Abstract
Prostate cancer that recurs following androgen-deprivation therapy is termed castration-resistant, which is incurable and is marked by reactivation of androgen receptor (AR) signaling. KIF20A, a kinesin with unique structural features, is overexpressed in human castration-resistant prostate cancer (CRPC) compared to androgen-dependent PC and benign tissue. KIF20A has well-described roles in mitotic processes, but it has a less characterized function in vesicle fission and trafficking within Golgi-driven secretory pathways. Stable expression of KIF20A in androgen-dependent PC cells promoted progression to CRPC through the activation of AR signaling in vitro and in vivo. KIF20A expression resulted in the secretion of autocrine factors in the conditioned media that activated AR and caused castration-resistant proliferation of naïve androgen-dependent cells. Pharmacologic disruption of vesicle biogenesis blocked KIF20A-driven castration-resistant proliferation of androgen-dependent PC. KIF20A depletion or treatment with the KIF20A-specific inhibitor, paprotrain, reduced CRPC . These data are the first to establish KIF20A as a driver of CRPC progression through AR activation and as a promising therapeutic target against CRPC.
Introduction
Men with advanced prostate cancer (PC) undergo androgen deprivation therapy (ADT) resulting in tumor remission. Unfortunately, PC typically progresses to incurable castration-resistance and in most cases, is due to the reactivation of androgen receptor (AR) signaling [1-3].
Using an unbiased systems biology approach designed to identify drivers of CRPC and ligand-independent AR signaling, we documented a clinically relevant seven gene network that is highly expressed in localized and metastatic CRPC patient samples and contains promising therapeutic targets [4]. The kinesin-6 family protein, KIF20A (or MKLP2), is a member of this gene set [4]. Compared to other kinesins, KIF20A, is under-studied in mammalian cells. KIF20A uses its ATPase hydrolysis domain to regulate microtubule bundling and protein transport during mitosis [5, 6]. KIF20A has a less-characterized role in secretion at the Golgi apparatus by acting as a stabilizer and anchor for Rab6-positive (GTPase-driven) vesicle trafficking [7, 8]. Unlike other kinesins, KIF20A contains an additional loop motif in its motor domain that may affect ATP and/or microtubule binding [9]. This structural distinction has permitted the specific targeting of KIF20A with small molecule inhibitors such as paprotrain [10].
Although there is limited understanding of KIF20A’s role in cancer, KIF20A has been identified in gene signatures associated with poorer prognosis in a variety of cancers including breast, gastric, pancreatic, renal cell and glioma [11-14]. In patients with advanced PC, increased expression of KIF20A is associated with worse tumor grade, higher Gleason score (indicative of greater malignancy) and with more rapid recurrence as measured by increasing circulating prostate specific antigen (PSA) [15].
Human tumor dataset analysis showed that KIF20A expression is elevated in advanced PC, and we demonstrated experimentally that KIF20A was sufficient for progression from androgen-dependence to CRPC both in vitro and in vivo. This KIF20A-driven castration-resistant tumor growth was dependent on functional AR, and KIF20A promoted the secretion of factors that stimulated AR transcriptional activity and castration-resistance of multiple androgen-dependent PC cell lines. KIF20A depletion or treatment with paprotrain, a KIF20A-specific antagonist, inhibited CRPC (but not non-tumorigenic prostate epithelial cell proliferation) at submicromolar doses. These data highlight a novel role for KIF20A in promoting CRPC via secreted factor activation of AR and support KIF20A as a therapeutic target for CRPC.
Materials and Methods
Tissue Culture
Human prostate cancer cell lines 22Rv1 (or CWR-22Rv1; #CRL-2505), RWPE-1 (#CRL-11609), and LNCaP (#CRL-1740) were obtained from ATCC (American Type Culture Collection). The following cell lines were provided by colleagues: VCaP (K. Pienta [Johns Hopkins University]), C4-2B (the late L. Chung [Cedar Sinai Medical Center]), and LAPC4 (R. Reiter [University of California: Los Angeles]). GP2-293 and Lenti-X cells were purchased from Clontech.
For maintenance, 22Rv1 and LNCaP cells were cultured in RPMI (Corning, #15-040-CV), 1% penicillin/streptomycin (ThermoFisher, #15140122), 2 mM L-glutamine (ThermoFisher, #25030024), and 10% FBS (Atlanta Biologicals). VCaP cells were cultured in DMEM GlutaMAX (Gibco, #10569-044), 1% Anti-Anti (ThermoFisher, #15240062), and 10% FBS. LAPC4 cells were cultured in IMDM (Gibco, #12440061), 1% penicillin/streptomycin, and 10% FBS. GP2-293, Lenti-X and C4-2B cells were cultured in DMEM (Corning, #10-013-CV), 1% penicillin/streptomycin, and 10% FBS. Cell images were captured using an inverted microscope (Leica Microsystems, DMi1) using default brightfield settings with a 10x lens. Cells are routinely checked for mycoplasma contamination using the LookOut Mycoplasma PCR Detection Kit (SigmaAldrich, #MP0035).
Experimental details of KIF20A stable expression in PC cells, cell proliferation assays, and collection of conditioned media are described in Supplemental Data.
Plasmids and Subcloning
The KIF20A-expression plasmid, p7130 pHAGE-P-CMVt-N-HA-GAW-KIF20A, was a gift from Peter Howley (Addgene plasmid #100156; http://n2t.net/addgene:100156; RRID:Addgene_100156)[16]. The HA-tagged KIF20A region of this plasmid was PCR amplified and subcloned into the retroviral vector, pQCXIN (BD Biosciences), using the restriction sites NotI and EcoRI. Successful subcloning was verified by Sanger sequencing (Genewiz). The MMTV-LUC and ΔGRE-LUC luciferase plasmids were provided by Mona Nemer (University of Ottawa). The viral packaging plasmids, VSVG and Δ8.2, were purchased from Clontech. The pLKO.1 shGFP plasmid was a gift from Priyamvada Rai (University of Miami). The pLKO.1 shKIF20A was purchased from SigmaAldrich (#TRCN0000290348).
