolidation with or without additional amsacrine and mitoxantrone in acute myeloid leukemia: results of the prospective randomized AML2003 trial. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31(17):2094–2102. doi: 10.1200/JCO.2012.46.4743. [DOI] [PubMed] [Google Scholar]
- 14.Röllig C, Kramer M, Gabrecht M, Hänel M, Herbst R, Kaiser U, et al. Intermediate-dose cytarabine plus mitoxantrone versus standard-dose cytarabine plus daunorubicin for acute myeloid leukemia in elderly patients. Ann Oncol Off J Eur Soc Med Oncol. 2018;29(4):973–8. [DOI] [PubMed]
- 15.Röllig C, Serve H, Hüttmann A, Noppeney R, Müller-Tidow C, Krug U, et al. Addition of sorafenib versus placebo to standard therapy in patients aged 60 years or younger with newly diagnosed acute myeloid leukaemia (SORAML): a multicentre, phase 2, randomised controlled trial. Lancet Oncol. 2015;16(16):1691–1699. doi: 10.1016/S1470-2045(15)00362-9. [DOI] [PubMed] [Google Scholar]
- 16.Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99(12):4326–4335. doi: 10.1182/blood.V99.12.4326. [DOI] [PubMed] [Google Scholar]
- 17.Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107(10):4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 18.Taube F, Georgi JA, Kramer M, Stasik S, Middeke JM, Röllig C, et al. CEBPA mutations in 4708 patients with acute myeloid leukemia: differential impact of bZIP and TAD mutations on outcome. Blood. 2022;139(1):87–103. doi: 10.1182/blood.2020009680. [DOI] [PubMed] [Google Scholar]
- 19.Stasik S, Schuster C, Ortlepp C, Platzbecker U, Bornhäuser M, Schetelig J, et al. An optimized targeted Next-Generation Sequencing approach for sensitive detection of single nucleotide variants. Biomol Detect Quantif. 2018;15:6–12. doi: 10.1016/j.bdq.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebhard C, Glatz D, Schwarzfischer L, Wimmer J, Stasik S, Nuetzel M, et al. Profiling of aberrant DNA methylation in acute myeloid leukemia reveals subclasses of CG-rich regions with epigenetic or genetic association. Leukemia. 2019;33(1):26–36. doi: 10.1038/s41375-018-0165-2. [DOI] [PubMed] [Google Scholar]
- 21.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33(4):451–8. [DOI] [PubMed]
- 22.Byrd JC, Edenfield WJ, Shields DJ, Dawson NA. Extramedullary myeloid cell tumors in acute nonlymphocytic leukemia: a clinical review. J Clin Oncol. 1995;13(7):1800–1816. doi: 10.1200/JCO.1995.13.7.1800. [DOI] [PubMed] [Google Scholar]
- 23.Schwyzer R, Sherman GG, Cohn RJ, Poole JE, Willem P. Granulocytic sarcoma in children with acute myeloblastic leukemia and t(8;21) Med Pediatr Oncol. 1998;31(3):144–149. doi: 10.1002/(SICI)1096-911X(199809)31:3<144::AID-MPO3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 24.Tallman MS, Hakimian D, Shaw JM, Lissner GS, Russell EJ, Variakojis D. Granulocytic sarcoma is associated with the 8;21 translocation in acute myeloid leukemia. J Clin Oncol Off J Am Soc Clin Oncol. 1993;11(4):690–697. doi: 10.1200/JCO.1993.11.4.690. [DOI] [PubMed] [Google Scholar]
- 25.Sen F, Zhang XX, Prieto VG, Shea CR, Qumsiyeh MB. Increased incidence of trisomy 8 in acute myeloid leukemia with skin infiltration (leukemia cutis) Diagn Mol Pathol Am J Surg Pathol Part B. 2000;9(4):190–194. doi: 10.1097/00019606-200012000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Shvartsbeyn M, Meehan SM, Gu P, Nierodzik ML, Perle MA. Trisomy 8 in myeloid leukemia cutis confirmed by fluorescence in situ hybridization analysis. J Cutan Pathol. 2012;39(11):1026–1029. doi: 10.1111/j.1600-0560.2012.01981.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X-H, Zhang R, Li Y. Granulocytic sarcoma of abdomen in acute myeloid leukemia patient with inv(16) and t(6;17) abnormal chromosome: case report and review of literature. Leuk Res. 2010;34(7):958–961. doi: 10.1016/j.leukres.