Abstract
Hematopoiesis is the process by which mature blood and immune cells are produced from hematopoietic stem and progenitor cells (HSCs and HSPCs). The last several decades of research have shed light on the origin of HSCs, as well as the heterogeneous pools of fetal progenitors that contribute to lifelong hematopoiesis. The overarching concept that hematopoiesis occurs in dynamic, overlapping waves throughout development, with each wave contributing to both continuous and developmentally limited cell types, has been solidified over the years. However, recent advances in our ability to track the production of hematopoietic cells in vivo has challenged several long-held dogmas on the origin and persistence of distinct hematopoietic cell types. In this review, we highlight emerging concepts in hematopoietic development and identify unanswered questions.
Keywords: Hematopoiesis, blood and immune cells, hematopoietic stem cells, hematopoietic progenitor cells, development, specification, self-renewal, differentiation, immune development, tissue-resident cells, innate immune cells, adaptive immune cells, blood development
Introduction
Hematopoietic stem cells (HSCs) are blood stem cells capable of self-renewal and differentiation into lineage-restricted progenitor cells that ultimately make mature cells types with highly specialized functions. HSCs were first described in the 1960’s as cells capable of giving rise to spleen colonies containing clonally multipotent cells (Becker, McCulloch, & Till, 1963; Wu, Till, Siminovitch, & McCulloch, 1968). These studies laid the groundwork for our initial understanding of blood stem cells and defined them as cells with combined self-renewal and multilineage hematopoietic differentiation capability upon transplantation into a suitable host. Subsequently, Spangrude et al. demonstrated that 30 HSCs was sufficient to rescue half a population of lethally irradiated mice, indicating the potent reconstitution capacity of these cells (Spangrude et al., 1988). Since then, the field of molecular and cellular biology has advanced rapidly, allowing for extremely sensitive detection of donor-derived cells to assess the clonal capacity of distinct subsets of hematopoietic stem and progenitor cells (HSPCs).
While transplantation of HSPCs into conditioned hosts has long been the “gold standard” in the field, in situ fate mapping, barcoding, and RNA sequencing (RNA-seq) have become increasingly common to interrogate HSC identity and function. There are pros and cons to each of these techniques. Transplantation assays test the inherent potential of stem and progenitor cells to produce different cell types. However, the conditioning regimens typically used to ensure robust reconstitution may affect the lineage capacity and/or preference of the incoming cells as well as the clearance of resident cell types, thus altering the perceived potential of the transplanted cell. Although reconstitution assays remain an essential tool to assess HSPC in vivo lineage potential and the therapeutic potential of cell subpopulations for regenerative medicine, HSPC transplantation might not reflect the cell ontogeny of normal hematopoiesis. Fate mapping using permanent genetic marking allows for tracking of HSPC differentiation in situ without disrupting normal hematopoiesis. Yet, it is difficult to reliably pinpoint the initiation of labeling to only HSCs, since different progenitors may transiently express the gene responsible for the labeling. In situ barcoding assays allow us to determine contributions of single clones to the hematopoietic system (Busch et al., 2015; Pei et al., 2017; Sun et al., 2014) (reviewed in Dharampuriya et al., 2017). However, interpretation of the outcomes is complex and rely heavily on mathematical modeling, and the conclusions have been challenged (Sawai et al., 2016). In addition to these functional assays, RNA-seq of bulk populations or single cells is heavily utilized. This is an extremely useful, yet relatively simple, tool to identify differences between cells. Although the data collected from RNA-seq is descriptive and not without caveats (Arzalluz-Luque et al., 2017; Griffiths et al., 2018; Herring et al., 2018), they provide a valuable database of genes to further investigate. Regardless of caveats, these assays have collectively proven extremely beneficial in furthering our understanding of hematopoiesis.
Increasingly sophisticated versions of these techniques have led to reconsideration of some long-held dogmas of hematopoiesis. Vast heterogeneity in stem and progenitor populations throughout development and in adulthood has been revealed, and the idea that the adult HSC pool is derived from one small pool of cells, as previously believed, has been challenged (Ganuza et al., 2017b; Pei et al., 2017; Yu et al., 2016). Accumulating evidence suggests that embryonic progenitors that precede lifelong, engraftable HSCs contribute to distinct cell types that persist with little replenishment from the adult HSC pool (Perdiguero et al., 2015; Hoeffel & Ginhoux, 2015). The extent to which different HSCs, and their fetal precursors, contribute to hematopoiesis during development and over the lifespan is under debate (Sawai et al., 2016). Even the concept that definitive, repopulation-capable HSCs persist for life has been challenged (Beaudin et al., 2016). In order to solidify emerging views and continue to gain deeper insights into the biology of HSCs, many labs focus their attention to the developmental origin of HSCs and the lifelong contribution of discrete waves of hematopoiesis. Important goals include gaining an actionable understanding of the biology of hematopoietic disease, and how to generate and culture HSCs in vitro for therapeutic use. In this review, with emphasis on lessons learned from the mouse, we aim to summarize the current perspective on the contributions of developmental waves of HSPCs to the specification and maintenance of lifelong hematopoiesis, and to pinpoint questions that remain unanswered.
HSC emergence: where, when, and how?
Cellular origins of HSCs: hemangioblast vs hemogenic endothelium
The mechanisms of HSC emergence have been under debate for decades. One hypothesis posited that hematopoietic and endothelial lineages arise through a specialized bi-potent progenitor known as the hemangioblast (Figure 1). This term was initially used to describe a group of mesodermal cells from the primitive streak that contained endothelial and hematopoietic cells (Murray, 1932). One study demonstrated that a subset of embryonic stem cell (ESC)-derived progenitors were capable of producing both endothelial and hematopoietic lineages (Lacaud et al., 2001). The authors concluded that this progenitor population was an in vitro equivalent of the hemangioblast. Using a brachyury-GFP transgene, Huber and colleagues identified a subset of Flk1+brachyury+ cells that modeled the mesodermal-to-hemangioblast transition (Huber et al., 2004) providing further ex vivo support for the existence of a hemangioblast.
Figure 1. Hemangioblast versus hemogenic endothelium schematic.
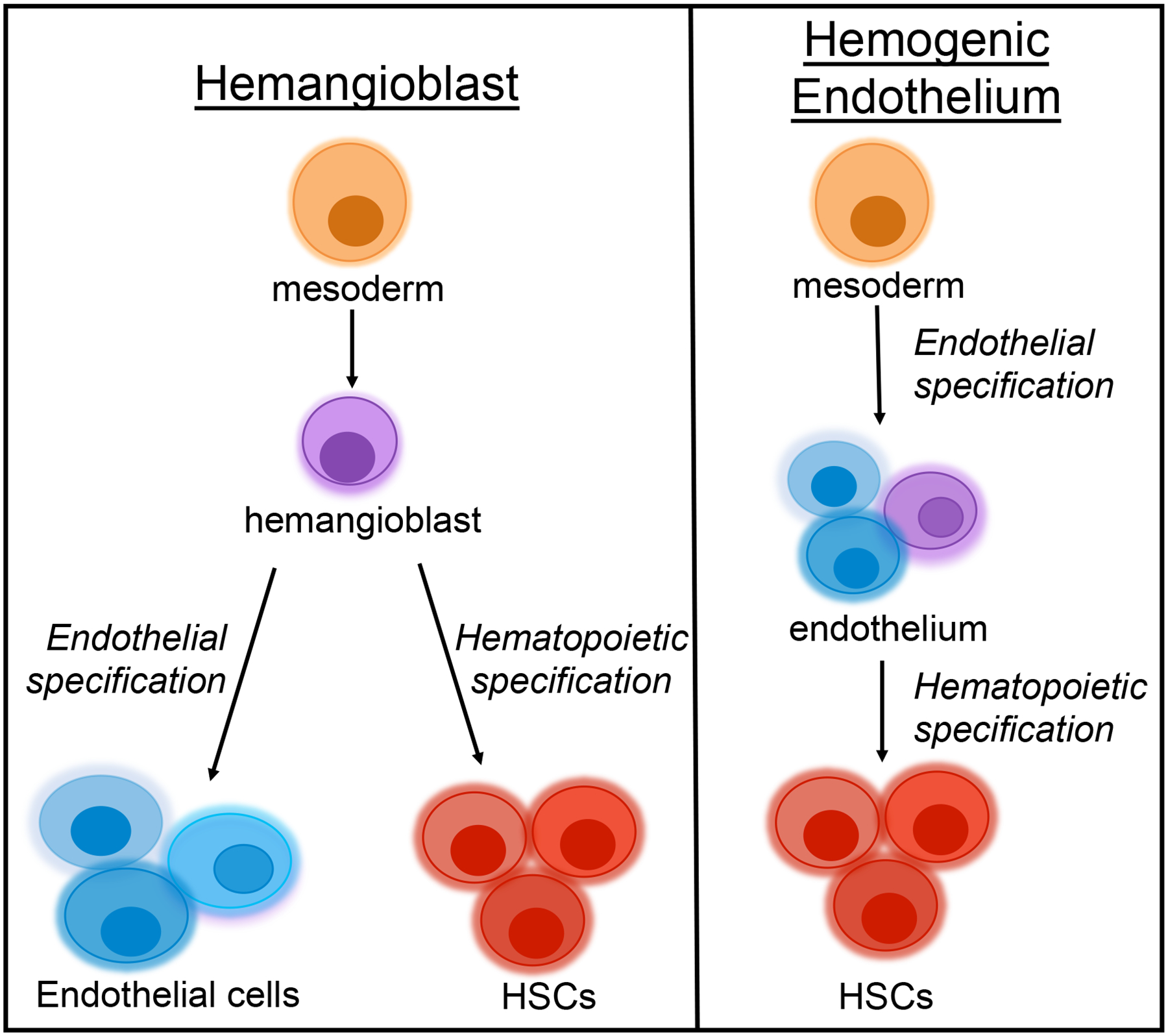
Two proposed models of HSC emergence in the embryo. One model proposes that a specialized bi-potent precursor named the hemangioblast gives rise to both endothelial cells and HSCs. An alternative model proposes that mesoderm gives rise first to endothelium, a small proportion of which can give rise to HSCs. In this schematic, the purple cell represents the subset of hemogenic endothelial cells. The different shades represent heterogeneity within both lineages.