RNA Isolation and RTqPCR
Total RNA was extracted using TRIzol (ThermoFisher, #15596018) and purified with the Direct-zol RNA Miniprep Plus kit (Zymo Research, #R2071) according to the manufacturer’s instructions. cDNA synthesis used 500 ng of total RNA with the High-Capacity cDNA Reverse Transcription kit (ThermoFisher, #4368814) and RNase Inhibitor (ThermoFisher, #N8080119). The qPCR reaction was carried out using the QuantStudio 3 (Applied Biosciences) with default settings, using Taqman Fast Universal PCR Master Mix (ThermoFisher, #4352042), TaqMan probes, and cDNA. The following TaqMan probes were used: GAPDH (Hs99999905_m1), FKBP5 (Hs01561006_m1), and KLK3 (Hs00377590_s1). Relative expression was calculated using comparative threshold cycle method (ΔΔCt).
Immunoblotting
The denatured protein lysates were electrophoresed through 10% polyacrylamide gels (Biorad, #4561034) and transferred to nitrocellulose membranes (Amersham, #10600003). The membranes were incubated in primary antibody with 5% Milk/TBS-T overnight at 4 °C. The following antibodies were used: KIF20A (1:1000, Bethyl Laboratories, #A300-879A); AR (1:1500, Millipore, #06-680); Actin (1:1000, SantaCruz, #SC-1616); HA (1:1000, SantaCruz, #SC-805); GAPDH (1:1000, Invitrogen, #MA5-15738). Membranes were washed three times for 10 minutes in TBS-T and incubated with the secondary antibody (Kindle Biosciences, [anti-mouse (#R1005), anti-rabbit (#R1006), anti-goat (#R1007)]) at 1:1000 in 5% Milk/TBS-T for 2 hours at room temperature. Membranes were rinsed with water and incubated in Ultra Digital-ECL Substrate Solution (Kindle Biosciences, #R1002) for 2 minutes. The blots were imaged at specific timed exposures, using the KwikQuant Imager (Kindle Biosciences, #D1001).
Reporter Gene Assay
We used a pair of luciferase-based MMTV (mouse mammary tumor virus) reporter plasmids to measure AR transcriptional activity [17]. MMTV-LUC contains the endogenous promoter/enhancer regions, which includes native glucocorticoid/androgen response elements to measure AR activity, and ΔGRE-LUC lacks the glucocorticoid/androgen response elements to quantify basal transcriptional activity. LNCaP EV or KIF20A cells were plated into 6-well plates with a seeding density of 3.0*105 cells per well. After 24 hours, the cells were androgen deprived (as previously described) and media containing 5% CSS was added to the cells. 1.6 μg of MMTV-LUC or ΔGRE-LUC plasmid was transiently transfected using the X-tremeGENE 9 DNA Transfection Reagent (Sigma-Aldrich, #6365787001) at a 3:1 (reagent to plasmid) ratio. After 24 hours of transfection, cells were harvested in RIPA lysis buffer (Rockland, #MB-030-0050). Luciferase signal was measured using the Luciferase Assay System (Promega, #E1501) according to the manufacturer’s instructions. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (ThermoFisher, #23225). Luminescence and absorbance signals were measured with the GloMax Explorer (Promega) using an acquisition time of 7.5 seconds.
Subcutaneous Xenografts
All animal studies were conducted under an approved IACUC (Institutional Animal Care and Use Committee) protocol with the University of Miami and adhered to the NIH (National Institutes of Health) Guide for the Care and Use of Laboratory Animals. Sample size justification is in the Supplemental data [18]. Pre-castrated 5-to 6-week-old SCID male mice (Envigo, C.B-17/IcrHsd-Prkdcscid) were subcutaneously injected with 2*106 LNCaP or VCaP cells expressing either Empty Vector (LNCaP, N = 16; VCaP, N = 8) or KIF20A (LNCaP, N = 16; VCaP, N = 20) into each hind flank in a 1:1 volume ratio of cells resuspended in DPBS and Matrigel (Corning, 356234). Bilateral injections were with the same cell line (either EV or KIF20A). Tumor volume was measured by caliper (without blinding) twice a week and calculated using the ellipsoid equation (0.52 * Length * Width * Depth). The endpoint for measuring castration-resistant growth was defined as the days after one KIF20A tumor bearing mouse reached the maximum tumor burden, 1000 mm3. Drug treatments were staggered such that treatment was initiated when average tumor volume for the bilateral xenografts reached 150 mm3 (LNCaP) or 100 mm3 (VCaP). Mice were randomized into the Enzalutamide (10 mg/kg; Cayman Chemical, #11596) or Vehicle (0.3% Methylcellulose; SigmaAldrich, #M7140) treatment arm and dosed daily via oral gavage for 28 (LNCaP) or 31 (VCaP) days. The mice were weighed twice a week. Circulating PSA (Prostate Specific Antigen) was measured by enzyme-linked immunosorbent assay (BioCheck Inc., #BC-1019) from blood plasma extracted after mice were humanely euthanized. Immunohistochemistry was performed by the Cancer Modeling Shared Resource, using human Ki67 and mouse CD31 antibodies. Images were captured using the OlyVIA (Olympus Viewer for Imaging Applications) software. Semi-quantification of positive signal was analyzed in ImageJ, using a protocol published by Crowe and Yue [19, 20].
Bioinformatic Data Analysis
KIF20A differential expression data and progression free survival calculations from TCGA-PRAD (The Cancer Genoma Atlas – Prostate Adenocarcinoma) were generated through the Xena Browser [21]. The KIF20A expression data from the MSKCC (Memorial Sloan Kettering Cancer Center, Cancer Cell 2010) were accessed through cBioPortal and analyzed after using the p-adjusted method, Benjamini-Hochberg correction [22-24]. The microarray datasets (Varambally, S et al. 2005 [GSE3325] [25]; Aryee, MJ et al. 2013 [GSE38241] [26]; Ross-Adams, H et al. 2015 [GSE70770] [27]; Rayford, W et al. 2021 [GSE169038] [28]) were accessed through the GEO (Gene Expression Omnibus) Accession database and differential gene expression was analyzed with False Discovery Rate (Benjamini-Hochberg) (p-adjusted < 0.05) correction in GEO2R.
