Abstract
Background
Yield-related traits including thousand grain weight (TGW), grain number per spike (GNS), grain width (GW), grain length (GL), plant height (PH), spike length (SL), and spikelet number per spike (SNS) are greatly associated with grain yield of wheat (Triticum aestivum L.). To detect quantitative trait loci (QTL) associated with them, 193 recombinant inbred lines derived from two elite winter wheat varieties Chuanmai42 and Chuanmai39 were employed to perform QTL mapping in six/eight environments.
Results
A total of 30 QTLs on chromosomes 1A, 1B, 1D, 2A, 2B, 2D, 3A, 4A, 5A, 5B, 6A, 6D, 7A, 7B and 7D were identified. Among them, six major QTLs QTgw.cib-6A.1, QTgw.cib-6A.2, QGw.cib-6A, QGl.cib-3A, QGl.cib-6A, and QSl.cib-2D explaining 5.96-23.75% of the phenotypic variance were detected in multi-environments and showed strong and stable effects on corresponding traits. Three QTL clusters on chromosomes 2D and 6A containing 10 QTLs were also detected, which showed significant pleiotropic effects on multiple traits. Additionally, three Kompetitive Allele Specific PCR (KASP) markers linked with five of these major QTLs were developed. Candidate genes of QTgw.cib-6A.1/QGl.cib-6A and QGl.cib-3A were analyzed based on the spatiotemporal expression patterns, gene annotation, and orthologous search.
Conclusions
Six major QTLs for TGW, GL, GW and SL were detected. Three KASP markers linked with five of these major QTLs were developed. These QTLs and KASP markers will be useful for elucidating the genetic architecture of grain yield and developing new wheat varieties with high and stable yield in wheat.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12863-022-01050-0.
Keywords: Wheat, Yield, Yield-related traits, Specific-locus amplified fragment (SLAF), Linkage analysis
Background
Common wheat (Triticum aestivum L.) is one of the three major crops worldwide and provides approximately 30% of global grain production and 20% of the calories consumed for humans [1]. Due to ongoing decrease of the global arable cultivated land area and increase of the population, the current rate of wheat yield increase will be insufficient to meet the future demand. Thus, breeding of high-yield wheat varieties to ensure future global food and nutrition security is an important target of the modern wheat breeding programs [2].
Wheat yield is a complex quantitative trait controlled by multiple genes and significantly influenced by interacting genetic and environmental factors [3, 4]. By contrast, yield components including thousand grain weight (TGW), grain number per spike (GNS), grain width (GW), grain length (GL), plant height (PH), spike length (SL) and spikelet number per spike (SNS) typically show higher heritability than that of the yield [5–7]. Therefore, targeting these traits and identifying the related genes or quantitative trait loci (QTL) is an important approach to improve grain yield potential in wheat.
The molecular cloning of genes associated with wheat yield is difficult owing to wheat’s huge and complicated genome. To date, only a few genes associated with grain yield have been cloned in wheat. For example, the application of semi-dwarfing genes Rht-B1b and Rht-D1b not only effectively improve the lodging resistance but also improve the harvest index, resulting in increasing yield since the 1970s [8–10]. The vernalization insensitive alleles of Vrn-1 (Vrn-A1, Vrn-B1, and Vrn-D1) shorten both the vegetative and the reproductive stages and have considerable impact on spike morphological traits [11, 12]. The grain-shape gene TasgD1 encoding a Ser/Thr protein kinase glycogen synthase kinase3 and independently control semispherical grain trait [13]. A jasmonic acid synthetic gene keto-acyl thiolase 2B was cloned in a TGW mutant, showing significant effects on TGW and GW [14]. Additionally, homologous cloning is an effective approach to characterize gene in wheat. As of today more than 20 genes related to yield have been isolated through homologous cloning approach, including WFZP, WAPO1, TaGW7, TaGW2, TaCKX6-D1, TaTGW6, TaGASR7, TaGL3 and TaGS-D1 et al [15–23].
Quantitative trait loci (QTL) mapping provides an effective approach to dissect the genetic architecture of complex quantitative traits. Over the past decades, numerous QTLs associated with yield or yield-related traits have been identified on all wheat chromosomes [3, 4, 11, 24–30]. For example, Rht8 located on chromosome 2DS was closely linked with marker xfdc53 and reduced plant height by 10% [31]; Rht25 on wheat chromosome arm 6AS showed pleiotropic effects on coleoptile length, heading date, SL, SNS and grain weight [32]. Two major QTLs for grain size and weight were detected on chromosome 4B, which together explained 46.3% of the phenotypic variance [33, 34]. Five stable QTLs for PH, SL and HD on chromosomes 1A, 2A, 2D and 6A were detected in an introgression line population [35]. Twelve major genomic regions with stable QTL controlling yield-related traits were detected on chromosomes 1B, 2A, 2B, 2D, 3A, 4A, 4B, 4D, 5A, 6A, and 7A [1]. However, among these QTLs reported previously, few of them were stably detected in multi–environments, which greatly restrict their potential utilization in marker-assisted selection (MAS) in breeding programs.
With the development of high-throughput sequencing technology, Single nucleotide polymorphisms (SNP) markers have been widely applied to construct high-density genetic maps for QTL mapping, due to their extensive and intensive distribution throughout genomes in many crop s[3, 36–38]. Specific-locus amplified fragment sequencing (SLAF-seq) was developed for economic and efficient high-throughput SNP discovery through restriction-site associated DNA tag sequencing (RAD-seq), which can provide abundant InDel and SNP markers to construct high-density genetic map [39–41].
In the present study, a high-resolution genetic map was constructed in a recombinant inbred line (RIL) population derived from two elite winter wheat varieties Chuanmai42 (CM42) and Chuanmai39 (CM39) based on SLAF-seq (Table S1, S2) [42]. Seven traits including TGW, GW, GL, PH, GNS, SL and SNS were assessed in multi-environments to detect potential major and stable QTL, which will lay out a foundation for further study on fine mapping and cloning of the underlying key genes for wheat yield.
Results
Phenotypic variation
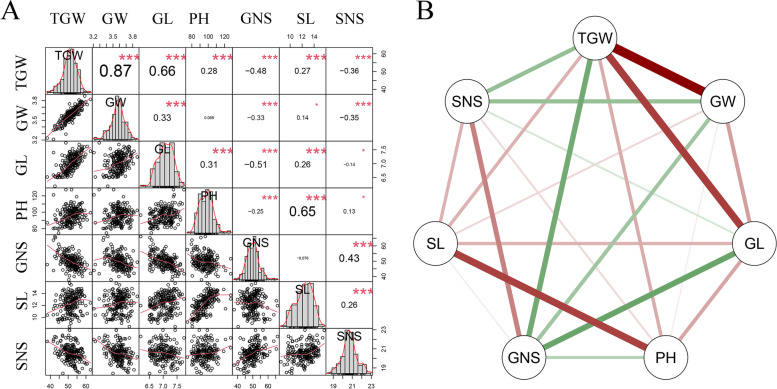
The phenotypic analysis showed that CM42 had higher trait values for TGW, GW, GL, GNS, PH and SL than those of CM39 in each of environments and the best linear unbiased prediction (BLUP) datasets (Table 1). In the RIL population, seven yield-related traits showed wide and significant variations in all environments and the BLUP datasets (Table 1). Of them, the TGW ranged from 20.81 to 72.7 gram (g), the GW ranged from 2.6 to 4.21 millimeter (mm), the GL ranged from 5.88 to 8.81 mm, the PH ranged from 65.08 to 148.3 centimeter (cm), the GNS ranged from 24 to 84.6, the SL ranged from 6.65 to 18.17 cm, and the SNS ranged from 15.83 to 27, respectively (Table 1). The BLUP datasets of all traits showed normal distributions in the RIL lines, which suggested polygenic inheritance of these traits (Fig. 1A). Additionally, the TGW, GL, PH, GNS and SL showed high across-environment broad-sense heritability of 0.54, 0.6, 0.91, 0.66 and 0.88, respectively (Table 1). Significant and positive correlations (P < 0.01) of the seven yield-related traits among all environments and the BLUP datasets were detected, which suggested that these traits were environmentally stable and mainly controlled by genetic factors (Table S3).
Table 1.