Ueno and colleagues tested the hemangioblast concept in vivo by making tetrachimeric mouse embryos that clonally expressed different fluorescent proteins (Ueno and Weissman, 2006). The dissimilar proportions of colored endothelial and hematopoietic cells within individual blood islands argued against a shared bi-potent progenitor that contributes equally to both lineages. Another group used a temporally-restricted fate mapping model, VEcadherin-CreERT2, to selectively label endothelial cells and thereby distinguish between HSC emergence from the endothelium or underlying mesenchyme (Zovein et al., 2008). By injecting pregnant mothers at E9.5 with tamoxifen, they aimed to label aorta-gonad-mesenepheros (AGM)-derived HSCs, while evading the earlier VEcadherin expression of the yolk sac (YS). By surveying additional models (SM22α-Cre and Myocardin-Cre) that labeled mesenchymal cells and their progeny, they ruled out a direct mesenchyme-to-HSC transition and concluded that only the endothelial lineage was capable of generating HSCs. Similarly supporting an endothelial ancestry, Oberlin et al. demonstrated that the majority of adult BM HSCs are derived from a VE-cadherin positive precursor (Oberlin et al., 2010). However, VEcadherin may not be entirely restricted to the endothelium. One study demonstrated that fetal HSCs express VEcadherin at E13.5, but lose expression by E16.5 (Kim et al., 2005); thus, the observed labeling of hematopoietic cells in this model may not signify an endothelial origin.
An alternative hypothesis to the bipotent hemangioblast proposed that mesoderm first gives rise to endothelial cells, some of which possess the capacity to generate HSCs (Figure 1). In support of HSC emergence after endothelial specification, Lancrin and colleagues identified a hemogenic endothelial precursor intermediate that was capable of giving rise to the first hematopoietic cells (Lancrin et al., 2009). Another group, using in vitro time lapse imaging, concluded that both ESC-derived mesodermal cells and mesodermal cells isolated from E7.5 embryos were able to give rise to hematopoietic cells through an endothelial intermediate, supporting the endothelial origin of HSCs (Eilken et al., 2009). Boisset and colleagues used in vivo time-lapse imaging to visualize HSC emergence from endothelium in the AGM region (Boisset et al., 2010). Similar findings were made simultaneously in zebrafish: two groups concluded that HSCs emerge from ventral aortic hemogenic endothelial cells prior to the onset of blood circulation (Bertrand et al., 2010; Kissa and Herbomel, 2010). Thus, HSC emergence appears similar in zebrafish and mouse, although de Brujin et al. concluded that HSC activity was limited to a few cells within the endothelial layer of the dorsal aorta in mouse, as opposed to the ventral aorta in fish (de Bruijn et al., 2002).
More recently, a group used single cell isolation and RNA-seq to identify molecular signatures of pre-HSCs (F. Zhou et al., 2016). They concluded that Rictor, a component of the mTORC pathway, is required for the emergence of HSCs from endothelial cells; a finding that supports the notion that HSCs arise from endothelial cells (Figures 1, 2). Ganuza and colleagues performed ex vivo cultures of murine cells from the midgestation aorta, vitelline artery, and umbilical artery, and found that only a very small percentage of the cells had in vitro functional properties of hemogenic endothelium (Ganuza et al., 2017a). Further, they suggested that heterogeneity within the hemogenic endothelial population might contribute to distinct populations with differential capacities to specify HSCs (Figure 2). This finding suggests that several types of precursors contribute to HSPCs. This idea is novel and striking given that some labs have believed for decades that one specific type of precursor contributes to the first HSCs. While no definitive conclusion has been made regarding the exact properties of the in vivo precursors to HSCs, it is clear that endothelial and hematopoietic lineages are closely linked (Figures 1, 2). The concepts of the hemangioblast versus the endothelial-to-hematopoietic transition are quite similar and may not be mutually exclusive: in both cases, mesoderm cells give rise to endothelial and hematopoietic lineages (Figures 1, 2). Several reviews discuss the hemangioblast versus hemogenic endothelial origin of HSCs in greater detail (Boisset and Robin, 2012; de Bruijn, 2014; Lacaud and Kouskoff, 2017; Nishikawa, 2012).
Figure 2. HSC emergence.

It is currently believed that definitive HSCs emerge in the AGM region of E10.5 murine embryos. There has been a Substantial evidence supports the notion that a subset of mesenchyme-derived endothelial cells (ECs), hemogenic endothelium (HE), has the potential to give rise to HSCs. Such de novo HSC specification in the AGM may contribute to lifelong hematopoiesis in parallel with YS-derived hematopoietic cells.
Are HSCs specified multiple times in distinct locations?
Tracking the ontogeny of HSCs has proven challenging due to the different temporal and spatial emergence of blood cells. It has been proposed that HSCs emerge de novo in either the YS, placenta, and/or (AGM), and eventually seed the fetal liver (FL) and adult bone marrow (BM). Early in vitro and in vivo colony forming assays (Huang and Auerbach, 1993; Liu and Auerbach, 1991; Moore and Metcalf, 1970; Perah and Feldman, 1977; Toles et al., 1989) suggested that HSCs emerge in the YS, migrate to the FL, and finally to the BM, where they reside in adulthood and contribute to lifelong hematopoiesis. In support of this model, Samokhvalov et al. demonstrated that Runx1+ cells, labeled with a single tamoxifen injection at E7.5 in a MER-cre-MER model, were able to develop into fetal myeloid and lymphoid progenitors and contribute to adult hematopoiesis (Samokhvalov et al., 2007). They concluded that these cells were able to colonize the umbilical cord, AGM, and ultimately the FL, suggesting that at least some definitive HSCs might be descendants of hematopoietic cells first specified in the YS. More recent data from their group substantiate these findings by providing evidence that a subset of extraembryonic Runx1+Gata1− cells in the E7.5 embryo migrate to intraembryonic sites prior to the onset of circulation (initiation of a heartbeat) (Tanaka et al., 2014). Similar findings were made using a Lyve1-cre mouse model (Lee et al., 2016). These results are quite striking as they suggest that systemic circulation is not required for HSC migration through the embryo.
However, it has also been suggested that HSCs are generated de novo in the AGM, challenging an exclusive YS origin of definitive HSCs (Medvinsky and Dzierzak, 1996). Using an in vitro organ culture system to tease apart the hematopoietic potential of cells residing in the YS and AGM region, the authors concluded that the AGM is the source of definitive HSCs that ultimately seed the adult BM. Taoudi and colleagues reported that VEcadherin+CD45+ pre-definitive HSCs in the AGM region give rise to the first definitive HSC (Taoudi et al., 2008). This finding contradicted the YS origin of HSCs and suggested that HSCs are generated de novo in the AGM rather than migrating from the YS. Several other studies have also reported de novo generation of HSCs in the AGM region (de Bruijn et al., 2000; Durand and Dzierzak, 2005; Peeters et al., 2009) or in the placenta, independent of circulation (Gekas et al., 2010; Rhodes et al., 2008).
Many labs have utilized Cre lineage tracing to directly assess the emergence of HSCs. Using an inducible Stem Cell Leukemia (SCL)-specific Cre line, one group addressed whether HSCs are generated de novo at each site during fetal development (Gothert et al., 2005). They injected tamoxifen into pregnant mothers at E10.5 in order to label AGM HSCs and analyzed the BM of adult progeny for labeled HSCs. As ~10% of the adult BM HSCs were labeled, at least some of the fetal HSCs appear to seed the adult BM. However, this result also revealed either inefficient HSC labeling (at this time point), or that additional HSCs may be specified after this time point. To address the possibility of de novo generation of HSCs at later timepoints, they compared the percent EYFP+ cells in the adult BM of mice dosed at E10.5 and E11.5 with the percent EYFP+ cells in the BM of mice transplanted with previously marked E14.5 FL cells. The authors reasoned that de novo generation of HSCs would have led to a decrease in EYFP+ cells in the BM of the transplanted mice. They found that mice transplanted with E14.5 FL HSCs had a similar percentage of labeled HSCs in their adult BM compared to the embryos treated with tamoxifen at E10.5, thus arguing against de novo generation beyond E10.5. However, it is important to note that the labeling efficiency of CreER models during embryonic development is frequently incomplete, and that the origins of the remaining 90% of unlabeled cells in this model remain unknown.