Statistical Analysis
Graphpad Prism 8 was used to produce graphs and for statistical analyses. The data underwent tests for normality (quantile-quantile and Shapiro-Wilk tests) and if conditions were met, the data used t-tests or ANOVAs to calculate significant differences. If normality conditions were not met, non-parametric tests were used. Sample size (N) designates biological experimental replicates. For proliferation assays, all measured cell count values from each biological replicate were used for statistical testing. Area Under the Curve values were calculated for experiments with datapoints measured over time.
Results
KIF20A expression is highest in CRPC and is prognostic for aggressive disease
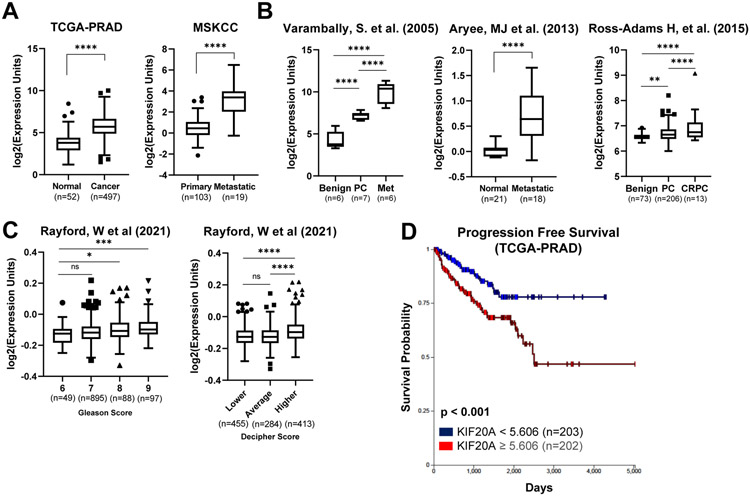
Analysis of PC patient transcriptomic datasets show KIF20A mRNA levels are elevated in PC compared to normal/benign prostate tissue (Figure 1A-B). Furthermore, KIF20A expression is higher in CRPC and metastatic samples compared to primary PC (Figure 1A-B). KIF20A expression increases with higher Gleason scores, the pathological grading system for PC with higher score indicative of more advanced disease (Figure 1C). The Decipher Score is a prognostic genomic test to predict a patient’s likelihood of progression to castration-resistance and metastasis [29]. Increased KIF20A expression is associated with a higher Decipher score (Figure 1C). Additionally, patient tumors with KIF20A expression values greater than the median expression (5.606) of all TCGA-PRAD (The Cancer Genome Atlas- Prostate Adenocarcinoma) samples were associated with lower progression-free survival (Figure 1D). Together, these data are consistent with KIF20A having a potential role in PC progression to deadly stages.
Figure 1. KIF20A expression increases with prostate cancer progression and is prognostic for poor outcome.
(A) KIF20A expression in normal or benign prostate, PC, CRPC and metastatic sites was obtained from RNA-seq datasets through cBioPortal and the Xena Browser. Significance was tested using the p-adjusted method, and expression values are plotted for visualization. TCGA-PRAD (The Cancer Genome Atlas-Prostate Adenocarcinoma); MSKCC (Memorial Sloan Kettering Cancer Center) (B) Microarray datasets (Varambally = GSE3325; Aryee = GSE38241; Ross-Adams = GSE70770) of prostate cancer patient samples. Significance was calculated using the p-adjusted method in GEO2R, and expression values are plotted for visualization. (C) KIF20A expression from microarray data (GSE169038) correlated with Gleason and Decipher score. (D) KIF20A expression correlated with Progression Free Survival was analyzed in the Xena Browser. 5.606 is the median expression for all PC samples. NS = Not significant; * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. Error bars = SEM
KIF20A promotes castration-resistant growth of androgen-dependent prostate cancer cell lines in vitro and in vivo
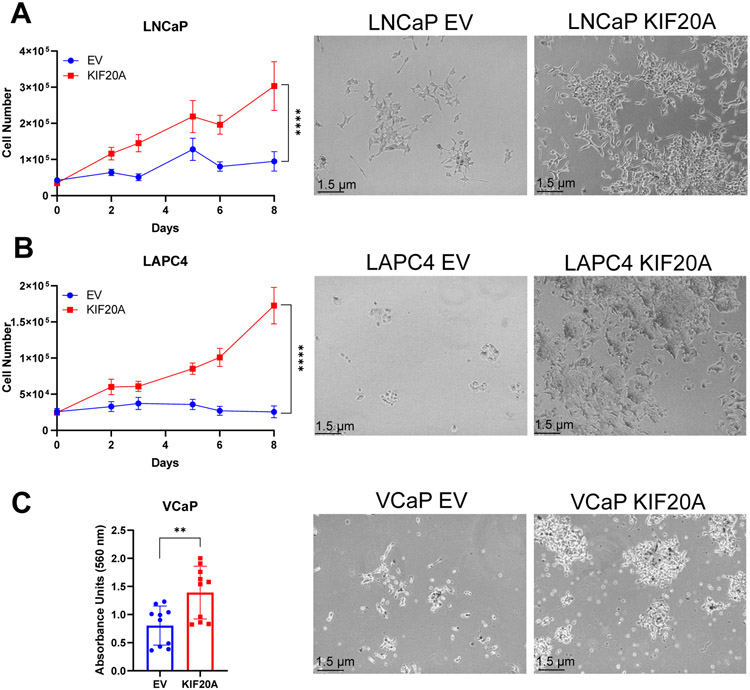
To determine whether KIF20A alone had the capacity to promote castration-resistance, we utilized three androgen-dependent cell lines LNCaP, LAPC4, and VCaP, which have significantly impeded or no growth in androgen-depleted media. We stably expressed KIF20A or Empty Vector (EV) control at protein levels approximately 1.5-2 times higher than endogenous KIF20A (Supplemental Figure 1) in each of these androgen-dependent cell lines and cultured the cells in media lacking androgens and other steroids. While androgen-dependent PC cells expressing EV failed to grow appreciably in androgen-depleted media, each of the androgen-dependent cell lines expressing ectopic KIF20A grew rapidly, indicating that KIF20A conferred castration resistance in androgen-dependent cells in vitro (Figure 2A-C). VCaP cell proliferation was quantified by colony formation (crystal violet staining) because these cells were difficult to dissociate and enumerate (Figure 2C). In contrast, KIF20A expression compared to the EV controls, did not provide a growth advantage to androgen-dependent LNCaP cells in androgen-replete media (10% Fetal Bovine Serum, FBS) (Supplemental Figure 2). Thus, the proliferative advantage conferred by KIF20A was specific to castration conditions.