Phenotypic variation of the seven yield-related traits, including thousand grain weight (TGW), grain number per spike (GNS), grain width (GW), grain length (GL), plant height (PH), spike length (SL) and spikelet number per spike (SNS), for the parents and the CM42×CM39 RIL lines in different environments
| Traits | Environments | Parents | The CM42×CM39 RIL lines | |||||
|---|---|---|---|---|---|---|---|---|
| CM42 | CM39 | Range | Mean | SD | CV (%) | H2 | ||
| TGW | 2017SHF | 54 | 52.94 | 38.34-70.88 | 58.57 | 5.84 | 9.98 | 0.54 |
| 2017SHL | 50.64 | 41.83 | 20.81-68.14 | 43.76 | 9.17 | 20.95 | ||
| 2018SHF | 54.79 | 53.47 | 40.44-72.7 | 54.67 | 5.58 | 10.2 | ||
| 2018SHL | 53.06 | 51.29 | 37.89-67.33 | 54.51 | 5.57 | 10.22 | ||
| 2019SHF | 52.4 | 42.42 | 32.59-66.54 | 51.27 | 5.94 | 11.59 | ||
| 2019SHL | 51.05 | 47.38 | 23.4-62.74 | 46.9 | 6.22 | 13.27 | ||
| BLUP | 52.36 | 50.44 | 38.24-62.56 | 51.65 | 3.98 | 7.7 | ||
| GW | 2017SHF | 3.68 | 3.42 | 3.19-4.21 | 3.82 | 0.16 | 4.28 | 0.49 |
| 2017SHL | 3.54 | 3.31 | 2.6-4.01 | 3.38 | 0.29 | 8.57 | ||
| 2018SHF | 3.58 | 3.53 | 3.19-4.04 | 3.69 | 0.16 | 4.35 | ||
| 2018SHL | 3.63 | 3.61 | 3.15-3.96 | 3.65 | 0.15 | 4.14 | ||
| 2019SHF | 3.6 | 3.16 | 3-3.9 | 3.5 | 0.18 | 5.21 | ||
| 2019SHL | 3.56 | 3.49 | 2.84-3.99 | 3.49 | 0.19 | 5.37 | ||
| BLUP | 3.59 | 3.51 | 3.21-3.87 | 3.59 | 0.11 | 3.06 | ||
| GL | 2017SHF | 7.73 | 7.17 | 6.78-8.81 | 7.76 | 0.41 | 5.26 | 0.6 |
| 2017SHL | 6.95 | 6.53 | 5.94-7.89 | 6.86 | 0.37 | 5.39 | ||
| 2018SHF | 6.87 | 6.72 | 5.89-7.92 | 6.95 | 0.37 | 5.3 | ||
| 2018SHL | 7.64 | 6.55 | 5.88-7.81 | 6.85 | 0.37 | 5.45 | ||
| 2019SHF | 7.32 | 6.43 | 6-7.71 | 6.86 | 0.33 | 4.84 | ||
| 2019SHL | 7.22 | 6.67 | 6.03-7.71 | 6.94 | 0.36 | 5.15 | ||
| BLUP | 7.27 | 6.98 | 6.19-7.75 | 7.04 | 0.3 | 4.26 | ||
| PH | 2016SHF | 90.34 | 89.5 | 66.5-120.3 | 91.53 | 9.5 | 10.38 | 0.91 |
| 2016SHL | 89.8 | 87.2 | 76-148.3 | 95.97 | 10.49 | 10.93 | ||
| 2017SHF | 97.67 | 96.33 | 81.33-143 | 103.3 | 10.65 | 10.31 | ||
| 2017SHL | 99 | 98.8 | 66.63-121.2 | 91.39 | 9.73 | 10.65 | ||
| 2018SHF | 91.7 | 87.08 | 65.08-131.9 | 93.9 | 11.82 | 12.59 | ||
| 2018SHL | 94.61 | 90 | 70.8-135.4 | 95.57 | 11.32 | 11.84 | ||
| 2019SHF | 90.05 | 85.9 | 69.45-126.8 | 98.74 | 9.89 | 10.02 | ||
| 2019SHL | 93.33 | 89.3 | 78.5-127.4 | 97.58 | 8.98 | 9.21 | ||
| BLUP | 93.24 | 91.91 | 74.65-127.5 | 96 | 9.14 | 9.52 | ||
| GNS | 2017SHF | 54 | 52 | 24-81.2 | 51.01 | 10.39 | 20.38 | 0.66 |
| 2017SHL | 44.5 | 43.6 | 26-77 | 41.94 | 8.08 | 19.27 | ||
| 2018SHF | 54.6 | 49.9 | 31.6-70.8 | 45.62 | 6.11 | 13.4 | ||
| 2018SHL | 54.5 | 54.1 | 35.3-70.8 | 52.07 | 7.18 | 13.78 | ||
| 2019SHF | 55.7 | 53.7 | 35.2-84.6 | 53.66 | 8.18 | 15.24 | ||
| 2019SHL | 56.5 | 56.2 | 35.5-75.8 | 53.77 | 7.07 | 13.15 | ||
| BLUP | 53.17 | 52.44 | 37.76-66.18 | 49.85 | 4.62 | 9.26 | ||
| SL | 2016SHF | 12.18 | 9.96 | 8.67-18 | 13.09 | 1.75 | 13.37 | 0.88 |
| 2016SHL | 12.1 | 9 | 6.65-14 | 10.53 | 1.61 | 15.33 | ||
| 2017SHF | 13.5 | 11.5 | 8.5-17.88 | 13.04 | 1.73 | 13.23 | ||
| 2017SHL | 13 | 11.5 | 8.33-17.67 | 12.93 | 1.88 | 14.51 | ||
| 2018SHF | 11.85 | 9.26 | 7.63-14.93 | 11.82 | 1.84 | 15.54 | ||
| 2018SHL | 13.02 | 10.9 | 7.55-15.7 | 11.3 | 1.72 | 15.18 | ||
| 2019SHF | 13.71 | 11.2 | 8.89-18.17 | 13.25 | 1.87 | 14.15 | ||
| 2019SHL | 12.4 | 10.5 | 8.5-16.3 | 12.51 | 1.56 | 12.51 | ||
| BLUP | 12.71 | 11.6 | 8.45-15.69 | 12.31 | 1.5 | 12.22 | ||
| SNS | 2017SHF | 18.6 | 19.6 | 16.2-25 | 19.58 | 1.39 | 7.08 | 0.4 |
| 2017SHL | 21.2 | 21.2 | 18-27 | 21.4 | 1.63 | 7.63 | ||
| 2018SHF | 21.9 | 21.5 | 17.7-24.5 | 21.66 | 1.13 | 5.2 | ||
| 2018SHL | 20.9 | 20.7 | 17.9-25.2 | 21.02 | 1.22 | 5.81 | ||
| 2019SHF | 21.7 | 21.2 | 17.9-25 | 21.29 | 1.2 | 5.63 | ||
| 2019SHL | 17.2 | 18.1 | 15.83-21.2 | 18.35 | 1.04 | 5.67 | ||
| BLUP | 20.3 | 20.35 | 18.42-22.96 | 20.55 | 0.84 | 4.1 | ||
SHF Shifang, SHL Shuangliu, BLUP best linear unbiased prediction, CV coefficient of variation, H2 broad-sense heritability
Fig. 1.
Phenotypic performances, distribution, and correlation coefficients of thousand grain weight (TGW), grain number per spike (GNS), grain width (GW), grain length (GL), plant height (PH), spike length (SL) and spikelet number per spike (SNS) in the CM42×CM39 RIL lines based on the BLUP datasets (A). B Visualization of correlations among investigated traits; Red and green lines represent positive and negative correlation, respectively; The line weight represent the size of correlation coefficient; *, ** and *** represent significant at P < 0.05, P < 0.01 and P < 0.001, respectively
Correlation analyses among different traits
The BLUP datasets of each trait was employed to assess their correlations in the CM42×CM39 RIL population. TGW had significantly positive correlation with GW, GL, PH and SL, and significantly negative correlation with GNS and SNS (P < 0.001) (Fig. 1). GW was significantly and positively correlated with GL (P < 0.001), weakly and positively correlated with SL (P < 0.05), significantly and negatively correlated with GNS and SNS (P < 0.001), and not correlated with PH, respectively (Fig. 1). GL had significantly positive correlation with PH and SL (P < 0.001), significantly negative correlation with GNS (P < 0.001), and weakly negative correlation with SNS (P < 0.05) (Fig. 1). Significantly positive correlations between PH and SL, GNS and SNS, and SL and SNS (P < 0.001), weakly positive correlations between PH and SNS (P < 0.05), significantly negative correlations between PH and GNS (P < 0.001), and no correlations between GNS and SL were detected, respectively (Fig. 1). Grain weight per spike (GWS) is comprised by TGW and GNS in wheat. Thus, we further analyzed the correlation between the seven yield-related traits and the GWS. The results showed that GWS was significantly positive and positively correlated with TGW, GW, GL, GNS, SNS and SL (P < 0.05), and no correlated with PH (Table S4).
QTL detection
Phenotypic data of the seven yield-related traits in each environment and the BLUP datasets were used for QTL detection, in which the BLUP datasets were treated as an additional environment. A total of 30 QTLs were identified in multi-environments and located on all chromosomes excepting 3B, 3D, 4B, 4D, 5D and 6B (Table 2).
Table 2.
