SUMMARY
Regulatory B cells (Bregs) suppress immune responses through the secretion of interleukin-10 (IL-10). This immunomodulatory capacity holds therapeutic potential, yet a definitional immunophenotype for enumeration and prospective isolation of B cells capable of IL-10 production remains elusive. Here, we simultaneously quantify cytokine production and immunophenotype in human peripheral B cells across a range of stimulatory conditions and time points using mass cytometry. Our analysis shows that multiple functional B cell subsets produce IL-10 and that no phenotype uniquely identifies IL-10+ B cells. Further, a significant portion of IL-10+ B cells co-express the pro-inflammatory cytokines IL-6 and tumor necrosis factor alpha (TNFα). Despite this heterogeneity, operationally tolerant liver transplant recipients have a unique enrichment of IL-10+, but not TNFα+ or IL-6+, B cells compared with transplant recipients receiving immunosuppression. Thus, human IL-10-producing B cells constitute an induced, transient state arising from a diversity of B cell subsets that may contribute to maintenance of immune homeostasis.
In brief
Through multiplexed screening of activating conditions and time points, Glass et al. find that human IL-10+ B cells lack a unifying cellular phenotype and frequently co-express pro-inflammatory cytokines. Mass-cytometry analyses reveal the breadth of phenotypic and polyfunctional profiles of IL-10+ B cells and their associations with liver-allograft operational tolerance.
Graphical Abstract
INTRODUCTION
In addition to their established roles in antibody production and antigen presentation, B cells can regulate inflammation. B cells with immunosuppressive properties, defined by the expression of the hallmark immunoregulatory cytokine interleukin-10 (IL-10), have been termed regulatory B cells (Bregs) (Fillatreau et al., 2002; Iwata et al., 2011; Mauri et al., 2003; Mizoguchi et al., 2002). Granzyme B (Zhu et al., 2017), IL-35 (Shen et al., 2014; Wang et al., 2014), and other secreted molecules (Lee et al., 2014; Parekh et al., 2003), as well as surface receptors (Catalán et al., 2021), have also been implicated in Breg immunomodulatory activity. Identification of an IL-10-producing B cell population with immunomodulatory activity was first reported in mice, where it was demonstrated that IL-10+ B cells were critical for the prevention or control of autoimmune disease and chronic inflammation (Wolf et al., 1996). A myriad of phenotypes and regulatory functions of Bregs have since been established using murine models of disease (Jansen et al., 2021; Ran et al., 2020), yet the relationship of murine IL-10+ B cells to the human immune system requires further study (Baba et al., 2020; Lighaam et al., 2018; Rosser et al., 2014).
Human IL-10+ B cells have been implicated in autoimmunity (Flores-Borja et al., 2013; Iwata et al., 2011; Matsumoto et al., 2014), alloimmunity (Cherukuri et al., 2021; Chesneau et al., 2014; Newell et al., 2015; Nova-Lamperti et al., 2016; Silva et al., 2012), allergy (Oliveria et al., 2018; van de Veen et al., 2013), cancer (Budczies et al., 2021; Mehdipour et al., 2019; Michaud et al., 2021; Wang et al., 2015), and infection (Das et al., 2012; Lopez-Abente et al., 2018). A wide variety of Breg surface phenotypes were described in these studies, some of which are distinct from established murine Breg phenotypes (Jansen et al., 2021). Thus, a significant barrier in understanding and defining human Bregs has been the lack of an encompassing immunophenotypic signature to identify cells with IL-10 production capacity. Additionally, human IL-10-producing Bregs have been primarily studied in disease states characterized by immune alterations through a single mode of ex vivo stimulation or without consideration of the substantial changes in B cell surface phenotype that occur upon activation. The low level of IL-10 expression by B cell subsets in vivo further complicates their detection (Fillatreau et al., 2002; Mauri and Menon, 2017; Yanaba et al., 2009). Nevertheless, CD24hiCD38hi transitional, CD9+, CD24hiCD27+ memory B cells and CD27intCD38+ plasmablasts have been shown to produce IL-10, and in some cases to possess IL-10-dependent regulatory properties in healthy humans, yet were diminished or absent in disease states (Blair et al., 2010; Brosseau et al., 2018; Iwata et al., 2011; Matsumoto et al., 2014; Menon et al., 2016). This suggests that human IL-10-producing Bregs participate in maintenance and regulation of immune homeostasis. However, the dynamics of IL-10 versus pro-inflammatory cytokine production by functional B cell subsets across healthy individuals and the impact of different activating stimuli remains to be verified by a systemic approach.
In clinical transplantation, the term operational tolerance describes a state whereby organ transplant recipients maintain stable graft function over an extended period in the absence of immunosuppression (IS). It remains unclear if operational tolerance involves an active immunoregulatory process. Nevertheless, spontaneously tolerant renal transplant recipients demonstrated an increase in total B cells, particularly CD38+CD24+IgD+ transitional and naive subsets, as well as an upregulated expression of B cell-associated genes by peripheral blood mononuclear cells (PBMCs), relative to their immunosuppressed counterparts (Newell et al., 2015; Pallier et al., 2010). Operationally tolerant kidney recipients have increased proportions of CD20+CD24hiCD38hi transitional B cells and CD20+CD38loCD24lo naive B cells and increased IL-10 production from activated B cells compared with healthy subjects and stable transplant recipients (Chesneau et al., 2014). Functionally, human IL-10+ B cells from healthy individuals and stable kidney transplant recipients on IS have been shown to decrease effector CD4+ T cell proliferation and type I pro-inflammatory cytokine or type II interferon production (Cherukuri et al., 2014; Nova-Lamperti et al., 2016). Granzyme B+ (GzmB+) B cells from stable immunosuppressed and operationally tolerant renal transplant recipients also inhibited CD4+ T cell proliferation (Chesneau et al., 2015). The role of Bregs in transplantation has focused extensively on kidney allograft recipients, with limited published data on the contribution of these cells to operational tolerance of other transplanted organs. A comparison of operationally tolerant kidney and liver recipients indicated an enrichment and expansion of B cell-associated genes in kidney recipients (Lozano et al., 2011). The relevance of B cells to tolerance in non-kidney transplant recipients remains unknown.
In homeostasis, B cells express only low or transient levels of IL-10, which obligates the use of ex vivo stimulation to clearly distinguish which B cells have IL-10-producing capacity. Moreover, a wide variety of stimuli have been reported for elucidating this across Breg studies (Baba et al., 2015; Iwata et al., 2011; Mauri and Bosma, 2012; Mauri and Menon, 2017; Tedder, 2015). These studies have, however, illuminated the importance of Toll-like receptor (TLR) engagement and CD40 activation in potent IL-10 induction (Iwata et al., 2011; Lighaam et al., 2018; Nova-Lamperti et al., 2016; van de Veen et al., 2013) and the augmentation of IL-10 expression by exogenous cytokines, such as IL-2, IL-35, and IL-21 (Dambuza et al., 2017; Tedder and Leonard, 2014; Yoshizaki et al., 2012). The role of cognate stimulation via the B cell receptor (BCR) is less clear and, in some cases, has been shown to interfere with IL-10 production (Bankó et al., 2017; Iwata et al., 2011; Maseda et al., 2012; Nova-Lamperti et al., 2016; Parekh et al., 2003; Yanaba et al., 2009).
Previously, we defined the spectrum of human B cell subsets across multiple tissues and related their phenotypic, metabolic, and immune signaling profiles (Glass et al., 2020). Here, we focus specifically on human Bregs and apply mass cytometry to achieve high-dimensional characterization of IL-10+ B cells, with simultaneous quantification of 38 cell-identity and state-defining molecules, from healthy individuals and operationally tolerant liver allograft recipients. We evaluate the impact of TLR ligands, CD40 stimulation, and exogenous cytokines on the generation of IL-10+ B cells among total PBMCs and determine the potential for discrete B cell subsets to give rise to IL-10+ populations. Our results challenge the paradigm that Bregs constitute a distinct, homogeneous B cell subset that is identifiable by a single phenotypic profile. Instead, using prospective isolation and a live-cell tracing assay, our findings indicate that IL-10+ B cells can emerge from numerous previously defined B cell subsets. Moreover, the predominate IL-10+ B cell subset may vary depending on the stimuli and the immune status of the individual. Our study highlights the diversity of IL-10+ B cell states and suggests phenotypic heterogeneity within activated B cell compartments may help maintain a balance between pro-inflammatory and immunoregulatory responses.
RESULTS
Emergence of human IL-10-producing B cells depends on cytokine and activation environment
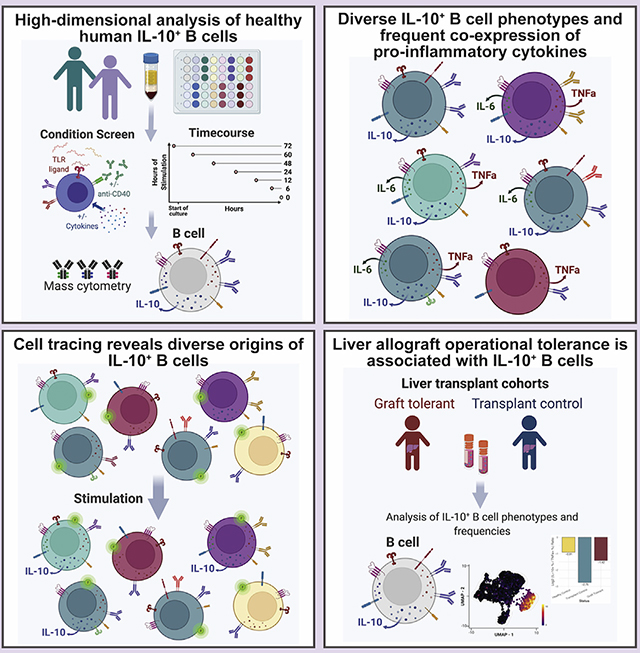
To identify stimuli that robustly induce IL-10-producing human peripheral B cells and assess their phenotypes, we activated human PBMCs from healthy individuals for 72 h with TLR ligands, anti-CD40, and exogenous cytokines, individually and in combination, and then applied mass cytometry to assess the expression of 24 surface and 14 intracellular proteins on total B cells (Figure 1A). We first compared the various stimulatory conditions (Figure 1B) for the ability to elicit IL-10 production from B cells in PBMCs from three healthy individuals (Figure 1C). TLR7/8 (R848) and TLR9 (CpG) stimulation, either alone or in combination with CD40 activation, yielded higher proportions of IL-10-producing B cells than did TLR4-based (lipopolysaccharide [LPS]) stimulation (Figure 1D). Of the 11 stimulation conditions tested, the cocktail of CpG, anti-CD40, IL-2, IL-21, and IL-35 (stimulation 10) was most effective in eliciting IL-10 production from B cells of healthy individuals (mean of 34.1% IL-10+ B cells) (Figure 1D).
Figure 1. IL-10+ B cell immunophenotype and cytokine production vary by activation microenvironment.
(A) Experimental workflow; n = 3 healthy individuals.
(B) Summary of in vitro stimulatory conditions.
(C) Exemplative biaxial plots demonstrating differential IL-10 expression by B cells from pooled donors under varying stimulation conditions. Unstimulated cells are depicted as condition 0.
(D) Percentage of IL-10+ B cells by condition and by individual (dots). Unstimulated B cell data are depicted as condition 0. Line depicts median, box depicts interquartile range (IQR), and whiskers depict IQR + −1.5*IQR.
(E) UMAP projection of donor-pooled B cells colored by stimulatory condition (top panel) and by positive status of the indicated cytokine (bottom panels). Mass-cytometry data were subsampled equally by condition. Manifold was derived using only expression of phenotypic markers.
(F) Top bar graph indicates the percentage of stimulated B cells by cytokine and GzmB expression status, as determined by Boolean gating of cytokine-positive cells. Co-expression status of populations are defined in the bottom dot-plot panel. Left bottom bar graph depicts percentage of cytokine/GzmB-positive B cells, regardless of co-expression, across conditions. Data were subsampled equally by condition.
(G) Median surface-marker expression for unstimulated B cells and IL-10+- and IL-10−-stimulated B cell subsets. Boxes indicate non-lineage markers with significant upregulation in IL-10+ B cells in 4 or more (of 11) stimulatory conditions.
(H) Relative difference in median expression of surface markers and cytokines in IL-10+- versus IL-10−-stimulated B cells by activating condition (1–11). Expression only shown for markers and cytokines with a difference greater than or equal to 0.1 and a p value less than 0.05. p values denote result of KS test with Bonferroni correction.
(I) Expression levels of CD38 and CD39 for IL-10− (blue) versus IL-10+ (red) B cells by stimulation condition. p values denote results of KS test with Bonferroni multiple-hypothesis correcting. Line depicts median, box depicts IQR, and whiskers depict IQR + − 1.5*IQR. *p < 0.05, **p < 0.01, and ***p < 0.001, and expression difference >0.1.