Figure 2. Stable expression of KIF20A in androgen-dependent prostate cancer cells confers castration resistant growth in vitro.
(A) LNCaP (N = 5), (B) LAPC4 (N = 4), and (C) VCaP (N = 2) cells were stably transduced with KIF20A or Empty Vector (EV) control. Androgen-deprived cells were cultured in 5% charcoal-stripped serum (CSS). Numbers of live (trypan blue excluded) LNCaP and LAPC4 cells were plotted over time. Because VCaP cells were difficult to dissociate, cell proliferation was determined by crystal violet staining after 12 days of culture. Eluted stain was measured by absorbance at 560 nm. Unpaired t-tests measured statistical significance. Representative cell images at 10X magnification were taken on Day 8 of proliferation. (Image contrast was increased for better visualization after image capture for LAPC4 or VCaP EV and KIF20A cells.) ** p < 0.01; **** p < 0.0001. Error bars: SEM
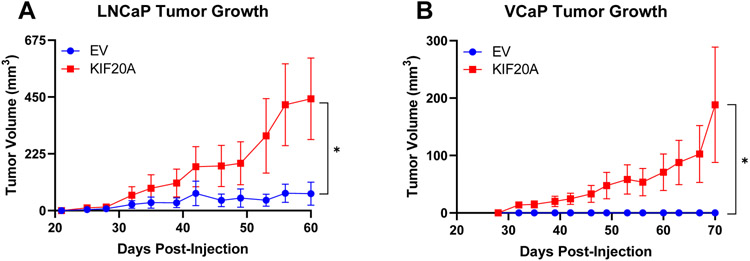
To investigate if KIF20A conferred castration resistance in vivo, we tested the capacity of KIF20A-expressing androgen-dependent PC cells to grow in castrated SCID mice. EV or ectopic KIF20A-expressing LNCaP or VCaP cells, were xenografted subcutaneously in castrated mice. LNCaP and VCaP KIF20A tumors grew robustly while their EV counterpart tumors grew little or not at all, indicating that KIF20A promoted castration resistance in vivo (Figure 3A-B). Thus, KIF20A expression in androgen-dependent PC cells was sufficient for tumor establishment and aggressive CRPC growth.
Figure 3. Stable expression of KIF20A in androgen-dependent prostate cancer cell xenografts confers castration resistant growth in vivo.
(A) LNCaP KIF20A (N = 7) or EV (N = 14) and (B) VCaP KIF20A (N = 16) or EV (N = 8) cells were xenografted into pre-castrated SCID mice. Area Under the Curve values were calculated for each xenograft. Unpaired t-tests determined statistical significance. * p < 0.05 Error bars: SEM
KIF20A-driven castration-resistant cell proliferation and tumor growth requires AR
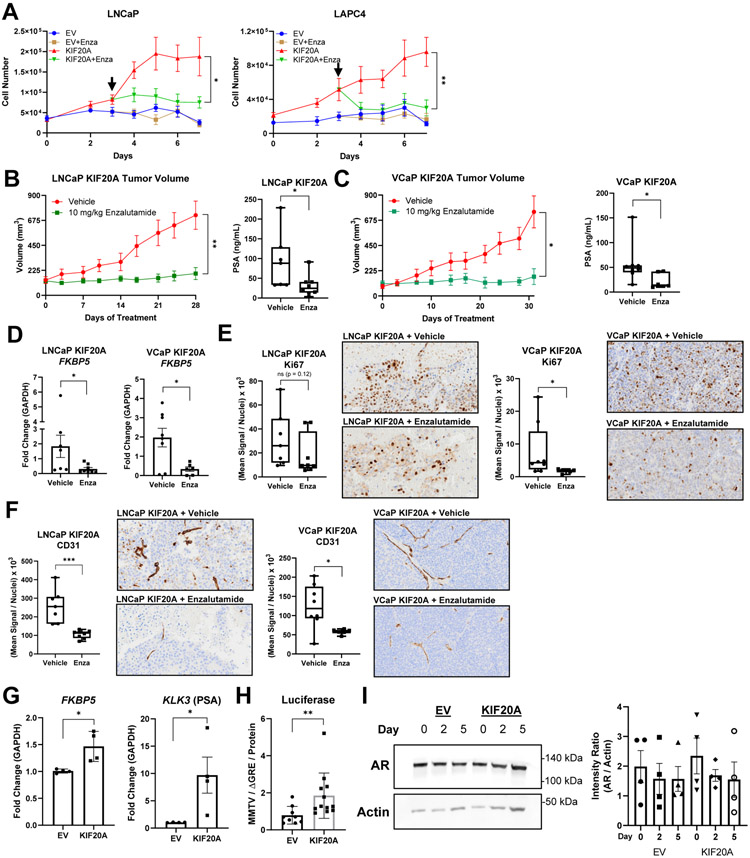
To determine whether castration-resistant growth of KIF20A-expressing androgen-dependent cells required AR activity, we tested the effects of the AR antagonist enzalutamide. Enzalutamide inhibited KIF20A-driven castration-resistant proliferation of LNCaP and LAPC4 cells in vitro (Figure 4A). Additionally, enzalutamide treatment of mice bearing LNCaP KIF20A or VCaP KIF20A xenografts significantly reduced tumor volumes and circulating PSA compared to mice receiving vehicle (Figure 4B-C). Enzalutamide treatment of LNCaP or VCaP KIF20A xenografts decreased AR target gene (FKBP5) expression (Figure 4D). Immunohistochemical analysis for Ki67 (a marker of proliferation) and CD31 (a marker of vascularization) revealed that enzalutamide significantly decreased Ki67 in VCaP KIF20A xenografts, and there was a trend indicative of decreased Ki67 in LNCaP KIF20A xenografts. Enzalutamide significantly decreased CD31 in both LNCaP and VCaP KIF20A xenografts (Figure 4E-F).