Quantitative trait loci (QTLs) for thousand grain weight (TGW), grain number per spike (GNS), grain width (GW), grain length (GL), plant height (PH), spike length (SL) and spikelet number per spike (SNS) identified across multi-environments in the CM42×CM39 RIL population
| Trait | QTL | Env. | Chr. | Interval (cM) |
|---|---|---|---|---|
| TGW | QTgw.cib-6A.1 | 18SHF/18SHL/BLUP | 6A | 41.3-42.46 |
| QTgw.cib-6A.2 | 17SHF/18SHL/19SHF/19SHL/BLUP | 6A | 52.98-59.52 | |
| GW | QGw.cib-2A | 18SHF/19SHF | 2A | 14.86-17.08 |
| QGw.cib-2B.1 | 19SHF/19SHL | 2B | 39.6-43.07 | |
| QGw.cib-2B.2 | 17SHF/BLUP | 2B | 121.67-121.93 | |
| QGw.cib-5A | 17SHF/BLUP | 5A | 27.76-27.97 | |
| QGw.cib-6A | 17SHF/17SHL/18SHL/19SHF/19SHL/BLUP | 6A | 49.98-58.87 | |
| QGw.cib-7B | 18SHF/19SHL | 7B | 179.93-180.13 | |
| GL | QGl.cib-3A | 17SHF/17SHL/18SHL/19SHF/19SHL/BLUP | 3A | 64.7-66.41 |
| QGl.cib-5A.1 | 17SHL/BLUP | 5A | 3.46-7.55 | |
| QGl.cib-5A.2 | 18SHL/19SHF | 5A | 86.87-87.49 | |
| QGl.cib-6A | 17SHF/18SHF/18SHL/19SHF/19SHL/BLUP | 6A | 42.36-43.4 | |
| QGl.cib-6D | 18SHL/19SHF/BLUP | 6D | 76.06-83.69 | |
| QGl.cib-7D | 17SHF/17SHL/BLUP | 7D | 32.68-38.76 | |
| PH | QPh.cib-1A | 16SHF/17SHL/19SHF | 1A | 28.34-30.95 |
| QPh.cib-2D | 16SHF/17SHL/18SHF/18SHL/19SHF/BLUP | 2D | 1.48-5.16 | |
| QPh.cib-4A | 16SHF/17SHL | 4A | 82.78-83.05 | |
| QPh.cib-5A | 17SHF/19SHL/BLUP | 5A | 126.27-126.52 | |
| QPh.cib-5B | 16SHF/17SHL | 5B | 134.43-134.74 | |
| QPh.cib-6A | 16SHF/17SHL/19SHF | 6A | 54.61-54.76 | |
| GNS | QGns.cib-2D | 18SHF/19SHF/BLUP | 2D | 0-5.16 |
| QGns.cib-6A | 18SHL/19SHF/BLUP | 6A | 56.45-59.52 | |
| SL | QSl.cib-2D | 16SHF/16SHL/17SHF/17SHL/18SHF/18SHL/19SHF/19SHL/BLUP | 2D | 1.48-5.16 |
| QSl.cib-5A | 18SHL/19SHF/BLUP | 5A | 17.71-21.48 | |
| QSl.cib-5B | 16SHF/16SHL/17SHF/BLUP | 5B | 40.07-40.38 | |
| QSl.cib-6A | 19SHF/BLUP | 6A | 58.87-64.37 | |
| SNS | QSns.cib-1B | 17SHF/18SHL/19SHF/BLUP | 1B | 28.8-33.3 |
| QSns.cib-1D | 17SHL/BLUP | 1D | 146.67-148.82 | |
| QSns.cib-4A | 17SHF/18SHL/19SHF/BLUP | 4A | 72.98-81.71 | |
| QSns.cib-7A | 19SHF/19SHL | 7A | 81.76-85.42 |
| Trait | Flanking Markers | LOD | PVE(%) | Add |
|---|---|---|---|---|
| TGW | Marker87546-Marker87736 | 6.17/8.03/4.44 | 13.49/16.38/9.89 | -1.89/-2/-1.04 |
| Marker90290-Marker91587 | 9.48/7.95/7.27/10.52/9.62 | 20.39/16.68/15.31/20.51/23.75 | -2.58/-2.06/-2.36/-2.88/-1.65 | |
| GW | Marker26336-Marker26958 | 5.48/2.76 | 9.51/5.81 | 0.05/0.04 |
| Marker29502-Marker29525 | 2.66/4.09 | 5.46/6.62 | -0.04/-0.05 | |
| Marker34419-Marker34417 | 3.83/3.7 | 5.2/5.2 | -0.04/-0.03 | |
| Marker70243-Marker70216 | 3.91/4.73 | 5.29/6.72 | 0.04/0.03 | |
| Marker90210-Marker91133 | 13.09/4.95/8.93/4.02/5.8/14.53 | 19.87/8.92/19.17/8.6/10.1/23.31 | -0.08/-0.09/-0.07/-0.05/-0.06/-0.06 | |
| Marker111000-Marker110965 | 5.36/5.98 | 9.07/9.89 | -0.05/-0.06 | |
| GL | Marker40793-Marker40901 | 5.31/2.97/6.1/5.69/3.87/5.68 | 11.86/6.55/10.17/10.31/7.37/9.54 | -0.13/-0.08/-0.12/-0.1/-0.09/-0.09 |
| Marker69377-Marker69395 | 2.83/3.66 | 6.28/6.26 | -0.08/-0.07 | |
| Marker71923-Marker71919 | 3.79/3.82 | 6.13/6.76 | -0.09/-0.08 | |
| Marker87807-Marker87738 | 5.32/4.62/7.72/3.39/5.37/7.41 | 11.85/10.15/13.11/5.96/10.37/12.7 | -0.13/-0.12/-0.13/-0.08/-0.11/-0.1 | |
| Marker99119-Marker99140 | 4.17/5/3.17 | 6.95/8.98/5.17 | 0.1/0.1/0.06 | |
| Marker111521-Marker111597 | 3/4.64/6.19 | 6.63/11.34/10.49 | -0.09/-0.11/-0.09 | |
| PH | Marker5758-Marker6328 | 5.58/6.07/3.64 | 7.62/7.53/5.87 | 2.96/3.03/2.65 |
| Marker35344-Marker35422 | 3.42/4.31/3/4.7/3.14/2.56 | 4.54/5.23/6.73/9.38/5.03/6.2 | 2.29/2.53/3/3.72/2.46/2.31 | |
| Marker57956-Marker57959 | 4.76/5.71 | 6.43/7.05 | -2.73/-2.95 | |
| Marker72631-Marker72950 | 3.02/2.91/2.52 | 7.18/7.03/5.62 | -2.8/-2.32/-2.2 | |
| Marker83905-Marker83879 | 3.07/3.18 | 4.07/3.8 | 2.17/2.16 | |
| Marker90459-Marker90388 | 6.13/8.86/5.6 | 8.42/11.37/9.25 | -3.24/-3.88/-3.46 | |
| GNS | Marker35164-Marker35422 | 2.57/2.63/4.73 | 5.73/4.97/6.46 | -1.44/-1.93/-1.27 |
| Marker90628-Marker91587 | 3.35/3.83/4.91 | 7.73/7.46/6.56 | 2.07/2.46/1.33 | |
| SL | Marker35344-Marker35422 | 3.42/6.86/6.82/8.05/8.05/8.83/4.82/4.43/7.43 | 6.84/11.15/9.46/10.91/14.89/13.41/6.18/8.31/13.51 | 0.46/0.6/0.66/0.76/0.75/0.7/0.53/0.48/0.58 |
| Marker69427-Marker69525 | 2.59/6/2.61 | 3.47/7.8/4.22 | -0.36/-0.6/-0.32 | |
| Marker81580-Marker81513 | 2.99/3.06/2.87/2.51 | 5.96/4.76/3.79/4.04 | 0.43/0.39/0.42/0.32 | |
| Marker91133-Marker91933 | 4.62/3 | 5.9/5.63 | -0.54/-0.39 | |
| SNS | Marker15740-Marker17413 | 16.86/4.13/6.11/5.4 | 16.18/7.47/9.85/8.21 | 0.82/0.35/0.4/0.25 |
| Marker23471-Marker23475 | 5.65/4.49 | 8.39/6.77 | -0.5/-0.22 | |
| Marker57882-Marker57915 | 2.51/4.94/5.3/5.63 | 2.34/10.46/9.69/9.68 | -0.31/-0.41/-0.39/-0.27 | |
| Marker103527-Marker103903 | 3.25/4.47 | 5.06/8.18 | 0.28/0.3 |
PVE mean of phenotypic variation explained, LOD logarithm of the odd, Add additive effect (Positive values indicate that the alleles from CM39 increases the trait scores, and negative values indicate that the allele from CM42 increases the trait scores), BLUP best linear unbiased prediction, Chr. chromosome, Env. environment
For TGW, two QTLs were detected on chromosomes 6A. QTgw.cib-6A.1 was detected in two environments and the BLUP datasets, explaining 9.89-16.38% of the phenotypic variance. QTgw.cib-6A.2 was a major QTL detected in four environments and the BLUP datasets and explained 15.31-23.75% of the phenotypic variance. Alleles of CM42 for the two QTLs contributed to higher TGW (Table 2).
For GW, six QTLs were identified on chromosomes 2A, 2B, 5A, 6A and 7B. Of them, a major QTL QGw.cib-6A was identified in five environments and the BLUP datasets, explaining 8.6-23.31% of the GW variation. The allele of CM42 contributed positively to the GW. The rest five minor QTLs were identified in two environments and explained 5.2-9.89% of the GW variation. The favorable alleles of QGw.cib-2A and QGw.cib-5A were contributed by CM39, and that of QGw.cib-2B.1, QGw.cib-2B.2 and QGw.cib-7B were contributed by CM42 (Table 2).
Among the six QTLs for GL, two major QTL QGl.cib-3A and QGl.cib-6A were identified in five environments and the BLUP datasets, explaining 6.55-11.86% and 5.96-13.11% of the GL variation, respectively. The positive additive effects of the two QTLs on GL were contributed by CM42. The rest four minor QTLs were identified in two or three environments on chromosome 5A, 6D and 7D, explaining 5.17-11.34% of the GL variation. The favorable alleles of QGl.cib-5A.1, QGl.cib-5A.2, and QGl.cib-7D were derived from CM42, and that of QGl.cib-6D was derived from CM39 (Table 2).
Among the six QTLs for PH, QPh.cib-2D on chromosome 2D was a stable QTL and detected in five environments and the BLUP datasets, explaining 4.54-9.38% of the PH variation. The allele of CM39 contributed to higher PH. The rest five minor QTLs on chromosomes 1A, 4A, 5A, 5B and 6A were detected in two or three environments, explaining 3.8-11.37% of the PH variation. The positive alleles of QPh.cib-1A and QPh.cib-5B were from CM39, and that of QPh.cib-4A, QPh.cib-5A and QPh.cib-6A were from CM42 (Table 2).
Two minor QTLs for GNS on chromosomes 2D and 6A were detected in two environments and the BLUP datasets and explained 4.97-6.46% and 6.56-7.73% of the GNS variation, respectively. Alleles from CM42 and CM39 at QGns.cib-2D and QGns.cib-6A, respectively, contributed to positive effects on GNS (Table 2).
For SL, four QTLs were detected on chromosomes 2D, 5A, 5B and 6A. A major QTL QSl.cib-2D was detected in eight environments and the BLUP datasets, explaining 6.18-14.89% of the SL variation. QSl.cib-5B was a stable QTL and detected in three environments and the BLUP datasets, explaining 3.79-5.96% of the SL variation. Alleles of CM39 for the two QTLs contributed to increase of SL. Two minor QTLs QSl.cib-5A and QSl.cib-6A were detected in two or three environments, explaining 3.47-7.8% and 5.63-5.9% of the SL variation, respectively. The positive alleles of the two QTLs were contributed by CM42 (Table 2).
Four QTLs for SNS were identified on chromosomes 1B, 1D, 4A and 7A. Of them, QSns.cib-1B and QSns.cib-4A were detected in three environments and the BLUP datasets, explaining 7.47-16.18% and 2.34-10.46% of the SNS variation, respectively. QSns.cib-1D and QSns.cib-7A were detected in two environments, explaining 6.77-8.39% and 5.06-8.18% of the SNS variation, respectively. The favorable alleles of QSns.cib-1B and QSns.cib-7A were contributed by CM39, and that of QSns.cib-1D and QSns.cib-4A were contributed by CM42 (Table 2).