The lack of consensus on the anatomical location(s) of the emergence of the first HSC(s) is likely, in part, due to models and methods being used. One issue is the length of time tamoxifen persists in the system. For example, Nishikawa’s group suggested that YS cells migrate to colonize the embryo; however, labeling of intraembryonic cells could potentially be explained by tamoxifen remaining in the system and labeling cells within the embryo that express Runx1 at a later time point. In addition, most groups use different Cre models, independently, that undoubtedly label divergent cell types. A thorough side by side comparison of the Cre lines that are specific to early hematopoietic development may help in our data analysis and interpretations, and ultimately better define hematopoietic development (Abram et al., 2014). Additionally, transplantation assays may not be sufficient to define which “HSCs” persist in situ (Figure 3). It has been demonstrated that embryonic HSCs engraft better in neonates compared to adults (Arora et al., 2014; Yoder et al., 1997). Additionally, evidence is accumulating that cells that do not engraft long term upon transplantation can persist for long periods of time in situ (Busch et al., 2015; Schoedel et al., 2016; Sun et al., 2014). Models that do not rely on niche perturbations or culturing of cells may provide us with better insights to the development of the blood system in situ (Ganuza et al., 2017b).
Figure 3. Fetal hematopoiesis occurs in waves.
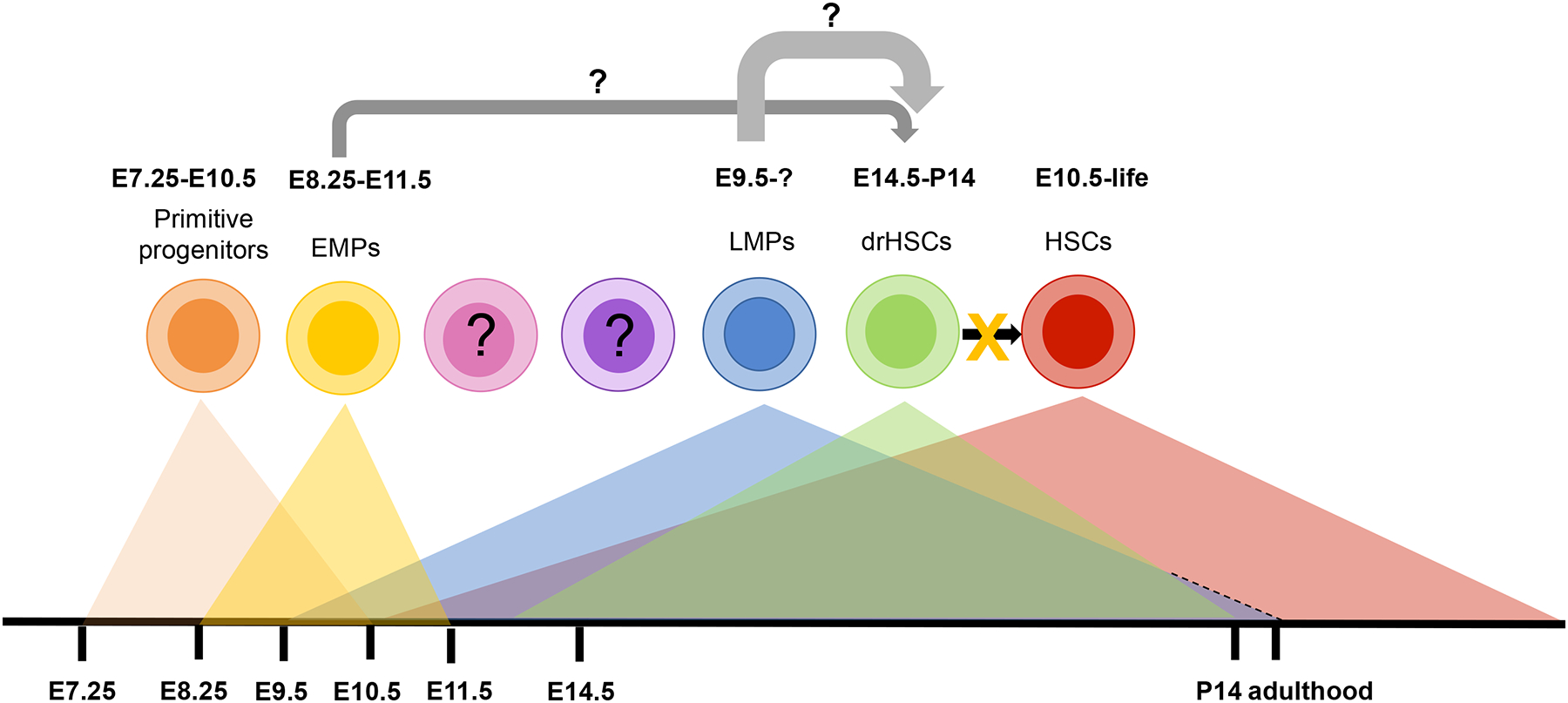
Several distinct progenitors exist at different developmental stages in situ. Primitive erythroid precursors can be isolated from E7.25 until E10.5. Erythro-myeloid progenitors (EMPs) exist from E8.25 until E11.5. Lympho-myeloid progenitors (LMPs) appear around E9.5. Engraftable, developmentally restricted (dr)HSCs can be detected from E12.5 until postnatal day 14 (P14). Definitive HSCs that contribute to lifelong hematopoiesis are identifiable from E10.5 onwards. It remains to be determined whether the drHSC are derived from EMPs, LMPs, or an independent precursor. It is clear that drHSCs do not give rise to definitive HSCs. Whether other previously unidentified HSPCs exist during this timeframe remains to be determined.
Molecular regulation of HSC specification
What are the molecular drivers of HSC specification and maintenance?
The molecular drivers of hematopoiesis change several times from a developing embryo to a fully formed vertebrate (Table 1). Efforts focused on understanding the different genetic programs that regulate HSC specification, expansion, and maintenance have identified distinct transcription factors involved in primitive erythropoiesis. Several studies have demonstrated that SCL/TAL1 is required for primitive hematopoiesis and specification of HSCs (Robb et al., 1995; Shivdasani, Mayer, & Orkin, 1995; Visvader, Fujiwara, & Orkin, 1998) and divergence of cardiac and hematopoietic lineages during development (Org et al., 2015). SCL is required for the initial specification of the hematopoietic system, but not for long-term HSC colonization in the FL (Schlaeger et al., 2005), and conditional deletion of SCL in adult mice showed that SCL is dispensable for adult HSC maintenance (Mikkola et al., 2003). Similarly, the transcriptional regulators Rbtn2 (LMO2) (Warren et al., 1994) and CBP (Tanaka et al., 2000) appear to be required for primitive hematopoiesis and specification of HSCs, but not for HSC maintenance.
Table 1. Molecular regulators of HSC specification.
A summary of the different genes involved in HSC specification and hematopoietic maintenance throughout development and the lifespan.
| Stage | Molecular Regulator | Deficit when deleted | Required for primitive hematopoiesis? | Required for definitive hematopoiesis? | References |
|---|---|---|---|---|---|
| Primitive hematopoiesis | Scl | No YS hematopoiesis. Disrupted capillary formation. |
Yes | No | Visvader,Fujiwara, & Orkin, 1998; Robb et al., 1995; Shivdasani,Mayer, & Orkin, 1995; Y Tanaka et al., 2000; Mikkola et al., 2003; Schlaeger, Mikkola, Gekas, Helgadottir, & Orkin, 2005; Org et al., 2015 |
| Primitive hematopoiesis | Cpb | Defective blood vessel formation. | Yes | ? | Tanaka et al., 2000 |
| Primitive hematopoiesis | Runx1 | Absence of fetal liver hematopoiesis. | Yes | No | Okuda, van Deursen, Hiebert, Grosveld, & Downing, 1996; Chen, Yokomizo, Zeigler, Dzierzak, & Speck, 2009; Tober, Yzaguirre, Piwarzyk, & Speck, 2013 |
| Primitive | Sox17 | Reduced hemogenic endothelium and HSCs in AGM region. | Yes | No | Clarke et al., 2013; Injune Kim, Saunders, & Morrison, 2007 |
| Primitive to definitive transition | ATF4 | FL hematopoiesis impaired. HSC self-renewal impaired. |
Yes | Yes? | Masuoka & Townes, 2002; Zhao et al., 2015 |
| Primitive to definitive transition | Cbfb | Nascent HSCs stuck in AGM (cannot enter circulation) | No | Yes | Bresciani et al., 2014; Sasaki et al., 1996 |
| Primitive to definitive transition | Gata-2 | Blocked HSC formation in AGM. Adult HSCs have impaired proliferation. |
No | Yes | Ling et al., 2004; Mehta et al., 2017; Rodrigues et al., 2005; Tsai et al., 1994 |
| Primitive to definitive transition | C-myb | Blocked migration from AGM. Conditional deletion in adults leads to impaired self-renewal. |
Yes | Yes | Emambokus et al., 2003; Lieu & Reddy, 2009; Mucenski et al., 1991; Zhang, Jin, Li, Qin, & Wen, 2011 |
| Primitive to definitive transition | C/EBPa | Impaired granulopoiesis. Enhanced repopulation activity of FL HSCs. Conditional deletion in adults leads to increased proliferation of HSCs. |
Yes | Yes | Ye et al., 2013 |
| Definitive hematopoiesis | Tet/Etv6 | Impaired megakrypoiesis | No | Yes | Hock et al., 2004 |
| Definitive hematopoiesis | E2A | Blocked B cell development leading to postnatal death. HSC pool maintenance perturbed. Impaired myelolymphoid/myloerythroid progenitor maturation. |
No | Yes | Semerad, Mercer, Inlay,Weissman, & Murre, 2009; Zhuang, Soriano, &Weintraub, 1994 |
| Definitive hematopoiesis | Bmi-1 | Impaired hematopoiesis postnatally. Reduced hematopoietic numbers. Increased adipocytes in bone ma rrow. |
No | Yes |
van der Lugt et al., 1994 Park et al., 2003 |
Like SCL, Runx1 is a transcription factor and an essential regulator of hematopoietic specification. Okuda et al. showed that Runx1 is required for FL hematopoiesis (Okuda et al., 1996). Mice with a homozygous mutation of Runx1 exhibited normal YS erythropoiesis, but eventually died around E12.5 due to failed FL hematopoiesis. Importantly, endothelial-specific loss of Runx1 precluded HSC formation (Chen et al., 2009), whereas hematopoietic deletion of Runx1 allowed normal EMP and HSC formation (Ichikawa et al., 2004). This supports HSC derivation from endothelial cells in a Runx1-dependent process. To further define the window of Runx1 dependency, Tober et al. performed timed deletions of Runx1 from E7.5 until E13.5 (Tober et al., 2013). When Runx1 was deleted between E9.5 and E11.5, recipients had no engraftment, while deletion after E11.5 led to high levels of engraftment. The authors concluded that Runx1 is required for EMP and HSC formation up until E10.5 and E11.5, respectively. In addition to Runx1, ATF4 (Masuoka and Townes, 2002; Zhao et al., 2015), Cbfb (Bresciani et al., 2014; Sasaki et al., 1996), Gata2 (Ling et al., 2004; Mehta et al., 2017; Rodrigues et al., 2005; Tsai et al., 1994), Sox17 (Clarke et al., 2013; Kim et al., 2007), and C-Myb (Emambokus et al., 2003; Lieu and Reddy, 2009; Mucenski et al., 1991; Zhang et al., 2011) have also been shown to be indispensable for normal mouse FL hematopoiesis (Table 1). Some of these factors also regulate adult HSC maintenance and function, while some are dispensable for adult hematopoiesis (Table 1). Likewise, a unique set of genes are required for the maintenance of adult HSCs (Hock et al., 2004; Semerad et al., 2009; van der Lugt et al., 1994; Ye et al., 2013; Zhuang et al., 1994). Interestingly, Sox17 and Lin28 are highly expressed in FL, but not adult, HSCs, and can be used to reprogram adult HSCs into cells that have regained at least some of the properties of fetal HSCs (He et al., 2011; Lang et al., 2013; Yuan et al., 2012). These studies have identified several key regulators of primitive and definitive hematopoiesis and have given us insight into the development and maintenance of distinct stages of HSCs (Table 1). However, the intricate network of genes being regulated by these factors remains under investigation, with the hopes of understanding the coordinated actions of key hematopoietic pathways that contribute to lifelong hematopoiesis.
