Abstract
Methanotrophic bacteria have significant potential for bioremediation, which would require methods for monitoring the presence and activity of these organisms in environmental samples. In this study, PCR was used to detect methanotrophic bacteria. Primers were designed on the basis of a partial sequence of pmoA, which encodes one of the proteins of the particulate methane monooxygenase. Specific amplification of a portion of pmoA was obtained with template DNA isolated from lab strains of methanotrophs. A pmoA product was also obtained by using DNA from groundwater. The identity of the PCR product was confirmed by sequencing or by amplification with a nested primer. Reverse transcriptase PCR detected pmoA mRNA.
Methanotrophic bacteria use methane as a source of carbon and energy. Methane is oxidized to CO2 or is incorporated into biomass. The first enzyme of the methane oxidation pathway is methane monooxygenase. All known methanotrophs produce a membrane-associated particulate methane monooxygenase (pMMO). A few strains produce a soluble methane monooxygenase (sMMO) in response to copper limitation. Because they are capable of cometabolic oxidation of trichloroethylene (TCE) and other persistent compounds, methanotrophs have potential for bioremediation (1, 4, 9, 13, 20). The initial interest in this potential has focused on sMMO because the rate of TCE oxidation with this enzyme is very high (20). However, because pMMO can also catalyze TCE oxidation (9, 30) with a low Ks for TCE (14, 25) and because methanotrophs that lack sMMO may be widely distributed, it is essential to assess the distribution and activity of pMMO when sites are evaluated prior to and during bioremediation.
The genes encoding sMMO (mmo) have been cloned and sequenced (6, 7, 26). The sequence information obtained has been used to design primers for PCR that can be used to detect a portion of mmoX in environmental samples (15, 17, 19). Primers for amplification of a portion of the gene encoding methanol dehydrogenase have been designed and used with environmental samples, but sequencing is required to distinguish products amplified from methanotrophs from products amplified from other methylotrophs (16).
Recently, the sequence of the genes encoding pMMO, pmoCAB, was determined and was shown to exhibit no significant homology to the mmo sequence (23). PCR primers that amplify a fragment of either pmoA or the gene for a related enzyme, amoA, have been designed (11). Because amo encodes the ammonia monooxygenase of ammonia-oxidizing bacteria, these primers cannot be used to distinguish between methanotrophs and ammonia oxidizers. This study was undertaken to develop approaches for specifically detecting pmo in groundwater. To do this, primers were designed on the basis of regions that are different in pmoA and amoA. These primers amplified a 330-bp sequence when DNA from a methanotroph was used as the template; no product was amplified when DNA from an ammonia oxidizer was used. Our methods should have applications for sites being assessed for bioremediation, as well as in other environmental studies on the distribution and activity of methanotrophs.
(A preliminary report of this work has been presented previously [8].)
MATERIALS AND METHODS
Bacteria and growth conditions.
Methylomicrobium album BG8, Methylococcus capsulatus (Bath), and Methylosinus trichosporium OB3b were grown in nitrate minimal salts medium (29) supplemented with 5 μM CuSO4 under an atmosphere containing 50% methane with shaking at 200 rpm at 30°C. To prepare enrichment cultures, cells from 150 ml of groundwater were collected on filters, and the filters were incubated in the same medium. When the medium became turbid, the cultures were streaked onto plates. An isolate from Yellowstone Lake was obtained from an enrichment culture inoculated with mat material collected near a fumarole in Sedge Bay. An isolate from Lake Michigan was isolated from an enrichment culture inoculated with Green Bay sediment (5). Escherichia coli JM109 was grown in Luria-Bertani broth (21), and Nitrosomonas europaea ATCC 19718 was grown in ATCC medium 221 (2).
Molecular techniques.