We applied uniform manifold approximation and projection (UMAP) (Becht et al., 2019) to visualize the range of phenotypes and cytokine profiles induced by the 11 different stimulation conditions (Figure 1E, top panel). IL-10+ cells were largely, though not exclusively, located in one island of the UMAP projection, but IL-35+ (p35), tumor necrosis factor alpha (TNFα)+, IL-6+, and IL-4+ B cells also occupied the same island indicating the polyfunctionality of IL-10+ B cells (Figure 1E, bottom panel). To quantify this polyfunctionality, we equally subsampled total B cells by stimulatory condition in silico and determined the proportion of those B cells that were able to produce IL-10 alone or in combination with IL-4, TNFα, IL-6, and/or the cytotoxic mediator GzmB (Figures 1F and S1A). While B cells that were exclusively IL-10 producers could be detected (blue bar), the greatest proportion of IL-10+ B cells also produced TNFa and IL-6 (purple bar) with lesser proportions of IL-10+ B cells also producing IL-4, GzmB, and the p35 subunit of IL-35.
To identify features that differentiate IL-10+ B cells from IL-10− B cells across the 11 different stimulation conditions, we compared expression of 24 surface markers and four cytokines between the two subsets (Figures 1G and 1H). IL-10+ B cells had significantly higher expression levels of IL-6 and/or TNFα compared with IL-10− B cells in most stimulatory conditions (p < 0.05, Kolmogorov-Smirnov [KS] test with Bonferroni correction). IL-10 and TNFα co-induction in ex-vivo-stimulated human B cells is consistent with reports from other groups (Cherukuri et al., 2014, 2021; Lighaam et al., 2018).Co-expression of IL-10 and IL-4 was significantly elevated with TLR4 and CD40 stimulation and with TLR7/8 ± CD40 stimulation (conditions 1, 2, and 5) (Figure 1H). In all stimulatory conditions, IL-10+ B cells show a highly activated B cell phenotype (Figures 1H, S1B, and S1C), which included upregulation of HLA-DR, CD38, CD183 (CXCR3), and CD197 (CCR7) and increased protein synthesis as indicated by puromycin labeling (Kimmey et al., 2019). While the median expression of these molecules was generally higher in IL-10+ B cells (Figure 1H), IL-10+- and IL-10−-stimulated B cells had similar expression profiles (Figure 1G).
Among individual surface markers, CD39, an immunomodulatory ectonucleotidase, most frequently demonstrated significant association with IL-10 expression by B cells (within conditions 1, 2, 4, 5, and 8) and generally had elevated median expression in IL-10+-stimulated B cells compared with IL-10−-stimulated B cells (Figures 1H, 1I, S1D, and S1E). CD38, an activation molecule and marker of rapidly proliferating cells, was the second most frequent non-lineage surface molecule to be significantly upregulated in IL-10+ B cells within the screen of stimulatory conditions. Notably, condition 11 induced a unique IL-10+ B cell population with significantly upregulated CD72, a BCR co-receptor with regulatory activity, and downregulated CD95, a memory-associated marker (Figure 1H). CD9 was also upregulated in IL-10+-stimulated B cells compared with IL-10− B cells in all but one stimulatory condition (9), but this expression change was not significant (Figures 1G and S1E). We also noted distinct CD23 upregulation and greater induction of CD23+ IL-10+ B cells upon R848-or LPS-based stimulation with CD40 activation and exogenous IL-4 and IL-10 compared with all other conditions (Figures S1B and S1C). Interestingly, CD23 is a negative regulator of BCR signaling as well as immunoglobulin G (IgG)/IgE antibody responses (Liu et al., 2016). Collectively, we observed that IL-10+ B cell surface phenotypes and cytokine profiles vary according to the specific stimulation environment. While CD39 most significantly identified exclusive IL-10-producing cells, no single phenotype captured these populations across all modes of activation.
Pro-inflammatory cytokines expression precedes B cell IL-10 induction
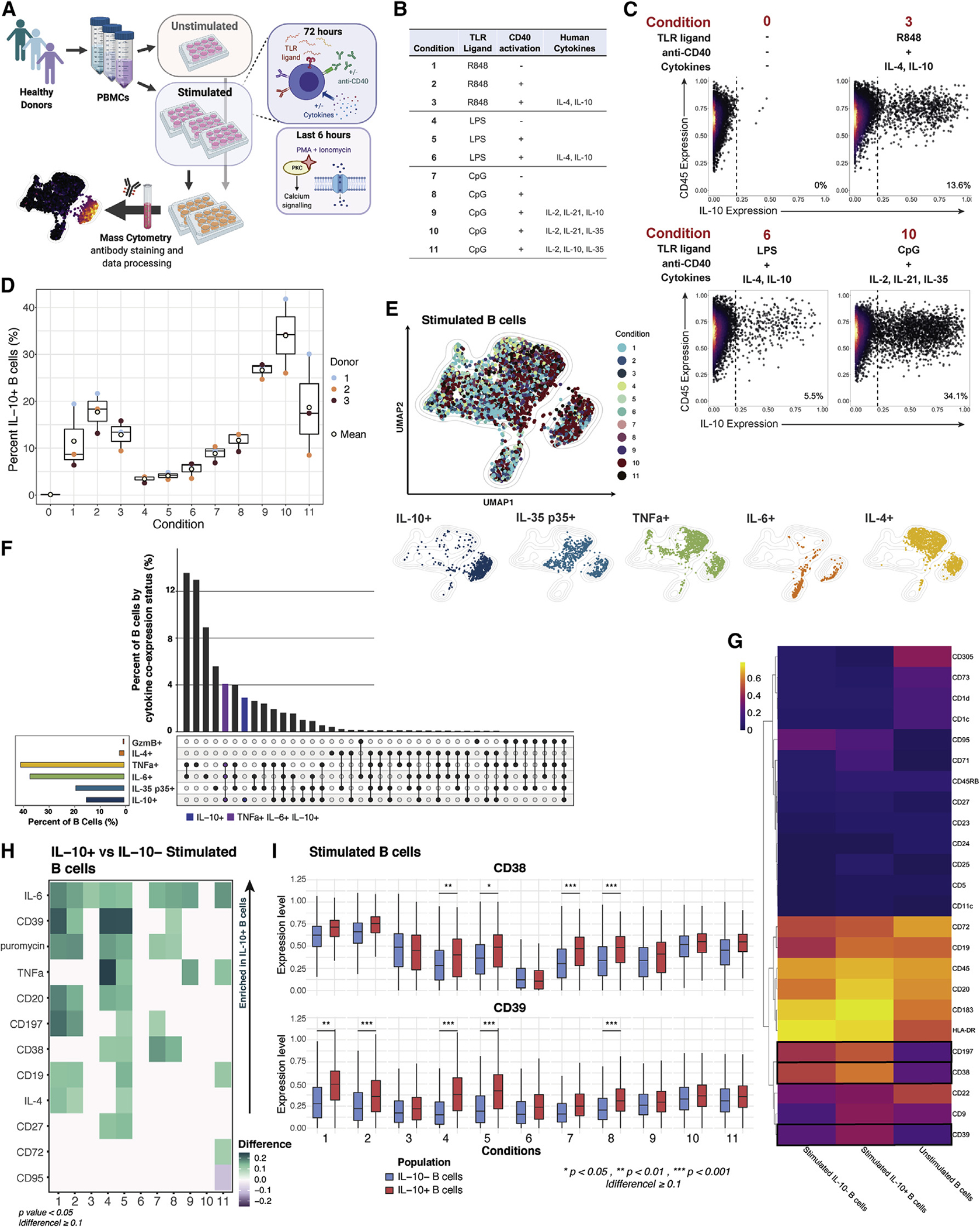
In our stimulation screen, the culture of PBMCs in the presence of CpG, anti-CD40, and exogenous recombinant human (rh) IL-2, IL-21, and IL-35 (condition 10) yielded maximal IL-10 expression by peripheral B cells. To map the temporal dynamics of cytokine production and marker expression by B cells, we recovered total PBMCs after 6, 12, 24, 48, 60, and 72 h of stimulation and applied mass-cytometry and high-dimensional analysis (Figure 2A). The frequency of IL-10+ B cells peaked at 48 h of ex vivo stimulation, and this observation was consistent across individuals (Figure 2B). Notably, high expression of the pro-inflammatory cytokines TNFα and IL-6 preceded IL-10 induction (Figures 2C, 2D, S2A, and S2B). This sequence of cytokine expression was consistent across individuals (Figures S2C and S2D). At the earliest time point analyzed (6 h), most B cells expressed both TNFα and IL-6, while <1% of cells expressed IL-10 (Figures 2C and S2B). TNFα+IL-10+ polyfunctional B cells began to emerge at 12–24 h, and by 48 h post-stimulation, a significant population of B cells that expressed only IL-10 was present, while the TNFα+ B cell population substantially contracted (Figures 2C, 2D, and S2A). The TNFα and IL-10 co-expression profile of total B cells remained largely stable between 48 and 72 h post-stimulation. These data suggest that in the early phase following activation, B cells predominantly provide inflammatory signals that are subsequently replaced by a more Breg state, including a peak of IL-10 production at 48 h of stimulation.
Figure 2. Overlapping expression of pro-inflammatory cytokines precedes IL-10 in activated B cells.
(A) Experimental workflow; n = 2 healthy individuals.
(B) Percentage of IL-10+ B cells by stimulation time point and individual (dots). Cross bar indicates mean percentage.
(C) Biaxial plots of donor-pooled total B cells by time point. Red box within the plot for time point 6 (h) highlights increasing TNFα expression. Purple boxes within the plots for time points 12, 24, and 48 (h) highlight increasing proportion of B cells co-expressing TNFα and IL-10. Blue box within plot for time point 48 (h) highlights increased proportion of B cells producing IL-10 alone. Unstimulated B cell data are depicted as time point 0.
(D) Percentage of stimulated B cells producing TNFα and IL-10 by time point (h).
(E) Difference in median expression of surface markers in unstimulated versus stimulated donor-pooled B cells by time point. Boxes in heatmap indicate markers with an absolute value of difference greater than 0.1. Expression difference only shown for markers and cytokines with a p value less than 0.05 (gray fill indicates p > 0.05). p values denote results of KS test with Bonferroni correction.
(F) Median expression level of markers with diverging patterns between donor-pooled IL-10+ and IL-10− B cells by time point. p values denote results of KS test with Bonferroni multiple hypothesis correcting. *p < 0.05. ***p < 0.001.
(G) UMAP plots of donor-pooled B cells colored by stimulation time point (large panel), by positive status of indicated cytokine expression, and by expression level of indicated surface markers (small panels). Light blue outline highlights IL-10+ B cell groupings. Mass-cytometry data were subsampled equally by time point. Unstimulated B cell data are depicted as time point 0. Manifold was derived using expression of only phenotypic markers.
To broadly characterize the features of ex-vivo-activated B cells at the key point of maximal IL-10 expression, we compared stimulated and unstimulated cell states. Total stimulated B cells recovered at 48 h indicated significant upregulation of the activation molecules CD38, CD197, and CD183 and the transferrin receptor CD71, as well as CD9, a multifunctional surface tetraspanin that impacts cellular proliferation, activation, and migration (Figures 2E, S2E, and S2F). In contrast, stimulated B cells exhibited significant downregulation of the inhibitory surface proteins CD72 and CD305 followed by the immune inhibitory control molecule CD73 and the lipid-antigen-presenting molecule CD1d (Figures 2E, S2E, and S2F). Memory markers CD27 and CD45RB and the B cell lineage marker CD19 were also downregulated upon stimulation (Figures 2E, S2E, and S2F). These results highlight the dynamics and complexity of B cell surface-molecule expression following activation and the associated challenges of applying classical gating strategies to isolate subsets of activated B cells.
Activation-induced IL-10+ B cell phenotypes mirror the overall B lymphocyte population
To determine if IL-10+ B cells have a unique and stable phenotype that persists following activation, we compared expression of 24 surface markers on IL-10+ and IL-10− stimulated B cells over a 72 h period. The transferrin receptor CD71, the immunoregulatory markers CD9 and CD25, and the activation molecules CD39 and CD197 were significantly upregulated (p < 0.05) on IL-10+ B cells compared with IL-10− B cells at 48 h or later after activation (Figures 2F and S2G). However, none of these markers were enriched in IL-10+ B cells across all time points. The activation molecule CD38 was enriched among IL-10+ B cell populations of some 72 h stimulation conditions of our screen (Figure 1H). In the time course, which assessed a single stimulation condition, CD38 was upregulated on IL-10+ cells only at late-activation time points, and this difference was not statistically significant compared with IL-10−-stimulated B cells (Figures 2F and S2G). Embedding B cells from all stimulated time points by UMAP highlighted that among the transiently IL-10-associated B cell markers, CD9, CD38, and CD39 were more widely expressed across the IL-10+ compartment than were CD25 and CD71 (Figure 2G, blue outlines). IL-10+ B cell populations were also enriched for higher protein-synthesis levels (indicated by puromycin) but were phenotypically indistinguishable from TNFα+ B cells (Figure 2G). In summary, no single surface marker uniquely defines IL-10+ B cells over time.
