Fig. 5. CRISPR/SaCas9–mediated disruption of Kcnq2/3 genes in Hcrt neurons leads to NREM sleep fragmentation in young mice.
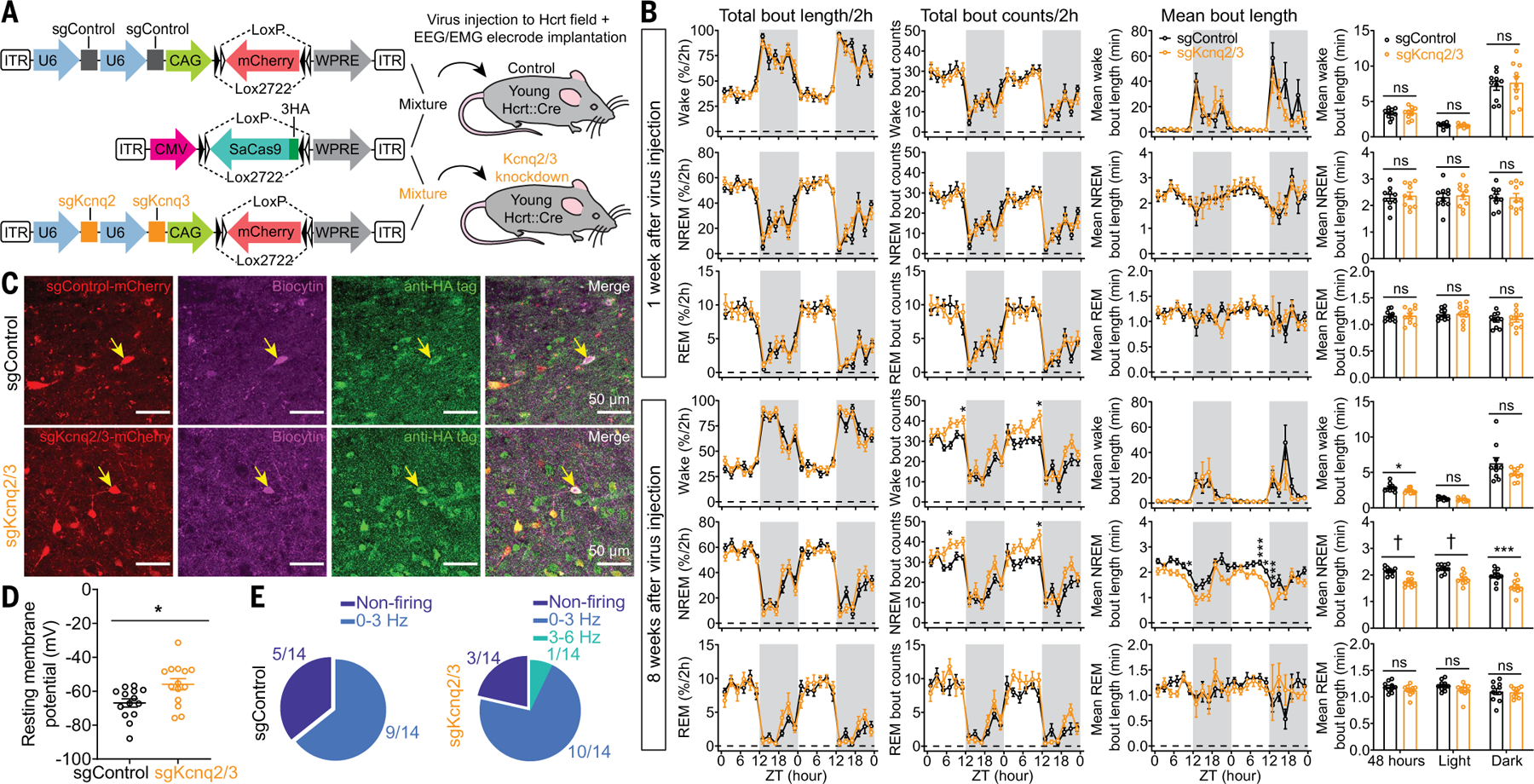
(A) Schematic of AAV sgControl, AAV SaCas9, AAV sgKcnq2/3 vector design, and bilateral viral infection of Hcrt neurons in young Hcrt∷Cre mice. (B) Two-hour (left) binned percentage, (middle left) bout counts, (middle right) mean bout length and (right) mean bout length based on circadian phase for wake, NREM, and REM sleep at 1 week (top) and 8 weeks (bottom) after injection of a virus mixture, as illustrated in (A) (n = 10 mice/group, dark phase indicated by gray shielding). (C) Representative slices expressing sgRNA with fluorescent mCherry flag and SaCas9–3HA for sgControl and sgKcnq2/3 group, respectively. Patch clamp recorded cells were labeled with biocytin, and post hoc antibody staining against HA tag confirmed the cells expressing SaCas9 for data analyses. (D) Comparison of RMPs between sgControl and sgKcnq2/3 group (n = 14 neurons from three mice each group). (E) Fractions of neurons with different firing frequencies in the (left) sgControl and (right) sgKcnq2/3 groups. Data indicate mean ± SEM [(B) left to middle right, two-way RM ANOVA followed by Šidák’s multiple comparisons; (B) right, Holm-Šidák; (D) Mann-Whitney U test; *P < 0.05, ***P < 0.005, †P < 0.0005; statistical details are avialable in the supplementary text].