Phenotypic and functional heterogeneity within the HSPC pool
Extensive transcriptome heterogeneity of cell populations
For several decades, the field has believed that one small pool of HSCs gives rise to a relatively homogeneous HSC pool and ultimately the blood system (Kumaravelu et al., 2002; Medvinsky et al., 2011; Medvinsky and Dzierzak, 1996; Morrison et al., 1995; Spangrude et al., 1988). Numerous studies have identified subsets of hematopoietic precursors and HSPCs using single-cell (sc)RNA-seq (Guo et al., 2013; Kowalczyk et al., 2015; Macaulay et al., 2016; Moignard et al., 2013; Wilson et al., 2015). In an impressive analyses of early mesoderm specification, one group assayed over 3,900 cells from developing embryos spanning E7.0 and E8.5 (Moignard et al., 2015). This revealed three cell clusters within the embryo: primitive streak/neural plate associated cells, a mixed group containing primitive streak/neural plate and hemogenic endothelial associated cells, and a final group consisting of hematopoietic associated cells. Computational analysis identified Sox7 as a potential regulator of early blood divergence. An inducible Sox7 overexpression model validated this prediction. Embryos overexpressing Sox7 displayed a clear reduction in primitive erythroid cells, indicating that downregulation of Sox7 is required for early blood cell commitment. Similarly, Scialdone et al. used bulk and scRNA-seq to interrogate the endothelial and hematopoietic specification from E6.5-E8.5 embryos (Scialdone et al., 2016). Gene expression data from wild type embryos and SCL mutants indicated that SCL regulates several genes associated with early blood specification, including Sox7.
In parallel to these embryonic studies, several groups have addressed heterogeneity within the adult HSPC pool using RNA-seq (Kowalczyk et al., 2015; Wilson et al., 2015). One group performed scRNA-seq of adult HSCs and their direct descendants, MPP1 cells, and identified subsets of quiescent and “active” HSCs, based on differential expression of cell cycle genes, DNA replication genes, and myeloid associated genes (Yang et al., 2016). The vast heterogeneity identified by these studies has prompted investigation by our lab and others as to whether other distinct stem and progenitor cells exist throughout the lifespan. Analogous scRNA-seq analysis of fetal stem and progenitor populations may shed light on previously unidentified populations in the developing embryo.
Functional heterogeneity of hematopoietic progenitor populations
To address the functional consequences of the revealed heterogeneity, many labs have utilized transplantation, barcoding, and lineage tracing with complex reporter systems. Interestingly, experiments that investigated the capacity of HSCs isolated from different developmental time points and tissues revealed extensive heterogeneity that changes throughout development and the lifespan (Benz and Eaves, 2012). This revelation suggested the existence of several distinct clones of HSCs and that pools are clonally selected based on supporting niches and the needs of the developing mouse. It also prompted the creation of more complex systems to address the clonal capacity of individual HSCs. Ganuza et al. utilized the Confetti system with several Cre lines to conclude, with the help of mathematical modeling, that several hundred developmental precursors contribute to long-term hematopoiesis (Ganuza et al., 2017). Unlike previous estimations of HSC numbers in the developing embryo, this model did not rely on ex vivo culture or transplantation of embryonic cells. Also using a multicolor reporter strategy, Yu et al. reported that the murine hematopoietic system is composed of persistent and non-persistent clones, with a small population of “dominant” clones that persist and give rise to most of the blood system while others completely disappear (Yu et al., 2016). Such a non-persistent, developmentally restricted (dr)HSC that retain serial, multilineage reconstitution capability was recently identified in FL (Beaudin et al., 2016). Pei et al, using an artificial DNA recombination locus, “polylox”, were able to tag thousands of single progenitor cells in the embryo and attribute the cell production capacity of individual clones (Pei et al., 2017). Like Ganuza and Yu, they concluded that the embryo is made up of many clones of HSCs and that some clones are more productive than others. Similar results have been made in the zebrafish (Henninger et al., 2017). While the extent of heterogeneity within the embryonic and adult HSC pools is still unclear, there is mounting consensus that many more clones than previously believed contribute to hematopoiesis during development and throughout the lifespan.
Do all HSCs persist?
There is conflicting evidence regarding the persistence of HSCs. One hypothesis is that all HSCs, endowed with self-renewal capability by definition, persist throughout the lifespan. As discussed above, the Nishikawa and Inlay/Mikkola groups have provided evidence that hematopoietic cells specified in the YS can persist for life (Lee et al., 2016; Samokhvalov et al., 2007). However, there is also accumulating evidence for the converse model, with some “HSC” populations extinguishing over time (Yu et al., 2016). Recently, our lab identified a developmentally-restricted HSC (drHSC) that does not persist into adulthood in unperturbed mice, supporting the notion that not all pools of HSCs endure for life (Beaudin et al., 2016). We identified this drHSC via use of the Flk2Cre/RosamTmG, also known as ”FlkSwitch”, mice (Boyer, Beaudin, & Forsberg, 2012; Boyer, Schroeder, Smith-Berdan, & Forsberg, 2011). Although functional HSCs by serial transplantation assays, the drHSCs only exist perinatally (Figure 3, 4). The irreversible, Cre-mediated genetic deletion that marks the drHSCs (but not adult HSCs) means that drHSCs cannot be precursors of adult HSCs, thereby questioning the idea that hematopoiesis is one linear progression through development. The temporal restriction of drHSCs to a perinatal window also defies the principle that engraftable HSCs persist and repopulate the entire blood system throughout the lifespan. The drHSCs efficiently gave rise to unique subsets of cells, including innate-like B and T lymphocytes, supporting the concept of developmental waves that comprise both unique lineage capacities and varying degrees of overlap in lineage potential (Figure 3). The ability to prospectively isolate drHSCs will enable new insights to the mechanisms regulating HSC persistence and lineage potential and extend recent work in the field that has highlighted the vast heterogeneity of hematopoietic progenitors (Table 2) (Eaves, 2015).
Figure 4. Progenitors at different time points give rise to both overlapping and distinct subsets of cells.

A simplified model depicting key features of developmental hematopoiesis. Around E7, cells of the yolk sac blood islands give rise to primitive macrophages and to primitive erythrocytes that are nucleated and express fetal globins. At E8.5, EMPs contribute to the first wave of tissue resident macrophages, including microglia in the developing brain. By E10.5, the first engraftable HSCs emerge in the AGM region of the embryo proper and initiate production of traditional mature blood cells (erythrocytes, platelets, granulocytes, monocytes/macrophages, B cells, T cells, and NK cells. At E14.5, developmentally restricted (dr)HSCs and persisting HSCs coexist in the fetal liver; drHSCs efficiently produce innate-like and traditional lymphoid cells. After birth, definitive HSCs primarily reside in the bone marrow and contribute to ”traditional” mature blood cells for the lifespan of the animal.
Table 2.