Effects of major QTL in mapping populations
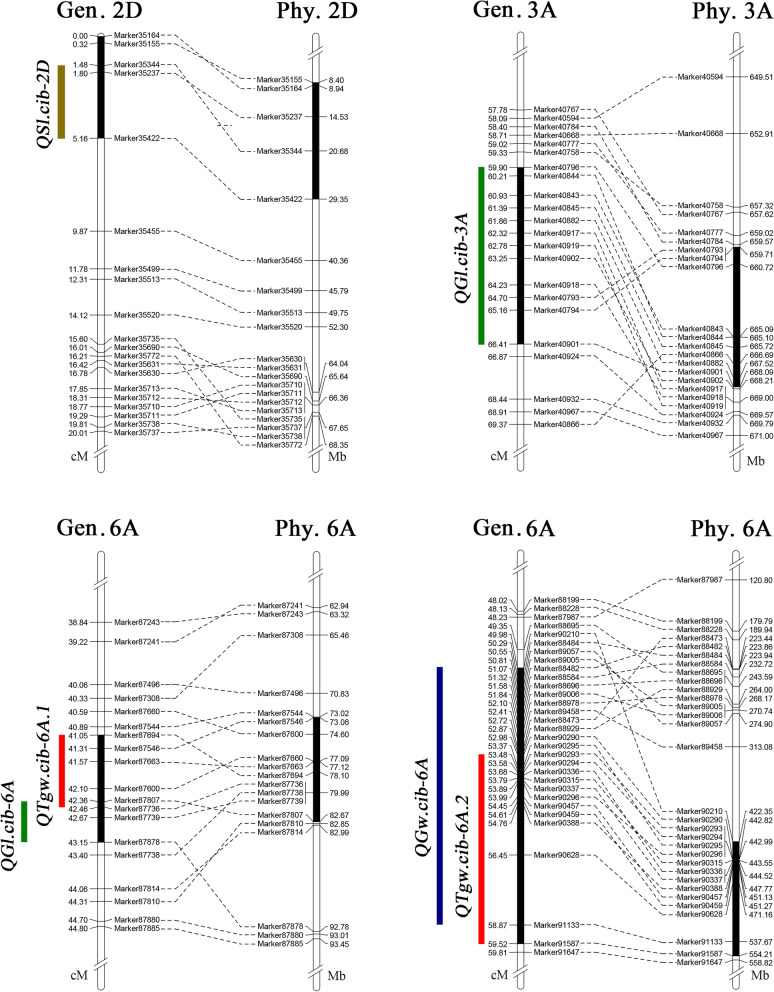
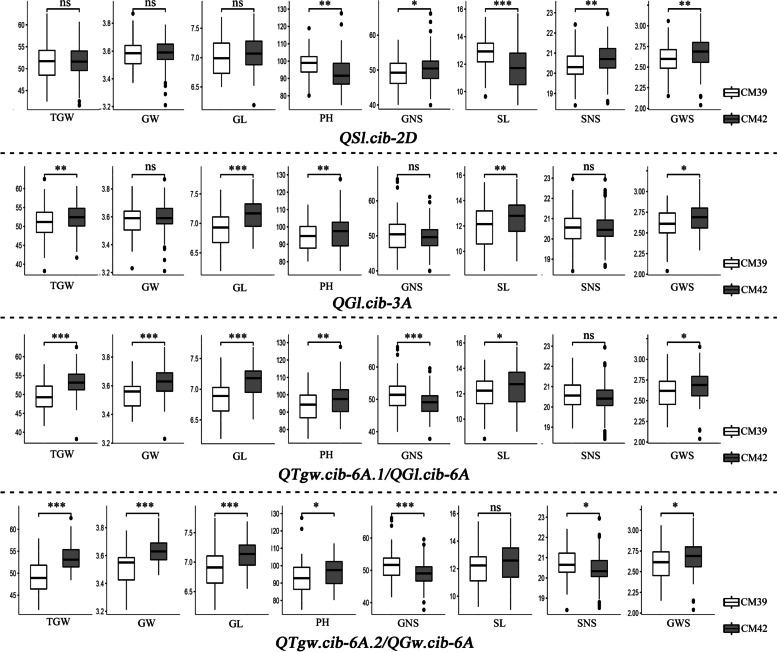
Six major QTLs QSl.cib-2D, QGl.cib-3A, QTgw.cib-6A.1, QTgw.cib-6A.2, QGw.cib-6A, and QGl.cib-6A were stably identified in multi-environments and the BLUP datasets (Table 2, Fig. 2). Based on the physical position of the flanking markers of them, three Kompetitive Allele Specific PCR (KASP) markers, K_2D-20925377, K_6A-83647812, and K_6A-54337781, tightly linked to QSl.cib-2D, QTgw.cib-6A.1/QGl.cib-6A, and QTgw.cib-6A.2/QGw.cib-6A, respectively, were successfully developed (Table S5, Fig. S1). We further analyzed the effects of these major QTLs on the seven yield-related trait and GWS using the three KASP markers and the flanking markers of QGl.cib-3A in the CM42×CM39 RIL population. The results showed that QSl.cib-2D significantly affected PH, GNS, SL, SNS and GWS, QGl.cib-3A significantly affected TGW, GL, PH, SL and GWS, QTgw.cib-6A.1/QGl.cib-6A significantly affected TGW, GW, GL, PH, GNS, SL and GWS, and QTgw.cib-6A.2/QGw.cib-6A significantly affected TGW, GW, GL, PH, GNS, SNS and GWS (Fig. 3).
Fig. 2.
The genetic and physical position of six major QTLs, QSl.cib-2D, QGl.cib-3A, QTgw.cib-6A.1, QTgw.cib-6A.2, QGw.cib-6A, and QGl.cib-6A detected in the CM42 ×CM39 RIL population; Chr., genetic position; Phy., physical position
Fig. 3.
Effects of major QTLs, QSl.cib-2D, QGl.cib-3A, QTgw.cib-6A.1, QGl.cib-6A, QTgw.cib-6A.2, and QGw.cib-6A, on seven yield-related traits and grain weight per spike (GWS) in the CM42×CM39 RIL population. CM42 and CM39 indicate the lines with the alleles from CM42 and CM39, respectively; *, ** and *** represent significance at P < 0.05, P < 0.01, and P < 0.001, respectively; ns represents non-significance
QTL clusters on chromosome 2D and 6A
The QTL cluster on 2D, including three QTLs QSl.cib-2D, QPh.cib-2D and QGns.cib-2D, was co-located between Marker35164 and Marker35422 (Table 2). Two QTL clusters were identified on chromosome 6A. One comprised two QTLs, QTgw.cib-6A.1 and QGl.cib-6A, was located between Marker87546 and Marker87738 (Table 2). The other one contained five QTLs, QTgw.cib-6A.2, QGw.cib-6A, QPh.cib-6A, QGns.cib-6A and QSl.cib-6A, was located between Marker90210 and Marker91587 (Table 2).
Discussion
QTL analysis and comparison with previous studies
Wheat yield-related traits are significantly associated with yield and typically show higher heritability than the yield itself, and thus, mining the genes or QTLs related to yield-related traits will be help for elucidating the genetic basis of wheat yield and facilitating the genetic improvement of varieties with high yield [5–7]. In the present study, a RIL population derived from two elite winter wheat varieties were used to dissect the genetic basis of variation for seven yield-related traits, including TGW, GNS, GW, GL, PH, SL and SNS. A total of 30 QTLs were identified in multiple environments, explaining 2.34-23.75% of the phenotypic variance (Table 2).
Fourteen QTLs were identified for grain size and weight, including two for TGW, six for GW and six for GL. Among them, QTgw.cib-6A.1 and QGl.cib-6A were co-located on chromosome arm 6AS, which was near to QTkw-6A.1 and QTgw.cau-6A.4 [1, 43]. QTgw.cib-6A.2 was located on chromosome arm 6AL and near to QTKW.caas-6AL and QTKW-6A.1 [44, 45]. The QTL QGw.cib-6A for GW was located in a large interval on chromosome 6A. This interval was near to a known gene TaGW2 controlling TGW and GW [46, 47]. QGw.cib-2A on chromosome 2A was overlapped with QGwt.crc-2A detected by McCartney et al [48]. QGw.cib-2B.1 on chromosome 2B was overlapped with qKW2B-1 detected by Xin et al [30]. QGw.cib-7B on chromosome 7B was located near to a QTL for TGW QTgw.wa-7BL [6]. Two QTLs for GL QGl.cib-3A and QGl.cib-5A.1 on chromosomes 3A and 5A, respectively, were overlapped with two QTLs for GL detected by Mohler et al [49]. QGl.cib-5A.2 was near to a QTL for TGW QTKW.ndsu.5A.1 reported previously [47]. QGl.cib-7D was overlapped with QGl.cau-7D detected by Yan et al [50]. For the rest three QTLs QGw.cib-2B.2, QGw.cib-5A and QGl.cib-6D, no stable QTL for grain size reported previously was overlapped with them, indicating they are likely novel QTL (Table 3).
Table 3.