Conventional Breg immunophenotypes capture few IL-10-producing B cells and enrich for pro-inflammatory cytokines
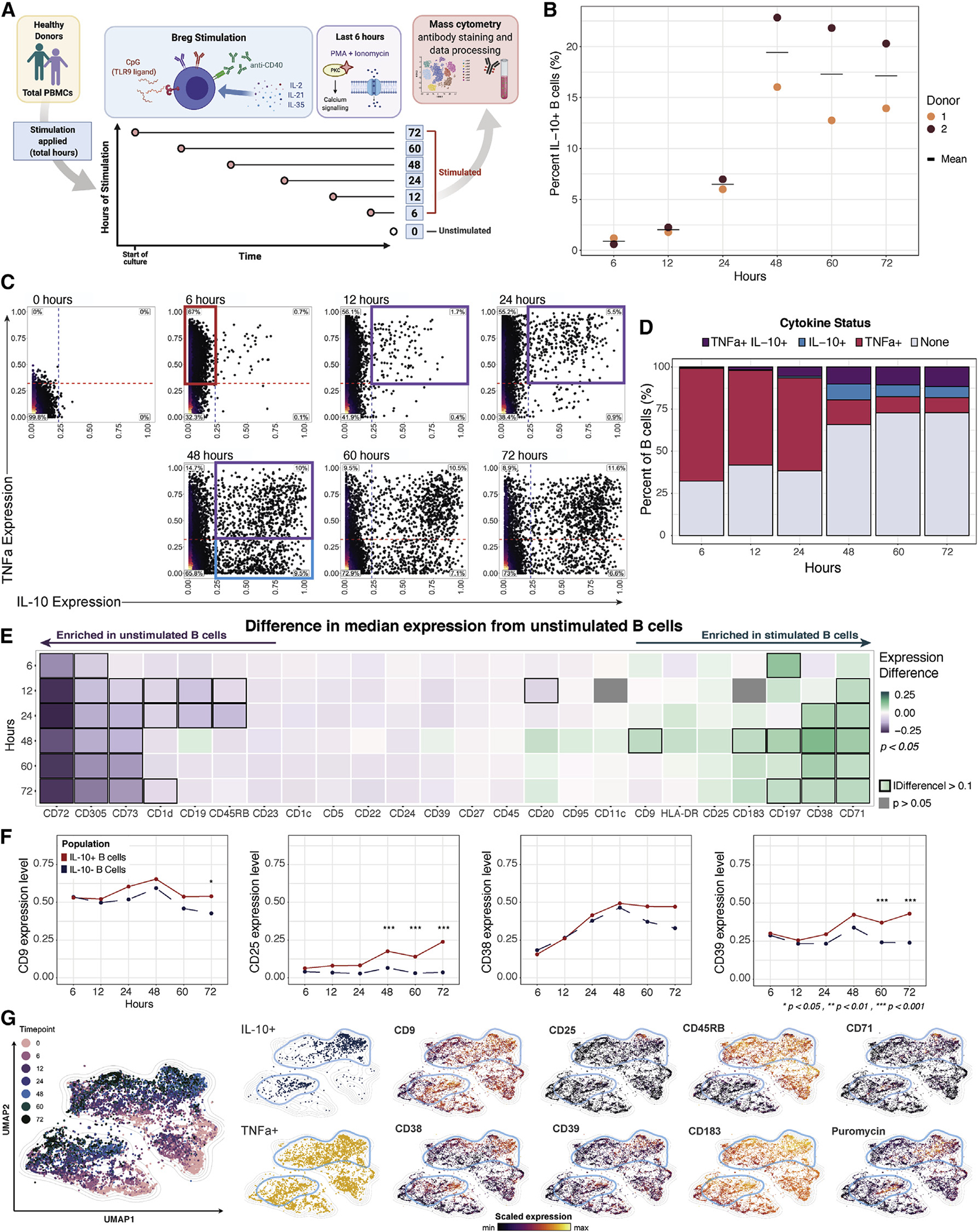
To reconcile our results with previous observations (Wortel and Heidt, 2017), we superimposed seven previously defined human Breg immunophenotypes onto our stimulation and time-course datasets and determined the fraction of total CD19+ B cells that they represent (Figure 3A). While each of these reported phenotypes were captured in our data, four out of seven collectively represented <2% of stimulated B cells as well as <2% of IL-10+ B cells (Figures 3A and 3B). The remaining three phenotypes, marked by CD1dhi, CD25+, and CD39hi, defined approximately 3%, 14%, and 12% of stimulated B cells as well as 3%, 25%, and 18% of IL-10+ B cells, respectively, in the pooled datasets (Figure 3A). Undefined or “other” phenotypes represented the majority (52%) of the IL-10+ B cell compartment (Figure 3B). Additionally, “other” B cells were the predominant population among IL-10+ B cells in 10 of the 11 B cell stimulatory conditions in the screen and in 4 of the 6 time points in the CpG, anti-CD40, and rhIL-2-, IL-21-, and IL-35-based stimulation analysis (Figure 3C). Thus, the majority of IL-10+ B cells were not accounted for by previously described Breg subsets. CD25+ and CD39hi B cells were the only subsets enriched in the IL-10+ B cell population compared with stimulated B cells (Figures 3B and 3C). Additionally, the relative proportions of the Breg phenotypes among total IL-10+ B cells varied substantially depending upon the condition and duration of stimulation and varied slightly by individual (Figures 3C and S3A).
Figure 3. Conventional Breg immunophenotypes associated with pro-inflammatory cytokines and a limited overlap with IL-10+ cells.
(A) Representative biaxial plots demonstrating gating of previously defined Breg surface phenotypes from the pooled stimulation-condition screen and time-course dataset; n = 5 healthy individuals. Dashed lines and solid boxes indicate thresholds of surface-marker expression level within gating of B cell subsets. Gating strategy for Breg subsets defined by >2 surface markers are indicated with 2 biaxial plots. Percentage of subset out of total donor-pooled B cells is indicated in the top right panel of each corresponding plot.
(B) UMAP projection of donor-pooled IL-10+ B cells colored by phenotype (left panel) and with overlay of unique B cell phenotypes (right panels). Frequency of B cell phenotypes among donor-pooled IL-10+ B cells is indicated in the top right corner of each corresponding plot. Other phenotype represents all remaining B cells not classified with specific surface-marker-based phenotypes. Mass-cytometry data were subsampled equally by condition and time point. Manifold was derived using expression of only phenotypic markers.
(C) Percentage of B cell phenotypes among total donor-pooled IL-10+ B cells by condition (duration of 72 h only) from stimulation screen (left panel) and by time point (condition 10 only; in h) from stimulation time course (right panel).
(D) Biaxial plots of TNFα and IL-10 expression for CD25+, CD39hi, and other B cells from stimulation screen. Dashed lines indicating threshold for TNFα+ status (red) and IL-10+ status (blue) are based on unstimulated B cell expression. Percentage of B cells in each quadrant is indicated.
(E)TNFα and IL-6 expression levels for total unstimulated B cells versus stimulated CD25+, CD39hi, and other phenotype B cells (left panels) and for stimulated IL-10+ CD25+, IL-10+ CD39hi, and IL-10+ other phenotype B cells (right panels). Diamond indicates median; dashed line indicates threshold for cytokine positivity based on unstimulated B cell expression. p values denote results of KS test with Bonferroni multiple-hypothesis correcting. *p < 0.05, and expression difference >0.1.
As CD25+ and CD39hi B cells were enriched within some IL-10-producing B cell populations, we further investigated their pro-inflammatory and immunoregulatory cytokine balance. We pooled B cell data from the stimulation screen and determined the co-expression of IL-10 and TNFα. Among CD25+ and CD39hi B cell populations, TNFα+IL-10− cells were highly abundant (37% and 34%, respectively), followed by TNFα+IL-10+ cells (20% and 15%, respectively) (Figures 3D and S3B). IL-10+TNFα− cells comprised 11% and 13% of the total CD25+ and CD39hi populations, respectively. The high proportion of TNFα+ cells among these phenotypes indicates that they are strongly activated B cells with pro-inflammatory features in addition to immunoregulatory cytokine secretion. Other phenotype B cells had a lower proportion of TNFα+IL-10− cells (27%) and TNFα+IL-10+ cells (7%), indicating a less pro-inflammatory bias (Figures 3D and S3B). Both the total CD25+ and CD39hi populations, as well as their IL-10+ fractions, expressed higher median levels of TNFα and IL-6 than the undefined or other B cell populations in all individuals (Figures 3E and S3C). Jointly, these data suggest that the IL-10-expressing CD25+ and CD39hi B cells have a comparatively high pro-inflammatory profile relative to other IL-10+ B cell subsets.
The expression of several surface molecules that have been reported as IL-10 associated in murine and human B cells (CD1c, CD1d, and CD5) decreased upon B cell-specific stimulation and were not correlated with peripheral B cell IL-10 expression, regardless of stimulation (Figure S4). Proposed Breg marker TIM-1 (CD365) was not detected on total unstimulated, stimulated, or IL-10+ human B cells, although TIM-1 expression was detected on in-vitro-activated CD4+ T cells (Figure S5). Collectively, previously observed Breg phenotypes comprise a limited proportion of IL-10+ B cell populations.
Multiple B cell subsets can produce IL-10, and distinct polyfunctional profiles arise from each subset
While stimulation of B cells is required for robust production of IL-10, the immunophenotype of these cells is severely perturbed and not reflective of the resting state. To understand the origins of IL-10-producing B cells, we fluorescent-activated cell sorted (FACS) six canonical B cell populations from PBMCs: CD38+CD24+ transitional, CD45RB−CD27−CD38−CD24− naive, CD45RB+CD27−memory, CD45RB+CD27+ memory, CD95+ memory, and CD19hi CD11c+ effector cells (Glass et al., 2020). The purified populations were labeled with carboxyfluorescein succinimidyl ester (CFSE), co-cultured with a 10:1 mixture of autologous PBMCs, stimulated for 48 h, and analyzed by mass cytometry (Figure 4A). CFSE+ cells were then selected in silico to associate the resulting stimulated B cells to their original phenotypes (Good et al., 2019).
Figure 4. Live-cell tracing reveals emergence of IL-10-producing B cells from multiple subsets and stimulation-induced phenotypic changes.
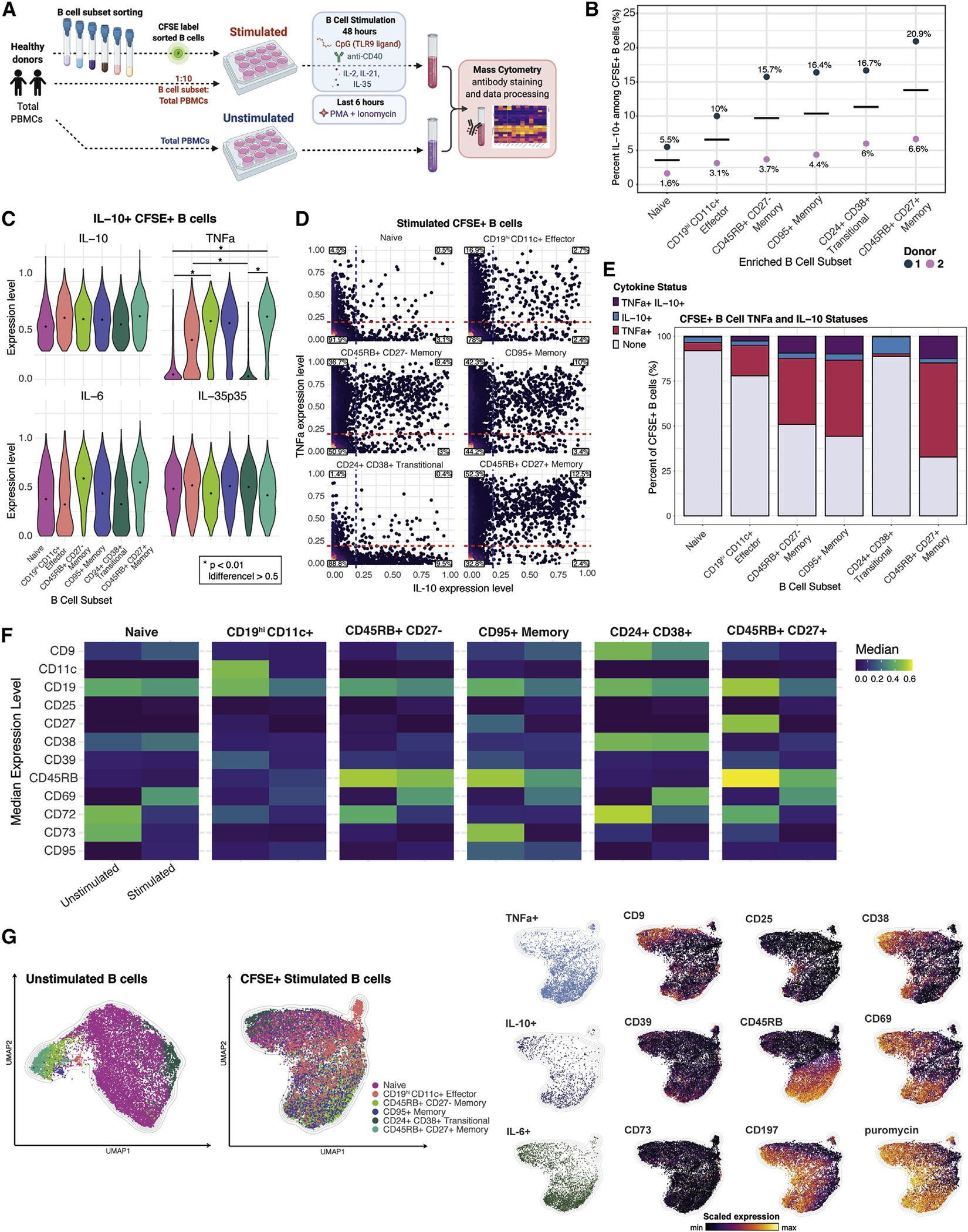
(A) Experimental workflow; n = 2 healthy individuals.