Figure 4. KIF20A-driven cell proliferation and tumor growth depends on AR signaling.
(A) Cell proliferation of LNCaP (N = 3) and LAPC4 (N = 2) EV or KIF20A cells cultured in 5% CSS. On day 3 (denoted by the black arrow), 10 μM Enzalutamide was added to the media. (B) LNCaP KIF20A or (C) VCaP KIF20A xenografts were randomly assigned to daily treatment arms, Vehicle (LNCaP, N = 7; VCaP, N = 8) or 10 mg/kg Enzalutamide (LNCaP, N = 8; VCaP, N = 6). Prostate Specific Antigen (PSA) was measured from blood plasma at the end of treatment. (D) RTqPCR of the AR target gene (FKBP5) was analyzed from LNCaP or VCaP KIF20A tumors from mice treated with enzalutamide or vehicle. (E) Ki67 and (F) CD31 immunohistochemistry quantification and representative 10x images exported from the OlyVIA software are shown. (G) RTqPCR (N = 4) of AR target genes (FKBP5, KLK3) was performed after LNCaP EV or KIF20A cells were deprived of androgens and cultured in 5% CSS for 3 hrs. (H) Luciferase assays (N = 3) with MMTV (containing androgen or glucocorticoid response elements) or ΔGRE (lacking response elements) plasmids were conducted in LNCaP EV or KIF20A cells cultured in 5% CSS. The ratios of luciferase values (MMTV/ΔGRE) were normalized to protein concentrations. (I) AR protein in LNCaP EV or KIF20A cells was quantified (N = 4) and normalized to Actin. T tests were performed. ns = not significant * p < 0.05; ** p < 0.01. Error bars = SEM
Since enzalutamide decreased proliferation of LNCaP KIF20A cells in vitro and in vivo, we examined the effect of KIF20A expression on AR target gene (FKBP5 and KLK3 [encodes PSA]) expression. LNCaP KIF20A cells (cultured in media lacking androgens) expressed higher levels of FKBP5 and KLK3 mRNA compared to EV controls (Figure 4G). We also used a dual luciferase reporter assay system to measure AR activity in media lacking androgens and found that ARE-driven luciferase was significantly higher in LNCaP KIF20A cells compared to EV controls (Figure 4H). The increased AR transcriptional activity observed in KIF20A-expressing cells was not due to increased AR protein levels (Figure 4I, Supplemental Figure 3).
Because AR often exhibits hypersensitivity to androgens in CRPC [30, 31], we tested the effects of androgen stimulation in KIF20A and EV-expressing androgen-dependent PC cells. Treatment of LNCaP KIF20A and EV cells with the synthetic androgen, R1881 (0.1 nM), rescued proliferation of LNCaP EV cells and increased proliferation of LNCaP KIF20A cells (Supplemental Figure 4A). Similarly, analysis of AR activity by luciferase assays revealed that KIF20A cells treated with R1881 exhibited a greater androgen-mediated response than EV cells (Supplemental Figure 4B). These data indicate that AR in KIF20A-expressing cells exhibits hypersensitivity to androgen stimulation.
KIF20A promotes secretion of factors sufficient for castration resistant proliferation
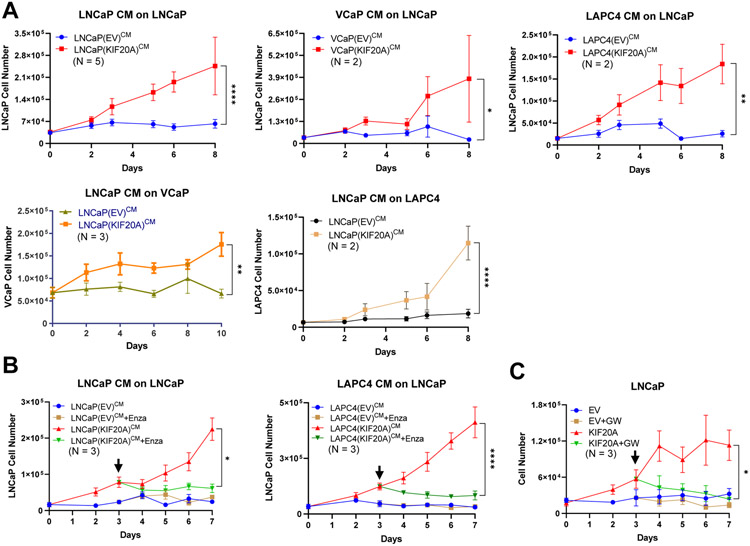
Because KIF20A participates in secretion at the trans-Golgi apparatus, stable expression of KIF20A may enhance secretory functions of PC cells. We examined the capacity of conditioned media from EV or KIF20A cells cultured without androgens to confer castration-resistant growth of parental (naïve) androgen-dependent PC cells. The conditioned media from each of the KIF20A-expressing androgen-dependent cell lines (LNCaP, VCaP, LAPC4) was sufficient for castration-resistant proliferation of parental LNCaP cells. Similarly, the conditioned media from LNCaP KIF20A cells was sufficient for castration-resistant proliferation of parental (naïve) VCaP and LAPC4 cells (Figure 5A). These data in multiple cell lines indicates that KIF20A promoted the secretion of factors sufficient for castration-resistant cell proliferation. Inhibition of AR using enzalutamide halted the castration-resistant proliferation that was mediated by conditioned media from KIF20A-expressing cells, suggesting that factors in the conditioned media signal through AR (Figure 5B).
Figure 5. KIF20A-expressing androgen-dependent prostate cancer cells secrete factors that drive AR-dependent castration-resistant cell proliferation.