Summary of different hematopoietic stem and progenitor cells (HSPCs) that contribute to development of the blood system.
| Progenitor | Stage | Persists throughout life? | Lineage bias | Lineage bias References |
|---|---|---|---|---|
| Primitive progenitors | E7.25-E10.5 | No | Myeloid (primitive red blood cells) | Haar & Ackerman. 1971 |
| Erythromyeloid progenitors (EMPs) | E8.25-E11.5 | No | Myeloid (erythrocytes, megakaryocytes, macrophages) | Palis et al. 1999 |
| Lymphomyeloid progenitors (LMPs) | E9.5s life | ND | Lymphoid, GM | Boiers et al. 2013 |
| Developmentally-restricted HSCs (drHSCs) | E12.5-P14 | No | Lymphoid (innate-like lymphocytes) | Beaudin et al. 2016 |
| Definitive HSCs | E10.5-life | Yes | Clone dependent? |
Medvinsky et al. 2011 Dzierzak. 2005 |
Purpose of developmental waves
Immune system layering
The very first hematopoietic cells produced during embryogenesis, prior to the emergence of reconstituting HSCs, are nucleated erythrocytes and macrophages (Table 2, Figure 3, 4). Erythrocytes are responsible for carrying oxygen throughout the developing embryo, and macrophages play important roles in tissue development and homeostasis (Blander, 2017; Gomez Perdiguero and Geissmann, 2013; Kierdorf et al., 2015; Nicolás-Ávila et al., 2018). The specification of these cells is intriguing since they do not appear to derive from the classical hierarchy that defines later stages of hematopoiesis. Similarly, tissue resident macrophages (TrMacs) have been the topic of substantial interest in recent years due to their reported pre-HSC origin (Lichanska and Hume, 2000; Mizoguchi et al., 1992; Sorokin et al., 1992). TrMacs have been widely implicated in disease, from neurodegeneration and autoimmune disorders to cancer (Honold and Nahrendorf, 2018; Nicolás-Ávila et al., 2018; Sevenich, 2018; Yin et al., 2017; Z. Zhou et al., 2016). Understanding how TrMacs develop and how they are maintained long-term will provide a better understanding of several diseases and may uncover potential therapeutic targets.
In the last decade, several groups have utilized fate mapping to identify distinct subsets of TrMacs that arise from YS progenitors and “self-renew” in their respective tissues (Epelman et al., 2014; Ghigo et al., 2013; Ginhoux et al., 2010; Hashimoto et al., 2013; Hoeffel et al., 2015, 2012; Kierdorf et al., 2013; Schulz et al., 2012; Yona et al., 2013)reviewed in Hoeffel and Ginhoux, 2018; Lavin et al., 2015). Neither resident brain macrophages (microglia) nor skin macrophages (Langerhans cells) appear to be maintained by BM-derived progenitors (Ajami et al., 2007; Merad et al., 2002). Instead, microglia and Langerhans cells are likely of fetal origin, without significant contribution from adult HSCs at steady state (Figure 4). Several groups have since reported BM-independent maintenance of additional TrMacs, including Kupffer cells of the liver, alveolar macrophages of the lung, cardiac macrophages of the heart, and red pulp macrophages of the spleen (Figure 4) (Epelman et al., 2014; Hashimoto et al., 2013; Yona et al., 2013). While most groups agree that many TrMacs have a fetal origin, the mechanisms that regulate seeding of these cells in distinct developmental waves and tissues requires further investigation. Surprisingly, we have found that tissue monocytes express IL7rα and contribute to TrMacs in several tissues (Beaudin, McCann, Leung, Worthington, & Forsberg, unpublished data). This finding was unexpected, as IL7rα is restricted to the lymphoid lineage in canonical hematopoiesis (Schlenner et al., 2010). Additional unanticipated regulators of tissue resident immune cells will likely be uncovered in the future.
Mast cells have also been suggested to have fetal origins. In 1983, one group injected YS- or FL-derived mast precursor cells into adult mice that lack mast cells to determine the site of precursor activity (Hayashi et al., 1983). They demonstrated that the precursor activity was limited to the YS, arguing that these likely do not originate from definitive HSCs. Very recently, Gentek et al. used a VEcadherin fate mapping model to assess the ontogeny of mast cells (Gentek et al., 2018). They concluded that mast cells largely originate from YS progenitors, but that adult definitive HSCs also contribute. They also demonstrated that YS-derived mast cells are transcriptionally distinct from their adult-derived counterparts. This finding is supportive of multiple waves of HSPCs contributing to tissue resident myeloid cells.
In addition to TrMacs and mast cells, several types of innate-like lymphocytes have been shown to have primarily fetal origins. One such cell type, B1a cells that predominantly exist in serous cavities, has become highly scrutinized (Beaudin and Forsberg, 2016). The Herzenberg group showed that fetal progenitor cells have a unique capability of generating Ly-1+ B cells (B1 cells) (Hayakawa et al., 1985). They concluded that only neonatal, not adult, cells had the capacity to generate B1a cells upon transplantation into irradiated recipients. This finding was further corroborated by additional studies (Barber et al., 2011; Ghosn et al., 2012). However, the conclusion that FL HSCs are incapable of contributing to B1a cells in an irradiated adult host (Ghosn et al., 2016) has been convincingly contested (Beaudin et al., 2016). The source of the divergent conclusions likely stem from strict phenotypic (Ghosn) versus functional (Beaudin and Kristensen) definitions of fetal HSCs. Several recent reviews provide more in depth discussion (Beaudin and Forsberg, 2016; Hadland and Yoshimoto, 2018; Hardy and Hayakawa, 2015; Herzenberg, 2015; Yoshimoto, 2015). While the cellular precursors of B1a cells remain under debate, there is general consensus on a primarily fetal origin of the B1a lineage.
Although the exact contribution of each wave to the developing blood system remains under investigation, there is clearly substantial overlap in hematopoietic cell production between waves. It appears that the earlier waves contribute to innate immune cells that provide basic immunity and aid in tissue formation and organization, while later perinatal waves contribute to innate and adaptive immune cells that prepare the fetus for exposure to environmental pathogens after birth. How early life exposures affect the emergence and expansion of different immune cells, and how that influence immune cell function in later life, is an important avenue of investigation.
Conclusion and Future directions
As highlighted in this review, hematopoiesis undergoes continual molecular and functional changes throughout development and the lifespan. It seems likely that several clones of HSCs arise independently, in temporally and spatially regulated waves (Figure 4, Table 2). Exactly how many HSCs emerge and persist in the embryo, and their relative lifelong contribution to the blood system remains under debate. Cre-lox, barcoding, and transplantation tracking systems have, together with RNA-seq, improved our understanding of blood development, yet some questions remain difficult to address. It is unclear which waves are essential and how much functional overlap exists between them. Rigorous testing would require specifically deleting one lineage without perturbing the others, but that will depend on development of more precise and efficient tools. The novel genetic regulators identified by RNA-seq, in conjunction with reconstitution assays, fate mapping models, and innovative microscopy techniques, promise to provide an increasingly comprehensive understanding of blood system development. Quantitative assessment of individual HSC output, in situ and upon transplantation, is also needed to provide deeper insight into the lifespan and differentiation capacity of HSCs. Additionally, understanding the amazing epigenetic memory of cells, such as the drHSCs that maintain lineage biased output upon serial transplantation, may allow us to better utilize, and potentially engineer, HSCs for therapeutic use, and will likely also shed light on the drivers of blood and immune disease. While our understanding of hematopoiesis has expanded immensely, the spatial and temporal regulation of hematopoietic development warrant continued investigation.
Acknowledgements
We thank Dr. Smrithi Rajendiran, Atesh Worthington, Eric Martin, and Stephanie Smith-Berdan for insightful comments on the manuscript. ECF is supported by NIH/NIDDK (R01DK100917), NIH/NHLBI (R01HL115158), by American Cancer Society Research Scholar Award (RSG-13-193-01-DDC), and an American Asthma Foundation Research Scholar award.
Footnotes
Competing interests: There are no conflicts of interest.