(B) Percentage of IL-10+ cells within stimulated CFSE+ B cells according to resting B cell phenotype and individual (dots). Cross bar indicates mean percentage for each subset.
(C) Expression levels of indicated cytokines by each of the CFSE+ IL-10+ donor-pooled B cell subsets. Diamond indicates median expression. p values denote results of KS test with Bonferroni multiple-hypothesis correcting. *p < 0.01, and expression difference >0.5.
(D) Biaxial plots of TNFα and IL-10 for stimulated CFSE+ donor-pooled B cell subsets. Dashed lines indicating threshold for TNFα+ status (red) and IL-10+ status (blue) are based on unstimulated B cell expression. Percentage of B cells in each quadrant is indicated.
(E) Percentage of B cells by TNFα and IL-10 expression for stimulated CFSE+ donor-pooled B cell subsets.
(F) Median expression level of surface markers with differential expression in unstimulated versus stimulated B cells in each of the six donor-pooled B cell subsets.
(G) Separately generated UMAP manifolds of unstimulated B cells (left panel) and stimulated CFSE+ B cells (right panels) colored by B cell subset, by positive status of indicated cytokine expression, and by expression level of indicated surface markers. Equal subsampling of unstimulated and stimulated B cells from mass-cytometry data. Each manifold contains 18,000 cells and was derived using expression of only phenotypic markers.
The proportion of IL-10+ cells that emerged from specific B cell subsets varied by individual, but the rank order of IL-10 positivity was consistent (Figure 4B). CD45RB+CD27+ memory and CD24+CD38+ transitional B cell subsets yielded the two highest proportions of IL-10+ B cells relative to the naive, CD19hiCD11c+ effector, CD45RB+CD27− memory, and CD95+ memory subsets (Figure 4B). Naive B cells yielded the lowest proportion of IL-10+ cells (Figure 4B). Notably, CD19hiCD11c+ effector B cells produce IL-10 cytokine (Figure 4B). The CD19hiCD11c+ effector memory subset corresponds to the CD21lo, T-bet+, and PD-1+ effector or atypical memory B cell populations described in prior studies (Glass et al., 2020).
To investigate how polyfunctionality varies by subset, we compared the expression levels of cytokines produced by IL-10+ B cells that originated from each of the sorted B cell subsets. The median IL-10 expression level of IL-10+ cells within each B cell maturation stage were similar, with slightly higher expression in CD45RB+CD27+ memory cells (Figures 4C and S6A). Whereas memory B cell subsets had increased TNFa expression levels, in comparison, the expression levels by IL-10+ cells from the naive and transitional subsets were markedly lower. IL-10+ CD19hiCD11c+ effector cells also had a moderately reduced TNFα expression level (Figure 4C). The median IL-6 expression levels in IL-10+ cells were also higher in the memory cell subsets compared with the effector, naive, and transitional subsets, while IL-35 p35 subunit expression levels were comparable among all IL-10+ B cell subsets, with some variation by individual (Figures 4C and S6A).
To gain further insight into the cytokine features of these B cell subsets, we specifically quantified the proportion of IL-10- and TNFα-producing cells within each population. Activated CD24+CD38+ transitional B cells yielded minimal TNFα+ IL-10− cells (<0.5%) but gave rise to the highest proportion of IL-10+ TNFα− cells (~10%) among all B cell subsets and across individuals (Figures 4D, 4E, S6B, and S6C). This aligns with prior reports indicating transitional B cells had greater regulatory effects on conventional CD4+ T cells than naive or memory B cells (Cherukuri et al., 2014, 2017, 2021; Hasan et al., 2019; Laguna-Goya et al., 2020; Newell et al., 2010; Nova-Lamperti et al., 2016;Shabir et al., 2015). Activation of naive B cells yielded relatively few TNFα+ or IL-10+ cells, such that most cells did not produce either cytokine (~92%) (Figures 4D, 4E, S6B, and S6C). This suggests that naive B cell have a higher activation threshold and/or lower responsiveness to Breg-specific stimulatory conditions, mirroring our recent findings that naive cells are more anergic than other B cell subsets (Glass et al., 2020). CD45RB+CD27+ memory B cells had the highest proportion of TNFα+ IL-10− cells (~52%) and TNFα+ IL-10+ cells (~13%) and the lowest proportion of TNFα− IL-10− cells (~33%) (Figures 4D, 4E, S6B, and S6C). Memory B cells, particularly the CD45RB+CD27+ B cell subset, are more skewed toward pro-inflammatory responses than other B cell maturation states under the same activation conditions.
The activation markers, CD69, and, to a lesser extent, CD25 were upregulated by all B cell subsets, confirming their responsiveness to the stimuli utilized (Figure 4F). CD9, which has been associated with IL-10+ Bregs, was upregulated within the naive, CD24+ CD38+ transitional, CD45RB+ CD27− memory, and CD95+ memory subsets, with individual variation (Figures 4F and S6D). Median expression of the activation molecule CD38 was higher for the naive, CD45RB+ CD27− memory, and CD45RB+ CD27+ memory subsets. Interestingly, CD39 was downregulated, but still detectable, on all B cell subsets. The reduction in CD39 expression was surprising given previously reported correlations between IL-10 and CD39 expression (Hasan et al., 2019, 2021). The canonical memory marker CD27 was downregulated by all memory B cell subsets, while the median expression levels of CD45RB decreased only slightly within 3 of the 4 memory B cell subsets. CD72 and CD73 were markedly downregulated by all B cell subsets, resulting in a complete loss of expression in the case of CD73 (Figures 4F and S6D).
The requirement of Breg-specific stimulation for the detection of IL-10-producing B cells led to substantial phenotypic changes within all B cell subsets analyzed (Figure 4F). Upon stimulation, surface-marker expression changes within distinct B cell subsets caused these populations to become phenotypically similar and largely indistinguishable, thereby precluding the isolation of the subsets by resting B cell phenotype gating strategies. UMAP visualizations of unstimulated and stimulated B cell subsets reveal the phenotypic changes and loss of distinct surface features (i.e., cells colored by resting B cell subset become highly overlapping within the plot) among the subsets upon stimulation (Figure 4G). Furthermore, stimulated IL-10+ B cells within all subsets showed moderate positive correlations with expression of the surface markers CD9, CD39, CD38, CD69, and CD197 as well as the pro-inflammatory cytokines and protein-synthesis activity (Figures 4G and S6E). Together, tracking of B cell subsets through ex vivo stimulation showed that each B cell subset can potentially produce IL-10, yet each subset’s IL-10+ population bears unique, concurrent pro-inflammatory cytokine profiles.
IL-10-producing B cells potential is increased in operationally tolerant liver allograft recipients
To assess the IL-10-producing B cells in transplantation, we stimulated the PBMCs of liver transplant recipients receiving maintenance IS, operationally allograft tolerant liver transplant recipients off IS for >1 year, and healthy controls and assessed the immunosuppressive IL-10 versus pro-inflammatory cytokine production by their B cell compartments (Table S1; Figure 5A). Importantly, the proportions of unstimulated B cell subsets among total peripheral B cells did not significantly differ between the operationally graft-tolerant liver transplant recipients and transplant-recipient controls (Figure S7A). We determined that operationally graft-tolerant liver transplant recipients who had maintained their allograft in the absence of IS had a significantly higher proportion of B cells capable of IL-10 production (p = 0.02) and overall elevated expression levels of IL-10 compared with liver transplant-recipient controls (Figures 5B and S7B; Table S2). In contrast, there was no significant difference in the proportion of TNFα+ or IL-6+ B cells between the groups (Figures 5B and S7C). Operationally graft-tolerant patients and healthy control subjects had higher proportions of IL-10+ TNFα+ B cells (16% and 18%, respectively) and IL-10+ TNFα− B cells (8% and 10%, respectively) compared with control transplant recipients (6% IL-10+ TNFα+ and 2% IL-10+ TNFα−) (Figure 5C). The proportions of IL-10− TNFα+ B cells were similar between the two liver transplant-recipient groups and lower in the healthy control group. We quantified the log2 ratio of IL10+:TNFα+ total B cells and found the operationally tolerant cohort had an approximately 2-fold higher ratio than the transplant-control cohort, suggesting a greater immunoregulatory bias of their B cell compartment (Figures 5D and S7D).
Figure 5. Operationally graft-tolerant liver transplant recipients are enriched for IL-10-producing B cells.
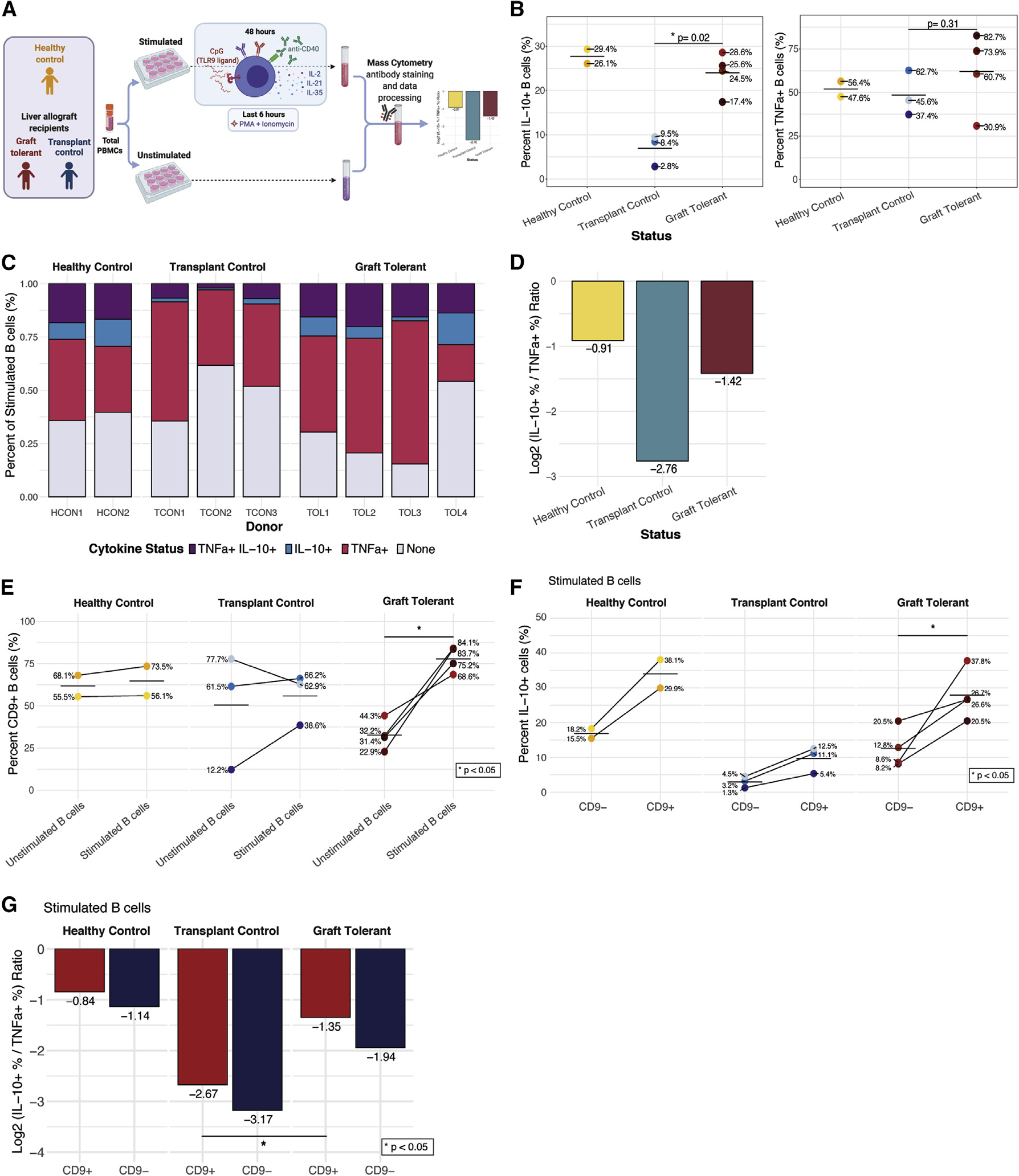
(A) Experimental workflow to analyze B cells of liver transplant recipients on IS (n = 3), operationally allograft-tolerant liver transplant recipients off IS for >1 year(n = 4), and healthy controls (no transplant; n = 2).
(B) Percentage of IL-10+ and TNFα+ B cells by group and individual (dots). Cross bar indicates mean percentage for each group. p values denote results of Wilcoxon rank-sum test. *p < 0.05.
(C) Percentage of B cells with each TNFα and/or IL-10 expression status among total stimulated B cells by group and individual.
(D) Log2 ratio of percentage of IL-10+ to TNFα+ donor-pooled B cells by group.
