Abstract
Objectives:
We aimed to review and describe antimicrobial resistance (AMR) prevalence in humans, animals, and the environment in Ethiopia.
Methods:
We conducted a structured review of literature on AMR in humans, animals, and the environment in Ethiopia from 2016–2020. We reported the pooled prevalence of AMR of bacterial pathogens in all 3 sectors.
Results:
We included 43 articles in our review. Only 5 studies evaluated AMR across multiple sectors. The most common bacteria in humans were Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus. High prevalence of resistance to third-generation cephalosporins, fluoroquinolones, and sulfamethoxazole-trimethoprim were seen in gram-negative organisms, often with >50% prevalence of resistance. Highest resistance rates were seen in humans, followed by environmental isolates. Salmonella spp. exhibited higher rates of resistance than previously reported in the literature. We found methicillin-resistant S. aureus (MRSA) in approximately half of S. aureus from the environment and a third from human isolates. Few studies evaluated AMR across all 3 sectors.
Conclusion:
Our review demonstrated high prevalence of AMR among bacteria in humans, animals, and the environment in Ethiopia. Integrating a One Health approach into AMR surveillance as part of Ethiopia’s national surveillance program will inform future implementation of One Health interventions.
Keywords: antimicrobial resistance, One Health, Ethiopia
Introduction
Antimicrobial resistance (AMR) is widely recognized as a global problem, including in sub-Saharan African countries (Elton et al., 2020, Gebretekle et al., 2020). Increasing rates of AMR render many antibiotics ineffective and result in increased morbidity and mortality due to bacterial infections (Global Action Plan on Antimicrobial Resistance 2015a, Report to the Secretary-General of the United Nations IACG, 2019). Antimicrobial misuse and overuse are attributed as drivers of increasing AMR worldwide, compounded by additional challenges in low- and middle-income countries (LMICs). In resource-limited areas, insufficient diagnostic infrastructure and laboratory capacity, inconsistent AMR surveillance, and inadequately resourced infection prevention and control contribute to empiric antibiotic use on the basis of syndromic approaches rather than microbiological data (Escher et al., 2021, Gebretekle et al., 2020, Gebretekle et al., 2018). This has led to high rates of antibiotic consumption in LMICs (Gebretekle et al., 2020), which create high selection pressure for resistant organisms.
In addition to human consumption, antibiotic use in food animals and agricultural crops are recognized as likely drivers of AMR in low-resource settings (Rousham et al., 2018). Globally, >70% of all antimicrobials are used in food animals, not only for treatment of diseases but also for infection prophylaxis and growth promotion (Van Boeckel et al., 2019). Transmission between humans and animals can occur through consumption of contaminated food of animal origin or direct contact with livestock (Rousham et al., 2018, White and Hughes, 2019). Antibiotic resistance genes are now considered an environmental pollutant, with exposure occurring through human and animal waste released into the soil and water, which are then used in agriculture (Manyi-Loh et al., 2018, Zalewska et al., 2021). In LMICs, healthcare waste combined with inadequately disinfected drinking water contribute to water contaminated with drug-resistant bacteria (Rousham et al., 2018, Talukdar et al., 2013). This complex interplay of AMR transmission between humans, animals, and ecosystems underscores the need for a One Health approach to better understand the mechanism of transmission and mitigate its spread.
A One Health approach to AMR, which uses an interdisciplinary approach to surveillance and implementation of programs, policies, and research, is increasingly recognized as a vital component to national and global AMR strategies (One Health Basics, 2018a)). In 2015, the World Health Organization (WHO) launched the Global Antimicrobial Resistance Surveillance System (GLASS), a collaborative effort to standardize AMR surveillance with the aim to inform policies and infection prevention strategies (Global Action Plan on Antimicrobial Resistance, 2015a). In Ethiopia, the Ethiopian Food, Medicines, and Healthcare Administration and Control Authority developed the “Strategy for the Prevention and Containment of Antimicrobial Resistance” plan in 2015 (Strategy for the Prevention and Containment of Antimicrobial Resistance for Ethiopia, 2015b). Then in 2017, they launched the Ethiopian Antimicrobial Resistance Surveillance System, a standardized, laboratory-based surveillance system and one of the first national efforts to combat AMR (Ethiopia Antimicrobial Resistance Surveillance Annual Report, 2020). More recently in December 2020, the Strategic Plan was revised with particular attention to a One Health platform (Ministry of Health MoA, 2020).
Since the implementation of Ethiopia’s AMR surveillance system, substantial achievements have been made, including an expanded surveillance network, collation of AMR surveillance data, and increased laboratory capacity (Ethiopia Antimicrobial Resistance Surveillance Annual Report, 2020). However, national AMR surveillance in Ethiopia is currently primarily focused on humans, and there remains a knowledge gap of AMR trends across animals and the environment. Despite extensive interaction between the 3 sectors, few research studies have evaluated AMR through the lens of One Health. Here, we provide a detailed, structured review of the AMR literature published during 2016–2020 in Ethiopia to describe AMR rates across the One Health sectors.
Methods
Search Strategy
A structured literature search was performed using PubMed, CINAHL, Global Health Database, AgriCOLA, Embase, and MEDLINE online databases. We included all articles on AMR in Ethiopia published in English from January 2016–October 2020. The literature search was conducted from October 6th–November 30th, 2020, by 1 author (KW). The search strategy used the following search string: (“antimicrobial resistance” OR “antibiotic resistance” OR “drug resistance” OR “gram-negative” OR “gram-positive”) AND (“Escherichia coli “OR “E. coli” OR “Salmonella” OR “Staphylococcus aureus “OR “Enterobacter cloacae” OR “Shigella “OR “Methicillin-resistant Staphylococcus aureus” OR “Klebsiella pneumoniae “OR “Acinetobacter baumannii” OR “Streptococcus pneumoniae)” AND (“foodborne infections” OR “healthcare infections”) AND (“animal” OR “livestock” OR “cattle” OR “cows” OR “beef” OR “poultry” OR “chickens” OR “pig” OR “swine”) OR “human” OR “environment” OR “One Health”) AND (“Ethiopia”).
Selection Criteria
Articles were reviewed by a single reviewer according to PRISMA guidelines. Full-text articles on AMR prevalence among bacteria isolated from humans, animals, and animal products (cows, pigs, and poultry), or the environment (swabs of surfaces and objects in clinical settings, surfaces in community settings including slaughterhouses, and water sources) in Ethiopia were screened for inclusion. Publications were reviewed and included if they reported AMR prevalence and information about sample collection. Studies evaluating AMR from sources of bacterial colonization (eg, nares swabs and stool samples from asymptomatic individuals) were excluded. Additionally, we excluded environmental samples collected from nonanimal food products (eg, juice and fruit). After our initial literature review, we identified and added 3 additional environmental studies that were discussed and referenced in another study. Publications reporting AMR for Mycobacterium tuberculosis or nonbacterial pathogens were excluded from this review.
We assessed AMR in the following clinically relevant pathogenic bacteria identified by the Global Antimicrobial Resistance Surveillance System (GLASS organisms, Additional File 1): Escherichia coli, Klebsiella pneumoniae, Acinetobacter spp., Staphylococcus aureus/Methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pneumoniae, Salmonella spp., and Shigella spp. Additionally, we included Enterobacter spp., Serratia spp., Proteus spp., and Citrobacter spp. because there is concern of growing resistance among these gram-negative organisms but excluded Neisseria gonorrhea, which is limited to humans.
Data Extraction
Data extraction was performed by 1 author (KW) and reviewed and confirmed by a second author (AWF). Data extracted included: (i) article information (first author, year, city/region, and sample source/host), (ii) study design (study approach, sample size, and setting), and (iii) results (clinical syndrome/infection, sample site [humans], sample source [animal and environment], organisms, and rates of resistance).