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Ovcharenko D, Stölzel F, Poitz D, Fierro F, Schaich M, Neubauer A, et al. miR-10a overexpression is associated with NPM1 mutations and MDM4 downregulation in intermediate-risk acute myeloid leukemia. Exp Hematol. 2011;39(10):1030–1042.e7. doi: 10.1016/j.exphem.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Falini B, Lenze D, Hasserjian R, Coupland S, Jaehne D, Soupir C, et al. Cytoplasmic mutated nucleophosmin (NPM) defines the molecular status of a significant fraction of myeloid sarcomas. Leukemia. 2007;21(7):1566–1570. doi: 10.1038/sj.leu.2404699. [DOI] [PubMed] [Google Scholar]
- 30.Döhner K, Schlenk RF, Habdank M, Scholl C, Rücker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106(12):3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 31.Ansari-Lari MA, Yang C-F, Tinawi-Aljundi R, Cooper L, Long P, Allan RH, et al. FLT3 mutations in myeloid sarcoma. Br J Haematol. 2004;126(6):785–791. doi: 10.1111/j.1365-2141.2004.05124.x. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Stölzel F, Onel K, Sukhanova M, Mirza MK, Yap KL, et al. Next-generation sequencing reveals clinically actionable molecular markers in myeloid sarcoma. Leukemia. 2015;29(10):2113–2116. doi: 10.1038/leu.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kashofer K, Gornicec M, Lind K, Caraffini V, Schauer S, Beham-Schmid C, et al. Detection of prognostically relevant mutations and translocations in myeloid sarcoma by next generation sequencing. Leuk Lymphoma. 2018;59(2):501. doi: 10.1080/10428194.2017.1339879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stasik S, Eckardt J-N, Kramer M, Röllig C, Krämer A, Scholl S, et al. Impact of PTPN11 mutations on clinical outcome analyzed in 1529 patients with acute myeloid leukemia. Blood Adv. 2021;5(17):3279–3289. doi: 10.1182/bloodadvances.2021004631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alfayez M, Issa GC, Patel KP, Wang F, Wang X, Short NJ, et al. The Clinical impact of PTPN11 mutations in adults with acute myeloid leukemia. Leukemia. 2021;35(3):691–700. doi: 10.1038/s41375-020-0920-z. [DOI] [PubMed] [Google Scholar]
- 36.Fobare S, Kohlschmidt J, Ozer HG, Mrózek K, Nicolet D, Mims AS, et al. Molecular associations, clinical, and prognostic implications of PTPN11 mutations in acute myeloid leukemia (Alliance). Blood Adv. 2021;bloodadvances.2021006242. [DOI] [PMC free article] [PubMed]
- 37.Xu D, Qu C-K. Protein tyrosine phosphatases in the JAK/STAT pathway. Front Biosci J Virtual Libr. 2008;1(13):4925–4932. doi: 10.2741/3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandey R, Saxena M, Kapur R. Role of SHP2 in Hematopoiesis and Leukemogenesis. Curr Opin Hematol. 2017;24(4):307–313. doi: 10.1097/MOH.0000000000000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nabinger SC, Li XJ, Ramdas B, He Y, Zhang X, Zeng L, et al. The protein tyrosine phosphatase, Shp2, positively contributes to FLT3-ITD-induced hematopoietic progenitor hyperproliferation and malignant disease in vivo. Leukemia. 2013;27(2):398–408. doi: 10.1038/leu.2012.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivins Zito C, Kontaridis MI, Fornaro M, Feng G-S, Bennett AM. SHP-2 regulates the phosphatidylinositide 3’-kinase/Akt pathway and suppresses caspase 3-mediated apoptosis. J Cell Physiol. 2004;199(2):227–236. doi: 10.1002/jcp.10446. [DOI] [PubMed] [Google Scholar]