(E) Percentage of CD9+ B cells among unstimulated and stimulated B cells by group and individual. p value denotes results of Wilcoxon signed-rank test. *p < 0.05.
(F) Percentage of IL-10+ cells among CD9−- and CD9+-stimulated B cells by group and individual. p value denotes results of Wilcoxon signed-rank test. *p < 0.05.
(G) Log2 ratio of percentage of IL-10+ to TNFα+ cells for CD9+ (red) and CD9− (blue) donor-pooled B cells by group. p value denotes results of Wilcoxon signed-rank test. *p < 0.05.
CD9+ cells were uniquely and significantly (p < 0.05) increased within stimulated B cells of operationally tolerant liver allograft recipients (Figures 5E and S7E). The CD9+ B cell populations of operationally tolerant graft recipients had a significantly higher proportion of IL-10+ cells compared with CD9− populations (p < 0.05). Furthermore, the proportions of IL-10+ B cells in CD9+ and CD9− populations were greater in all operationally tolerant graft recipients than in all transplant controls (Figure 5F). CD9+ B cells also had a higher log2 ratio of IL-10+:TNFα+ B cells compared with CD9− populations (Figures 5G and S7F). The stimulated CD9+ B cell population of operationally tolerant transplant recipients had a significantly higher ratio of IL-10+:TNFα+ cells (i.e., was enriched for IL-10 production) compared with transplant controls (p < 0.05). However, CD9 expression was not detected on all IL-10+ B cells in any of the subjects, nor did all CD9+ B cells produce IL-10 (Figures 5F and S7G). In summary, operationally graft-tolerant liver transplant recipients exhibited a higher frequency and magnitude of IL-10 production in B cells with a corresponding increase in the trafficking molecule CD9.
DISCUSSION
High-dimensional immune profiling revealed human IL-10-producing peripheral B cells exhibit highly activated, yet heterogeneous, phenotypes. By screening across activating conditions and time points, we found that IL-10+ B cell surface and cytokine profiles were diverse and strongly influenced by the method and duration of activation. Cell tracing revealed that a variety of resting, canonical B cell subsets can give rise to IL-10+ B cells. In fact, no single, unifying phenotype captured the majority of IL-10+ B cells. Instead, we found that IL-10+ B cells extend well beyond formerly proposed Breg phenotypes, such as CD39hi and CD25+ B cells (Wortel and Heidt, 2017). The IL-2 receptor alpha chain (CD25) was upregulated on a minor subset of IL-10+ B cells yet was expressed by a higher frequency of B cells after longer in vitro stimulations, suggesting that this upregulation may be partially driven by autocrine IL-10 production (Fluckiger et al., 1993). In summary, IL-10+ B cells lack the features of a dedicated immunological subset and represent an impermanent state. These findings confirm and extend the work of Lighaam et al., who showed that ex vivo IL-10 production is the transient property of an activated B cell state (Lighaam et al., 2018).
Based on our data, we hypothesize that the regulatory capacity of B cells may be more accurately quantified by their cytokine co-expression profiles than solely by the detection of IL-10 expression. A significant portion of IL-10+ B cells co-expressed IL-6 and TNFα, suggesting that IL-10+ B cells are not a distinct, monofunctional population. It is possible that a feedforward effect of IL-6 and/or TNFα enhances IL-10 production by activated B cells. Prior studies established that human IL-10+ peripheral B cells co-express the pro-inflammatory cytokines TNFα and IL-6 (Cherukuri et al., 2014; Lighaam et al., 2018) and that transitional B cells have a higher ratio of IL-10:TNFα. These observations coincided with the finding that transitional B cells demonstrate significantly enhanced immunoregulatory activity on CD4+ T cells, suggesting that IL-10:TNFα ratios reflect a meaningful metric of immunoregulatory capacity (Cherukuri et al., 2014, 2017, 2021; Hasan et al., 2019; Laguna-Goya et al., 2020; Newell et al., 2010; Nova-Lamperti et al., 2016; Shabir et al., 2015). The immunoregulatory bias of transitional B cells may result from their recent emigration from the bone marrow, in which inflammation during B cell lymphopoiesis would be undesirable.
Our ability to understand the heterogeneity of the IL-10+ B cell compartment hinged on comprehensively assessing ex-vivo-stimulation conditions and durations. Both direct activation of B cells and indirect non-B cell signals likely contributed to the induction of IL-10 production by B cells, enabling analysis in a more physiologically relevant setting. We found that CpG stimulation combined with a CD40 agonist, IL-2, IL-21, and IL-35 was the strongest inducer of IL-10 expression in B cells, and the peak of this IL-10 production was at 48 h of ex vivo stimulation. In a related study, TLR9-based activation with CpG yielded the highest proportion of IL-10+ cells and the highest levels of IL-10 expression in adult B cells, as compared with all other TLR agonists, with CD40 activation (Iwata et al., 2011). Prior studies established that stimulation of human B cells with a CpG TLR9 agonist and CD40 activation induces IL-10 production and immunosuppressive function from specific IL-10+ B cell subsets (Cherukuri et al., 2014, 2021; Iwata et al., 2011). These immunosuppressive activities include inhibition of proliferation, activation, and pro-inflammatory cytokine (IFNγ, TNFα) expression from CD4+ T cell in humans (Bouaziz et al., 2010; Cherukuri et al., 2014, 2021; Iwata et al., 2011). The overlap of the focal activating cocktail used in our study, and in these prior works, reinforces the notion that IL-10+ B cell populations profiled here likely possess immunoregulatory activity. Importantly, these prior studies provide initial support that human peripheral B cell function can be interpreted through their cytokine expression profiles.
Similar to other studies in humans, we could not compare IL-10 production by B cells from multiple tissues in the same individual. Tissue-resident IL-10-producing B cells may share characteristics with those circulating in peripheral blood. In fact, a prior study established that B cells derived from newborn cord blood and adult tonsil and spleen tissues exhibited the highest IL-10 production after stimulation with CpG and CD40 activation for 48 h in a screen of ex vivo stimulations (Iwata et al., 2011). This previous work and the data from the current study highlight the necessity and potency of a TLR9 agonist and CD40 activation in induction of IL-10 from human B cells. A comprehensive and high-dimensional assessment of tissue-defined IL-10+ B cell phenotypes would be a useful resource and follow up to the present study.
Initially, we profiled healthy individuals to understand the dynamics of IL-10+ B cells in homeostasis. To extend these findings to a clinical scenario, we analyzed peripheral IL-10+ B cells in operationally tolerant liver transplant recipients, as these cells could contribute to the establishment or maintenance of allograft tolerance. Most studies of clinical transplantation have focused on the levels of Bregs as indicators of impending rejection or long-term outcome rather than on transplant tolerance. We analyzed liver transplant recipients, as it has been suggested that liver recipients include a larger proportion, compared with other solid-organ recipients, of individuals who are spontaneously tolerant and could maintain their allograft in the absence of IS (Dai et al., 2020; Feng, 2016; Lerut and Sanchez-Fueyo, 2006). We found that operationally allograft-tolerant liver transplant recipients have a higher proportion of IL-10-producing B cells and elevated expression levels of IL-10 by B cells (similar to healthy control subjects) compared with transplant-recipient controls. The same was not true of TNFα and IL-6. Thus, B cells from the operationally tolerant cohort are skewed toward an immunoregulatory state that may contribute to immune tolerance and allograft homeostasis in this cohort.
The tetraspanin CD9 is well-established as a marker of murine Bregs with immunosuppressive activity and IL-10 production (Braza et al., 2015; Brosseau et al., 2018; Matsushita et al., 2016; Sun et al., 2015). In humans, B cells of healthy individuals are known to co-express CD9 and IL-10 upon activation, and CD9 expression is enriched in transitional B cells (Hasan et al., 2019). Here, we reported a significant correlation between the expression of CD9 and IL-10 by stimulated B cells of operationally tolerant liver transplant recipients and a higher IL-10:TNFα ratio in CD9+ versus CD9- cells (Figure 5). However, not all CD9+ B cells produced IL-10 and, in fact, 38%–85% of IL-10−-stimulated B cells were CD9+. Interestingly, the positive correlation between IL-10 and CD9 expression by B cells was not significant within healthy individuals until 72 h of stimulation. CD9 is therefore associated with IL-10 production but is neither a requirement nor a fixed surface marker of IL-10+ B cells.
In conclusion, we have utilized high-dimensional cytometry to reveal the phenotypic diversity and origins of human IL-10+ B cells and demonstrate the absence of a single, canonical subset of IL-10+ B cells in the peripheral blood of healthy humans and liver transplant recipients. Our findings provide a comprehensive overview of phenotypic and cytokine expression profiles of IL-10+ human B cells across a range of ex-vivo-activating conditions and durations. This study emphasizes the importance of assessing B cell polyfunctionality alongside IL-10 expression to accurately distinguish between pro-inflammatory- and immunoregulatory-skewed populations. The dynamic and heterogeneous nature of IL-10-producing B cells, and their enrichment among transitional B cells, should be considered when analyzing total or sorted IL-10+ B cell populations. Despite the variable origins and potential impermanence of IL-10+ B cells, the impact of the IL-10+-activated B cell state has been clearly associated with immunoregulatory activity and outcomes in human disease and allograft responses. This work should serve as a resource to future studies on IL-10-producing B cells and their immunoregulatory contributions in humans.
Limitations of the study
In this work, we deeply profiled the polyfunctionality and immunophenotypic states of human IL-10-producing B cells from cohorts of healthy individuals and transplant recipients. Robust induction of IL-10 by B cells in vitro requires stimulations of greater than 24 h, which may not be representative of in vivo conditions. Furthermore, the molecular mechanism(s) of IL-10 induction remains a question for future work. To assay a large number of stimulation conditions, time points, and sorted cell populations, we multiplexed individual donors across these myriad experimental conditions, which necessitated using smaller numbers of individuals. Thus, the consistency of our findings should be tested across more individuals of varying demographics. In the clinical cohort, transplant controls were on IS, while graft-tolerant subjects were not, which is an unavoidable dilemma of transplant immunology studies due to standard of care. The absence of IS could contribute to some differences in B cell phenotypes and frequencies. Furthermore, while our analysis suggests CD9 expression and IL-10:TNFα ratios of B cells are associated with, and could serve as, biomarkers for allograft tolerance, it remains to be determined if these features are causative or merely correlated.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Olivia Martinez (omm@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human specimens
Deidentified human blood was obtained from healthy adult donors (Stanford Blood Center, n = 9). Healthy adult donors consisted of 3 female and 6 male individuals with an age range of 19–49 years old (mean age 31). Blood samples were also collected from pediatric and adult liver transplant recipients (n = 7) at Lucile Packard Children’s Hospital and Stanford Hospital. Liver allograft transplants recipient blood donors consisted of 4 female and 3 male individuals with an age range of 0.42–21 years old (mean age 14). All samples were obtained under informed consent and in accordance with Stanford’s Institutional Review Board.
METHOD DETAILS
Study design
The purpose of this study was to provide comprehensive and high-dimensional analyses of the phenotypic heterogeneity and polyfunctionality of IL-10+ B cells in the context of healthy individuals and operationally graft tolerant liver transplant recipients. To this end, we tested in vitro stimulation conditions and durations of human PBMC samples for optimal B cell IL-10 expression. We assessed the origins of IL-10-producing cells by sorting six defined B cell subsets and tracked these cells through the Breg-inducing activation process with CFSE labeling. Finally, we applied optimum in vitro conditions identified from prior experiments to PBMCs from immunosuppressed organ transplant recipients and operationally graft tolerant transplant recipients to investigate the IL-10+ B cell profile in these patient groups.
Peripheral blood processing
PBMCs (lymphocytes) were purified from fresh whole blood samples using Ficoll-Paque (GE Healthcare) gradients. Healthy blood donor and transplant recipient patient cells were processed immediately for in vitro stimulations or cryopreserved in pure fetal bovine serum with 10% dimethyl sulfoxide at −80°C overnight and −196°C long-term.
Cell culture of peripheral blood mononuclear cells
Total PBMCs were suspended in RPMI 1640 supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, L-glutamine, HEPES, sodium pyruvate, glucose and 2-Mercaptoethanol (Gibco; 50 μM) at 1 × 106 cells/mL and rested for 30 minutes. In the stimulation screen experiment, rested PBMCs were stimulated for 72 hours with TLR ligand, with or without CD40 activation and with or without a variety of exogenous recombinant human cytokines, as outlined in Figures 1A and 1B. Stimulation reagents and concentrations are outlined in Table S3. In all stimulations, phorbol 12-myristate 13-acetate (PMA; 50 ng/mL), ionomycin (1 μg/mL), and Brefeldin A (BFA; 1x) were added for the final 5 hours of culture. In the stimulation timecourse experiment, rested PBMCs were stimulated with CpG ODN2006 (10 μg/mL) and anti-CD40 (500 ng/mL) as well as exogenous recombinant human IL-2 (600 IU/mL), IL-21 (100 ng/mL) and IL-35 (20 ng/mL) for 6, 12, 24, 48, 60 and 72 hours (outlined in Figure 2A), with addition of PMA, ionomycin, and BFA in the final 5 hours. In the B cell sorting and transplant recipient cohort experiments, the same Breg-specific stimulation with CpG, anti-CD40 and exogenous cytokines as well as PMA/ionomycin and BFA was applied for a total of 48 hours (outlined in Figures 4A and 5A). All PBMC cultures were maintained at 37°C and 5% CO2.