Statistical analysis
Data were extracted by organism and sector (humans, animals, and environment), and descriptive statistics were used for summarizing frequencies and proportions. For calculating the prevalence of AMR, we focused on resistance to antimicrobials prioritized by GLASS (Additional File 1) (2015a). Confidence intervals for proportions were used to estimate the pooled prevalence of AMR of each organism-antibiotic combination, and this was reported separately for humans, animals, and the environment, as well as overall. We used oxacillin or cefoxitin resistance to determine the prevalence of methicillin-resistant Staphylococcus aureus (MRSA). In our results and main tables, we report the pooled prevalence of AMR among the bacterial pathogens included. Resistance rates reported by individual studies are reported in Additional Files 2–4.
Results
Initial literature search yielded a total of 1534 articles. After excluding 515 duplicates and 671 articles by screening titles and abstracts that were not pertinent to AMR, 348 articles were reviewed in full. An additional 308 articles were excluded because they did not meet our selection criteria (Figure 1), leaving 40 articles in the final data extraction. A total of 3 additional studies were later identified after reviewing references from included studies; thus, a total of 43 studies were included in this review.
Figure 1.
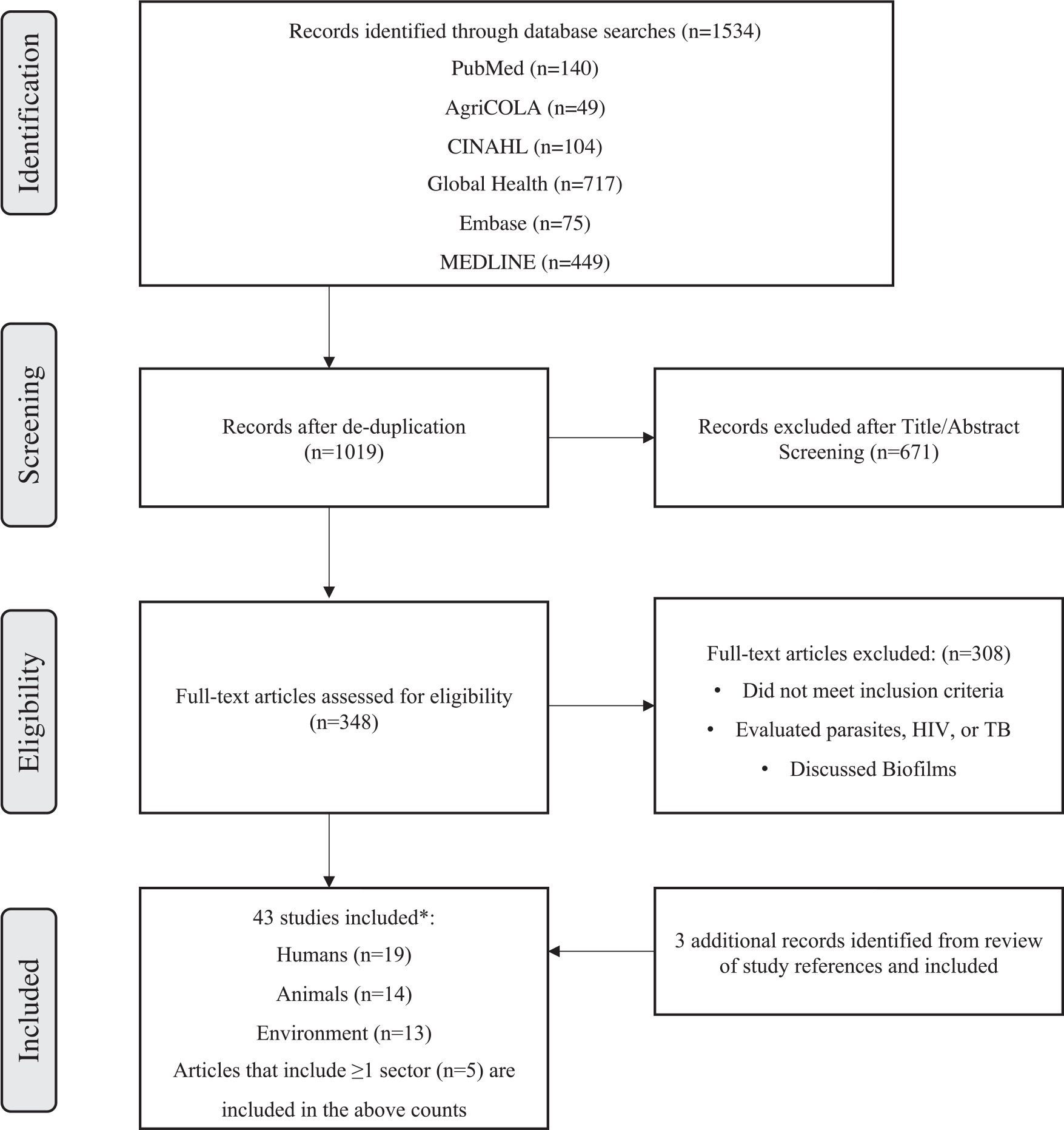
Study selection process for literature review of AMR in humans, animals, and the environment in Ethiopia
Study Characteristics
Of the 43 full-text articles included for review, all were cross-sectional, most of which were retrospective. Studies were conducted in 17 cities and 6 regions in Ethiopia, representing urban and periurban areas (Figure 2). A total of 19 studies evaluated AMR in humans, 14 studies in animals, and 13 studies in environmental samples. Only 5 studies evaluated AMR across multiple sectors, all of which were conducted in slaughterhouses or dairy farms (Abdi et al., 2017, Abunna, 2017, Beyene et al., 2017, Garedew et al., 2016, Takele et al., 2018).
Figure 2.