(A) Conditioned media was collected from LNCaP, VCaP, LAPC4 KIF20A or EV cells growing in androgen-depleted media. Parental or naïve androgen-dependent cells (LNCaP, VCaP, or LAPC4) were androgen-depleted and cultured in conditioned media harvested from KIF20A or EV cell lines. (B) LNCaP cultured in the designated conditioned media were treated with Enzalutamide on day 3 (black arrow). (C) LNCaP KIF20A or EV cells were cultured in 5% CSS and treated on day 3 (black arrow) with an inhibitor of vesicle biogenesis, GW4869. Statistical t tests tested for significant differences. Biological replicates, N, are located inside each graph. * p < 0.05; ** p < 0.01; **** p < 0.0001. Error bars = SEM
Because KIF20A promoted the secretion of factors that were sufficient to confer castration-resistent proliferation to androgen dependent PC cells, we used GW4869 [32] to inhibit vesicle biogenesis. Consistent with KIF20A’s secretory functions underlying release of CRPC-promoting factors, the proliferation of KIF20A-expressing cells was blocked by GW4869 (Figure 5C).
KIF20A is a therapeutic target for castration-resistant prostate cancer
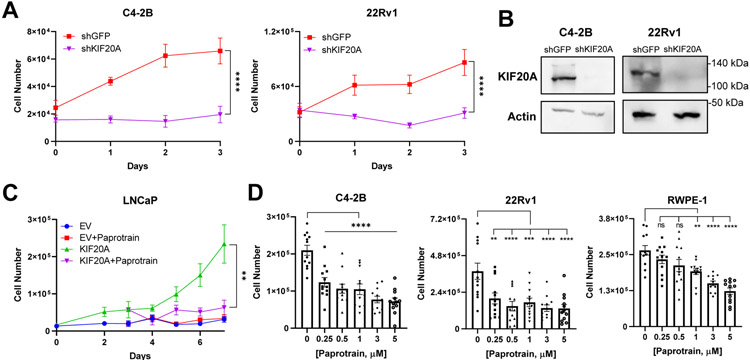
Depletion of KIF20A with an shRNA halted CRPC cell proliferation, supporting the potential for KIF20A to serve as a therapeutic target in PC (Figure 6A-B). Paprotrain (PAssenger PROteins TRAnsport INhibitor), is a cell permeable, reversible, ATP-uncompetitive inhibitor of the ATPase region of KIF20A. This inhibitor is selective for KIF20A and does not bind to its closely related kinesin, KIF23, or other superfamily kinesins [10, 33]. Submicromolar concentrations of paprotrain significantly inhibited proliferation of LNCaP KIF20A (Figure 6C) and CRPC 22Rv1 and C4-2B cells (Figure 6D). In contrast, non-tumorigenic prostate epithelial cells, RWPE-1, were not inhibited by submicromolar concentrations of paprotrain but required 1 μM and higher doses (Figure 6D).
Figure 6. KIF20A inhibition blocks CRPC proliferation.
(A) Proliferation of C4-2B (N = 3) and 22Rv1 (N = 3) CRPC cells in 5% CSS was analyzed after stable transduction of shRNAs against KIF20A or GFP control. (B) Western blotting was conducted to evaluate KIF20A knockdown. (C) LNCaP KIF20A or EV cells (N = 3) were treated wtih vehicle or 500 nM paprotrain. (D) C4-2B (N = 3), 22Rv1 (N = 4), and RWPE-1 (N = 3) cells were treated with the indicated doses of paprotrain or vehicle control for 48 hours. Dunnett’s multiple comparisons test was used to measure significance between paprotrain compared to the vehicle. NS = not significant; ** p < 0.01; *** p < 0.001; **** p < 0.0001. Error bars = SEM
Discussion
We found that KIF20A expression was sufficient to drive PC progression from androgen-dependence to castration-resistance in cell-based assays and in subcutaneous xenografts in castrated mice. Because enzalutamide, a potent AR antagonist, blocked castration-resistant growth of KIF20A-expressing androgen-dependent cells both in vitro and in vivo, AR must be required. However, AR levels were not increased by ectopic expression of KIF20A in androgen-dependent cells suggesting that AR transcriptional activity is increased by other mechanisms. Evaluation of the autocrine effects of conditioned media from KIF20A-expressing androgen-dependent cells indicated that expression of KIF20A resulted in secretion of factors that promoted AR activity. GW4869, which blocks vesicle biogenesis, inhibited proliferation of KIF20A-expressing cells suggesting that KIF20A drives CRPC proliferation through promoting secretion. This finding is consistent with KIF20A’s known functions in Golgi-mediated vesicle fission and secretion.
KIF20A may promote castration-resistance through activation of both full length-AR and AR variants. However, in the case of LNCaP and LAPC4 KIF20A or EV cells, AR variants were not detectable (Figure 4D, Supplemental Figure 3) [34, 35]. Additionally, KIF20A expression in VCaP cells did not affect AR-V7 protein levels (Supplemental Figure 3). Castration-resistant LNCaP xenografts can have increased expression of androgen biosynthetic enzymes [36-38]; thus, KIF20A may increase AR activity in androgen-dependent PC cells by increasing androgen synthesis. Although it is not known whether KIF20A interacts with AR, our data examining conditioned media from KIF20A cells indicate that KIF20A enhances AR activity by promoting secretion of autocrine factors.
Depletion of KIF20A in CRPC cells decreases proliferation in vitro (Figure 6) and xenograft growth in vivo [15]. Conversely, we show that ectopic expression of KIF20A promoted progression from androgen-dependence to castration-resistance in vitro and in vivo by promoting AR activity. In an ovarian cancer cell line, KIF20A expression increases proliferation in vitro [39]. Interestingly, we observed that KIF20A expression promoted proliferation of androgen-dependent PC cells only in castration conditions but not in androgen-replete media. Thus, KIF20A, in the context of androgen-dependent PC cells, provided a survival and proliferative advantage selectively by enhancing AR activity in the absence of androgens.
Exploring KIF20A’s function at the Golgi apparatus could uncover mechanisms of secretion that promote CRPC. Such processes have received little attention in PC. In VCaP and LNCaP cells, androgen treatment promotes Endoplasmic-Reticulum-to-Golgi vesicle trafficking by increasing transcription of protein transport genes (including CREB3L2, a transcription factor that co-localizes with AR) [40]. Additionally, CRPC cells release paracrine-acting factors that stimulate bone-derived mesenchymal stem cells and stromal cells of fibroblast lineage [41, 42], suggesting that secretion may be advantageous for CRPC progression and tumorigenesis. Although KIF20A serves dual functions in mitosis and secretion, our data are most consistent with KIF20A’s role in secretion of factors that in the context of PC are important for CRPC progression. We show that inhibition of vesicle biogenesis halted castration-resistant proliferation of KIF20A-expressing cells, indicating that this secretory process is important for KIF20A-driven growth in androgen-depleted conditions. Therefore, expression of KIF20A could promote CRPC progression through enhancing or accelerating the secretion of factors that promote AR-driven cell proliferation in castration conditions.