Mass cytometry processing and acquisition
Antibody conjugation, staining, and data acquisition were performed as previously described (Hartmann et al., 2018). Briefly, metal-isotope labeled antibodies used in this study were conjugated using the MaxPar X8 Antibody Labeling kit per manufacturer instruction (Fluidigm) or were purchased from Fluidigm pre-conjugated. Each conjugated antibody was quality checked and titrated to optimal staining concentration using a combination of primary human cells (Figures S4 and S5). PBMCs from cultures were spiked with a tri-molecular biosynthesis cocktail, consisting of 5-Bromouridine (BRU; Sigma, 5 mM), 5-Iodo-2-deoxyuridine (IdU; Sigma, 100 uM) and puromycin (Sigma, 10 μg/mL), and incubated for 30 minutes (Kimmey et al., 2019), then washed in cell staining media (CSM; PBS with 0.5% BSA and 0.02% sodium azide and benzonase 25 × 108 U/mL (Sigma)) and resuspended in 0.5μM cisplatin in phosphate-buffered saline (PBS) for 5 minutes to label non-viable cells (Sigma). Cells were washed in CSM and fixed with 1.6% PFA in PBS for 10 minutes at room temperature (RT). Fixed cells were Palladium barcoded (Fluidigm) as previously described (94). Barcoded cells were then suspended in TruStain FC blocker (Biolegend) for 10 minutes at RT and washed in CSM prior to staining. All surface staining was performed in CSM for 30 minutes at RT (antibodies listed in Table S4). Cells were washed in CSM and permeabilized with 0.02% saponin (Sigma) in CSM for 30 minutes on ice. Intracellular and anti-biotin staining was performed in 0.02% saponin in CSM for 1 hour at RT. Before acquisition, samples were washed in CSM and resuspended in intercalation solution (1.6% PFA in PBS, 0.02% saponin (Sigma) and 0.5 μM iridium-intercalator (Fluidigm)) for 1 hour at RT or overnight at 4°C to label DNA. Before acquisition, samples were washed once in CSM and twice in ddH2O. All samples were filtered through a 35μm nylon mesh cell strainer, resuspended at 1 × 106 cells/mL in ddH2O supplemented with 1x EQ four element calibration beads (Fluidigm), and acquired on a CyTOF2 mass cytometer (Fluidigm) using the Super Sampler injection system (Victorian Airship).
CD365/TIM-1 mass cytometry antibody validation
CD4+ T cell stimulation method consisted of total PBMCs treated with 3 days with plate-bound anti-CD3 (10 ug/mL) and anti-CD28 (5 ug/mL) and soluble human IL-2 (50 IU/mL) and IL-4 (12.5 ug/mL) and addition of Brefeldin A (1x) in the final 5 hours. Breg stimulation method consisted of total PBMCs treated with 2 days with CpG ODN2006 (10 ug/mL) and anti-CD40 (500 ng/mL) as well as exogenous recombinant human IL-2 (600 IU/mL), IL-21 (100 ng/mL) and IL-35 (20 ng/mL) with addition of PMA (50 ng/mL), ionomycin (1 ug/mL), and Brefeldin A (1x) in the final 5 hours.
Sorting of B cell subsets and dye-based cell tracking
Total PBMCs were isolated from Trima Accel leukocyte reduction system (LRS) chambers as described above and then Fc blocked and subjected to magnetic lineage depletion according to the manufacturer’s instructions using BD Streptavidin Particles Plus and the BD IMag Cell Separation Magnet (BD Biosciences) with a cocktail of biotinylated antibodies consisting of CD3, CD7, CD15, CD33, CD56, CD61, and CD235ab (Biolegend). The cells bound by the biotinylated antibody cocktail were captured by streptavidin particles for depletion. The remaining B cell-enriched suspensions were stained for surface antigens (antibodies listed in Table S4) in CSM in the dark for 30 minutes on ice and then washed in CSM. Stained cell suspensions were incubated with eFluor 780 viability stain (eBioscience; 1:1000 dilution) in the dark for 5 minutes on ice to label non-viable cells, then immediately washed in PBS and transferred for cell sorting on a BD Biosciences FACS Aria II equipped with violet, blue and red lasers (Stanford Blood Center, Flow Cytometry Core). Within the cell sorting phase, total B cells were defined by surface expression as CD20+ and B cell subsets were identified as CD24−CD38− naïve, CD24+CD38+ transitional, CD45RB+CD27+ memory, CD45RB+CD27− memory, CD95+ memory, and CD11c+ effector memory cells. 200,000 cells from each B cell subset were sorted into pure FBS on ice and immediately washed 3 times in 37°C PBS and labeled with CellTrace carboxyfluorescein succinimidyl ester (CFSE) Cell Proliferation Kit (Invitrogen; 1μM) according to the manufacturer’s instructions, followed by an RPMI wash and final resuspension in 37°C complete RPMI (as described above). The sort gating strategies for individual B cell subsets and validation of CFSE-labeling of these subsets are shown in Figure S8. Each CFSE-labeled B cell subset was added to two replicate cultures with autologous donor total PBMCs at a 1:10 final concentration (sorted B cell subsets:PBMCs) and 2 × 106 cells/mL. A Breg-specific stimulation, as described above, was applied to final cultures for 48 hours (outlined in Figure 4A).
QUANTIFICATION AND STATISTICAL ANALYSIS
Mass cytometry data pre-processing
Data processing were performed as previously described (Glass et al., 2020). Briefly, acquired samples were bead-normalized using MATLAB-based software as previously described (Finck et al., 2013) and debarcoded using MATLAB-based software (Zunder et al., 2015). Normalized data was then uploaded onto CellEngine (cellengine.com, Primity Bio, Fremont, CA) for singlet (Tsai et al., 2020) and B cell gating (Figure S9). Gated data was downloaded and further processed with the R programming language (http://www.r-project.org) and Bioconductor (http://www.bioconductor.org) software. Data was transformed with an inverse hyperbolic sine (asinh) transformation with a cofactor of 5. Each marker was scaled to the 99.9th percentile of expression of all cells in that experiment. Cytokine positive thresholds for B cells were defined by the 99.9th percentile of expression for each cytokine in unstimulated B cells within each experiment. Batch correction was not applied as all samples within an experiment were barcoded and acquired in the same mass cytometry run, except for individual samples in the B cell subset sorting experiment. For the B cell sorting experiment, batch effect was minimal, so markers were 99.9th percentile scaled by batch and then combined for analysis.
Data visualization
To visualize co-expression of molecules measured on different B cell populations in the stimulation screen, timecourse and B cell sorting experiments, cells labeled with the same mass cytometry panels were subsampled (equally by B cell population, for the screen analysis) and plotted on a UMAP plot using the umap package in R (Figures 1E, 2E, 3B, 4G). The plots were generated based on expression of all phenotypic markers expressed on B cells. In the stimulation screen and B cell phenotype analysis, the UMAP plots were additionally based on cytokine expression (Figures 1E and 3B). Isotype was not used to generate maps to prevent artificial separation of phenotypically similar cells. Subsampling by B cell population facilitated visualization of heterogeneity without the map being dominated by the most abundant populations.
For activated B cell state analysis, stimulated B cells were over-clustered into 100 clusters using FlowSOM with all detectable surface molecules as input. Clusters were then hierarchically clustered based on expression of B cell surface molecules and isotype. Unstimulated B cells were over-clustered with FlowSOM and manually assigned to cell subsets, as described previously (Glass et al., 2020).
Statistics
All statistical tests for differences in distribution of markers and cytokines between B cell populations from mass cytometry data were performed on equally subsampled populations using the KS test. This non-parametric test determines the equality of two continuous distributions. All p values were corrected by the Bonferroni method, the most conservative of multiple hypothesis correction approaches. For comparisons of single marker expression levels between B cell populations and frequencies of single B cell populations between conditions, timepoints or clinical groups, the Wilcoxon rank sum test was performed with Bonferroni correction. The details of statistical tests and p value thresholds for individual experimental data analyses are described in relevant main and supplementary figures and figure legends.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-CD45 (clone HI30 ) - 89 Y | Biolegend | Cat # 3089003B |
| anti-CD9 (clone HI9a) - 141 Pr | Biolegend | Cat # 312102 |
| anti-CD39/ENTPD1 (clone A1) - 142 Nd | Biolegend | Cat # 328202 |
| anti-CD5 (clone UCHT2) - 143 Nd | Biolegend | Cat # 300602 |
| anti-CD45RB (clone MEM-55) - 145 Nd | Fluidigm | Cat # 3145009B |
| anti-CD20 (clone 2H7) - 147 Sm | Fluidigm | Cat # 3147001B |
| anti-CD25 (clone BC96) - 149 Sm | Biolegend | Cat # 3149010B |
| anti-CD11c (clone Bu15) - 150 Nd | Biolegend | Cat # 337221 |
| anti-CD71 (clone CY1G4) - 151 Eu | Biolegend | Cat # 334102 |
| anti-IgK (clone A8B5) - 153 Eu | Invitrogen | Cat # MA110385 |
| anti-CD27 (clone M-T271) - 155 Gd | Biolegend | Cat # 3155001B |
| anti-CD183/CXCR3 (clone G025H7) - 156 Gd | Fluidigm | Cat # 3156004B |
| anti-CD72 (clone 3F3) - 157 Gd | Biolegend | Cat # 316202 |
| anti-CD23 (clone EBVCS-5) - 160 Gd | Biolegend | Cat # 338502 |
| anti-CD365/TIM-1 (clone 1D12) - 161 Dy | Biolegend | Cat # 353902 |
| anti-CD95/FASR (clone DX2) - 164 Dy | Biolegend | Cat # 3164008B |
| anti-CD19 (clone HIB19) - 165 Ho | Biolegend | Cat # 302202 |
| anti-CD197/CCR7 (clone G043H7) - 167 Er | Fluidigm | Cat # 3167009A |
| anti-CD73/NT5E1 (clone AD2) - 168 Er | Biolegend | Cat # 3168015B |
| anti-CD24 (clone ML5) - 169 Tm | Biolegend | Cat # 3169004B |
| anti-HLA-DR (clone L243) - 170 Er | Fluidigm | Cat # 3170013B |
| anti-CD1d (clone 51.1) - 171 Yb | Biolegend | Cat # 350302 |
| anti-CD38 (clone HIT2) - 172 Yb | Biolegend | Cat # 3172007B |
| anti-CD1c (clone L161) - 173 Yb | Biolegend | Cat # 331502 |
| anti-IgL (clone MHL-38) - 174 Yb | BD Biosciences | Cat # 316602 |
| anti-CD305/LAIR1 (clone NKTA255) - 176 Yb | Genetex | Cat # GTX00485 |
| anti-CD22 (clone HIB22) - 209 Bi | Biolegend | Cat # 302511 |
| anti-Biotin (clone 1D4-C5) - 113 ln | Biolegend | Cat # 409002 |
| anti-IgM (polyclonal) - 140 Ce | Invitrogen | Cat # H15000 |
| anti-IL-4 (clone MP4-25D2) - 144 Nd | Fluidigm | Cat # 3144010B |
| anti-IgD (clone IA6-2) - 146 Nd | Biolegend | Cat # 3146005B |
| anti-IgA (polyclonal) - 148 Nd | Fluidigm | Cat # 3148007B |
| anti-TNFa (clone Mab11) - 152 Sm | Fluidigm | Cat # 3152002B |
| anti-IL-6 (clone MQ2-13A5) - 154 Sm | Biolegend | Cat # 501102 |
| anti-Puromycin (clone 12D10) - 158 Gd | Miltenyi Biotec | Cat # MABE343 |
| anti-IL-35/IL-12 p35 (clone 27537) - 159 Tb | RD Systems | Cat # MAB1570-100 |
| anti-IgG (clone M1310G05) - 162 Dy | Biolegend | Cat # 410701 |
| anti-Granzyme B (clone 351927) - 163 Dy | RD Systems | Cat # MAB2906 |
| anti-IL-10 (clone JES3-9D7) - 166 Er | Biolegend | Cat # 501423 |
| anti-BrU (clone 3D4) - 175 Lu | BD Biosciences | Cat # BDB555627 |
| anti-CD3 (clone OKT3) - Biotin | Biolegend | Cat # 317320 |
| anti-CD7 (clone CD7-6B6) - Biotin | Miltenyi Biotec | Cat # 130-105-839 |
| anti-CD15 (clone HI98) - Biotin | Biolegend | Cat # 301914 |
| anti-CD33 (clone WM53) - Biotin | Biolegend | Cat # 303426 |
| anti-CD56 (clone 5.1H11) - Biotin | Biolegend | Cat # 362536 |
| anti-CD61 (clone Y2/51) - Biotin | Miltenyi Biotec | Cat # 130-098-522 |
| anti-CD235ab (clone HIR2) - Biotin | Biolegend | Cat # 306618 |
| anti-CD20 (clone 2H7) - PerCP-Cy5.5 | Biolegend | Cat # 302319 |
| anti-CD38 (clone HIT2) - FITC | Biolegend | Cat # 303527 |