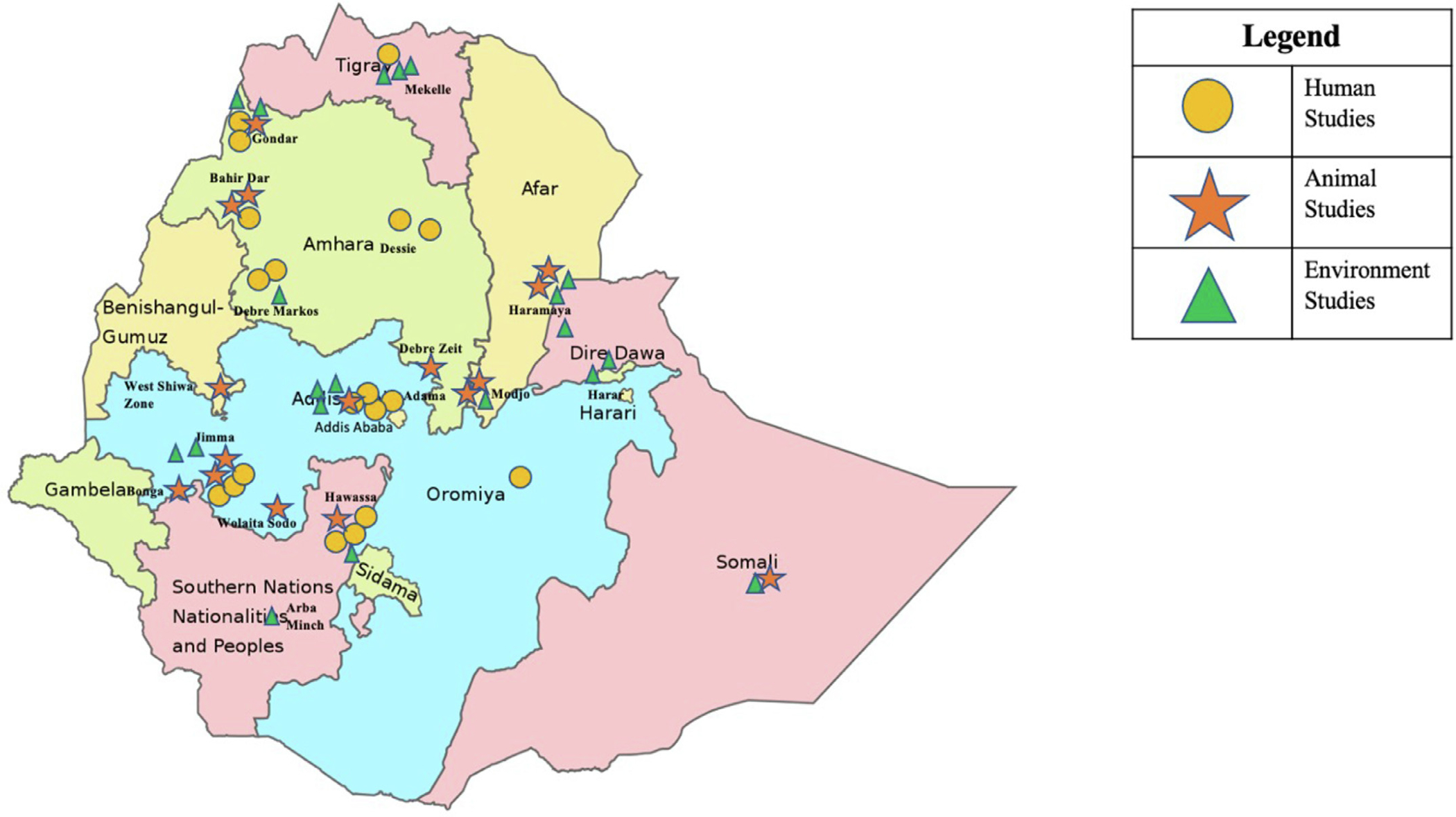
Geographical locations of AMR studies conducted in Ethiopia between January 2016 and October 2020
Antimicrobial resistance rates in humans
A total of 19 studies evaluated bacterial AMR in humans. Most studies were conducted in urban cities, predominantly in Addis Ababa, Jimma, and Hawassa (Figure 2) and described AMR of bacteria in a single, specific infectious syndrome, such as surgical site infections, urinary tract infections, otitis media, or diarrhea (Additional File 2) (Argaw-Denboba et al., 2016, Bitew Kifilie et al., 2018, Deyno et al., 2017b, Gorems et al., 2018, Hailu, 2018, Lamboro et al., 2016, Mamuye, 2016, Nigussie and Amsalu, 2017, Shimekaw et al., 2020, Tadesse et al., 2018, Terfassa and Jida, 2018, Teshome et al., 2019, Tsige et al., 2020). Only 1 study evaluated AMR specifically in bloodstream infections (Arega et al., 2018). The most frequently sampled sites for culture were urine (n = 1664, 29%), ear swabs (n = 1521, 25%), wounds (n = 1420, 25%), and stool (n = 752, 13%). Only 305 (5%) of all samples obtained were blood cultures.
Most bacteria isolated were gram-negative organisms (80%), most frequently E. coli (n = 676), Klebsiella spp. (n = 347), Proteus spp. (n = 422), and Salmonella spp. (n = 97). Susceptibility against broad-spectrum gram-negative antimicrobial agents such as cefepime, piperacillin/tazobactam, and meropenem were infrequently tested for susceptibility, and only 20% of human samples were tested for carbapenems. However, among the bacteria that were tested, 20% (117/582) were carbapenem-resistant. When carbapenem susceptibility was assessed, resistance was observed in Serratia spp. (n = 3, 60%), Enterobacter spp. (n = 20, 53%), Proteus spp. (n = 3, 43%), Citrobacter spp. (n = 19, 38%), Klebsiella spp. (n = 30, 18%), and E. coli (n = 19, 13%).
E. coli had high pooled prevalence of resistance to ciprofloxacin (77%; 95% CI: 74%–80%), sulfamethoxazole/trimethoprim (SMX/TMP) (54%; 95% CI: 50%–58%), ceftriaxone (46%; 95% CI: 42%–50%), and ceftazidime (29%; 95% CI: 26%–33%) (Table 1). Compared with E. coli, Klebsiella spp. had higher rates of resistance to SMX/TMP (74%; 95% CI: 69%–79%), ceftriaxone (66%; 95% CI: 61%–71%), and ceftazidime (52%; 95% CI: 47%–58%) but lower rates of resistance to ciprofloxacin (35%; 95% CI 30%–40%).
Table 1.
Pooled prevalence of AMR among Enterobacterales from humans, animals, and the environment.
| Organism | Sector | Total Number Positive Cultures | Ceftriaxone (%, 95% CI) | Ceftazidime (%, 95% CI) | Cefepime (%, 95% CI) | Meropenem (%, 95% CI) | Ciprofloxacin (%, 95% CI) | SMX/TMP (%, 95% CI) | Gentamicin (%, 95% CI) |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| E. coli | Humans | 676 | 46 (42–50) | 29 (26–33) | 1 (0–1) | 6 (4–8) | 77 (74–80) | 54 (50–58) | 36 (32–39) |
| Animals | 297 | 2 (0.4–4) | 10 (7–14) | N/A | N/A | 1 (0–3) | 18 (13–22) | 3 (1–5) | |
| Environment | 254 | 13 (9–17) | 9 (5–12) | 31 (25–36) | 18 (13–22) | 35 (29–41) | 38 (32–44) | 26 (21–31) | |
| Total | 1227 | 28 (26–31) | 20 (18–23) | 7 (5–8) | 7 (6–9) | 50 (47–53) | 42 (39–45) | 26 (23–28) | |
| K. pneumoniae | Humans | 97 | 43 (37–49) | 44 (38–50) | 0 | 5 (2–8) | 38 (32–45) | 55 (49–62) | 36 (30–42) |
| Animals | 0 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Environment | 45 | 41 (27–55) | 19 (7–30) | 24 (11–36) | 9 (1–17) | 16 (6–27) | 26 (13–39) | 12 (3–21) | |
| Total | 142 | 42 (34–50) | 36 (28–44) | 7 (3–12) | 6 (2–10) | 31 (24–39) | 46 (38–54) | 28 (21–36) | |
| Klebsiella spp. (not K. pneumoniae) | Humans | 250 | 76 (71–81) | 57 (51–63) | 14 (9–18) | 14 (9–18) | 34 (28–40) | 83 (78–87) | 64 (58–70) |
| Animals | 0 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Environment | 49 | 26 (14–39) | 6 (−0.1–13) | 6 (−0.1–13) | 2 (−2–6) | 14 (4–24) | 37 (24–51) | 29 (16–42) | |
| Total | 299 | 67 (61–72) | 48 (42–53) | 7 (4–10) | 12 (8–15) | 30 (25–36) | 74 (69–79) | 57 (52–63) | |
| Proteus spp. | Humans | 422 | 82 (79–86) | 3 (1–5) | 4 (2–5) | 1 (0–1) | 11 (8–14) | 83 (79–86) | 21 (17–25) |
| Animals | 0 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Environment | 22 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Total | 444 | 82 (79–86) | 3 (1–5) | 4 (2–5) | 1 (0–1) | 11 (8–14) | 83 (79–86) | 21 (17–25) | |
| Salmonella spp. | Humans | 103 | 16 (9–23) | 9 (3–14) | N/A | N/A | 24 (15–32) | 27 (19–36) | 8 (3–14) |
| Animals | 268 | 12 (8–16) | N/A | N/A | N/A | 8 (5–11) | 46 (40–52) | 8 (4–11) | |
| Environment | 15 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Total | 386 | 9 (6–11) | 2 (1–4) | N/A | N/A | 10 (7–13) | 32 (28–37) | 6 (4–9) | |
| Citrobacter spp. | Humans | 158 | 44 (36–52) | 19 (13–25) | 5 (2–9) | 12 (7–17) | 16 (10–22) | 58 (50–66) | 26 (19–33) |
| Animals | 0 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Environment | 37 | 19 (6–32) | 5 (−2–13) | 11 (1–21) | 0 | 27 (13–41) | 30 (15–44) | 14 (2–25) | |
| Total | 195 | 39 (32–47) | 16 (10–22) | 6 (2–10) | 10 (5–14) | 18 (12–24) | 53 (45–60) | 23 (17–30) | |
| Enterobacter spp. | Humans | 113 | 57 (48–66) | 24 (16–32) | 3 (0–6) | 18 (11–25) | 15 (9–22) | 52 (42–61) | 34 (25–42) |
| Animals | 0 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Environment | 13 | N/A | N/A | N/A | N/A | N/A | N/A | ||
| Total | 126 | 53 (45–62) | 22 (14–29) | 2 (0–5) | 16 (10–22) | 17 (10–23) | 50 (41–58) | 31 (23–39) | |
| Shigella spp. | Humans | 55 | 20 (9–31) | 22 (11–33) | N/A | N/A | 29 (17–41) | 37 (25–50) | 9 (1–16) |
| Animals | 10 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Environment | 15 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Total | 80 | 23 (14–33) | 15 (7–23) | N/A | N/A | 20 (11–29) | 26 (16–35) | 6 (1–11) | |
The pooled prevalence of AMR and 95% confidence intervals were calculated for each organism and antibiotic by sector and as an aggregated total. Pooled prevalence of AMR was only calculated when the total number of positive cultures from a sector was ≥50. When susceptibility testing was not performed or when the sample size was too small to calculate pooled prevalence, “N/A” was used to designate non-applicability. Acinetobacter spp. (n=39), Serratia spp. (n=15), and Streptococcus pneumoniae (n=38) were excluded from the table, as their aggregated totals were <50. For full details of each study in our review, please see Additional Files 2–4.