References
- Abram CL, Roberge GL, Hu Y, Lowell CA, 2014. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408, 89–100. 10.1016/j.jim.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FMV, 2007. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci 10, 1538–43. 10.1038/nn2014 [DOI] [PubMed] [Google Scholar]
- Arora N, Wenzel PL, McKinney-Freeman SL, Ross SJ, Kim PG, Chou SS, Yoshimoto M, Yoder MC, Daley GQ, 2014. Effect of Developmental Stage of HSC and Recipient on Transplant Outcomes. Dev. Cell 29, 621–628. 10.1016/j.devcel.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzalluz-Luque Á, Devailly G, Mantsoki A, Joshi A, 2017. Delineating biological and technical variance in single cell expression data. Int. J. Biochem. Cell Biol 90, 161–166. 10.1016/j.biocel.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber CL, Montecino-Rodriguez E, Dorshkind K, 2011. Reduced production of B-1-specified common lymphoid progenitors results in diminished potential of adult marrow to generate B-1 cells. Proc. Natl. Acad. Sci. U. S. A 108, 13700–4. 10.1073/pnas.1107172108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin AE, Boyer SW, Perez-Cunningham J, Hernandez GE, Derderian SC, Jujjavarapu C, Aaserude E, MacKenzie T, Forsberg EC, 2016. A Transient Developmental Hematopoietic Stem Cell Gives Rise to Innate-like B and T Cells. Cell Stem Cell 19, 768–783. 10.1016/j.stem.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin AE, Forsberg EC, 2016. To B1a or not to B1a: do hematopoietic stem cells contribute to tissue-resident immune cells? Blood 128, 2765–2769. 10.1182/blood-2016-10-697813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BECKER AJ, McCULLOCH EA, TILL JE, 1963. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 197, 452–4. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DYR, Traver D, 2010. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111. 10.1038/nature08738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander JM, 2017. The many ways tissue phagocytes respond to dying cells. Immunol. Rev 277, 158–173. 10.1111/imr.12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset J-C, Robin C, 2012. On the origin of hematopoietic stem cells: Progress and controversy. Stem Cell Res. 8, 1–13. 10.1016/j.scr.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Boisset J-C, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C, 2010. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120. 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Boyer SW, Beaudin AE, Forsberg EC, 2012. Mapping differentiation pathways from hematopoietic stem cells using Flk2/Flt3 lineage tracing. Cell Cycle 11, 3180–8. 10.4161/cc.21279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer SW, Schroeder AV, Smith-Berdan S, Forsberg EC, 2011. All Hematopoietic Cells Develop from Hematopoietic Stem Cells through Flk2/Flt3-Positive Progenitor Cells. Cell Stem Cell 9, 64–73. 10.1016/j.stem.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresciani E, Carrington B, Wincovitch S, Jones M, Gore AV, Weinstein BM, Sood R, Liu PP, 2014. CBF and RUNX1 are required at 2 different steps during the development of hematopoietic stem cells in zebrafish. Blood 124, 70–78. 10.1182/blood-2013-10-531988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, Rodewald H-R, 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546. 10.1038/nature14242 [DOI] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA, 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891. 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RL, Yzaguirre AD, Yashiro-Ohtani Y, Bondue A, Blanpain C, Pear WS, Speck NA, Keller G, 2013. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat. Cell Biol 15, 502–510. 10.1038/ncb2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies LC, Jenkins SJ, Allen JE, Taylor PR, 2013. Tissue-resident macrophages. Nat. Immunol 14, 986–995. 10.1038/ni.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn M, 2014. The hemangioblast revisited. Blood 124, 2472–2473. 10.1182/blood-2014-09-597674 [DOI] [PubMed] [Google Scholar]
- de Bruijn MFTR, Ma X, Robin C, Ottersbach K, Sanchez M-J, Dzierzak E, 2002. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity 16, 673–83. [DOI] [PubMed] [Google Scholar]
- de Bruijn MFTR, Speck NA, Peeters MCE, Dzierzak E, 2000. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19, 2465–2474. 10.1093/emboj/19.11.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharampuriya PR, Scapin G, Wong C, John Wagner K, Cillis JL, Shah DI, 2017. Tracking the origin, development, and differentiation of hematopoietic stem cells. Curr. Opin. Cell Biol 49, 108–115. 10.1016/j.ceb.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand C, Dzierzak E, 2005. Embryonic beginnings of adult hematopoietic stem cells. Haematologica 90, 100–8. [PubMed] [Google Scholar]
- Eaves CJ, 2015. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood 125, 2605–2613. 10.1182/blood-2014-12-570200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilken HM, Nishikawa S-I, Schroeder T, 2009. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 457, 896–900. 10.1038/nature07760 [DOI] [PubMed] [Google Scholar]
- Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J, 2003. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 22, 4478–88. 10.1093/emboj/cdg434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, Schilling JD, Schwendener R, Sergin I, Razani B, Forsberg EC, Yokoyama WM, Unanue ER, Colonna M, Randolph GJ, Mann DL, 2014. Embryonic and Adult-Derived Resident Cardiac Macrophages Are Maintained through Distinct Mechanisms at Steady State and during Inflammation. Immunity 40, 91–104. 10.1016/j.immuni.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganuza M, Hadland B, Chabot A, Li C, Kang G, Bernstein I, McKinney-Freeman S, 2017a. Murine hemogenic endothelial precursors display heterogeneous hematopoietic potential ex vivo. Exp. Hematol 51, 25–35.e6. 10.1016/j.exphem.2017.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganuza M, Hall T, Finkelstein D, Chabot A, Kang G, McKinney-Freeman S, 2017b. Lifelong haematopoiesis is established by hundreds of precursors throughout mammalian ontogeny. Nat. Cell Biol 19, 1153–1163. 10.1038/ncb3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekas C, Rhodes KE, Vanhandel B, Chhabra A, Ueno M, Mikkola HKA, 2010. Hematopoietic stem cell development in the placenta. Int. J. Dev. Biol 54, 1089–1098. 10.1387/ijdb.103070cg [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentek R, Ghigo C, Hoeffel G, Bulle MJ, Msallam R, Gautier G, Launay P, Chen J, Ginhoux F, Bajénoff M, 2018. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity 48, 1160–1171.e5. 10.1016/j.immuni.2018.04.025 [DOI] [PubMed] [Google Scholar]
- Ghigo C, Mondor I, Jorquera A, Nowak J, Wienert S, Zahner SP, Clausen BE, Luche H, Malissen B, Klauschen F, Bajénoff M, 2013. Multicolor fate mapping of Langerhans cell homeostasis. J. Exp. Med 210, 1657–1664. 10.1084/jem.20130403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn EEB, Waters J, Phillips M, Yamamoto R, Long BR, Yang Y, Gerstein R, Stoddart CA, Nakauchi H, Herzenberg LA, 2016. Fetal Hematopoietic Stem Cell Transplantation Fails to Fully Regenerate the B-Lymphocyte Compartment. Stem Cell Reports 6, 137–149. 10.1016/j.stemcr.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn EEB, Yamamoto R, Hamanaka S, Yang Y, Herzenberg LA, Nakauchi H, Herzenberg LA, 2012. Distinct B-cell lineage commitment distinguishes adult bone marrow hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A 109, 5394–8. 10.1073/pnas.1121632109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M, 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–5. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, de Bruijn M, Rodewald H-R, Geissmann F, 2015. The Origin of Tissue-Resident Macrophages: When an Erythro-myeloid Progenitor Is an Erythro-myeloid Progenitor. Immunity 43, 1023–1024. 10.1016/j.immuni.2015.11.022 [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Geissmann F, 2013. Myb-Independent Macrophages: A Family of Cells That Develops with Their Tissue of Residence and Is Involved in Its Homeostasis. Cold Spring Harb. Symp. Quant. Biol 78, 91–100. 10.1101/sqb.2013.78.020032 [DOI] [PubMed] [Google Scholar]
- Gothert JR, Gustin SE, Hall MA, Green AR, Göttgens B, Izon DJ, Begley CG, 2005. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood 105, 2724–2732. 10.1182/blood-2004-08-3037 [DOI] [PubMed] [Google Scholar]
- Griffiths JA, Scialdone A, Marioni JC, 2018. Using single‐cell genomics to understand developmental processes and cell fate decisions. Mol. Syst. Biol 14, e8046. 10.15252/msb.20178046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Luc S, Marco E, Lin T-W, Peng C, Kerenyi MA, Beyaz S, Kim W, Xu J, Das PP, Neff T, Zou K, Yuan G-C, Orkin SH, 2013. Mapping Cellular Hierarchy by Single-Cell Analysis of the Cell Surface Repertoire. Cell Stem Cell 13, 492–505. 10.1016/j.stem.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland B, Yoshimoto M, 2018. Many layers of embryonic hematopoiesis: new insights into B-cell ontogeny and the origin of hematopoietic stem cells. Exp. Hematol 60, 1–9. 10.1016/j.exphem.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Hayakawa K, 2015. Perspectives on fetal derived CD5 + B1 B cells. Eur. J. Immunol 45, 2978–2984. 10.1002/eji.201445146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, Greter M, Mortha A, Boyer SW, Forsberg EC, Tanaka M, van Rooijen N, García-Sastre A, Stanley ER, Ginhoux F, Frenette PS, Merad M, 2013. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804. 10.1016/j.immuni.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K, Hardy RR, Herzenberg LA, Herzenberg LA, 1985. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J. Exp. Med 161, 1554–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi C, Sonoda T, Kitamura Y, 1983. Bone marrow origin of mast cell precursors in mesenteric lymph nodes of mice. Exp. Hematol 11, 772–8. [PubMed] [Google Scholar]