The physical interval of QTL detected in the present study and comparison with previously studies.
| Trait | QTL | Chromosome | Physical position (Mb) | Nearby known locus | Reference |
|---|---|---|---|---|---|
| TGW | QTgw.cib-6A.1 | 6A | 73.08-82.67 | QTkw-6A.1, QTgw.cau-6A.4 | [1, 43] |
| QTgw.cib-6A.2 | 6A | 442.82-554.21 | QTKW.caas-6AL, QTKW-6A.1 | [44, 45] | |
| GW | QGw.cib-2A | 2A | 517.02-581.44 | QGwt.crc-2A | [48] |
| QGw.cib-2B.1 | 2B | 150.75-151.74 | qKW2B-1 | [30] | |
| QGw.cib-2B.2 | 2B | 734.72-734.72 | |||
| QGw.cib-5A | 5A | 202.92-212.92 | |||
| QGw.cib-6A | 6A | 422.35-537.67 | TaGW2 | [46, 47] | |
| QGw.cib-7B | 7B | 735.93-740.06 | QTgw.wa-7BL | [6] | |
| GL | QGl.cib-3A | 3A | 659.71-668.09 | IWA4298-IWB11347 | [49] |
| QGl.cib-5A.1 | 5A | 26.14-29.28 | IWA4871-IWB34408 | [49] | |
| QGl.cib-5A.2 | 5A | 453.5-453.6 | QTKW.ndsu.5A.1 | [47] | |
| QGl.cib-6A | 6A | 79.99-82.67 | |||
| QGl.cib-6D | 6D | 75.08-83.92 | |||
| QGl.cib-7D | 7D | 66.19-107.61 | QGl.cau-7D | [50] | |
| PH | QPh.cib-1A | 1A | 345.37-443.28 | ||
| QPh.cib-2D | 2D | 20.68-29.35 | Rht8, QPLH-2D | [31, 51] | |
| QPh.cib-4A | 4A | 704.53-704.58 | Chr4A-B57-Hap6 | [52] | |
| QPh.cib-5A | 5A | 501.62-523.22 | Chr5A-B54-Hap3 | [52] | |
| QPh.cib-5B | 5B | 607.07-608.06 | |||
| QPh.cib-6A | 6A | 447.77-451.27 | Rht18 | [53] | |
| GNS | QGns.cib-2D | 2D | 8.4-29.35 | Rht8 | [31, 51] |
| QGns.cib-6A | 6A | 471.16-554.21 | QTKW.caas-6AL, QTKW-6A.1 | [44, 45] | |
| SL | QSl.cib-2D | 2D | 20.68-29.35 | Rht8, QPLH-2D | [31, 51] |
| QSl.cib-5A | 5A | 35.84-45.91 | QSL5A.3 | [54] | |
| QSl.cib-5B | 5B | 404.42-406.31 | |||
| QSl.cib-6A | 6A | 537.67-584 | |||
| SNS | QSns.cib-1B | 1B | 381.92-439.8 | QSn.sau-1BL | [5] |
| QSns.cib-1D | 1D | 482.32-485.76 | |||
| QSns.cib-4A | 4A | 691.53-703.17 | |||
| QSns.cib-7A | 7A | 524.95-562.63 | QSn-7A.2 | [55] |
PH and SL are important traits related to plant architecture and yield potential in wheat [12, 56]. In the present study, six and four QTLs for PH and SL were identified, respectively. Among them, QPh.cib-2D and QSl.cib-2D were co-located in the same interval on chromosome arm 2DS, which was overlapped with the dwarfing gene Rht8 [31, 51]. QPh.cib-4A and QPh.cib-5A were located near to two loci for PH reported by Luján Basile et al [52]. QPh.cib-6A on chromosome 6A was overlapped with the dwarfing gene Rht18 [53]. QSl.cib-5A on chromosome 5A was located near to QSL5A.3 detected by Liu et al [54]. For the rest four QTLs QPh.cib-1A, QPh.cib-5B, QSl.cib-5B and QSl.cib-6A, no stable QTL for PH and SL reported previously was overlapped with them, indicating they are likely novel (Table 3).
Two QTLs for GNS and four QTLs for SNS were identified in the present study. Of them, QGns.cib-2D were co-located with QPh.cib-2D and QSl.cib-2D on chromosome 2D and overlapped with the dwarfing gene Rht8 [31, 51]. QGns.cib-6A was co-located with QTgw.cib-6A.2 and near to two QTLs for TGW QTKW.caas-6AL and QTKW-6A.1 [44, 45]. QSns.cib-1B for SNS on chromosome 1B was overlapped with the QSn.sau-1BL reported recently [5]. QSns.cib-7A for SNS on chromosome 7A was overlapped with QSn-7A.2 detected by Cao et al [55]. For the rest two QTLs QSns.cib-1D and QSns.cib-4A, no stable QTL for SNS reported previously was overlapped with them, indicating they are likely novel (Table 3).
QTL cluster on chromosomes 2D and 6A
Numerous co-located QTLs associated with multiple traits have been reported in the previous studies [2, 5, 24, 57, 58], which are beneficial to improve breeding efficiency for multiple elite traits, and thus is favorable for pyramiding breeding. In the present study, three QTLs QSl.cib-2D, QPh.cib-2D and QGns.cib-2D were co-located in the interval of 8.4-29.35 Mb on chromosome arm 2DS (Table 2). The allele of CM42 at the locus decreases SL and PH while increasing GNS. Additionally, the locus was overlapped with the dwarfing gene Rht8, which has been reported to associated with QTLs for PH, SL, SNS, GNS, spikelet compactness, TGW, and grain yield [12, 51, 59–61]. Interestingly, no QTL for grain size and weight detected in the present study was overlapped with the locus, indicating it had no effect on grain size and weight. Given CM42 was bred by utilizing synthetic wheat germplasm [62], further studies, such as fine-mapping and map-based cloning are needed to future reveal the relationship between the locus and Rht8. However, the results in this study showed that the locus could be utilized in optimization PH with no penalty for grain size and weight in MAS.
Two QTL clusters were detected on chromosome 6A in the present study. One comprised two QTLs, QTgw.cib-6A.1 and QGl.cib-6A, was located on chromosome arm 6AS (Table 2, Fig. 2). The other one comprised five QTLs, QTgw.cib-6A.2, QGw.cib-6A, QGns.cib-6A, QPh.cib-6A, and QSl.cib-6A, was located on chromosome arm 6AL (Table 2, Fig. 2). The QTL cluster on chromosome 6AL was overlapped with the haplotype block encompassing TaGW2 and additional 2167 genes which was located between 187 Mb and 455 Mb on chromosome 6A and defined by Brinton et al [63]. Therefore, fine-mapping and map-based cloning is needed to dissect the relationships between TaGW2 and the QTL cluster on chromosome 6AL in the future study. For the QTL cluster on chromosome 6AS, which was located between 73.08 Mb and 82.67 Mb and far apart the haplotype block of TaGW2 [63], indicating that they are different loci for grain weight.
Additive effects of three major QTLs on TGW and GNS
Due to there is a trade-off between TGW and GNS, increasing one of them may not contribute to an increase in grain yield of wheat. We further analyzed the additive effects of three major QTLs, QPh/Sl.cib-2D, QGl.cib-3A and QTgw.cib-6A.2, on the TGW and GNS. As showed in the Table 4, lines possessing the allele from CM42 at the three loci had relatively higher TGW and GNS, which might partly explain the high yield of CM42. Additionally, lines possessing the alleles from CM42 at QPh/Sl.cib-2D and QTgw.cib-6A.2 and the allele from CM39 at QGl.cib-3A also had relatively higher TGW and GNS. However, for the other combination schemes, either the higher TGW but lower GNS, or higher GNS but lower TGW, or both lower TGW and GNS were harvested. Overall, the QTLs and KASP markers in this study will be useful for elucidating the genetic architecture of grain yield and developing new wheat varieties with high and stable yield in wheat.
Table 4.
Analyses of additive effects on TGW and GNS of three major QTLs QPh/Sl.cib-2D, QGl.cib-3A and QTgw.cib-6A.2
| QTL | Lines | TGW(g) ** | GNS * |
|---|---|---|---|
| aabbcc | 20 | 46.97±2.85 a | 52.25±5.25 cd |
| AABBcc | 14 | 47.63±3.29 a | 49.78±5.12 abc |
| AAbbcc | 7 | 49.75±3.76 ab | 55.2±5.51 d |
| aaBBcc | 16 | 51.53±2.42 bc | 50.58±3.48 bc |
| aabbCC | 20 | 53.13±3.49 cd | 47.74±4.1 a |
| AAbbCC | 18 | 53.17±2.17 cd | 50.58±4 bc |
| aaBBCC | 21 | 54.25±2.94 d | 48.84±3.47 ab |
| AABBCC | 31 | 53.32±2.88 d | 50.07±2.86 bc |
aa, bb and cc represent the allele from CM39 at QPh/Sl.cib-2D, QGl.cib-3A and QTgw.cib-6A.2, respectively; AA, BB and CC represent the allele from CM42 at QPh/Sl.cib-2D, QGl.cib-3A and QTgw.cib-6A.2, respectively; Lines represent the number of different haplotypes; * and ** represent significance at P < 0.05 and P < 0.01, respectively; The superscript letter indicates significant difference among groups
Potential candidate genes for QTgw.cib-6A.1/QGl.cib-6A and QGl.cib-3A
Among these major QTL, QSl.cib-2D is likely allele with Rht8. In the previous study, TraesCS2D01G055700 was reported by Chai et al [64] as a possible candidate gene of Rht8. QTgw.cib-6A.2/QGw.cib-6A was needed additional populations to narrow their physical interval. Therefore, we mainly analyzed possible candidates for QTgw.cib-6A.1/QGl.cib-6A and QGl.cib-3A in the present study.
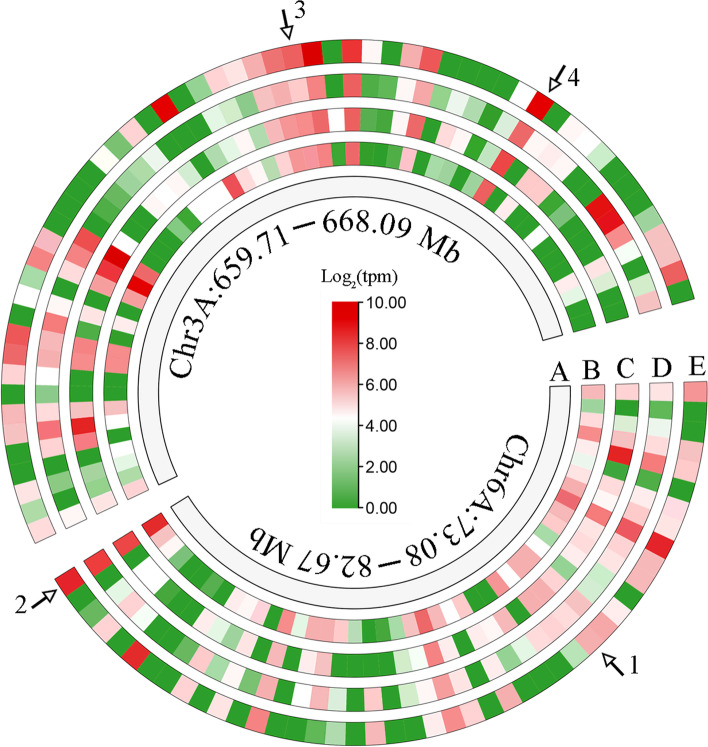
QTgw.cib-6A.1 and QGl.cib-6A were co-located between 73.08 and 82.67 Mb on Chinese Spring (CS) chromosome arm 6AS, and QGl.cib-3A was located between 659.71 and 668.09 Mb on CS chromosome arm 3AL (Table 3, Fig. 2). In the interval of QTgw.cib-6A.1/QGl.cib-6A and QGl.cib-3A, there were 81 and 85 predicted genes in the CS genome, respectively (Table S6, S7). Expression pattern analyses showed that 45 and 57 genes in the interval of QTgw.cib-6A.1/QGl.cib-6A and Gl.cib-3A expressed in various tissue, respectively (Fig. 4) [65, 66]. Among them, several were abundantly expressed in grain, indicating they are likely associated with grain growth and development (Fig. 4). For example, TraesCS6A02G107800 is an ortholog of the rice RGG2 and encodes a guanine nucleotide-binding protein subunit gamma 2 (Table S6). Miao et al previously reported that RGG2 played a negative role in plant growth and yield production and that manipulation of RGG2 can increase the plant biomass, grain weight, length and yield in rice [67]. TraesCS6A02G112400 and TraesCS3A02G424000 encode polyubiquitin and small ubiquitin-related modifier, respectively (Table S6, S7). TraesCS3A02G421900 encodes a 26S proteasome regulatory subunit (Table S7), which participates in the ubiquitin/26S proteasome pathway and mediate the degradation of the complex of ubiquitin receptor and poly-ubiquitinated protein [68, 69]. Previous studies revealed that the ubiquitin pathway play an important role in regulation grain size and weight in rice [70, 71]. These results indicated that the four genes may be closely related to grain size and weight in wheat and useful for fine mapping and cloning of QTgw.cib-6A.1/QGl.cib-6A and QGl.cib-3A in our following work.