Most Salmonella species (97%) were obtained from stool specimens, and only 2 were found in blood cultures. Data on serovars were not available in most studies. Pooled estimates of Salmonella spp. resistance to ciprofloxacin were 25% (95% CI 16%–34%) and ceftriaxone 17% (95% CI 10%–25%).
Typical hospital-acquired gram-negative organisms, such as Citrobacter spp., Enterobacter spp., and Proteus spp., also demonstrated high rates of AMR, especially to SMX/TMP, ceftriaxone, and aminoglycosides (Table 1).
A total of 15 human studies identified Staphylococcus aureus with a total of 1062 isolates, and we determined the pooled prevalence of MRSA to be 34% (95% CI 31%–36%) (Table 2). The pooled prevalence of S. aureus resistance to SMX/TMP was 49% (95% CI 46%–52%) and ceftriaxone 28% (95% CI 25%–30%). Overall, few studies tested S. aureus against vancomycin, daptomycin, linezolid, doxycycline, or clindamycin.
Table 2.
Pooled prevalence of AMR among Staphylococcus aureus isolates from humans, animals, and the environment.
| Staphylococcus aureus | Total Number Positive Cultures | Pooled Cefoxitin Resistance (n, %) | 95% Confidence Intervals |
|---|---|---|---|
|
| |||
| Humans | 1062 | 357, 34% | 31–36% |
| Animals | 120 | Not tested | N/A |
| Environmental | 240 | 128, 53% | 47–60% |
| Community settings | 61 | 21, 34% | 22–46% |
| Hospital settings | 179 | 107, 60% | 53–67% |
| Total | 1422 | 485, 37%* | 35–40% |
The pooled prevalence of MRSA and 95% confidence intervals were calculated for S. aureus isolates from humans, animals, and the environment. Cefoxitin or oxacillin resistance were used as surrogates to determine prevalence of MRSA. Environmental samples were further stratified by samples obtained from the community (e.g., slaughterhouses and dairy farms) versus from hospital settings. We could not determine MRSA rates in animal sectors as these studies did not report susceptibility testing for MRSA. For this reason, only human and environmental samples were used to calculate the total pooled prevalence of MRSA.
Animicrobial resistance rates in animals
A total of 14 studies evaluated AMR in animals or food of animal origin, including chickens (n = 5), cattle (n = 8), or both (n = 1). We did not find any studies in Ethiopia assessing AMR in pigs. Most studies evaluated bacteria isolated from food of animal origin, such as milk, raw or cooked meat, and eggs. Animal studies were conducted in 15 urban or periurban cities rather than rural or pastoral regions. From a One Health perspective, 4 studies tested for AMR in pathogens isolated from all 3 sectors (Abdi et al., 2017, Abunna, 2017, Beyene et al., 2017, Garedew et al., 2016), and 1 study evaluated AMR in Salmonella isolated from both cattle and human fecal samples (Takele et al., 2018).
In total 5237 samples were collected from animals or animal products, and 700 samples (13%) tested positive for pathogenic bacteria. Fewer types of bacteria were isolated and tested for AMR, focusing primarily on E. coli/E. coli O157:H7 (n = 297, 42%) and Salmonella species (n = 274, 39%). Staphylococcus aureus was isolated in 129 (18%) samples. Prevalence of carbapenem-resistant Enterobacterales could not be calculated owing to lack of carbapenem susceptibility testing performed in animal samples.
E. coli was the most common organism isolated from animals and food of animal origin. High rates of resistance to SMX/TMP were observed; however, lower resistance rates to fluoroquinolones and third-generation cephalosporins were seen compared with E. coli isolated from human clinical samples. The pooled prevalence of resistance to SMX/TMP was 18% (95% CI 13%–22%), ceftazidime 10% (95% CI 7%–14%), ciprofloxacin 1.4% (95% CI 0%–3%), and ceftriaxone 2% (95% CI 0%–4%) (Table 1).
Salmonella species were identified in 8 studies with a total of 274 isolates. Pooled prevalence of resistance against SMX/TMP was 34% (95% CI 28%–39%) with lower rates of resistance observed for ceftriaxone 6% (95% CI 3%–9%) and ciprofloxacin 5% (95% CI 3%–8%). However, we noted that susceptibilities against ceftriaxone were only tested in 3 studies despite being a common alternative to fluoroquinolones for the treatment of severe Salmonella disease.
Staphylococcus aureus (n = 129) was identified in 2 studies and all samples were collected from milk of dairy cattle. One of these studies found that 62% of S. aureus isolates were resistant to cefoxitin (Sileshi and Munees, 2016). However, susceptibility testing against oxacillin or cefoxitin was not consistently performed; thus, pooled estimates of the prevalence of MRSA could not be calculated in animals. The study characteristics and AMR rates for individual studies are shown in Additional File 3.
Antimicrobial resistance rates in the environment
Thirteen studies evaluated AMR in environmental samples. We included water sources, surfaces of clinical settings, and surfaces of community settings and excluded swabs of human hands or nonanimal food products (Additional File 4). Five studies used a One Health approach by assessing AMR among bacteria from both animal and environmental sources (Abdi et al., 2017, Abunna, 2017, Beyene et al., 2017, Garedew et al., 2016, Takele et al., 2018). Four studies included swabs of human hands in abattoir settings, which offered a unique One Health perspective of AMR across the human-animal-ecosystem interface.
A total of 1657 samples were collected, of which 906 (55%) positive cultures yielded 1713 bacterial isolates. The most common pathogens isolated were E. coli (n = 254), followed by S. aureus (n = 240) and Klebsiella spp. (n = 94) (Additional file 4). Susceptibility to carbapenems was tested in fewer than half (44%) of all gram-negative isolates; of these, carbapenem resistance was identified in 38% of gram-negative bacterial isolates. Carbapenem resistance was observed in Acinetobacter spp. (n = 29, 74%), Klebsiella spp. (n = 5, 50%), E. coli (n = 45, 28%), and Serratia spp. (n = 1, 25%).