- He S, Kim I, Lim MS, Morrison SJ, 2011. Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev. 25, 1613–1627. 10.1101/gad.2052911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninger J, Santoso B, Hans S, Durand E, Moore J, Mosimann C, Brand M, Traver D, Zon L, 2017. Clonal fate mapping quantifies the number of haematopoietic stem cells that arise during development. Nat. Cell Biol 19, 17–27. 10.1038/ncb3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring CA, Chen B, McKinley ET, Lau KS, 2018. Single-Cell Computational Strategies for Lineage Reconstruction in Tissue Systems. Cell. Mol. Gastroenterol. Hepatol 5, 539–548. 10.1016/j.jcmgh.2018.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzenberg LA, 2015. Layered evolution in the immune system: a view from history. Ann. N. Y. Acad. Sci 1362, 1–5. 10.1111/nyas.12795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Meade E, Medeiros S, Schindler JW, Valk PJM, Fujiwara Y, Orkin SH, 2004. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 18, 2336–2341. 10.1101/gad.1239604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, Poidinger M, Zolezzi F, Larbi A, Ng LG, Chan JKY, Greter M, Becher B, Samokhvalov IM, Merad M, Ginhoux F, 2015. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–78. 10.1016/j.immuni.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Ginhoux F, 2018. Fetal monocytes and the origins of tissue-resident macrophages. Cell. Immunol 10.1016/j.cellimm.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Hoeffel G, Ginhoux F, 2015. Ontogeny of Tissue-Resident Macrophages. Front. Immunol 6, 486. 10.3389/fimmu.2015.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, Leboeuf M, Low D, Oller G, Almeida F, Choy SHY, Grisotto M, Renia L, Conway SJ, Stanley ER, Chan JKY, Ng LG, Samokhvalov IM, Merad M, Ginhoux F, 2012. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med 209, 1167–81. 10.1084/jem.20120340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honold L, Nahrendorf M, 2018. Resident and Monocyte-Derived Macrophages in Cardiovascular Disease. Circ. Res 122, 113–127. 10.1161/CIRCRESAHA.117.311071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Auerbach R, 1993. Identification and characterization of hematopoietic stem cells from the yolk sac of the early mouse embryo. Proc. Natl. Acad. Sci. U. S. A 90, 10110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G, 2004. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432, 625–30. 10.1038/nature03122 [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Hirai H, Ogawa S, Kurokawa M, 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med 10, 299–304. 10.1038/nm997 [DOI] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Hölscher C, Müller DN, Luckow B, Brocker T, Debowski K, Fritz G, Opdenakker G, Diefenbach A, Biber K, Heikenwalder M, Geissmann F, Rosenbauer F, Prinz M, 2013. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci 16, 273–80. 10.1038/nn.3318 [DOI] [PubMed] [Google Scholar]
- Kierdorf K, Prinz M, Geissmann F, Gomez Perdiguero E, 2015. Development and function of tissue resident macrophages in mice. Semin. Immunol 27, 369–378. 10.1016/j.smim.2016.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Saunders TL, Morrison SJ, 2007. Sox17 Dependence Distinguishes the Transcriptional Regulation of Fetal from Adult Hematopoietic Stem Cells. Cell 130, 470–483. 10.1016/j.cell.2007.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Yilmaz OH, Morrison SJ, 2005. CD144 (VE-cadherin) is transiently expressed by fetal liver hematopoietic stem cells. Blood 106, 903–905. 10.1182/blood-2004-12-4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, Herbomel P, 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115. 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A, 2015. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872. 10.1101/gr.192237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaravelu P, Hook L, Morrison AM, Ure J, Zhao S, Zuyev S, Ansell J, Medvinsky A, 2002. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development 129, 4891–9. [DOI] [PubMed] [Google Scholar]
- Lacaud G, Kouskoff V, 2017. Hemangioblast, hemogenic endothelium, and primitive versus definitive hematopoiesis. Exp. Hematol 49, 19–24. 10.1016/J.EXPHEM.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Lacaud G, Robertson S, Palis J, Kennedy M, Keller G, 2001. Regulation of hemangioblast development. Ann. N. Y. Acad. Sci 938, 96–107; discussion 108. [DOI] [PubMed] [Google Scholar]
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G, 2009. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457, 892–895. 10.1038/nature07679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J-Y, Shi Y, Chin YE, 2013. Reprogramming cancer cells: back to the future. Oncogene 32, 2247–2248. 10.1038/onc.2012.349 [DOI] [PubMed] [Google Scholar]
- Lavin Y, Merad M, 2013. Macrophages: gatekeepers of tissue integrity. Cancer Immunol. Res 1, 201–9. 10.1158/2326-6066.CIR-13-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Mortha A, Rahman A, Merad M, 2015. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol 15, 731–744. 10.1038/nri3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LK, Ghorbanian Y, Wang W, Wang Y, Kim YJ, Weissman IL, Inlay MA, Mikkola HKA, 2016. LYVE1 Marks the Divergence of Yolk Sac Definitive Hemogenic Endothelium from the Primitive Erythroid Lineage. Cell Rep. 10.1016/j.celrep.2016.10.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichanska AM, Hume DA, 2000. Origins and functions of phagocytes in the embryo. Exp. Hematol 28, 601–11. [DOI] [PubMed] [Google Scholar]
- Lieu YK, Reddy EP, 2009. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc. Natl. Acad. Sci 106, 21689–21694. 10.1073/pnas.0907623106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K-W, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai F-Y, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E, 2004. GATA-2 Plays Two Functionally Distinct Roles during the Ontogeny of Hematopoietic Stem Cells. J. Exp. Med 200, 871–882. 10.1084/jem.20031556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CP, Auerbach R, 1991. In vitro development of murine T cells from prethymic and preliver embryonic yolk sac hematopoietic stem cells. Development 113, 1315–23. [DOI] [PubMed] [Google Scholar]
- Macaulay IC, Svensson V, Labalette C, Ferreira L, Hamey F, Voet T, Teichmann SA, Cvejic A, 2016. Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep. 14, 966–977. 10.1016/j.celrep.2015.12.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuoka HC, Townes TM, 2002. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99, 736–45. [DOI] [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E, 1996. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86, 897–906. [DOI] [PubMed] [Google Scholar]
- Medvinsky A, Rybtsov S, Taoudi S, 2011. Embryonic origin of the adult hematopoietic system: advances and questions. Development 138, 1017–31. 10.1242/dev.040998 [DOI] [PubMed] [Google Scholar]
- Mehta C, Johnson KD, Gao X, Ong IM, Katsumura KR, McIver SC, Ranheim EA, Bresnick EH, 2017. Integrating Enhancer Mechanisms to Establish a Hierarchical Blood Development Program. Cell Rep. 20, 2966–2979. 10.1016/j.celrep.2017.08.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Manz MG, Karsunky H, Wagers A, Peters W, Charo I, Weissman IL, Cyster JG, Engleman EG, 2002. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat. Immunol 3, 1135–41. 10.1038/ni852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola HKA, Klintman J, Yang H, Hock H, Schlaeger TM, Fujiwara Y, Orkin SH, 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421, 547–551. 10.1038/nature01345 [DOI] [PubMed] [Google Scholar]
- Mizoguchi S, Takahashi K, Takeya M, Naito M, Morioka T, 1992. Development, differentiation, and proliferation of epidermal Langerhans cells in rat ontogeny studied by a novel monoclonal antibody against epidermal Langerhans cells, RED-1. J. Leukoc. Biol 52, 52–61. [DOI] [PubMed] [Google Scholar]
- Moignard V, Woodhouse S, Fisher J, Göttgens B, 2013. Transcriptional hierarchies regulating early blood cell development. Blood Cells, Mol. Dis 51, 239–247. 10.1016/j.bcmd.2013.07.007 [DOI] [PubMed] [Google Scholar]
- Moignard V, Woodhouse S, Haghverdi L, Lilly AJ, Tanaka Y, Wilkinson AC, Buettner F, Macaulay IC, Jawaid W, Diamanti E, Nishikawa S-I, Piterman N, Kouskoff V, Theis FJ, Fisher J, Göttgens B, 2015. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat. Biotechnol 33, 269–276. 10.1038/nbt.3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MA, Metcalf D, 1970. Ontogeny of the haemopoietic system: yolk sac origin of in vivo and in vitro colony forming cells in the developing mouse embryo. Br. J. Haematol 18, 279–96. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL, 1995. The purification and characterization of fetal liver hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A 92, 10302–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Potter SS, 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65, 677–89. [DOI] [PubMed] [Google Scholar]
- Murray PDF, 1932. The Development in vitro of the Blood of the Early Chick Embryo. Proc. R. Soc. B Biol. Sci 111, 497–521. 10.1098/rspb.1932.0070 [DOI] [Google Scholar]
- Nicolás-Ávila JA, Hidalgo A, Ballesteros I, 2018. Specialized functions of resident macrophages in brain and heart. J. Leukoc. Biol 10.1002/JLB.6MR0118-041R [DOI] [PubMed] [Google Scholar]
- Nishikawa S, 2012. Hemangioblast: an in vitro phantom. Wiley Interdiscip. Rev. Dev. Biol 1, 603–608. 10.1002/wdev.38 [DOI] [PubMed] [Google Scholar]
- Oberlin E, El Hafny B, Petit-Cocault L, Souyri M, 2010. Definitive human and mouse hematopoiesis originates from the embryonic endothelium: a new class of HSCs based on VE-cadherin expression. Int. J. Dev. Biol 54, 1165–73. 10.1387/ijdb.103121eo [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR, 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–30. [DOI] [PubMed] [Google Scholar]