- 41.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 42.Ganzel C, Manola J, Douer D, Rowe JM, Fernandez HF, Paietta EM, et al. Extramedullary disease in adult acute myeloid leukemia is common but lacks independent significance: analysis of patients in ECOG-ACRIN cancer research group trials, 1980–2008. J Clin Oncol Off J Am Soc Clin Oncol. 2016;34(29):3544–3553. doi: 10.1200/JCO.2016.67.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez HF, Sun Z, Litzow MR, Luger SM, Paietta E, Racevskis J, et al. Extramedullary acute myeloid leukemia presenting in young adults demonstrates sensitivity to high-dose anthracycline: a subset analysis from ECOG-ACRIN 1900. Haematologica. 2019;104(4):e147. doi: 10.3324/haematol.2018.197277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agis H, Weltermann A, Fonatsch C, Haas O, Mitterbauer G, Müllauer L, et al. A comparative study on demographic, hematological, and cytogenetic findings and prognosis in acute myeloid leukemia with and without leukemia cutis. Ann Hematol. 2002;81(2):90–95. doi: 10.1007/s00277-001-0412-9. [DOI] [PubMed] [Google Scholar]
- 45.Ganzel C, Lee J-W, Fernandez HF, Paietta EM, Luger SM, Lazarus HM, et al. CNS involvement in AML at diagnosis is rare and does not affect response or survival: data from 11 ECOG-ACRIN trials. Blood Adv. 2021;5(22):4560–4568. doi: 10.1182/bloodadvances.2021004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vairy S, Tran TH. IKZF1 alterations in acute lymphoblastic leukemia: The good, the bad and the ugly. Blood Rev. 2020;44:100677. doi: 10.1016/j.blre.2020.100677. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Zhang X, Li X, Lv Y, Zhu Y, Wang J, et al. The specific distribution pattern of IKZF1 mutation in acute myeloid leukemia. J Hematol OncolJ Hematol Oncol. 2020;13(1):140. doi: 10.1186/s13045-020-00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastidas Torres AN, Cats D, Mei H, Fanoni D, Gliozzo J, Corti L, et al. Whole-genome analysis uncovers recurrent IKZF1 inactivation and aberrant cell adhesion in blastic plasmacytoid dendritic cell neoplasm. Genes Chromosomes Cancer. 2020;59(5):295–308. doi: 10.1002/gcc.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cornelissen JJ, Gratwohl A, Schlenk RF, Sierra J, Bornhäuser M, Juliusson G, et al. The European LeukemiaNet AML Working Party consensus statement on allogeneic HSCT for patients with AML in remission: an integrated-risk adapted approach. Nat Rev Clin Oncol. 2012;9(10):579–590. doi: 10.1038/nrclinonc.2012.150. [DOI] [PubMed] [Google Scholar]
- 51.Shem-Tov N, Saraceni F, Danylesko I, Shouval R, Yerushalmi R, Nagler A, et al. Isolated extramedullary relapse of acute leukemia after allogeneic stem cell transplantation: different kinetics and better prognosis than systemic relapse. Biol Blood Marrow Transpl J Am Soc Blood Marrow Transpl. 2017;23(7):1087–1094. doi: 10.1016/j.bbmt.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 52.Shan M, Lu Y, Yang M, Wang P, Lu S, Zhang L, et al. Characteristics and transplant outcome of myeloid sarcoma: a single-institute study. Int J Hematol. 2021;113(5):682–692. doi: 10.1007/s12185-021-03081-2. [DOI] [PubMed] [Google Scholar]
- 53.Sakaguchi H, Miyamura T, Tomizawa D, Taga T, Ishida H, Okamoto Y, et al. Effect of extramedullary disease on allogeneic hematopoietic cell transplantation for pediatric acute myeloid leukemia: a nationwide retrospective study. Bone Marrow Transplant. 2021;56(8):1859–1865. doi: 10.1038/s41409-021-01250-9. [DOI] [PubMed] [Google Scholar]
- 54.Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJ, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med. 2018;379(24):2330–2341. doi: 10.1056/NEJMoa1808777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Impact of continuous variables on odds of EM manifestation and multivariable analysis for outcome.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.