Nearly all 184 samples from water sources had positive cultures, all of which grew gram-negative organisms. A total of 255 bacterial isolates were identified, and 75% were E. coli. Water samples were collected from hospital wastewater systems, as well as from abattoirs and downstream rivers in Addis Ababa (Belachew et al., 2018, Takele et al., 2018, Tesfaye et al., 2019, Teshome et al., 2020). Of 478 samples from surfaces in the community, 300 were swabs from handles of city buses, where most positive cultures (54/66) grew S. aureus. The remaining samples were obtained from surfaces from abattoirs or dairy farms, and Shigella (n = 15), S. aureus (n = 7), and Salmonella (n = 3) were isolated. From hospital settings, the most common organisms isolated were S. aureus (n = 179), Klebsiella spp. (n = 60), and E. coli (n = 54), followed by other nosocomial gram-negative organisms such as Acinetobacter spp, Citrobacter spp, and Serratia spp (Table 2).
Among 254 positive cultures with E. coli, pooled prevalence of resistance was highest for SMX/TMP (38%; 95% CI: 32%–44%), ciprofloxacin (35%; 95% CI: 29%–41%), and cefepime (31%; 95% CI 25%–36%). For Klebsiella spp. (n = 94), the pooled prevalence of resistance for ceftriaxone was 33% (95% CI 24%–43%) and SMX/TMP was 32% (95% CI 22%–41%).
In total, S. aureus was isolated from 240 positive cultures, and the pooled prevalence of MRSA was 53% (95% CI 47%–60%) (Table 2). When stratified by community versus hospital settings, MRSA prevalence was 34% among S. aureus isolated from the community versus 60% from hospital surfaces.
Salmonella was only identified in 15 bacterial isolates from water sources and a dairy farm. Susceptibility testing to antibiotics were inconsistent and low in frequency; however, when tested, there was no resistance reported to ciprofloxacin or ceftriaxone from these environmental samples.
Discussion
Our review of the AMR literature in Ethiopia revealed high prevalence of resistance to common and clinically important antimicrobials among GLASS priority pathogens (Global Action Plan on Antimicrobial Resistance, 2015a). Our broad overview included studies from diverse regions across Ethiopia and included a wide range of samples obtained from humans, animals, and the environment. We identified a notable gap in the AMR literature of studies with an integrated, One Health approach to surveillance in Ethiopia, with only 5 studies describing AMR across all 3 sectors (Abdi et al., 2017, Abunna, 2017, Beyene et al., 2017, Garedew et al., 2016, Takele et al., 2018). Previous studies in Ethiopia have focused on only a single pathogen, a particular clinical syndrome, or only 1 or 2 One Health sectors. More recently, a systematic review and meta-analysis of AMR was published in Ethiopia through a One Health lens (Gemeda et al., 2021). However, authors focused on bacteria in the animal-source food chain; thus, only food handlers were included for human samples. Our literature review is unique in that it included studies of human clinical samples along with animal and environmental studies in Ethiopia.
Nearly all animal studies were conducted in urban and periurban areas (Figure 2) and included animal husbandry systems, composed mostly of dairy cattle and poultry. The absence of studies in pigs is possibly because pork consumption is less common in Ethiopia. Although intensive dairy cattle constitute only a small portion of the nation’s cattle, this sector is important because it represents a population with better access to pharmacies and veterinary care, which may lead to greater exposure and risk to AMR.
For this review, we focused on priority antimicrobials identified by GLASS according to its Access, Watch, Research (AWaRe) classification system (Additional File 1) (Sharland et al., 2018). Antimicrobials classified as “Access” are those used to treat common, susceptible bacteria and are expected to have low rates of resistance. Those in the “Watch” group have higher rates of resistance and are recommended to be prioritized in surveillance and stewardship programs. Finally, the “Reserve” group of antimicrobials are those that should be reserved to treat multidrug-resistant organisms. We found high resistance rates among 5 antibiotics in the AWaRe “Access” group and 8 in the “Watch” group, emphasizing the importance of not only AMR surveillance programs but also of implementation of antibiotic stewardship programs.
Antimicrobial susceptibility testing appeared to be inconsistent and disproportionately low in animal and environmental isolates compared with humans. In many animal and environmental isolates, pooled prevalence of resistance could not be calculated due to lack of susceptibility data, highlighting a gap in AMR data in the animal and environmental sectors. We observed that in many cases, clinically irrelevant antibiotics were tested for susceptibility, whereas other clinically important antibiotics were not. In addition to increased laboratory capacity and support for susceptibility testing, AMR surveillance would benefit from standardized procedures or panels for susceptibility testing for different categories of pathogens.
Susceptibility testing against carbapenems was exceedingly low across all sectors, which is problematic given the growing concerns of carbapenemase-producing bacteria in sub-Saharan Africa (Manenzhe et al., 2015). True rates of carbapenem resistance in this region are difficult to ascertain owing to lack of carabapenem susceptibility testing. However, in isolates where testing was performed, the pooled prevalence of carbapenem resistance was as high as 20%. Increased and consistent susceptibility testing to carbapenems should be performed to identify prevalence and trends of carbapenem-resistant Enterobacterales.
Regarding distribution and types of culture samples, we found low numbers of blood cultures, with only 5% of all cultures consisting of blood. Cultures of sterile sites can offer important microbiological information because these typically represent true infections; whereas, cultures obtained from wounds and urine can represent colonization or contamination and are difficult to interpret in the absence of clinical data.
We found higher rates of MRSA among S. aureus isolates in the environment compared with humans (53% vs 34%). However, when environmental samples were stratified by hospital and community settings, we discovered higher rates of MRSA from hospital settings (60% vs 34%), mostly from hospital surfaces and equipment, suggesting a need for improved and thorough cleaning practices to reduce surface contamination with AMR organisms in healthcare settings. In a One Health study by Beyene, T et al, S. aureus was isolated from dairy milk, beef, human hand swabs, and equipment at dairy farms and abattoirs, demonstrating possible transmission of organisms between humans, animals, and the environment (Beyene et al., 2017). Both settings suggest that AMR transmission between One Health sectors may occur owing to inadequate hygiene during points of contact, such as touching hospital surfaces or during milking or slaughtering of animals. Our review found MRSA rates to be comparable with the pooled prevalence of methicillin resistance (47%) noted in a meta-analysis of S. aureus resistance in Ethiopia (Deyno et al., 2017a). High MRSA rates are concerning as infections caused by MRSA have limited treatment options and have been shown to have worse clinical outcomes, including longer hospitalizations and higher mortality (Bassetti et al., 2012, Cosgrove et al., 2005). To mitigate the spread of MRSA between the 3 sectors, we recommend improved cleaning protocols of hospital surfaces and increased education about hand hygiene in dairy farms and slaughterhouses.
Although the studies with a true One Health approach were few, they showed the interconnection of the 3 domains, primarily in abattoirs, dairy farms, and butcher shops. In these studies, samples were taken from human hands, animals or animal products, and environmental surfaces and showed similar organisms or resistance (Abdi et al., 2017, Abunna, 2017, Beyene et al., 2017, Garedew et al., 2016, Takele et al., 2018). This suggests the potential circulation of AMR isolates among the human-animal-environment domains, which may have serious impact on human and animal health. Risk factors for AMR organisms in animals varies depending on the type of production system, but prophylactic antibiotics in animal feed and water may contribute to the development of AMR, which could be transmitted to humans through consumption of animal products containing antibiotic residue. However, little research has been done in Ethiopia to assess the impact of prophylactic antibiotics on the development of AMR in farm settings. Additionally, studies that prospectively collect samples from multiple sectors simultaneously are needed to inform future areas for intervention to reduce AMR transmission.
Studies that identified drug-resistant organisms in hospital wastewater systems suggest that healthcare-acquired resistance could be transmitted into the community and environment through wastewater (Belachew et al., 2018, Tesfaye et al., 2019, Teshome et al., 2020). Environmental exposure to antimicrobials has adverse effects on environmental and human health, and a recent global study of pharmaceutical pollution in rivers across 104 countries revealed high concentrations in sub-Saharan Africa, South Asia, and South America (Wilkinson et al., 2022). Rivers with highest rates of pharmaceutical contamination were in LMICs where wastewater management infrastructure is poor (Wilkinson et al., 2022). In fact, Addis Ababa, Ethiopia had the third highest concentration of pharmaceutical pollution in rivers in the world (Wilkinson et al., 2022). Future studies should sample not only wastewater systems within the hospital but also in the community near or around the hospital and from rivers downstream from the hospital, which would substantiate the One Health concept of AMR transmission between humans in the healthcare setting, agricultural crops, and livestock through contaminated water.