The factors secreted into the conditioned media may be growth factors, cytokines, chemokines, microRNAs, androgens, extracellular vesicles (exosomes), or a combination of these components. For instance, KIF20A expression may increase the secretion of mature microRNAs or oncomiRs (oncogenic microRNAs) into the conditioned media, which could increase castration-resistant progression through AR signaling [43, 44]. Additionally, secreted cytokines may promote pro-inflammatory pathways resulting in progression to castration-resistance [45]. KIF20A may facilitate castration-resistance through increasing the release of extracellular vesicles or exosomes, which can include tumor-promoting proteins and/or miRNAs [46]. While the nature of these factors is currently unknown, our results show that conditioned media from multiple KIF20A-expressing androgen-dependent PC cell lines is sufficient to promote castration-resistant growth and AR signaling in naïve androgen-dependent PC cells.
While KIF20A inhibition may affect normal cellular processes, KIF20A expression increases with prostate cancer progression. This increased expression suggests that a therapeutic window exists permitting the selective targeting of KIF20A in lethal CRPC. KIF20A’s unique structure allowed for the generation of paprotrain a KIF20A-specific inhibitor that does not block other kinesins [9, 10]. Because paprotrain effectively blocked CRPC cell proliferation at submicromolar concentrations (with no significant effects on non-tumorigenic prostate epithelial cells), KIF20A may be an ideal target in CRPC.
In summary, ectopic KIF20A expression alone was sufficient to promote the progression of androgen-dependent to castration-resistant disease in vitro and in vivo. This process involved KIF20A enhancement of AR activity through the secretion of factors and occured in the absence of exogenous androgens. Based on these findings and the existence of KIF20A-selective inhibitors that are effective at low concentrations, further investigation of KIF20A as a therapeutic target in CRPC is warranted.
Supplementary Material
Acknowledgments
We thank Dr. Deukwoo Kwon (University of Miami) for assistance with statistical analyses, and the Cancer Modeling Shared Resource (Sylvester Comprehensive Cancer Center; University of Miami Miller School of Medicine), which is supported by the National Cancer Institute (NCI) Cancer Center Support Grant (CCSG) P30-CA240139, for the immunohistochemistry assistance. We thank Drs. M. Julia Martinez and Nahuel Peinetti for helpful comments on the manuscript. This work was supported by VA Merit Review 1I01BX002773 (to K.L.B.), the Mike Slive Foundation (to K.L.B.), and Developmental Funds from the Sylvester Comprehensive Cancer Center (to K.L.B.).
Footnotes
Conflicts of Interest: None
References
- 1.Einstein DJ, Arai S, Balk SP. Targeting the androgen receptor and overcoming resistance in prostate cancer. Curr Opin Oncol 2019; 31: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang YA, Sfakianos J, Tewari AK, Cordon-Cardo C, Kyprianou N. Molecular tracing of prostate cancer lethality. Oncogene 2020; 39: 7225–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai K, McManus JM, Sharifi N. Hormonal Therapy for Prostate Cancer. Endocr Rev 2021; 42: 354–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magani F, Bray ER, Martinez MJ, Zhao N, Copello VA, Heidman L et al. Identification of an oncogenic network with prognostic and therapeutic value in prostate cancer. Mol Syst Biol 2018; 14: e8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adriaans IE, Hooikaas PJ, Aher A, Vromans MJM, van Es RM, Grigoriev I et al. MKLP2 Is a Motile Kinesin that Transports the Chromosomal Passenger Complex during Anaphase. Curr Biol 2020; 30: 2628–2637.e2629. [DOI] [PubMed] [Google Scholar]
- 6.Terada Y, Uetake Y, Kuriyama R. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J Cell Biol 2003; 162: 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echard A, Jollivet F, Martinez O, Lacapère J-J, Rousselet A, Janoueix-Lerosey I et al. Interaction of a Golgi-Associated Kinesin-Like Protein with Rab6. Science 1998; 279: 580–585. [DOI] [PubMed] [Google Scholar]
- 8.Miserey-Lenkei S, Bousquet H, Pylypenko O, Bardin S, Dimitrov A, Bressanelli G et al. Coupling fission and exit of RAB6 vesicles at Golgi hotspots through kinesin-myosin interactions. Nature Communications 2017; 8: 1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atherton J, Yu IM, Cook A, Muretta JM, Joseph A, Major J et al. The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry. eLife 2017; 6: e27793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labrière C, Talapatra SK, Thoret S, Bougeret C, Kozielski F, Guillou C. New MKLP-2 inhibitors in the paprotrain series: Design, synthesis and biological evaluations. Bioorg Med Chem 2016; 24: 721–734. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura M, Takano A, Thang PM, Tsevegjav B, Zhu M, Yokose T et al. Characterization of KIF20A as a prognostic biomarker and therapeutic target for different subtypes of breast cancer. Int J Oncol 2020; 57: 277–288. [DOI] [PubMed] [Google Scholar]
- 12.Saito K, Ohta S, Kawakami Y, Yoshida K, Toda M. Functional analysis of KIF20A, a potential immunotherapeutic target for glioma. J Neurooncol 2017; 132: 63–74. [DOI] [PubMed] [Google Scholar]