The DNA used for PCR was prepared with a GenomicPrep Cell and Tissue DNA Isolation Kit (Amersham Pharmacia, Milwaukee, Wis.). Alternatively, DNA was prepared with an Amersham Pharmacia GFX kit with the following modifications: solution I included 2% hexadecyltrimethylammonium bromide, solution II was a 1% sodium N-laurylsarcosinate solution, and solution III was a 6 M guanidine thiocyanate solution. DNA was isolated from groundwater by using a protocol modified from the protocol of Fuhrman et al. (10). Cells were collected on Durapore series GV filters (Millipore Corp., Bedford, Mass.), and 0.3% hexadecyltrimethylammonium bromide and 0.9 M NaCl were added prior to phenol extraction.
RNA was prepared from cultured bacterial cells with TRIzol (Gibco BRL, Gaithersburg, Md.) by using the recommendations of the manufacturer. To prepare RNA from groundwater, cells were collected from groundwater on Duropore filters.
The PCR amplifications were performed in 100-μl (total volume) reaction mixtures in thin-wall tubes under a layer of mineral oil (Sigma Chemical Co., St. Louis, Mo.) by using a model 480 thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.). Each PCR was performed in a solution that contained each deoxynucleoside triphosphate at a concentration of 200 to 400 μM, 1.5 mM MgCl2, 10 mM NaCl, 0.01 mM EDTA, 0.1 mM dithiothreitol, 5 mM Tris HCl (pH 8.0), 2% dimethyl sulfoxide, 5% glycerol, 0.1% Triton X-100, and 2.5 U of Taq polymerase (Promega, Madison, Wis.). For PCR, the concentration of each member of the primer pair was as follows: 0.15 μM for the 16S ribosomal DNA (rDNA) primers, 0.075 μM for the mmoX primers, and 0.10 μM for the pmoA primers. The PCR conditions were as follows: incubation at 95°C for 10 min; Taq added; 29 cycles consisting of incubation at 94°C for 1 min, at 45°C for 1 min, at 72°C for 1 min, and (for the final cycle) at 94°C for 1 min, at 45°C for 1 min, and at 72°C for 5 min. For DNA purified from cultured cells, 20 ng of DNA was used as the template. For environmental samples the level of DNA was less than the level which could be directly quantified on an agarose gel.
To detect RNA, an uncoupled reverse transcriptase PCR (RT-PCR) was used. A DNA copy (cDNA) was reverse transcribed from the RNA by using the avian myeloblastosis virus RT and the AMV/Tfl buffer components of an Access RT-PCR kit (Promega). The cDNA was isolated by using a PCR Purification Kit (Qiagen Inc., Santa Clarita, Calif.) and was used as a template for PCR. PCR and RT-PCR products were analyzed on 1.5% agarose gels.
Environmental samples.
Groundwater was collected from three wells maintained by the Department of Geosciences of the University of Wisconsin-Milwaukee. Two of these wells (wells GLRF and CAP) are 3 to 5 m deep in a sand-gravel aquifer. Well LAPHAM is 83 to 105 m deep in a dolomite aquifer.
RESULTS
The sequences of the pmoA and amoA genes were aligned by Holmes et al. (11). These investigators used primers amo/pmof and amo/pmor (Table 1) to amplify a portion of either pmoA or amoA, sequenced the PCR products, and aligned the resulting sequences. On the basis of highly conserved regions of the pmoA sequences that are distinct from the amoA sequence, primers specific for pmoA (primers pmof1 and pmor) were designed in the present study (Table 1). Primer pmof1 binds to a site on the DNA 165 bp downstream from the primer amo/pmof binding site.
TABLE 1.