E. coli and Klebsiella spp. were the most common gram-negative organisms isolated, and both exhibited high rates of AMR to third-generation cephalosporins, fluoroquinolones, and SMX/TMP. A recent One Health review of AMR in Cameroon found similarly high rates of AMR in Enterobacterales isolated from hospital settings (Mouiche et al., 2019). There has been growing attention to drug-resistant gram-negative organisms, including those with extended-spectrum beta-lactamases (Abayneh and Worku, 2020), which render many commonly used antibiotics ineffective. For example, previous evidence showed that using piperacillin-tazobactam to treat patients with E. coli or K. pneumoniae bacteremia with ceftriaxone resistance had poorer outcomes than those treated with carbapenems (Harris et al., 2018). In LMICs, where broad-spectrum antibiotics such as carbapenems may be unavailable, options to effectively treat resistant gram-negative infections may be limited.
Multidrug-resistant Salmonella is an increasing global concern and has been reported in sub-Saharan Africa, including Ethiopia. In our review, Salmonella spp. were primarily isolated from fecal specimens, and only 2 of 305 blood cultures grew Salmonella. In the most recent Typhoid Fever Surveillance in Africa Program, blood cultures from 847 febrile patients from Butajira, Ethiopia over 2 years revealed only 3 cases of invasive Salmonella disease, all S. typhi with no resistance to cephalosporins, fluoroquinolones, or SMX-TMP (Marks et al., 2017). In contrast, our review showed higher pooled prevalence of resistance to antibiotics commonly used to treat Salmonella disease in humans, including ciprofloxacin, ceftriaxone, and SMX-TMP. Lower rates of resistance to ceftriaxone and fluoroquinolones were seen in animals; however, resistance to SMX-TMP remained high at >30% (Figure 3c). This suggests that there may be variance in AMR prevalence in Salmonella spp. reported in the literature and to be suspicious of single reports of pansusceptibility of Salmonella with small sample sizes per study and a wide range of resistance rates reported in Ethiopia.
Figure 3.
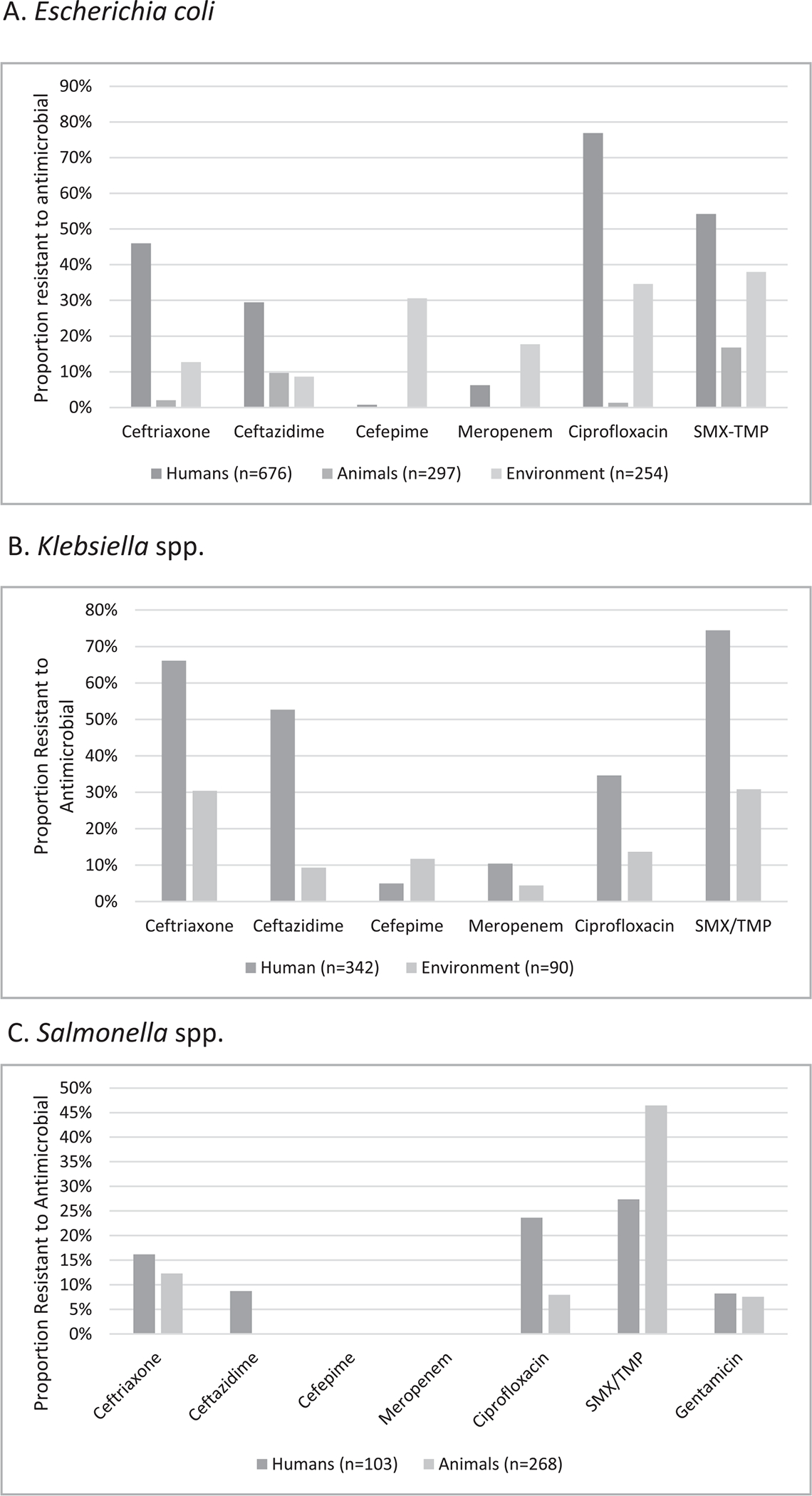
Pooled prevalence of AMR for select gram-negative pathogens from studies of human, animal, and environmental samples. Escherichia coli (A), Klebsiella spp. (B), and Salmonella spp. (C) are included in the graphs. Klebsiella spp. were not isolated from animal studies. Salmonella spp. were isolated from all 3 sectors; however, the sample size in environmental samples (n=15) was too small to calculate pooled prevalence of resistance.
Our review has several limitations. A known limitation of One Health AMR research is the lack of studies evaluating all 3 sectors simultaneously. Without integrated AMR data across all 3 sectors, it is difficult to assess the true prevalence of AMR, the directionality of transmission, and how to effectively combat resistance at the human-animal-environment interface (Rousham et al., 2018). Prospective studies sampling human, animal, and environments simultaneously are needed; however, this is resource-intensive and logistically challenging, especially in resource-limited settings.
Another limitation is that our initial search did not include small ruminants such as sheep and goats. However, when re-examining the literature for AMR studies in Ethiopia in sheep and goats, only a few studies were carried out during our search period (Abreham et al., 2019, Messele et al., 2017).
Finally, all human samples were obtained from hospitalized patients in clinical settings, which may create bias toward including patients with nosocomial infections. In these settings, cultures may only be obtained after prolonged hospital courses when patients have not improved on empiric antibiotics, thus selecting out for patients with higher rates of drug-resistant organisms. However, this approach is difficult to avoid, as gathering AMR data in humans usually occurs in clinical settings, and we aimed to avoid collecting data from asymptomatic individuals with bacterial colonization.