Fig. 4.
Expression pattern of genes within the QTgw.cib-6A.1/QGl.cib-6A and QGl.cib-3A intervals. 1, 2, 3 and 4 marked by the arrow represent TraesCS6A02G107800, TraesCS6A02G112400, TraesCS3A02G421900 and TraesCS3A02G424000, respectively; A represents the physical interval of QTgw.cib-6A.1/QGl.cib-6A and QGl.cib-3A on chromosome 6A and 3A; B, C, D and E represent root, leaf/shoot, spike and, grain, respectively
Conclusion
In this study, a total of 30 QTLs for TGW, GNS, GW, GL, PH, SL, and SNS were identified, explaining 2.34-23.75% of the phenotypic variance. Among them, six major QTLs QTgw.cib-6A.1, QTgw.cib-6A.2, QGw.cib-6A, QGl.cib-3A, QGl.cib-6A, and QSl.cib-2D were detected. Three KASP markers linked with five of these major QTLs were developed. These QTLs and KASP markers will be useful for elucidating the genetic architecture of grain yield and developing new wheat varieties with high and stable yield in wheat. Additionally, candidate genes of QTgw.cib-6A.1/QGl.cib-6A and QGl.cib-3A were preliminary analyzed.
Methods
Plant materials and field trials
A RIL population (F10) comprising 193 lines derived from a cross CM42 and CM39 were used for QTL detection in the present study. CM42 is the first wheat elite variety in the world bred by using synthetic hexaploid wheat (Triticum turgidum×Aegilops tauschii) germplasm, and showed high yield potential in Sichuan and the Yangzi River region [62], while CM39 is an elite winter wheat variety with different genetic background to that of CM42. During four growing seasons of wheat from 2015-2016 to 2018-2019, the RIL population along with their parents were evaluated at two experimental sites in Sichuan province of China, including Shuangliu (SHL, 103° 52'E, 30°34'N) and Shifang (SHF, 104°11'E, 31°6'N). Randomized block design was adopted for all of the trials. Each line was planted in a one-row plot with 50 seeds per row, a row length of 2.0 m, and a row spacing of 0.3 m. Five replicates were performed under each environment. Nitrogen and superphosphate fertilizers were applied at a rate of 80 and 100 kg/ha, respectively, at sowing. Crop management and disease control were performed according to local cultivation practices.
Phenotyping and statistical analysis
At maturity, ten representative plants from middle row of each line were randomly selected to investigate agronomic traits including TGW, GL, GW, GNS, PH, SL, SNS and GWS. SL was measured as the length from the base of the rachis to the tip of the terminal spikelet, excluding the awns. SNS was determined by counting the number of spikelets in main spikes; PH was measured from the soil surface to the tip of the spike, excluding the awns. Subsequently, the main spike of all selected plants were harvested and manually threshed for evaluating GNS, TGW, GW, GL and GWS using SC-G software (Wseen Co., Ltd, Hangzhou, China). PH and SL were evaluated in eight environments, and the rest traits were evaluated in six environments.
Basic phenotypic statistical analyses, frequency distribution, correlation analyses and student’s t tests were performed with SPSS version 20.0 (Chicago, IL, USA). The phenotype distribution graph was drawn using the plugin “CorrPlot” in TBtools [72]. The relationships among measured traits were visualized using the R package “qgraph”. The BLUP data across evaluated environments was calculated using the “lmer” function implemented in R package “lme4”. ANOVA was performed over all trials which indicated statistically significant main effects for genotypes (G), environments (E), G × E interactions for all measured traits using the SAS software (SAS Institute Inc., North Carolina, USA). The broad sense heritability (H2) was estimated based on the following equation: , where is the variance of genotypes, is the variance of genotype by environmental effect, is the residual variance, n is the number of environments and r is the number of replicates [73].
Linkage map construction and QTL detection
A whole-genome genetic map constructed previously was adopted for QTL mapping [42]. The genetic map was constructed using the CM42×CM39 RIL population with SLAF markers. A total of 4996 Bin SLAFs were distributed in 21 linkage groups and covered a total genetic distance of 2,859.94 cM with an average interval of 0.57 cM between adjacent Bin marker (Table S1, S2) [42].
QTL analysis was conducted using the inclusive composite interval mapping (ICIM) function of IciMapping 4.1 (https://www.isbreeding.net) with the minimal LOD score was set at 2.5. The missing phenotype was deleted in QTL analysis. QTL was named according to the provision of Genetic Nomenclature (http://wheat.pw.usda.gov/ggpages/wgc/98/Intro.htm), where ‘CIB’ represents Chengdu Institute of Biology. QTLs consistently identified in at least three environments and in combined analysis with ≥10% of phenotypic variation explained were considered as major QTLs.
Development of Kompetitive Allele-Specific PCR Markers
On the basis of the preliminary QTL mapping results, the flanking markers of major QTL were blasted against the CS reference genome sequence (RefSeq v1.0; https://wheat-urgi.versailles.inra.fr/) to gain their physical positions [74]. The SNPs within the physical interval of major QTLs were used for developing KASP markers tight linked with them. The KASP marker primers were designed using the PrimerServer tool in Triticeae Multi-omics Center (http://202.194.139.32/) [75]. Standard FAM and HEX adapters were added to the allele-specific forward primers at the 5′ ends. The KASP assays were run in a Bio-Rad CFX96 real-time PCR system in 10μL reaction volumes with the following PCR cycling parameters: hot start enzyme activation at 94 °C for 15 min; a touchdown of 10 cycles (94 °C for 20 s, and touchdown starting at 61 °C and decreasing by 0.6 °C per 1-min cycle); then 26 cycles of regular PCR (94 °C for 20 s, 55 °C for 60 s, and rest at 37 °C for 1 min). If the clustering was not significant, further cycling was performed at 94 °C for 20 s and 55 °C for 60 s (3–10 cycles per step)
Prediction of candidate gene
Genes between the physical intervals of major QTLs were extracted from IWGSC RefSeq v1.1 annotation for CS [74]. The annotations and functions of a given gene were analyzed using UniProt (https://www.uniprot.org/). The expression pattern analysis was performed by using Wheat Expression Browser (http://www.wheat-expression.com/), and the circle graph of expression values was drawn using TBtools [72]. The orthologous gene analysis between wheat and rice was conducted using the Triticeae-Gene Tribe (http://wheat.cau.edu.cn/TGT/) [76].
Supplementary Information
Acknowledgement
Not applicable.
Abbreviations
- CM42
Chuanmai42
- CM39
Chuanmai39
- SHF
Shifang
- SHL
Shuangliu
- TGW
Thousand grain weight
- GNS
Grain number per spike
- GW
Grain width
- GL
Grain length
- PH
Plant height
- SL
Spike length
- SNS
Spikelet number per spike
- QTL
Quantitative trait loci
- SLAF
Specific-locus amplified fragment
- MAS
Marker-assisted selection
- KASP
Kompetitive Allele Specific PCR
- BLUP
Best linear unbiased prediction
- CS
Chinese Spring
Authors’ contributions
TL undertook the field trials and subsequent analysis of all available data including the phenotyping and population genotyping, and drafted this manuscript. QL and ZP undertook the genetic map constructed. JW, ZY, YT, YS, JZ, XQ and XP participated in phenotyping. ZL, WY and JL developed and provided us the CC population. MY, JL, YW and HZ discussed results. HL and GD designed the experiments, guided the entire study, participated in data analysis, discussed results and revised the manuscript. The authors read and approved the final manuscript.
Funding
This work is supported by National Key R&D Program of China (2016YFD0100102), Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA08020205), Science and Technology Support Project of Sichuan Province, China (2016NZ0103), Key Project of Crop Breeding of Sichuan Province (2016NYZ0030), and Science and technology projects of Sichuan Province (2020YFSY0049).
Availability of data and materials
All data used in this study was present in the manuscript and supporting materials.
Declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable
Competing interests
All authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guan P, Lu L, Jia L, Kabir MR, Zhang J, Lan T, et al. Global QTL Analysis Identifies Genomic Regions on Chromosomes 4A and 4B Harboring Stable Loci for Yield-Related Traits Across Different Environments in Wheat (Triticum aestivum L.) Front Plant Sci. 2018;9:1–14. doi: 10.3389/fpls.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isham K, Wang R, Zhao W, Wheeler J, Klassen N, Akhunov E, et al. QTL mapping for grain yield and three yield components in a population derived from two high-yielding spring wheat cultivars. Theor Appl Genet. 2021;134:2079–2095. doi: 10.1007/s00122-021-03806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li T, Deng G, Tang Y, Su Y, Wang J, Cheng J. Identification and Validation of a Novel Locus Controlling Spikelet Number in Bread Wheat ( Triticum aestivum L.) Front Plant Sci. 2021;12:1–14. doi: 10.3389/fpls.2021.611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui F, Ding A, Li J, Zhao C, Wang L, Wang X, et al. QTL detection of seven spike-related traits and their genetic correlations in wheat using two related RIL populations. Euphytica. 2012;186:177–192. doi: 10.1007/s10681-011-0550-7. [DOI] [Google Scholar]
- 5.Ren T, Fan T, Chen S, Li C, Chen Y, Ou X, et al. Utilization of a Wheat55K SNP array-derived high-density genetic map for high-resolution mapping of quantitative trait loci for important kernel-related traits in common wheat. Theor Appl Genet. 2021;134:807–821. doi: 10.1007/s00122-020-03732-8. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Liu W, Wang H, Li L, Wu J, Yang X, et al. QTL mapping of yield-related traits in the wheat germplasm 3228. Euphytica. 2011;177:277–292. doi: 10.1007/s10681-010-0267-z. [DOI] [Google Scholar]
- 7.Chu CG, Xu SS, Friesen TL, Faris JD. Whole genome mapping in a wheat doubled haploid population using SSRs and TRAPs and the identification of QTL for agronomic traits. Mol Breed. 2008;22:251–266. doi: 10.1007/s11032-008-9171-9. [DOI] [Google Scholar]
- 8.Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA. “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet. 2002;105:1038–1042. doi: 10.1007/s00122-002-1048-4. [DOI] [PubMed] [Google Scholar]
- 9.Lv C, Song Y, Gao L, Yao Q, Zhou R, Xu R, et al. Integration of QTL detection and marker assisted selection for improving resistance to Fusarium head blight and important agronomic traits in wheat. Crop J. 2014;2:70–78. doi: 10.1016/j.cj.2013.10.004. [DOI] [Google Scholar]
- 10.Du Y, Chen L, Wang Y, Yang Z, Saeed I, Daoura BG, et al. The combination of dwarfing genes Rht4 and Rht8 reduced plant height, improved yield traits of rainfed bread wheat (Triticum aestivum L.) F Crop Res. 2018;215:149–155. doi: 10.1016/j.fcr.2017.10.015. [DOI] [Google Scholar]
- 11.Fan X, Cui F, Ji J, Zhang W, Zhao X, Liu JJ, et al. Dissection of pleiotropic QTL regions controlling wheat spike characteristics under different nitrogen treatments using traditional and conditional QTL mapping. Front Plant Sci. 2019;10:1–13. doi: 10.3389/fpls.2019.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhai H, Feng Z, Li J, Liu X, Xiao S, Ni Z, et al. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front. Plant Sci. 2016;7:1–13. doi: 10.3389/fpls.2016.01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X, Xin M, Xu R, Chen Z, Cai W, Chai L, et al. A single amino acid substitution in STKc_GSK3 kinase conferring semispherical grains and its implications for the origin of triticum sphaerococcum. Plant Cell. 2020;32:923–934. doi: 10.1105/tpc.19.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Yan Y, Wu TT, Zhang GL, Yin H, Chen W, et al. Cloning of wheat keto-acyl thiolase 2B reveals a role of jasmonic acid in grain weight determination. Nat Commun. 2020;11:6266. doi: 10.1038/s41467-020-20133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrovolskaya O, Pont C, Sibout R, Martinek P, Badaeva E, Murat F, et al. Frizzy panicle drives supernumerary spikelets in bread wheat. Plant Physiol. 2015;167:189–199. doi: 10.1104/pp.114.250043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voss-Fels KP, Keeble-Gagnère G, Hickey LT, Tibbits J, Nagornyy S, Hayden MJ, et al. High-resolution mapping of rachis nodes per rachis, a critical determinant of grain yield components in wheat. Theor Appl Genet. 2019;132:2707–2719. doi: 10.1007/s00122-019-03383-4. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Bai Z, Li X, Wang P, Wu Q, Yang L, et al. SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor Appl Genet. 2012;125:1057–1068. doi: 10.1007/s00122-012-1895-6. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Zhao YL, Gao LF, Zhao GY, Zhou RH, Zhang BS, et al. TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 2012;195:574–584. doi: 10.1111/j.1469-8137.2012.04194.x. [DOI] [PubMed] [Google Scholar]
- 19.Hanif M, Gao F, Liu J, Wen W, Zhang Y, Rasheed A, et al. TaTGW6-A1, an ortholog of rice TGW6, is associated with grain weight and yield in bread wheat. Mol Breed. 2016;36:1–8. doi: 10.1007/s11032-015-0425-z. [DOI] [Google Scholar]
- 20.Dong L, Wang F, Liu T, Dong Z, Li A, Jing R, et al. Natural variation of TaGASR7-A1 affects grain length in common wheat under multiple cultivation conditions. Mol Breed. 2014;34:937–947. doi: 10.1007/s11032-014-0087-2. [DOI] [Google Scholar]
- 21.Yang J, Zhou Y, Wu Q, Chen Y, Zhang P, Zhang Y, et al. Molecular characterization of a novel TaGL3-5A allele and its association with grain length in wheat (Triticum aestivum L.) Theor Appl Genet. 2019;132:1799–1814. doi: 10.1007/s00122-019-03316-1. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Liu J, Xia X, He Z. TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol Breed. 2014;34:1097–1107. doi: 10.1007/s11032-014-0102-7. [DOI] [Google Scholar]
- 23.Wang W, Pan Q, Tian B, He F, Chen Y, Bai G, et al. Gene editing of the wheat homologs of TONNEAU1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J. 2019;100:251–264. doi: 10.1111/tpj.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao S, Xu D, Hanif M, Xia X, He Z. Genetic architecture underpinning yield component traits in wheat. Theor Appl Genet. 2020;133:1811–1823. doi: 10.1007/s00122-020-03562-8. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto Y, Nguyen AT, Yoshioka M, Iehisa JCM, Takumi S. Identification of quantitative trait loci controlling grain size and shape in the D genome of synthetic hexaploid wheat lines. Breed Sci. 2013;63:423–429. doi: 10.1270/jsbbs.63.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marza F, Bai GH, Carver BF, Zhou WC. Quantitative trait loci for yield and related traits in the wheat population Ning7840 x Clark. Theor Appl Genet. 2006;112:688–698. doi: 10.1007/s00122-005-0172-3. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Wu L, Gan X, Chen W, Liu B, Fedak G, et al. Mapping quantitative trait loci for 1000-grain weight in a double haploid population of common wheat. Int J Mol Sci. 2020;21:3960. doi: 10.3390/ijms21113960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Xia X, Xiao Y, He Z, Wang D, Trethowan R, et al. QTL mapping for plant height and yield components in common wheat under water-limited and full irrigation environments. Crop Pasture Sci. 2015;66:660–670. doi: 10.1071/CP14236. [DOI] [Google Scholar]
- 29.Su Q, Zhang X, Zhang W, Zhang N, Song L, Liu L, et al. QTL Detection for Kernel Size and Weight in Bread Wheat (Triticum aestivum L.) Using a High-Density SNP and SSR-Based Linkage Map. Front. Plant Sci. 2018;9:1–13. doi: 10.3389/fpls.2018.01484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xin F, Zhu T, Wei S, Han Y, Zhao Y, Zhang D, et al. QTL Mapping of Kernel Traits and Validation of a Major QTL for Kernel Length-Width Ratio Using SNP and Bulked Segregant Analysis in Wheat. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohan A, Grant NP, Schillinger WF, Gill KS. Characterizing reduced height wheat mutants for traits affecting abiotic stress and photosynthesis during seedling growth. Physiol Plant. 2021;172:233–246. doi: 10.1111/ppl.13321. [DOI] [PubMed] [Google Scholar]
- 32.Mo Y, Vanzetti LS, Hale I, Spagnolo EJ, Guidobaldi F, Al-Oboudi J, et al. Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor Appl Genet. 2018;131:2021–2035. doi: 10.1007/s00122-018-3130-6. [DOI] [PubMed] [Google Scholar]
- 33.Guan P, Shen X, Mu Q, Wang Y, Wang X, Chen Y, et al. Dissection and validation of a QTL cluster linked to Rht-B1 locus controlling grain weight in common wheat (Triticum aestivum L.) using near-isogenic lines. Theor Appl Genet. 2020;133:2639–2653. doi: 10.1007/s00122-020-03622-z. [DOI] [PubMed] [Google Scholar]
- 34.Duan X, Yu H, Ma W, Sun J, Zhao Y, Yang R, et al. A major and stable QTL controlling wheat thousand grain weight: identification, characterization, and CAPS marker development. Mol Breed. 2020;40:68. doi: 10.1007/s11032-020-01147-3. [DOI] [Google Scholar]
- 35.Chen W, Sun D, Li R, Wang S, Shi Y, Zhang W, et al. Mining the stable quantitative trait loci for agronomic traits in wheat (Triticum aestivum L.) based on an introgression line population. BMC Plant Biol. 2020;20:1–9. doi: 10.1186/s12870-019-2170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maccaferri M, Ricci A, Salvi S, Milner SG, Noli E, Martelli PL, et al. A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol J. 2015;13:648–663. doi: 10.1111/pbi.12288. [DOI] [PubMed] [Google Scholar]
- 37.Li T, Deng G, Su Y, Yang Z, Tang Y, Wang J, et al. Identification and validation of two major QTLs for spike compactness and length in bread wheat (Triticum aestivum L.) showing pleiotropic effects on yield-related traits. Theor Appl Genet. 2021;134:3625–3641. doi: 10.1007/s00122-021-03918-8. [DOI] [PubMed] [Google Scholar]