| anti-CD24 (clone ML5) - Brilliant Violet 421 | Biolegend | Cat # 311113 |
| anti-CD45RB (clone MEM-55) - PE | Biolegend | Cat # 310204 |
| anti-CD27 (clone O323) - Brilliant Violet 605 | Biolegend | Cat # 302829 |
| anti-CD95 (clone DX2) - Brilliant Violet 510 | Biolegend | Cat # 305639 |
| anti-CD11c (clone Bu15) - Alexa Fluor 647 | Biolegend | Cat # 337229 |
| anti-CD14 (clone M5E2) - 113 In | Biolegend | Cat # 301802 |
| anti-CD11b (clone ICRF44) - 115 In | Biolegend | Cat # 301302 |
| anti-CD16 (clone 3G8) - 143 Nd | Biolegend | Cat # 3209002B |
| anti-CD69 (clone FN50) - 144 Nd | Fluidigm | Cat # 3144018B |
| anti-CD4 (clone RPA-T4) - 151 Eu | Biolegend | Cat # 300502 |
| anti-TIGIT (clone MAB7898) - 153 Eu | Fluidigm | Cat # 3153019B |
| anti-CD3 (clone UCHT1) - 156 Gd | Biolegend | Cat # 300402 |
| anti-CD8a (clone RPA-T8) - 171 Yb | Biolegend | Cat # 301002 |
| anti-CD279/PD1 (clone EH12.2H7) - 174 Yb | Fluidigm | Cat # 3174020B |
| anti-CD127 (clone A019D5) - 176 Yb | Biolegend | Cat # 351302 |
| anti-CD5 (clone UCHT2) - 209 Bi | Biolegend | Cat # 300602 |
| anti-CD152/CTLA4 (clone 14D3) - 160 Gd | Invitrogen | Cat # 14-1529-82 |
| anti-IL17A (clone BL168) - 161 Dy | Fluidigm | Cat # 3161008B |
| anti-IFNg (clone B27) - 168 Er | Fluidigm | Cat # 3168005B |
| anti-CFSE/FITC (polyclonal) - 198 Pt | Southern Biotech | Cat # 6400-01 |
| Purified anti-human CD28 (clone CD28.2) | Biolegend | Cat # 302933 |
| Purified anti-human CD3 (clone OKT3) | Biolegend | Cat # 317325 |
| Soluble human anti-CD40/TNFRSF5 clone 82111 | RD Systems | Cat # MAB6321-100 |
|
| ||
| Biological samples | ||
|
| ||
| Leukocyte reduction chamber from adult human blood | Stanford Blood Center | stanfordbloodcenter.org |
| Whole blood from healthy adult donors | Obtained under informed consent with IRB approval | n/a |
| Whole blood from pediatric and adult liver transplant recipients | Obtained under informed consent with IRB approval | n/a |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Ficoll-Paque Plus | GE Healthcare | Cat # 300-25 |
| RPMI 1640 media (ATCC formulation) | Thermo Fisher Scientific | Cat # A1049101 |
| Fetal bovine serum USDA approved lot | Omega Scientific, Inc. | Cat # FB-01 |
| 2-mercaptoethanol | Gibco | Cat # 21-985-023 |
| Benzonase nuclease | Sigma-Aldrich | Cat # E1014-25KU |
| R848 | Mabtech | Cat # 3660-1 |
| Lipopolysaccharides (LPS) from Escherichia coli O127:B8 | Sigma | Cat # L4516-1MG |
| CpG ODN 2006 | Invivogen | Cat # tlrl-2006 |
| Recombinant human IL-4 | RD Systems | Cat # 204-IL-020 |
| Recombinant human IL-10 Biolegend Cat # 571002 | ||
| Recombinant human IL-2 | Mabtech | Cat # 3660-1 |
| Recombinant human IL-21 | RD Systems | Cat # 8879-IL-010 |
| Recombinant human IL-35 | Enzo Life | Cat # ALX-522-140-C010 |
| Phorbol 12-myristate 13-acetate | Sigma | Cat # P8139-1MG |
| Ionomycin calcium salt from Streptomyces conglobatus | Sigma | Cat # I0634-1 MG |
| Brefeldin A Solution | eBioscience | Cat # 00-4506-51 |
| Human TruStain FcX (Fc Receptor Blocking Solution) | Biolegend | Cat # 422302 |
| DPBS | Thermo Fisher Scientific | Cat # 14190250 |
| Bovine Serum Album (BSA) Heat-shock Treated | Thermo Fisher Scientific | Cat # BP1600-100 |
| Cell-ID Intercalator-Ir | Fluidigm | Cat # 201192A |
| Cell-ID Cisplatin | Fluidigm | Cat # 201064 |
| Saponin | Sigma-Aldrich | Cat # 84510-100G |
| Calibration Beads, EQ, Four Element | Fluidigm | Cat # 201078 |
| Fixable Viability Dye eFluor 780 | eBioscience | Cat # 65-0865-14 |
| Streptavidin Particles Plus | BD Biosciences | Cat # 557812 |
| FITC Streptavidin | Biolegend | Cat # 405201 |
| CellTrace carboxyfluorescein succinimidyl ester (CFSE) Cell Proliferation Kit | Invitrogen | Cat # C34554 |
| 5-Bromouridine | Sigma | Cat # 850187 |
| 5-Iodo-2-deoxyuridine | Sigma | Cat # I7125 |
| Puromycin, Dihydrochloride - CAS 58-58-2 | Sigma | Cat # P212121 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Cell-ID 20-Plex Pd Barcoding Kit | Fluidigm | Cat # 201060 |
| Maxpar X8 Antibody Labeling Kit | Fluidigm | Cat # 201176B |
| IMag Cell Separation Magnet | BD Biosciences | Cat # 552311 |
|
| ||
| Software and algorithms | ||
|
| ||
| RStudio | RStudio | rstudio.com |
| Cytobank | Cytobank | cytobank.org |
| CellEngine | Primity Bio | cellengine.com |
Highlights.
Mass-cytometry analysis reveals human IL-10+ B cells do not have a unique phenotype
Cell tracing shows that a diversity of human B cell subsets can produce IL-10
IL-10+ B cells often co-express the pro-inflammatory cytokines TNFα and IL-6
Liver-allograft operational tolerance is associated with IL-10+ B cell frequency
ACKNOWLEDGMENTS
We thank the flow cytometry core staff at Stanford Blood Center for guidance and assistance with cell sorting. We also thank P. Favaro, J. Pena, and J. Toh for guidance with B cell-sorting plans, F. Hartmann, D. Mrdjen, and J. Harden for guidance with mass-cytometry experiments, and T. Bruce, S. Greenbaum, S. Kimmey, E. McCaffery, and A. Calderon for experimental contributions. We thank B. Sahaf and K. Bruton for advice on optimization of B cell culture conditions. Figures were created using BioRender (http://www.biorender.com) and Illustrator (Adobe). The primary funding support of this work was NIH grant UO1 AI35947 (O.M.M.). O.M.M., S.M.K., and M.C.G were supported by NIH grant U01 AI35947-02. O.M.M. was supported by NIH grant RO1 AI113130. M.C.G. was supported by NIH Molecular and Cellular Immunology Training Grant institutional postdoctoral fellowship 5 T32 AI07290. D.R.G. was supported by the Bio-X SIGF. J.-P.O. was supported by the CIHR Postdoctoral Fellowship. B.M. was supported by the Transplant and Tissue Engineering Center at Lucile Packard Children’s Hospital postdoctoral fellowship and the Stanford MCHRI postdoctoral fellowship. S.C.B. was supported by NIH grants 1DP2OD022550-01, 1R01AG056287-01, 1R01AG057915-01, R01AG068279, 1U24CA224309-01, UH3 CA246633, and U19 AG065156-01 and The Bill and Melinda Gates Foundation.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110728.
REFERENCES
- Baba Y, Matsumoto M, and Kurosaki T (2015). Signals controlling the development and activity of regulatory B-lineage cells. Int. Immunol. 27, 487–493. [DOI] [PubMed] [Google Scholar]
- Baba Y, Saito Y, and Kotetsu Y (2020). Heterogeneous subsets of B-lineage regulatory cells (Breg cells). Int. Immunol. 32, 155–162. [DOI] [PubMed] [Google Scholar]
- Bankó Z, Pozsgay J, Szili D, Tóth M, Gáti T, Nagy G, Rojkovich B, and Sármay G (2017). Induction and differentiation of IL-10–producing regulatory B cells from healthy blood donors and Rheumatoid arthritis patients. J. Immunol. 198, 1512–1520. [DOI] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2019). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44. [DOI] [PubMed] [Google Scholar]
- Blair PA, Noreña LY, Flores-Borja F, Rawlings DJ, Isenberg DA, Ehrenstein MR, and Mauri C (2010). CD19+CD24hiCD38hi B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic Lupus Erythematosus patients. Immunity 32, 129–140. [DOI] [PubMed] [Google Scholar]
- Bouaziz J-D, Calbo S, Maho-Vaillant M, Saussine A, Bagot M, Bensussan A, and Musette P (2010). IL-10 produced by activated human B cells regulates CD4+ T-cell activation in vitro. Eur. J. Immunol. 40, 2686–2691. [DOI] [PubMed] [Google Scholar]
- Braza F, Chesne J, Durand M, Dirou S, Brosseau C, Mahay G, Cheminant MA, Magnan A, and Brouard S (2015). A regulatory CD9+ B-cell subset inhibits HDM-induced allergic airway inflammation. Allergy 70, 1421–1431. [DOI] [PubMed] [Google Scholar]
- Brosseau C, Durand M, Colas L, Durand E, Foureau A, Cheminant M-A, Bouchaud G, Castan L, Klein M, Magnan A, et al. (2018). CD9+ regulatory B cells induce T cell apoptosis via IL-10 and are reduced in severe asthmatic patients. Front. Immunol. 9, 3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budczies J, Kirchner M, Kluck K, Kazdal D, Glade J, Allgäuer M, Kriegsmann M, Heußel C-P, Herth FJ, Winter H, et al. (2021). A gene expression signature associated with B cells predicts benefit from immune checkpoint blockade in lung adenocarcinoma. Oncoimmunology 10, 1860586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán D, Mansilla MA, Ferrier A, Soto L, Oleinika K, Aguillón JC, and Aravena O (2021). Immunosuppressive mechanisms of regulatory B cells. Front. Immunol. 12, 611795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherukuri A, Rothstein DM, Clark B, Carter CR, Davison A, Hernandez-Fuentes M, Hewitt E, Salama AD, and Baker RJ (2014). Immunologic human renal allograft injury associates with an altered IL-10/TNF-α expression ratio in regulatory B cells. J. Am. Soc. Nephrol. 25, 1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherukuri A, Salama AD, Carter CR, Landsittel D, Arumugakani G, Clark B, Rothstein DM, and Baker RJ (2017). Reduced human transitional B cell T1/T2 ratio is associated with subsequent deterioration in renal allograft function. Kidney Int. 91, 183–195. [DOI] [PubMed] [Google Scholar]
- Cherukuri A, Salama AD, Mehta R, Mohib K, Zheng L, Magee C, Harber M, Stauss H, Baker RJ, Tevar A, et al. (2021). Transitional B cell cytokines predict renal allograft outcomes. Sci. Transl. Med. 13, eabe4929. [DOI] [PubMed] [Google Scholar]
- Chesneau M, Pallier A, Braza F, Lacombe G, Gallou SL, Baron D, Giral M, Danger R, Guerif P, Aubert-Wastiaux H, et al. (2014). Unique B cell differentiation profile in tolerant kidney transplant patients. Am. J. Transplant. 14, 144–155. [DOI] [PubMed] [Google Scholar]
- Chesneau M, Michel L, Dugast E, Chenouard A, Baron D, Pallier A, Durand J, Braza F, Guerif P, Laplaud D-A, et al. (2015). Tolerant kidney transplant patients produce B cells with regulatory properties. J. Am. Soc. Nephrol. 26, 2588–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Zheng Y, Thomson AW, and Rogers NM (2020). Transplant tolerance induction: insights from the liver. Front. Immunol. 11, 1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza IM, He C, Choi JK, Yu C-R, Wang R, Mattapallil MJ, Wingfield PT, Caspi RR, and Egwuagu CE (2017). IL-12p35 induces expansion of IL-10 and IL-35-expressing regulatory B cells and ameliorates autoimmune disease. Nat. Commun. 8, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Ellis G, Pallant C, Lopes AR, Khanna P, Peppa D, Chen A, Blair P, Dusheiko G, Gill U, et al. (2012). IL-10–Producing regulatory B cells in the pathogenesis of chronic Hepatitis B virus infection. J. Immunol. 189, 3925–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S (2016). Spontaneous and induced tolerance for liver transplant recipients. Curr. Opin. Organ Transplant. 21, 53–58. [DOI] [PubMed] [Google Scholar]
- Fillatreau S, Sweenie CH, McGeachy MJ, Gray D, and Anderton SM (2002). B cells regulate autoimmunity by provision of IL-10. Nat. Immunol. 3, 944–950. [DOI] [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, and Bendall SC (2013). Normalization of mass cytometry data with bead standards. Cytometry A 83A, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Borja F, Bosma A, Ng D, Reddy V, Ehrenstein MR, Isenberg DA, and Mauri C (2013). CD19+CD24hiCD38hi B cells maintain regulatory T cells while limiting TH1 and TH17 differentiation. Sci. Transl. Med. 5, 173ra23. [DOI] [PubMed] [Google Scholar]