Primers used for amplification of DNA from methanotrophic bacteria
| Target gene | Size of product (bp) | Primer | Primer sequence (5′–3′)a | Reference |
|---|---|---|---|---|
| amoA or pmoA | 525 | amo/pmof | GGNGACTGGGACTTCTGG | 11 |
| amo/pmor | GAASGCNGAGAAGAASGC | |||
| mmoX | 535 | mmof | GGCTCCAAGTTCAAGGTCGAGC | 15 |
| mmor | TGGCACTCGTAGCGCTCCGGCTCG | |||
| 16S rDNA | 1,507 | 16Sf | AGAGTTTGATCATGGCTCAG | 12 |
| 16Sr | TACGGYTACCTTGTTACGACTT | |||
| pmoA | 330 | pmof1 | GGGGGAACTTCTGGGGITGGAC | This study |
| pmor | GGGGGRCIACGTCITTACCGAA | |||
| pmoA | 178 | pmof2 | TTCTAYCCDRRCAACTGGCC | This study |
| pmor | GGGGGRCIACGTCITTACCGAA |
Y = C or T; D = G, A, or T; R = A or G; S = G or C; N = A, G, C, or T.
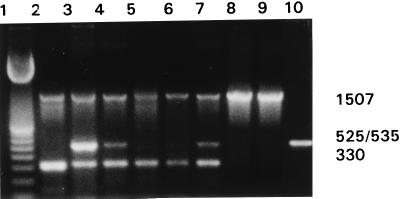
PCR was performed with three primer pairs to simultaneously detect pmoA, mmoX (primers designed by McDonald et al. [15]), and 16S rDNA. 16S rDNA, which was amplified by universal primers (12), was used as a control for the adequacy of the DNA as a template for PCR. The expected amplification products were obtained (Fig. 1). While sMMO is present in M. capsulatus (Bath) and M. trichosporium OB3b, it is not present in M. album BG8 (27, 28). The negative controls, E. coli and the ammonia oxidizer N. europaea, did not yield mmoX or pmoA products (Fig. 1, lanes 8 and 9). An amoA product was amplified from N. europaea with primers amo/pmof and amo/pmor (Fig. 1, lane 10).
FIG. 1.
Multiplex PCR. The 16Sf-16Sr, mmof-mmor, and pmof1-pmor primer pairs were used for lanes 2 through 9. The amo/pmof-amo/pmor primer pair was used for lane 10. DNA was obtained from M. album BG8 (lane 2), M. capsulatus (Bath) (lane 3), M. trichosporium OB3b (lane 4), a groundwater enrichment culture (lane 5), an isolate from sediment from Green Bay in Lake Michigan (lane 6), an isolate from Yellowstone Lake in Yellowstone National Park (lane 7), E. coli (lane 8), and N. europaea (lanes 9 and 10). Lane 1 contained a 100-bp ladder (Amersham Pharmacia). PCR products were identified by their sizes, as follows: 16S rDNA, 1,507 bp; mmoX, 535 bp; amo-pmo, 525 bp; and pmoA, 330 bp.
The identities of the PCR products were confirmed by sequencing the products obtained by using M. capsulatus (Bath), M. trichosporium OB3b, and M. album BG8 DNAs as templates. The sequences obtained were 97 to 98% identical to the sequence described previously (11).
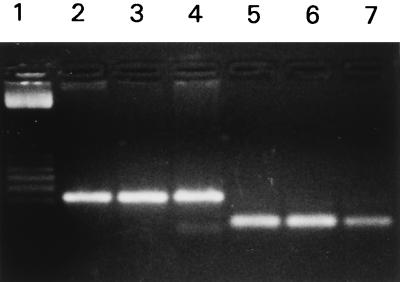
Another approach used to verify the identities of the PCR products was to use the pmoA product as a template for amplification of another product with a nested primer (primer pmof2) (Table 1). This resulted in a 178-bp product when the pmoA PCR products of M. capsulatus (Bath), M. trichosporium OB3b, and M. album BG8 were used as templates (Fig. 2). In addition, the identity of the product obtained from M. album BG8 with the nested primer was verified by sequencing (data not shown). Nested PCR was also used to confirm the identities of the pmoA products obtained with DNA from isolates from Yellowstone Lake, Green Bay, and a groundwater enrichment culture (Fig. 1); the identities of the latter two products were confirmed by sequencing (data not shown).
FIG. 2.