- 38.Li T, Deng G, Su Y, Yang Z, Tang Y, Wang J, et al. Genetic dissection of quantitative trait loci for grain size and weight by high-resolution genetic mapping in bread wheat (Triticum aestivum L.) Theor Appl Genet. 2022;135:257–271. doi: 10.1007/s00122-021-03964-2. [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Zhang J, Chen Y, Zhang C, Gong J, Song Z, et al. Identification of candidate genes for key fibre-related QTLs and derivation of favourable alleles in Gossypium hirsutum recombinant inbred lines with G. barbadense introgressions. 2020:707–20. [DOI] [PMC free article] [PubMed]
- 40.Ma J, Pei W, Ma Q, Geng Y, Liu G, Liu J, et al. QTL analysis and candidate gene identification for plant height in cotton based on an interspecific backcross inbred line population of Gossypium hirsutum × Gossypium barbadense. Theor Appl Genet. 2019;132:2663–2676. doi: 10.1007/s00122-019-03380-7. [DOI] [PubMed] [Google Scholar]
- 41.Han J, Han D, Guo Y, Yan H, Wei Z, Tian Y, et al. QTL mapping pod dehiscence resistance in soybean (Glycine max L. Merr.) using specific-locus amplified fragment sequencing. Theor Appl Genet. 2019;132:2253–2272. doi: 10.1007/s00122-019-03352-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Pan Z, Gao Y, Li T, Liang J, Zhang Z, et al. Quantitative Trait Locus (QTLs) Mapping for Quality Traits of Wheat Based on High Density Genetic Map Combined With Bulked Segregant Analysis RNA-seq (BSR-Seq) Indicates That the Basic 7S Globulin Gene Is Related to Falling Number. Front Plant Sci. 2020;11:1–21. doi: 10.3389/fpls.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui F, Zhao C, Ding A, Li J, Wang L, Li X, et al. Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor Appl Genet. 2014;127:659–675. doi: 10.1007/s00122-013-2249-8. [DOI] [PubMed] [Google Scholar]
- 44.Li F, Wen W, He Z, Liu J, Jin H, Cao S, et al. Genome - wide linkage mapping of yield - related traits in three Chinese bread wheat populations using high - density SNP markers. Theor Appl Genet. 2018;131:1903–1924. doi: 10.1007/s00122-018-3122-6. [DOI] [PubMed] [Google Scholar]
- 45.Lee HS, Jung JU, Kang CS, Heo HY, Park CS. Mapping of QTL for yield and its related traits in a doubled haploid population of Korean wheat. Plant Biotechnol Rep. 2014;8:443–454. doi: 10.1007/s11816-014-0337-0. [DOI] [Google Scholar]
- 46.Maphosa L, Langridge P, Taylor H, Parent B, Emebiri LC, Kuchel H, et al. Genetic control of grain yield and grain physical characteristics in a bread wheat population grown under a range of environmental conditions. Theor Appl Genet. 2014;127:1607–1624. doi: 10.1007/s00122-014-2322-y. [DOI] [PubMed] [Google Scholar]
- 47.Kumar A, Mantovani EE, Seetan R, Soltani A, Echeverry-Solarte M, Jain S, et al. Dissection of Genetic Factors underlying Wheat Kernel Shape and Size in an Elite x Nonadapted Cross using a High Density SNP Linkage Map. Plant. Genome. 2016;9. [DOI] [PubMed]
- 48.McCartney CA, Somers DJ, Humphreys DG, Lukow O, Ames N, Noll J, et al. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 x “AC Domain”. Genome. 2005;48:870–883. doi: 10.1139/g05-055. [DOI] [PubMed] [Google Scholar]
- 49.Mohler V, Albrecht T, Castell A, Diethelm M, Schweizer G, Hartl L. Considering causal genes in the genetic dissection of kernel traits in common wheat. J Appl Genet. 2016;57:467–476. doi: 10.1007/s13353-016-0349-2. [DOI] [PubMed] [Google Scholar]
- 50.Yan L, Liang F, Xu H, Zhang X, Zhai H, Sun Q, et al. Identification of QTL for grain size and shape on the D genome of natural and synthetic allohexaploid wheats with near-identical AABB genomes. Front Plant Sci. 2017;8:1–14. doi: 10.3389/fpls.2017.01705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daba SD, Tyagi P, Brown-Guedira G, Mohammadi M. Genome-wide association study in historical and contemporary U.S. winter wheats identifies height-reducing loci. Crop J. 2020;8:243–251. doi: 10.1016/j.cj.2019.09.005. [DOI] [Google Scholar]
- 52.Luján Basile SM, Ramírez IA, Crescente JM, Conde MB, Demichelis M, Abbate P, et al. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019;19:1–16. doi: 10.1186/s12870-019-2015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ford BA, Foo E, Sharwood R, Karafiatova M, Vrána J, MacMillan C, et al. Rht18 Semidwarfism in Wheat Is Due to Increased GA 2-oxidaseA9 Expression and Reduced GA Content. Plant Physiol. 2018;177:168–180. doi: 10.1104/pp.18.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu K, Sun X, Ning T, Duan X, Wang Q, Liu T, et al. Genetic dissection of wheat panicle traits using linkage analysis and a genome-wide association study. Theor Appl Genet. 2018;131:1073–1090. doi: 10.1007/s00122-018-3059-9. [DOI] [PubMed] [Google Scholar]
- 55.Cao P, Liang X, Zhao H, Feng B, Xu E, Wang L, et al. Identification of the quantitative trait loci controlling spike-related traits in hexaploid wheat (Triticum aestivum L.) Planta. 2019;250:1967–1981. doi: 10.1007/s00425-019-03278-0. [DOI] [PubMed] [Google Scholar]
- 56.Tian X, Wen W, Xie L, Fu L, Xu D, Fu C, et al. Molecular mapping of reduced plant height gene Rht24 in bread wheat. Front Plant Sci. 2017;8:1–9. doi: 10.3389/fpls.2017.01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma J, Tu Y, Zhu J, Luo W, Liu H, Li C, et al. Flag leaf size and posture of bread wheat: genetic dissection, QTL validation and their relationships with yield-related traits. Theor Appl Genet. 2020;133:297–315. doi: 10.1007/s00122-019-03458-2. [DOI] [PubMed] [Google Scholar]
- 58.Gao F, Wen W, Liu J, Rasheed A, Yin G, Xia X, et al. Genome-Wide Linkage Mapping of QTL for Yield Components, Plant Height and Yield-Related Physiological Traits in the Chinese Wheat Cross Zhou 8425B/Chinese Spring. Front Plant Sci. 2015;6:1099. doi: 10.3389/fpls.2015.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang K, Wang J, Qin H, Wei Z, Hang L, Zhang P, et al. Assessment of the individual and combined effects of Rht8 and Ppd-D1a on plant height, time to heading and yield traits in common wheat. Crop J. 2019;7:845–856. doi: 10.1016/j.cj.2019.06.008. [DOI] [Google Scholar]
- 60.Rebetzke GJ, Ellis MH, Bonnett DG, Mickelson B, Condon AG, Richards RA. Height reduction and agronomic performance for selected gibberellin-responsive dwarfing genes in bread wheat (Triticum aestivum L.) F Crop Res. 2012;126:87–96. doi: 10.1016/j.fcr.2011.09.022. [DOI] [Google Scholar]
- 61.Wang Y, Du Y, Yang Z, Chen L, Condon AG, Hu YG. Comparing the effects of GA-responsive dwarfing genes Rht13 and Rht8 on plant height and some agronomic traits in common wheat. F Crop Res. 2015;179:35–43. doi: 10.1016/j.fcr.2015.04.010. [DOI] [Google Scholar]
- 62.Yang W, Liu D, Li J, Zhang L, Wei H, Hu X, et al. Synthetic hexaploid wheat and its utilization for wheat genetic improvement in China. J Genet Genomics. 2009;36:539–546. doi: 10.1016/S1673-8527(08)60145-9. [DOI] [PubMed] [Google Scholar]
- 63.Brinton J, Ramirez-Gonzalez RH, Simmonds J, Wingen L, Orford S, Griffiths S, et al. A haplotype-led approach to increase the precision of wheat breeding. Commun Biol. 2020;3:1–11. doi: 10.1038/s42003-020-01413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chai L, Chen Z, Bian R, Zhai H, Cheng X, Peng H, et al. Correction to: Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.) Theor Appl Genet. 2019;132:3225. doi: 10.1007/s00122-019-03421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Borrill P, Ramirez-Gonzalez R, Uauy C. expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiol. 2016;170:2172–2186. doi: 10.1104/pp.15.01667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, et al. The transcriptional landscape of polyploid wheat. Science. 2018;361:eaar6089. doi: 10.1126/science.aar6089. [DOI] [PubMed] [Google Scholar]
- 67.Miao J, Yang Z, Zhang D, Wang Y, Xu M, Zhou L, et al. Mutation of RGG2, which encodes a type B heterotrimeric G protein γ subunit, increases grain size and yield production in rice. Plant Biotechnol J. 2019;17:650–664. doi: 10.1111/pbi.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li N, Li Y. Ubiquitin-mediated control of seed size in plants. Front Plant Sci. 2014;5:1–6. doi: 10.3389/fpls.2014.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, et al. The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell. 2003;15:965–980. doi: 10.1105/tpc.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li N, Xu R, Li Y. Molecular Networks of Seed Size Control in Plants. Annu Rev Plant Biol. 2019;70:435–463. doi: 10.1146/annurev-arplant-050718-095851. [DOI] [PubMed] [Google Scholar]
- 71.Li Q, Li L, Yang X, Warburton ML, Bai G, Dai J, et al. Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 2010;10:1–15. doi: 10.1186/1471-2229-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 73.Smith SE, Kuehl RO, Ray IM, Hui R, Soleri D. Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop Sci. 1998;38:1125–1129. doi: 10.2135/cropsci1998.0011183X003800050003x. [DOI] [Google Scholar]
- 74.Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:eaar7191. [DOI] [PubMed]
- 75.Ma S, Wang M, Wu J, Guo W, Chen Y, Li G, et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol Plant. 2021;14:1965–1968. doi: 10.1016/j.molp.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 76.Chen Y, Song W, Xie X, Wang Z, Guan P, Peng H, et al. A Collinearity-Incorporating Homology Inference Strategy for Connecting Emerging Assemblies in the Triticeae Tribe as a Pilot Practice in the Plant Pangenomic Era. Mol Plant. 2020;13:1694–1708. doi: 10.1016/j.molp.2020.09.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study was present in the manuscript and supporting materials.