- Fluckiger AC, Garrone P, Durand I, Galizzi JP, and Banchereau J (1993). Interleukin 10 (IL-10) upregulates functional high affinity IL-2 receptors on normal and leukemic B lymphocytes. J. Exp. Med. 178, 1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DR, Tsai AG, Oliveria JP, Hartmann FJ, Kimmey SC, Calderon AA, Borges L, Glass MC, Wagar LE, Davis MM, et al. (2020). An integrated multi-omic single-cell atlas of human B cell identity. Immunity 53, 217–232.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good Z, Borges L, Vivanco Gonzalez N, Sahaf B, Samusik N, Tibshirani R, Nolan GP, and Bendall SC (2019). Proliferation tracing with single-cell mass cytometry optimizes generation of stem cell memory-like T cells. Nat. Biotechnol. 37, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FJ, Simonds EF, and Bendall SC (2018). A universal live cell barcoding-platform for multiplexed human single cell analysis. Sci. Rep. 8, 10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MM, Thompson-Snipes L, Klintmalm G, Demetris AJ, O’Leary J, Oh S, and Joo H (2019). CD24hiCD38hi and CD24hiCD27+ human regulatory B cells display common and distinct functional characteristics. J. Immunol. 203, 2110–2120. [DOI] [PubMed] [Google Scholar]
- Hasan MM, Nair SS, O’Leary JG, Thompson-Snipes L, Nyarige V, Wang J, Park W, Stegall M, Heilman R, Klintmalm GB, et al. (2021). Implication of TIGIT+ human memory B cells in immune regulation. Nat. Commun. 12, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Matsushita T, Horikawa M, DiLillo DJ, Yanaba K, Venturi GM, Szabolcs PM, Bernstein SH, Magro CM, Williams AD, et al. (2011). Characterization of a rare IL-10–competent B-cell subset in humans that parallels mouse regulatory B10 cells. Blood 117, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen K, Cevhertas L, Ma S, Satitsuksanoa P, Akdis M, and Veen W (2021). Regulatory B cells. A. Z. Allergy 00, 1–17. [DOI] [PubMed] [Google Scholar]
- Kimmey SC, Borges L, Baskar R, and Bendall SC (2019). Parallel analysis of tri-molecular biosynthesis with cell identity and function in single cells. Nat. Commun. 10, 1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna-Goya R, Utrero-Rico A, Cano-Romero FL, Gómez-Massa E, González E, Andrés A, Mancebo-Sierra E, and Paz-Artal E (2020). Imbalance favoring follicular helper T cells over IL10+ regulatory B cells is detrimental for the kidney allograft. Kidney Int. 98, 732–743. [DOI] [PubMed] [Google Scholar]
- Lee KM, Stott RT, Zhao G, SooHoo J, Xiong W, Lian MM, Fitzgerald L, Shi S, Akrawi E, Lei J, et al. (2014). TGF-β-producing regulatory B cells induce regulatory T cells and promote transplantation tolerance. Eur. J. Immunol. 44, 1728–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerut J, and Sanchez-Fueyo A (2006). An appraisal of tolerance in liver transplantation. Am. J. Transplant. 6, 1774–1780. [DOI] [PubMed] [Google Scholar]
- Lighaam LC, Unger P-PA, Vredevoogd DW, Verhoeven D, Vermeulen E, Turksma AW, ten Brinke A, Rispens T, and van Ham SM (2018). In vitro-induced human IL-10+ B cells do not show a subset-defining marker signature and plastically co-express IL-10 with pro-inflammatory cytokines. Front. Immunol. 9, 1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Richard K, Wiggins M, Zhu X, Conrad DH, and Song W (2016). CD23 can negatively regulate B-cell receptor signaling. Sci. Rep. 6, 25629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Abente J, Prieto-Sanchez A, Munñoz-Fernandez M-Á, Correa-Rocha R, and Pion M (2018). Human immunodeficiency virus type-1 induces a regulatory B cell-like phenotype in vitro. Cell Mol. Immunol. 15, 917–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano JJ, Pallier A, Martinez-Llordella M, Danger R, López M, Giral M, Londoño MC, Rimola A, Soulillou JP, Brouard S, et al. (2011). Comparison of transcriptional and blood cell-phenotypic markers between operationally tolerant liver and kidney recipients. Am. J. Transplant. 11, 1916–1926. [DOI] [PubMed] [Google Scholar]
- Maseda D, Smith SH, DiLillo DJ, Bryant JM, Candando KM, Weaver CT, and Tedder TF (2012). Regulatory B10 cells differentiate into antibody-secreting cells after transient IL-10 production in vivo. J. Immunol. 188, 1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Baba A, Yokota T, Nishikawa H, Ohkawa Y, Kayama H, Kallies A, Nutt SL, Sakaguchi S, Takeda K, et al. (2014). Interleukin-10Producing plasmablasts exert regulatory function in autoimmune inflammation. Immunity 41, 1040–1051. [DOI] [PubMed] [Google Scholar]
- Matsushita T, Le Huu D, Kobayashi T, Hamaguchi Y, Hasegawa M, Naka K, Hirao A, Muramatsu M, Takehara K, and Fujimoto M (2016). A novel splenic B1 regulatory cell subset suppresses allergic disease through phosphatidylinositol 3-kinase–Akt pathway activation. J. Allergy Clin. Immunol. 138, 1170–1182.e9. [DOI] [PubMed] [Google Scholar]
- Mauri C, and Bosma A (2012). Immune regulatory function of B cells. Annu. Rev. Immunol. 30, 221–241. [DOI] [PubMed] [Google Scholar]
- Mauri C, and Menon M (2017). Human regulatory B cells in health and disease: therapeutic potential. J. Clin. Invest. 127, 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri C, Gray D, Mushtaq N, and Londei M (2003). Prevention of arthritis by interleukin 10–producing B cells. J. Exp. Med. 197, 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdipour F, Razmkhah M, Faghih Z, Bagheri M, Talei A-R, and Ghaderi A (2019). The significance of cytokine-producing B cells in breast tumor-draining lymph nodes. Cell Oncol. 42, 381–395. [DOI] [PubMed] [Google Scholar]
- Menon M, Blair PA, Isenberg DA, and Mauri C (2016). A regulatory feedback between plasmacytoid dendritic cells and regulatory B cells is aberrant in systemic Lupus Erythematosus. Immunity 44, 683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud D, Steward CR, Mirlekar B, and Pylayeva-Gupta Y (2021). Regulatory B cells in cancer. Immunol. Rev. 299, 74–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi A, Mizoguchi E, Takedatsu H, Blumberg RS, and Bhan AK (2002). Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity 16, 219–230. [DOI] [PubMed] [Google Scholar]
- Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, Burlingham WJ, Marks WH, Sanz I, Lechler RI, et al. (2010). Identification of a B cell signature associated with renal transplant tolerance in humans. J. Clin. Invest. 120, 1836–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell KA, Asare A, Sanz I, Wei C, Rosenberg A, Gao Z, Kanaparthi S, Asare S, Lim N, Stahly M, et al. (2015). Longitudinal studies of a B cell–derived signature of tolerance in renal transplant recipients. Am. J. Transplant. 15, 2908–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nova-Lamperti E, Fanelli G, Becker PD, Chana P, Elgueta R, Dodd PC, Lord GM, Lombardi G, and Hernandez-Fuentes MP (2016). IL-10-produced by human transitional B-cells down-regulates CD86 expression on B-cells leading to inhibition of CD4 + T-cell responses. Sci.Rep. 6, 20044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveria J-P, El-Gammal AI, Yee M, Obminski CD, Scime TX, Watson RM, Howie K, O’Byrne PM, Sehmi R, and Gauvreau GM (2018). Changes in regulatory B-cell levels in bone marrow, blood, and sputum of patients with asthma following inhaled allergen challenge. J. Allergy Clin. Immunol. 141, 1495–1498.e9. [DOI] [PubMed] [Google Scholar]
- Pallier A, Hillion S, Danger R, Giral M, Racapé M, Degauque N, Dugast E, Ashton-Chess J, Pettré S, Lozano JJ, et al. (2010). Patients with drug-free long-term graft function display increased numbers of peripheral B cells with a memory and inhibitory phenotype. Kidney Int. 78, 503–513. [DOI] [PubMed] [Google Scholar]
- Parekh VV, Prasad DVR, Banerjee PP, Joshi BN, Kumar A, and Mishra GC (2003). B cells activated by lipopolysaccharide, but not by anti-ig and anti-CD40 antibody, induce anergy in CD8+ T cells: role of TGF-b1. J. Immunol. 170, 5897–5911. [DOI] [PubMed] [Google Scholar]
- Ran Z, Yue-Bei L, Qiu-Ming Z, and Huan Y (2020). Regulatory B cells and its role in central nervous system inflammatory demyelinating diseases. Front. Immunol. 11, 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser EC, Blair PA, and Mauri C (2014). Cellular targets of regulatory B cell-mediated suppression. Mol. Immunol. 62, 296–304. [DOI] [PubMed] [Google Scholar]
- Shabir S, Girdlestone J, Briggs D, Kaul B, Smith H, Daga S, Chand S, Jham S, Navarrete C, Harper L, et al. (2015). Transitional B lymphocytes are associated with protection from kidney allograft rejection: a prospective study. Am. J. Transplant. 15, 1384–1391. [DOI] [PubMed] [Google Scholar]
- Shen P, Roch T, Lampropoulou V, O’Connor RA, Stervbo U, Hilgenberg E, Ries S, Dang VD, Jaimes Y, Daridon C, et al. (2014). IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature 507, 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva HM, Takenaka MCS, Moraes-Vieira PMM, Monteiro SM, Hernandez MO, Chaara W, Six A, Agena F, Sesterheim P, Barbé-Tuana FM, et al. (2012). Preserving the B-cell compartment favors operational tolerance in human renal transplantation. Mol. Med. 18, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Wang J, Pefanis E, Chao J, Rothschild G, Tachibana I, Chen JK, Ivanov II, Rabadan R, Takeda Y, et al. (2015). Transcriptomics identify CD9 as a marker of murine IL-10-competent regulatory B cells. Cell Rep. 13, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedder TF (2015). B10 cells: a functionally defined regulatory B cell subset. J. Immunol. 194, 1395–1401. [DOI] [PubMed] [Google Scholar]
- Tedder TF, and Leonard WJ (2014). Regulatory B cells—IL-35 and IL-21 regulate the regulators. Nat. Rev. Rheumatol. 10, 452–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AG, Glass DR, Juntilla M, Hartmann FJ, Oak JS, Fernandez-Pol S, Ohgami RS, and Bendall SC (2020). Multiplexed single-cell morphometry for hematopathology diagnostics. Nat. Med. 26, 408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veen W, Stanic B, Yaman G, Wawrzyniak M, Söllner S, Akdis DG, Rückert B, Akdis CA, and Akdis M (2013). IgG4 production is confined to human IL-10–producing regulatory B cells that suppress antigen-specific immune responses. J. Allergy Clin. Immunol. 131, 1204–1212. [DOI] [PubMed] [Google Scholar]
- Wang R-X, Yu C-R, Dambuza IM, Mahdi RM, Dolinska MB, Sergeev YV, Wingfield PT, Kim S-H, and Egwuagu CE (2014). Interleukin-35 induces regulatory B cells that suppress autoimmune disease. Nat. Med. 20, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yuan X, Chen H, Xie G, Ma Y, Zheng Y, Zhou Y, and Shen L (2015). CD19 + CD24 hi CD38 hi Bregs involved in downregulate helper T cells and upregulate regulatory T cells in gastric cancer. Oncotarget 6, 33486–33499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SD, Dittel BN, Hardardottir F, and Janeway CA Jr. (1996). Experimental autoimmune encephalomyelitis induction in genetically B cell–deficient mice. J. Exp. Med. 184, 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortel C, and Heidt S (2017). Regulatory B cells: phenotype, function and role in transplantation. Transpl. Immunol. 41, 1–9. [DOI] [PubMed] [Google Scholar]
- Yanaba K, Bouaziz J-D, Matsushita T, Tsubata T, and Tedder TF (2009). The development and function of regulatory B cells expressing IL-10 (B10 cells) requires antigen receptor diversity and TLR signals. J. Immunol. 182, 7459–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, Spolski R, Poe JC, Leonard WJ, and Tedder TF (2012). Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature 491, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zeng Y, Dolff S, Bienholz A, Lindemann M, Brinkhoff A, Schedlowski M, Xu S, Sun M, Guberina H, et al. (2017). Granzyme B producing B-cells in renal transplant patients. Clin. Immunol. 184, 48–53. [DOI] [PubMed] [Google Scholar]
- Zunder ER, Finck R, Behbehani GK, Amir ED, Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, et al. (2015). Palladium-based mass-tag cell barcoding with a doublet-filtering scheme and single cell deconvolution algorithm. Nat. Protoc. 10, 316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