Confirmation of the identify of the pmoA PCR product by nested PCR. The pmof1-pmor primer pair was used for lanes 2 through 4. The pmof2-pmor primer pair was used for lanes 5 through 7. The templates used for lanes 2 through 4 were DNA prepared from M. album BG8, M. capsulatus (Bath), and M. trichosporium OB3b, respectively. The templates used for lanes 5 through 7 were the products of the PCR in lanes 2 through 4, respectively. Lane 1 contained a 100-bp ladder (Amersham Pharmacia). PCR products of 330 bp were detected in lanes 2 through 4, and 178-bp PCR products were detected in lanes 5 through 7. No product was obtained with the pmof2-pmor primer pair when the amo product (Fig. 1, lane 10) was used as a template (data not shown).
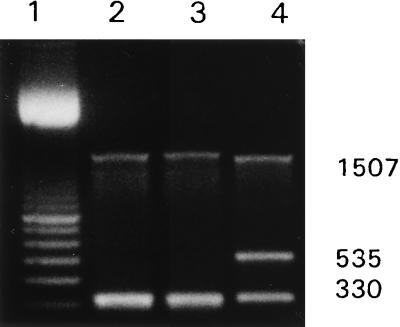
DNA was prepared from groundwater obtained at three sites. These DNAs were used as templates for multiplex PCR. A pmoA product was obtained with DNA from all three sites, while an mmoX product was obtained only with DNA from one site (Fig. 3). The identity of the nested product obtained with well GLRF DNA was confirmed by sequencing (data not shown).
FIG. 3.
Multiplex PCR analysis of groundwater. The 16Sf-16Sr, mmof-mmor, and pmof1-pmor primer pairs were used. DNA was prepared from groundwater obtained from the following sites: well CAP (lane 2), well GLRF (lane 3), and well LAPHAM (lane 4). Lane 1 contained a 100-bp ladder (Amersham Pharmacia). The 16S rDNA PCR product (1,507 bp) and the pmoA PCR product (330 bp) were detected in lanes 2 through 4; the mmoX PCR product (535 bp) was detected in lane 4. No product was obtained when distilled water was substituted for groundwater (data not shown).
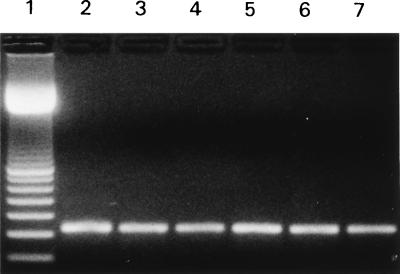
RNA was extracted from methanotroph lab strains and groundwater. Each RNA was reverse transcribed to cDNA, which was used as a template for PCR performed with pmoA primers. A pmoA product was obtained with cDNA that had been reverse transcribed from RNA prepared from lab strains, groundwater, and a groundwater enrichment culture (Fig. 4). The identities of the PCR products were confirmed by nested PCR (data not shown). No product was obtained when the RNA preparations were used directly without an RT step (data not shown).
FIG. 4.
RT-PCR performed with the pmof1-pmor primer pair. RNA was obtained from M. album BG8 (lane 2), M. capsulatus (Bath) (lane 3), M. trichosporium OB3b (lane 4), a groundwater enrichment culture (lane 5), well GLRF groundwater (lane 6), and well LAPHAM groundwater (lane 7). Lane 1 contained a 100-bp ladder (Amersham Pharmacia). A 330-bp product was detected in lanes 2 through 7.
DISCUSSION
The pmof1-pmor primer pair is specific for pMMO, as a product was not obtained with DNA prepared from E. coli or the ammonia oxidizer N. europaea. Using a second forward primer (pmof2) provided a simple way to confirm the identity of the pmoA PCR product. In addition, it is possible that using nested PCR could extend the application of this approach to detection of methanotrophs in environmental samples that contain PCR inhibitors; nested PCR has been used to detect Legionella spp. in such samples (18).