On the basis of our review, there are several opportunities for future AMR research with a One Health approach. First, we need to identify barriers to routine cultures and antibiotic susceptibility testing in not only human clinical settings but also in veterinary medicine and agricultural sectors. Susceptibility testing with appropriate antibiotics should be standardized with protocols and discussed with clinicians to test for the most clinically relevant antibiotics. Second, additional studies from animals and the environment (particularly agriculture, aquaculture, live animal markets, and small ruminants) are needed. These were under-represented in our review and would offer greater generalizability of the AMR data. Third, prospective studies with a One Health approach—integrating collection of AMR data from all 3 sectors simultaneously—would provide important surveillance information and help us to better understand AMR transmission across sectors.
Fourth, it is imperative that we gain additional knowledge about antibiotic-prescribing practices among physicians and veterinarians, as well as usage among livestock owners. Despite the limited AMR data in Ethiopia, the existing data show increasing AMR prevalence to commonly used empiric antibiotics. The WHO has published a methodology for conducting point prevalence surveys on antibiotic use in hospitals (WHO methodology for point prevalence survey on antibiotic use in hospitals, 2018b); however, additional information on antibiotic consumption in the animals would inform policies on antimicrobial stewardship across all sectors.
Ethiopia has already established a national surveillance program with increased support and funding for laboratory capacity, and the revised Antimicrobial Resistance Prevention and Containment Strategic Plan prioritizes a One Health approach. Our review is aligned with its second strategic objective, which is to strengthen the knowledge and evidence on antimicrobial use and resistance through surveillance (Ministry of Health MoA, 2020). We found high pooled prevalence of AMR in bacteria from humans, animals, and environmental samples in Ethiopia, but we identified gaps in AMR data from animal and environmental sectors. There is a noticeable lack of studies that use a One Health approach to collecting and reporting AMR data across all 3 sectors. Next steps to optimizing a One Health approach would be to develop standardized protocols for antimicrobial susceptibility testing in not only humans but also animals and environmental samples. This will support future AMR surveillance by making routine cultures and susceptibility testing more efficient and clinically relevant. Future AMR interventions and policies should prioritize representation from all stakeholders, including the environmental sector, which has historically been under-represented (Essack, 2018, Khan et al., 2018). As Ethiopia carries out its “Strategy for the Prevention and Containment of Antimicrobial Resistance,” a collaborative effort among all 3 sectors will be crucial to a One Health approach to AMR surveillance. Integrating AMR surveillance from humans, animals, and the environment is key to understanding mechanisms of transmission and will inform future implementation of One Health interventions to combat AMR across all 3 sectors.
Supplementary Material
Acknowledgments
We thank Hannah Rogers, the Head of Information Services at the Woodruff Health Sciences Center Library at Emory University, for her expertise and assistance with the initial literature search.
We also thank Shenita Peterson, Public Health Informationist for the Emory Libraries, for additional assistance with the literature search.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Data generated and analyzed during this review are included in this published article in the form of the main tables and additional figures. Additional details of our analysis are available from the corresponding author on reasonable request.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.03.041.
References
- Abayneh M, Worku T.. Prevalence of multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing gram-negative bacilli: A meta-analysis report in Ethiopia. Drug Target Insights 2020;14:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdi RD, Mengstie F, Beyi AF, Beyene T, Waktole H, Mammo B, et al. Determination of the sources and antimicrobial resistance patterns of Salmonella isolated from the poultry industry in Southern Ethiopia. BMC Infect Dis 2017;17(1):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreham S, Teklu A, Cox E, Sisay Tessema T. Escherichia coli O157:H7: distribution, molecular characterization, antimicrobial resistance patterns and source of contamination of sheep and goat carcasses at an export abattoir, Mojdo, Ethiopia. BMC Microbiol 2019;19(1):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fea Abunna. Isolation, identification and antimicrobial susceptibility profiles of Salmonella isolates from dairy farms in and around Modjo town. Ethiopia. Ethiopian Veterinary Journal 2017;21:92. [Google Scholar]
- Arega B, Woldeamanuel Y, Adane K, Sherif AA, Asrat D.. Microbial spectrum and drug-resistance profile of isolates causing bloodstream infections in febrile cancer patients at a referral hospital in Addis Ababa. Ethiopia. Infect Drug Resist 2018;11:1511–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaw-Denboba A, Abejew AA, Mekonnen AG.. Antibiotic-Resistant Bacteria Are Major Threats of Otitis Media in Wollo Area, Northeastern Ethiopia: A Ten-Year Retrospective Analysis. Int J Microbiol 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti M, Trecarichi EM, Mesini A, Spanu T, Giacobbe DR, Rossi M, et al. Risk factors and mortality of healthcare-associated and community-acquired Staphylococcus aureus bacteraemia. Clin Microbiol Infect 2012;18(9):862–9. [DOI] [PubMed] [Google Scholar]
- Belachew T, Mihret A, Legesse T, Million Y, Desta K.. High level of drug resistance by gram-negative bacteria from selected sewage polluted urban rivers in Addis Ababa, Ethiopia. BMC Res Notes 2018;11(1):524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene T, Hayishe H, Gizaw F, Beyi AF, Abunna F, Mammo B, et al. Prevalence and antimicrobial resistance profile of Staphylococcus in dairy farms, abattoir and humans in Addis Ababa, Ethiopia. BMC Res Notes 2017;10(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitew Kifilie A, Dagnew M, Tegenie B, Yeshitela B, Howe R, Abate E.. Bacterial Profile, Antibacterial Resistance Pattern, and Associated Factors from Women Attending Postnatal Health Service at University of Gondar Teaching Hospital, Northwest Ethiopia. Int J Microbiol 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y.. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect Control Hosp Epidemiol 2005;26(2):166–74. [DOI] [PubMed] [Google Scholar]
- Deyno S, Fekadu S, Astatkie A.. Resistance of Staphylococcus aureus to antimicrobial agents in Ethiopia: a meta-analysis. Antimicrob Resist Infect Control 2017a;6:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyno S, Toma A, Worku M, Bekele M.. Antimicrobial resistance profile of staphylococcus aureus isolates isolated from ear discharges of patients at University of Hawassa comprehensive specialized hospital. BMC Pharmacol Toxicol 2017b;18(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton L, Thomason MJ, Tembo J, Velavan TP, Pallerla SR, Arruda LB, et al. Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrob Resist Infect Control 2020;9(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher NA, Muhummed AM, Hattendorf J, Vonaesch P, Zinsstag J.. Systematic review and meta-analysis of integrated studies on antimicrobial resistance genes in Africa-A One Health perspective. Trop Med Int Health 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essack SY.. Environment: the neglected component of the One Health triad. Lancet Planet Health 2018;2(6):e238–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethiopia Antimicrobial Resistance Surveillance Annual Report (Second Year) September 2018-October 2019; 2020. Available from: http://repository.iifphc.org/handle/123456789/753?show=full. [Accessed 15 Nov 2021].