- 13.Ren X, Chen X, Ji Y, Li L, Li Y, Qin C et al. Upregulation of KIF20A promotes tumor proliferation and invasion in renal clear cell carcinoma and is associated with adverse clinical outcome. Aging (Albany NY) 2020; 12: 25878–25894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai K, Hirata S, Irie A, Senju S, Ikuta Y, Yokomine K et al. Identification of HLA-A2-restricted CTL epitopes of a novel tumour-associated antigen, KIF20A, overexpressed in pancreatic cancer. Br J Cancer 2011; 104: 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Chai C, Shen T, Li X, Ji J, Li C et al. Aberrant KIF20A Expression Is Associated with Adverse Clinical Outcome and Promotes Tumor Progression in Prostate Cancer. Dis Markers 2019; 2019: 4782730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galligan JT, Martinez-Noël G, Arndt V, Hayes S, Chittenden TW, Harper JW et al. Proteomic analysis and identification of cellular interactors of the giant ubiquitin ligase HERC2. J Proteome Res 2015; 14: 953–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magani F, Peacock SO, Rice MA, Martinez MJ, Greene AM, Magani PS et al. Targeting AR Variant-Coactivator Interactions to Exploit Prostate Cancer Vulnerabilities. Mol Cancer Res 2017; 15: 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Houghton PJ. Interval approach to assessing antitumor activity for tumor xenograft studies. Pharmaceutical Statistics 2010; 9: 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crowe AR, Yue W. Semi-quantitative Determination of Protein Expression using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Bio Protoc 2019; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landini G, Martinelli G, Piccinini F. Colour deconvolution: stain unmixing in histological imaging. Bioinformatics 2020; 37: 1485–1487. [DOI] [PubMed] [Google Scholar]
- 21.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnology 2020; 38: 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005; 8: 393–406. [DOI] [PubMed] [Google Scholar]
- 26.Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med 2013; 5: 169ra110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross-Adams H, Lamb AD, Dunning MJ, Halim S, Lindberg J, Massie CM et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine 2015; 2: 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayford W, Beksac AT, Alger J, Alshalalfa M, Ahmed M, Khan I et al. Comparative analysis of 1152 African-American and European-American men with prostate cancer identifies distinct genomic and immunological differences. Commun Biol 2021; 4: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalela D, Löppenberg B, Sood A, Sammon J, Abdollah F. Contemporary Role of the Decipher® Test in Prostate Cancer Management: Current Practice and Future Perspectives. Rev Urol 2016; 18: 1–9. [PMC free article] [PubMed] [Google Scholar]
- 30.Gregory CW, Johnson RT, Mohler JL, French FS, Wilson EM. Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Research 2001; 61: 2892–2898. [PubMed] [Google Scholar]
- 31.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinänen R, Palmberg C et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet 1995; 9: 401–406. [DOI] [PubMed] [Google Scholar]
- 32.Catalano M, O'Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell Vesicles 2020; 9: 1703244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tcherniuk S, Skoufias DA, Labriere C, Rath O, Gueritte F, Guillou C et al. Relocation of Aurora B and survivin from centromeres to the central spindle impaired by a kinesin-specific MKLP-2 inhibitor. Angew Chem Int Ed Engl 2010; 49: 8228–8231. [DOI] [PubMed] [Google Scholar]
- 34.Roggero CM, Jin L, Cao S, Sonavane R, Kopplin NG, Ta HQ et al. A detailed characterization of stepwise activation of the androgen receptor variant 7 in prostate cancer cells. Oncogene 2021; 40: 1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma T, Bai S, Qi Y, Zhan Y, Ungerleider N, Zhang DY et al. Increased transcription and high translation efficiency lead to accumulation of androgen receptor splice variant after androgen deprivation therapy. Cancer Lett 2021; 504: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Locke JA, Guns ES, Lubik AA, Adomat HH, Hendy SC, Wood CA et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res 2008; 68: 6407–6415. [DOI] [PubMed] [Google Scholar]
- 37.Chang K-H, Li R, Papari-Zareei M, Watumull L, Zhao YD, Auchus RJ et al. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proceedings of the National Academy of Sciences 2011; 108: 13728–13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auchus RJ, Sharifi N. Sex Hormones and Prostate Cancer. Annu Rev Med 2020; 71: 33–45. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Guo H, Wang Z, Bu H, Wang S, Wang H et al. Cyclin F and KIF20A, FOXM1 target genes, increase proliferation and invasion of ovarian cancer cells. Experimental Cell Research 2020; 395: 112212. [DOI] [PubMed] [Google Scholar]
- 40.Hu L, Chen X, Narwade N, Lim MGL, Chen Z, Tennakoon C et al. Single-cell analysis reveals androgen receptor regulates the ER-to-Golgi trafficking pathway with CREB3L2 to drive prostate cancer progression. Oncogene 2021. [DOI] [PubMed] [Google Scholar]
- 41.Kaur SP, Verma A, Lee HK, Barnett LM, Somanath PR, Cummings BS. Inhibition of glypican-1 expression induces an activated fibroblast phenotype in a human bone marrow-derived stromal cell-line. Sci Rep 2021; 11: 9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ridge SM, Bhattacharyya D, Dervan E, Naicker SD, Burke AJ, Murphy JM et al. Secreted factors from metastatic prostate cancer cells stimulate mesenchymal stem cell transition to a pro-tumourigenic 'activated' state that enhances prostate cancer cell migration. Int J Cancer 2018; 142: 2056–2067. [DOI] [PubMed] [Google Scholar]
- 43.Hizir MS, Balcioglu M, Rana M, Robertson NM, Yigit MV. Simultaneous detection of circulating oncomiRs from body fluids for prostate cancer staging using nanographene oxide. ACS Appl Mater Interfaces 2014; 6: 14772–14778. [DOI] [PubMed] [Google Scholar]
- 44.Fernandes RC, Toubia J, Townley S, Hanson AR, Dredge BK, Pillman KA et al. Post-transcriptional Gene Regulation by MicroRNA-194 Promotes Neuroendocrine Transdifferentiation in Prostate Cancer. Cell Rep 2021; 34: 108585. [DOI] [PubMed] [Google Scholar]
- 45.Archer M, Dogra N, Kyprianou N. Inflammation as a Driver of Prostate Cancer Metastasis and Therapeutic Resistance. Cancers 2020; 12: 2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaglani S, Gonzalez-Kozlova E, Lundon DJ, Tewari AK, Dogra N, Kyprianou N. Exosomes as A Next-Generation Diagnostic and Therapeutic Tool in Prostate Cancer. International Journal of Molecular Sciences 2021; 22: 10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.