The pmoA primers designed in this study could be used in multiplex PCR in conjunction with primers for 16S rDNA and mmoX. This allows the simultaneous detection of genes encoding pMMO and sMMO with amplification of 16S rDNA, providing a positive control.
A pmoA PCR product was amplified from DNA obtained from all three groundwater samples tested. In contrast to the pmoA product, an mmoX product was obtained only with DNA from one of these groundwater samples. This implies that either the levels of the methanotrophs with sMMO were below the level of detection or that the mmoX primers used could not detect the methanotrophs that were present. Detection of pmoA DNA from methanotrophs cultured from different environments (Fig. 1) and from Green Bay sediment (9a) suggested that this approach may be widely applicable.
In this study pmoA mRNA was detected by RT-PCR performed with RNA obtained from cultures and groundwater. This is significant because it has been suggested that detection of mRNA by RT-PCR is a better indicator of cell viability than is the detection of 16S rRNA (24). Moreover, detection of specific messages has potential applications for evaluation of activities of microbes at bioremediation sites or in bioreactors (3, 22). While in this study detection of pmoA DNA and RNA was qualitative, this work provides the basis for approaches that could be used to quantify methanotrophs and message encoding pMMO.
ACKNOWLEDGMENTS
This work was supported by a grant from the Water Resources Center through the UWS Groundwater Research Program. The isolate from Yellowstone Lake was obtained in a field study supported by grants to J. Maki (grant NA76RU0060 from the National Oceanic and Atmospheric Administration) and R. Cuhel (grant EAR9708501 from the National Science Foundation), and the sediment from Green Bay was obtained in a study supported by a grant to D. N. Edgington (grant R/MW-78 from the National Oceanic and Atmospheric Administration).
We thank C. Wimpee for helpful advice throughout this work. We are grateful to D. Cherkauer for providing access to sampling wells. The assistance of P. D. Anderson with some of this work is gratefully acknowledged.
Footnotes
This is publication no. 416 from the Center for Great Lakes Studies.
REFERENCES
- 1.Alvarez-Cohen L, McCarty P L, Boulygina E, Hanson R S, Brusseau G A, Tsien H C. Characterization of a methane-utilizing bacterium from a bacterial consortium that rapidly degrades trichloroethylene and chloroform. Appl Environ Microbiol. 1992;58:1886–1893. doi: 10.1128/aem.58.6.1886-1893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Type Culture Collection. Catalogue of bacteria and phages. 18th ed. Rockville, Md: American Type Culture Collection; 1992. p. 424. [Google Scholar]
- 3.Bogan B W, Schoenike B, Lamar R T, Cullen D. Manganese peroxidase mRNA and enzyme activity levels during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysporium. Appl Environ Microbiol. 1996;62:2381–2386. doi: 10.1128/aem.62.7.2381-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman J P, Jimenez L, Rosario I, Hazen T C, Sayler G S. Characterization of the methanotrophic bacterial community present in a trichloroethylene-contaminated subsurface groundwater site. Appl Environ Microbiol. 1993;59:2380–2387. doi: 10.1128/aem.59.8.2380-2387.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz L A, Klump J V, Collins M L P, Brantner C A, Remsen C C. Activity of methanotrophic bacteria in Green Bay sediments. FEMS Microbiol Lett. 1995;16:1–8. [Google Scholar]
- 6.Cardy D L N, Laidler V, Salmond G P C, Murrell J C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991;5:335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 7.Cardy D L N, Laidler V, Salmond G P C, Murrell J C. The methane monooxygenase gene cluster of Methylosinus trichosporium: cloning and sequencing of the mmoC gene. Arch Microbiol. 1991;156:477–483. doi: 10.1007/BF00245395. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Y S, Halsey J L, Anderson P D, Remsen C C, Collins M L P. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Use of PCR to detect particulate methane monooxygenase in groundwater; p. 377. [Google Scholar]
- 9.DiSpirito A A, Gulledge J, Shiemke A K, Murrell J C, Lidstrom M E, Krema C L. Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II, and type X methanotrophs. Biodegradation. 1992;2:151–164. [Google Scholar]
- 9a.Fode, K. A., and M. L. P. Collins. Unpublished data.