- Org T, Duan D, Ferrari R, Montel-Hagen A, Van Handel B, Kerenyi MA, Sasidharan R, Rubbi L, Fujiwara Y, Pellegrini M, Orkin SH, Kurdistani SK, Mikkola HK, 2015. Scl binds to primed enhancers in mesoderm to regulate hematopoietic and cardiac fate divergence. EMBO J. 34, 759–777. 10.15252/embj.201490542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M, Ottersbach K, Bollerot K, Orelio C, de Bruijn M, Wijgerde M, Dzierzak E, 2009. Ventral embryonic tissues and Hedgehog proteins induce early AGM hematopoietic stem cell development. Development 136, 2613–2621. 10.1242/dev.034728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei W, Feyerabend TB, Rössler J, Wang X, Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C, Chen W, Sauer S, Wolf S, Höfer T, Rodewald H-R, 2017. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460. 10.1038/nature23653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perah G, Feldman M, 1977. In vitro activation of the in vivo colony-forming units of the mouse yolk sac. J. Cell. Physiol 91, 193–9. 10.1002/jcp.1040910205 [DOI] [PubMed] [Google Scholar]
- Rhodes KE, Gekas C, Wang Y, Lux CT, Francis CS, Chan DN, Conway S, Orkin SH, Yoder MC, Mikkola HKA, 2008. The Emergence of Hematopoietic Stem Cells Is Initiated in the Placental Vasculature in the Absence of Circulation. Cell Stem Cell 2, 252–263. 10.1016/j.stem.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb L, Lyons I, Li R, Hartley L, Köntgen F, Harvey RP, Metcalf D, Begley CG, 1995. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. U. S. A 92, 7075–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, Enver T, Vyas P, Scadden DT, 2005. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 106, 477–484. 10.1182/blood-2004-08-2989 [DOI] [PubMed] [Google Scholar]
- Samokhvalov IM, Samokhvalova NI, Nishikawa S, 2007. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 446, 1056–1061. 10.1038/nature05725 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T, 1996. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc. Natl. Acad. Sci. U. S. A 93, 12359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai CM, Babovic S, Upadhaya S, Knapp DJHF, Lavin Y, Lau CM, Goloborodko A, Feng J, Fujisaki J, Ding L, Mirny LA, Merad M, Eaves CJ, Reizis B, 2016. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity 45, 597–609. 10.1016/j.immuni.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeger TM, Mikkola HKA, Gekas C, Helgadottir HB, Orkin SH, 2005. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood 105, 3871–4. 10.1182/blood-2004-11-4467 [DOI] [PubMed] [Google Scholar]
- Schlenner SM, Madan V, Busch K, Tietz A, Läufle C, Costa C, Blum C, Fehling HJ, Rodewald HR, 2010. Fate Mapping Reveals Separate Origins of T Cells and Myeloid Lineages in the Thymus. Immunity 32, 426–436. 10.1016/j.immuni.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Schoedel KB, Morcos MNF, Zerjatke T, Roeder I, Grinenko T, Voehringer D, Göthert JR, Waskow C, Roers A, Gerbaulet A, 2016. The bulk of the hematopoietic stem cell population is dispensable for murine steady-state and stress hematopoiesis. Blood 128, 2285–2296. 10.1182/blood-2016-03-706010 [DOI] [PubMed] [Google Scholar]
- Schulz C, Perdiguero EG, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SEW, Pollard JW, Frampton J, Liu KJ, Geissmann F, 2012. A Lineage of Myeloid Cells Independent of Myb and Hematopoietic Stem Cells. Science (80-.) 336, 86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Scialdone A, Tanaka Y, Jawaid W, Moignard V, Wilson NK, Macaulay IC, Marioni JC, Göttgens B, 2016. Resolving early mesoderm diversification through single-cell expression profiling. Nature 535, 289–293. 10.1038/nature18633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C, 2009. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci 106, 1930–1935. 10.1073/pnas.0808866106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenich L, 2018. Brain-Resident Microglia and Blood-Borne Macrophages Orchestrate Central Nervous System Inflammation in Neurodegenerative Disorders and Brain Cancer. Front. Immunol 9, 697. 10.3389/fimmu.2018.00697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Mayer EL, Orkin SH, 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373, 432–434. 10.1038/373432a0 [DOI] [PubMed] [Google Scholar]
- Sorokin SP, Hoyt RF, Blunt DG, McNelly NA, 1992. Macrophage development: II. Early ontogeny of macrophage populations in brain, liver, and lungs of rat embryos as revealed by a lectin marker. Anat. Rec 232, 527–550. 10.1002/ar.1092320410 [DOI] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL, 1988. Purification and characterization of mouse hematopoietic stem cells. Science 241, 58–62. [DOI] [PubMed] [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho Y-J, Klein A, Hofmann O, Camargo FD, 2014. Clonal dynamics of native haematopoiesis. Nature 514, 322–327. 10.1038/nature13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S, 2000. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech. Dev 95, 133–45. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sanchez V, Takata N, Yokomizo T, Yamanaka Y, Kataoka H, Hoppe PS, Schroeder T, Nishikawa S-I, 2014. Circulation-independent differentiation pathway from extraembryonic mesoderm toward hematopoietic stem cells via hemogenic angioblasts. Cell Rep. 8, 31–9. 10.1016/j.celrep.2014.05.055 [DOI] [PubMed] [Google Scholar]
- Taoudi S, Gonneau C, Moore K, Sheridan JM, Blackburn CC, Taylor E, Medvinsky A, 2008. Extensive Hematopoietic Stem Cell Generation in the AGM Region via Maturation of VE-Cadherin+CD45+ Pre-Definitive HSCs. Cell Stem Cell 3, 99–108. 10.1016/j.stem.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Tober J, Yzaguirre AD, Piwarzyk E, Speck NA, 2013. Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development 140, 3765–3776. 10.1242/dev.094961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toles JF, Chui DH, Belbeck LW, Starr E, Barker JE, 1989. Hemopoietic stem cells in murine embryonic yolk sac and peripheral blood. Proc. Natl. Acad. Sci. U. S. A 86, 7456–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH, 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–6. 10.1038/371221a0 [DOI] [PubMed] [Google Scholar]
- Ueno H, Weissman IL, 2006. Clonal Analysis of Mouse Development Reveals a Polyclonal Origin for Yolk Sac Blood Islands. Dev. Cell 11, 519–533. 10.1016/j.devcel.2006.08.001 [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8, 757–69. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Fujiwara Y, Orkin SH, 1998. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 12, 473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH, 1994. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell 78, 45–57. [DOI] [PubMed] [Google Scholar]
- Wilson NK, Kent DG, Buettner F, Shehata M, Macaulay IC, Calero-Nieto FJ, Sánchez Castillo M, Oedekoven CA, Diamanti E, Schulte R, Ponting CP, Voet T, Caldas C, Stingl J, Green AR, Theis FJ, Göttgens B, 2015. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell 16, 712–724. 10.1016/j.stem.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Till JE, Siminovitch L, McCulloch EA, 1968. Cytological evidence for a relationship between normal hemotopoietic colony-forming cells and cells of the lymphoid system. J. Exp. Med 127, 455–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tanaka Y, Seay M, Li Z, Jin J, Garmire LX, Zhu X, Taylor A, Li W, Euskirchen G, Halene S, Kluger Y, Snyder MP, Park I-H, Pan X, Weissman SM, 2016. Single cell transcriptomics reveals unanticipated features of early hematopoietic precursors. Nucleic Acids Res. 45, gkw1214. 10.1093/nar/gkw1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhang H, Amabile G, Yang H, Staber PB, Zhang P, Levantini E, Alberich-Jordà M, Zhang J, Kawasaki A, Tenen DG, 2013. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat. Cell Biol 15, 385–394. 10.1038/ncb2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Valin KL, Dixon ML, Leavenworth JW, 2017. The Role of Microglia and Macrophages in CNS Homeostasis, Autoimmunity, and Cancer. J. Immunol. Res 2017, 1–12. 10.1155/2017/5150678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder MC, Hiatt K, Mukherjee P, 1997. In vivo repopulating hematopoietic stem cells are present in the murine yolk sac at day 9.0 postcoitus. Proc. Natl. Acad. Sci. U. S. A 94, 6776–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, Hume DA, Perlman H, Malissen B, Zelzer E, Jung S, 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, 2015. The first wave of B lymphopoiesis develops independently of stem cells in the murine embryo. Ann. N. Y. Acad. Sci 1362, 16–22. 10.1111/nyas.12612 [DOI] [PubMed] [Google Scholar]
- Yu VWC, Yusuf RZ, Oki T, Wu J, Saez B, Wang X, Cook C, Baryawno N, Ziller MJ, Lee E, Gu H, Meissner A, Lin CP, Kharchenko PV, Scadden DT, 2016. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell 167, 1310–1322.e17. 10.1016/j.cell.2016.10.045 [DOI] [PubMed] [Google Scholar]
- Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA, 2012. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science 335, 1195–200. 10.1126/science.1216557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jin H, Li L, Qin FX-F, Wen Z, 2011. cMyb regulates hematopoietic stem/progenitor cell mobilization during zebrafish hematopoiesis. Blood 118, 4093–4101. 10.1182/blood-2011-03-342501 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhou J, Liu D, Dong F, Cheng H, Wang W, Pang Y, Wang Y, Mu X, Ni Y, Li Z, Xu H, Hao S, Wang X, Ma S, Wang Q.-f., Xiao G, Yuan W, Liu B, Cheng T, 2015. ATF4 plays a pivotal role in the development of functional hematopoietic stem cells in mouse fetal liver. Blood 126, 2383–2391. 10.1182/blood-2015-03-633354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Li X, Wang W, Zhu P, Zhou J, He W, Ding M, Xiong F, Zheng X, Li Z, Ni Y, Mu X, Wen L, Cheng T, Lan Y, Yuan W, Tang F, Liu B, 2016. Tracing haematopoietic stem cell formation at single-cell resolution. Nature 533, 487–492. 10.1038/nature17997 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Ding M, Huang L, Gilkeson G, Lang R, Jiang W, 2016. Toll-like receptor-mediated immune responses in intestinal macrophages; implications for mucosal immunity and autoimmune diseases. Clin. Immunol 173, 81–86. 10.1016/j.clim.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, Weintraub H, 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79, 875–84. [DOI] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, Tallquist MD, Iruela-Arispe ML, 2008. Fate Tracing Reveals the Endothelial Origin of Hematopoietic Stem Cells. Cell Stem Cell 3, 625–636. 10.1016/j.stem.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]