- Garedew L, Hagos Z, Zegeye B, Addis Z.. The detection and antimicrobial susceptibility profile of Shigella isolates from meat and swab samples at butchers’ shops in Gondar town, Northwest Ethiopia. J Infect Public Health 2016;9(3):348–55. [DOI] [PubMed] [Google Scholar]
- Gebretekle GB, Haile Mariam D, Abebe Taye W, Mulu Fentie A, Amogne Degu W, Alemayehu T, et al. Half of Prescribed Antibiotics Are Not Needed: A Pharmacist-Led Antimicrobial Stewardship Intervention and Clinical Outcomes in a Referral Hospital in Ethiopia. Front Public Health 2020;8:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebretekle GB, Haile Mariam D, Abebe W, Amogne W, Tenna A, Fenta TG, et al. Opportunities and barriers to implementing antibiotic stewardship in low and middle-income countries: Lessons from a mixed-methods study in a tertiary care hospital in Ethiopia. PLoS One 2018;13(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemeda BA, Assefa A, Jaleta MB, Amenu K, Wieland B.. Antimicrobial resistance in Ethiopia: A systematic review and meta-analysis of prevalence in foods, food handlers, animals, and the environment. One Health 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Action Plan on Antimicrobial Resistance. World Health Organization; 2015a. Available from: https://www.who.int/antimicrobial-resistance/global-action-plan/en/. [Accessed 1 Nov 2021]. [Google Scholar]
- Gorems K, Beyene G, Berhane M, Mekonnen Z.. Antimicrobial susceptibility patterns of bacteria isolated from patients with ear discharge in Jimma Town, Southwest, Ethiopia. BMC Ear Nose Throat Disord 2018;18:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailu DDAMDZYAYWSBF.. Drug resistance patterns of bacterial isolates from infected wounds at Bahir Dar Regional Health Research Laboratory Center, Northwest Ethiopia. African Journals Online 2018;30(3). [Google Scholar]
- Harris PNA, Tambyah PA, Lye DC, Mo Y, Lee TH, Yilmaz M, et al. Effect of Piperacillin-Tazobactam vs Meropenem on 30-Day Mortality for Patients With E coli or Klebsiella pneumoniae Bloodstream Infection and Ceftriaxone Resistance: A Randomized Clinical Trial. JAMA 2018;320(10):984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IACG. Report to the Secretary-General of the United Nations. Interagency Coordination Group on Antimicrobial Resistance; 2019.
- Khan MS, Rothman-Ostrow P, Spencer J, Hasan N, Sabirovic M, Rahman-Shepherd A, et al. The growth and strategic functioning of One Health networks: a systematic analysis. Lancet Planet Health 2018;2(6):e264–73. [DOI] [PubMed] [Google Scholar]
- Lamboro T, Ketema T, Bacha K.. Prevalence and Antimicrobial Resistance in Salmonella and Shigella Species Isolated from Outpatients. Jimma University Specialized Hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamuye Y. Antibiotic Resistance Patterns of Common Gramnegative Uropathogens in St. Paul’s Hospital Millennium Medical College. Ethiop J Health Sci 2016;26(2):93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenzhe RI, Zar HJ, Nicol MP, Kaba M.. The spread of carbapenemase-producing bacteria in Africa: a systematic review. J Antimicrob Chemother 2015;70(1):23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manyi-Loh C, Mamphweli S, Meyer E, Okoh A.. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018;23(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks F, von Kalckreuth V, Aaby P, Adu-Sarkodie Y, El Tayeb MA, Ali M, et al. Incidence of invasive salmonella disease in sub-Saharan Africa: a multicentre population-based surveillance study. Lancet Glob Health 2017;5(3):e310–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messele YE, Abdi RD, Yalew ST, Tegegne DT, Emeru BA, Werid GM.. Molecular determination of antimicrobial resistance in Escherichia coli isolated from raw meat in Addis Ababa and Bishoftu, Ethiopia. Ann Clin Microbiol Antimicrob 2017;16(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Health MoA, Environment Forest and Climate Change Commission. Antimicrobial Resistance Prevnetion and Containment Strategic Plan: “One-Health Approach”. 2020; Third edition. [Google Scholar]
- Mouiche MMM, Moffo F, Akoachere JTK, Okah-Nnane NH, Mapiefou NP, Ndze VN, et al. Antimicrobial resistance from a one health perspective in Cameroon: a systematic review and meta-analysis. BMC Public Health 2019;19(1):1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigussie D, Amsalu A.. Prevalence of uropathogen and their antibiotic resistance pattern among diabetic patients. Turk J Urol 2017;43(1):85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Health Basics. The Centers for Disease Control and Prevention NCEZID; 2018a. Available from: https://www.cdc.gov/onehealth/basics/index.html. [Accessed 25 Oct 2021].
- Rousham EK, Unicomb L, Islam MA.. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc Biol Sci 2018;285(1876). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharland M, Pulcini C, Harbarth S, Zeng M, Gandra S, Mathur S, et al. Classifying antibiotics in the WHO Essential Medicines List for optimal use-be AWaRe. Lancet Infect Dis 2018;18(1):18–20. [DOI] [PubMed] [Google Scholar]
- Shimekaw M, Tigabu A, Tessema B.. Bacterial Profile, Antimicrobial Susceptibility Pattern, and Associated Risk Factors Among Patients With Wound Infections at Debre Markos Referral Hospital, Northwest, Ethiopia. Int J Low Extrem Wounds 2020. [DOI] [PubMed] [Google Scholar]
- Sileshi S, Munees A.. Prevalence and antibiotic susceptibility of Staphylococcus aureus from lactating cow’s milk in Bahir Dar dairy farms. African Journal of Microbiology Research 2016;10(35):1444–54. [Google Scholar]
- Strategy for the Prevention and Containment of Antimicrobial Resistance for Ethiopia. Second ed: Ethiopian Food, Medicine and Healthcare Administration and Control Authority; 2015b. Available from: https://www.who.int/publications/m/item/ethiopia. [Accessed 20 Nov 2021]. [Google Scholar]
- Tadesse S, Alemayehu H, Tenna A, Tadesse G, Tessema TS, Shibeshi W, et al. Antimicrobial resistance profile of Staphylococcus aureus isolated from patients with infection at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. BMC Pharmacol Toxicol 2018;19(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takele S, Woldemichael K, Gashaw M, Tassew H, Yohannes M, Abdissa A.. Prevalence and drug susceptibility pattern of Salmonella isolates from apparently healthy slaughter cattle and personnel working at the Jimma municipal abattoir, south-West Ethiopia. Trop Dis Travel Med Vaccines 2018;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One 2013;8(4):e61090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terfassa A, Jida M.. Prevalence and Antibiotics Susceptibility Pattern of Salmonella and Shigella Species among Diarrheal Patients Attending Nekemte Referral Hospital, Oromia, Ethiopia. Int J Microbiol 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye H, Alemayehu H, Desta AF, Eguale T.. Antimicrobial susceptibility profile of selected Enterobacteriaceae in wastewater samples from health facilities, abattoir, downstream rivers and a WWTP in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control 2019;8:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshome A, Alemayehu T, Deriba W, Ayele Y.. Antibiotic Resistance Profile of Bacteria Isolated from Wastewater Systems in Eastern Ethiopia. J Environ Public Health 2020;2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshome B, Teklemariam Z, Admassu Ayana D, Marami D, Asaminew N. Salmonella and Shigella among patients with diarrhea at public health facilities in Adama, Ethiopia: Prevalence, antimicrobial susceptibility pattern, and associated factors. SAGE Open Med 2019;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsige Y, Tadesse S, GE T, Tefera MM, Amsalu A, Menberu MA, et al. Prevalence of Methicillin-Resistant Staphylococcus aureus and Associated Risk Factors among Patients with Wound Infection at Referral Hospital, Northeast Ethiopia. J Pathog 2020;2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel TP, Pires J, Silvester R, Zhao C, Song J, Criscuolo NG, et al. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019;365(6459):eaaw1944. [DOI] [PubMed] [Google Scholar]
- White A, Hughes JM.. Critical Importance of a One Health Approach to Antimicrobial Resistance. Ecohealth 2019;16(3):404–9. [DOI] [PubMed] [Google Scholar]
- WHO methodology for point prevalence survey on antibiotic use in hospitals. Geneva: World Health Organization; 2018b. Available from: https://www.who.int/publications/i/item/WHO-EMP-IAU-2018.01. [Accessed 1 Sept 2021]. [Google Scholar]
- Wilkinson JL, Boxall ABA, Kolpin DW, Leung KMY, Lai RWS, Galban-Malagon C, et al. Pharmaceutical pollution of the world’s rivers. Proc Natl Acad Sci U S A 2022;119(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalewska M, Blazejewska A, Czapko A, Popowska M. Antibiotics and Antibiotic Resistance Genes in Animal Manure - Consequences of Its Application in Agriculture. Front Microbiol 2021;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