- 10.Fuhrman J A, Comeau D E, Hagstrom A, Chan A M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol. 1988;54:1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes A J, Costello A, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 12.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. J. Chichester, United Kingdom: Wiley & Sons, Ltd.; 1991. pp. 132–175. [Google Scholar]
- 13.Little C D, Palumbo A V, Herbes S E, Lidstrom M E, Tyndall R L, Gilmer P J. Trichloroethylene biodegradation by a methane-oxidizing bacterium. Appl Environ Microbiol. 1988;54:951–956. doi: 10.1128/aem.54.4.951-956.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lontoh S, Semrau J D. Methane and trichloroethylene degradation by Methylosinus trichosporium OB3b expressing particulate methane monooxygenase. Appl Environ Microbiol. 1998;64:1106–1114. doi: 10.1128/aem.64.3.1106-1114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald I R, Kenna E M, Murrell J C. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl Environ Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald I R, Murrell J C. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs: Appl Environ Microbiol. 1997;63:3218–3224. doi: 10.1128/aem.63.8.3218-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miguez C B, Bourque D, Sealy J A, Greer C W, Groleau D. Detection and isolation of methanotrophic bacteria possessing soluble methane monoxygenase (sMMO) genes using the polymerase chain reaction (PCR) Microb Ecol. 1997;33:21–31. doi: 10.1007/s002489900004. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto H, Yamamoto H, Arima K, Fujii J, Maruta K, Izu K, Shiomori T, Yoshida S-I. Development of a new semi-nested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl Environ Microbiol. 1997;63:2489–2494. doi: 10.1128/aem.63.7.2489-2494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohan K S, Walia S K. Detection of soluble methane monooxygenase producing Methylosinus trichosporium OB3b by polymerase chain reaction. Can J Microbiol. 1994;40:969–973. doi: 10.1139/m94-154. [DOI] [PubMed] [Google Scholar]
- 20.Oldenhuis R, Oedzes J Y, van der Waarde J J, Janssen D B. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol. 1991;57:7–14. doi: 10.1128/aem.57.1.7-14.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. p. A.1. [Google Scholar]
- 22.Selvaratnam S, Schoedel B A, McFarland B L, Kulpa C F. Application of reverse transcriptase PCR for monitoring expression of the catabolic dmpN gene in a phenol-degrading sequencing batch reactor. Appl Environ Microbiol. 1995;61:3981–3985. doi: 10.1128/aem.61.11.3981-3985.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semrau J D, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, Holmes A J, Finch R, Murrell J C, Lidstrom M E. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheridan G E C, Masters C I, Shallcross J A, Mackey B M. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith K S, Costello A M, Lidstrom M E. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl Environ Microbiol. 1997;63:4617–4620. doi: 10.1128/aem.63.11.4617-4620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stainthorpe A C, Lees V, Salmond G P C, Dalton H, Murrell J C. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath) Gene. 1990;91:27–34. doi: 10.1016/0378-1119(90)90158-n. [DOI] [PubMed] [Google Scholar]
- 27.Stainthorpe A C, Salmond G P C, Dalton H, Murrell J C. Screening of obligate methanotrophs for soluble methane monooxygenase genes. FEMS Microbiol Lett. 1990;70:211–216. [Google Scholar]
- 28.Stanley S H, Prior S D, Leak D J, Dalton H. Copper stress underlies the fundamental change in intracellular location of methane-monoxygenase in methane-oxidizing organisms: studies in batch and continuous cultures. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- 29.Whittenbury R, Phillips K C, Wilkinson J F. Enrichment, isolation, and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 30.Zahn J A, DiSpirito A A. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]