Abstract
Emerging evidences have shown that long noncoding RNA (lncRNA) plays an important role in the immune escape of cancer cells. Our previous study has demonstrated that lncRNA MIAT is associated with the immune infiltration of hepatocellular carcinoma (HCC). However, the underlying mechanism of MIAT regulating the PD‐L1‐mediated immune escape of HCC is poorly understood. Quantitative real‐time PCR (qRT‐PCR) was used to detect the expression of MIAT and PD‐L1 mRNA in HCC. The relationship between MIAT, miR‐411‐5p, STAT3 and PD‐L1 was explored by dual‐luciferase reporter assay, cytotoxicity assay, Western blot and RNA immunoprecipitation (RIP). In addition, the xenograft model was established to determine the effect of MIAT on PD‐L1 expression in vivo. We found that MIAT and PD‐L1 were significantly upregulated in HCC tissues and the expression of PD‐L1 was regulated by MIAT. The knockdown of MIAT enhanced the cytotoxicity of T cells on HCC cells. MIAT negatively regulated miR‐411‐5p expression, upregulated STAT3 and ultimately increased PD‐L1 expression from the transcription level. The inhibition of miR‐411‐5p reversed STAT3 and PD‐L1 expression inhibited by MIAT knockdown in HCC cells. This study suggests a novel lncRNA‐mediated mechanism for HCC cells to evade the immune response; MIAT/miR‐411‐5p/STAT3/PD‐L1 may be a novel therapeutic target for HCC.
Keywords: hepatocellular carcinoma, lncRNA MIAT, miR‐411‐5p, PD‐L1, STAT3
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is the fourth most common cause of cancer‐related death in the world. 1 In September 2017, with the development of immunotherapy, Food and Drug Administration (FDA) approved anti‐PD‐1 antibody nivolumab as a second‐line treatment scheme for patients with advanced HCC who had previously been treated with sorafenib 2 and achieved good curative effect, while more patients who received checkpoint blocker therapy could not achieve the ideal results. 3 Therefore, exploring the reasons for the poor efficacy of immune checkpoint therapy has become a hot spot in clinical HCC treatment. The discovery of novel intervention targets is expected to promote the progress in this field. Studies have shown that the amplification or higher expression of PD‐L1 significantly and independently correlated with unfavourable survival in HCC patients, authenticating the PD‐1/PD‐L1 axis as rational immunotherapeutic targets for HCC. 4 PD‐L1 expression was associated with a poor clinical outcome and vascular formation in patients with HCC, so it might serve as predictive tissue biomarkers. 5
Long noncoding RNA (lncRNA) is a transcript longer than 200 nucleotides with no protein‐coding capacity. 6 In recent years, more and more studies have shown that lncRNA can participate in tumour immune escape or immune resistance through multiple modes, and affect tumour progression. For example, MEG3 suppresses regulatory T‐cell (Treg) differentiation through the P53/miR‐149‐3p/FOXP3 axis in oesophageal cancer 7 ; SNHG12 promotes IL‐6/miR‐21 crosstalk between ovarian cancer cells and M2 macrophages 8 ; FENDRR inhibits the Treg‐mediated immune escape of HCC cells by upregulating GADD45B through sponging miR‐423‐5p 9 ; and UCA1 protects PD‐L1 expression from the repression of anti‐tumour miRNAs in gastric cancer cells. 10
In our previous studies, we found that lncRNA MIAT was related to the immune infiltration of HCC, but its specific mechanism is unknown. 11 MIAT (myocardial infarction‐associated transcript) was related to myocardial infarction and is widely expressed in the nervous system and retinal tissue. 12 Recent studies have shown that MIAT plays a role similar to oncogenes in thyroid papillary carcinoma, 13 HCC, 14 cholangiocarcinoma 15 and non‐small‐cell lung cancer. 16 Therefore, this study focuses on the molecular mechanism of MIAT participating in the immune escape of HCC cells based on the results of bioinformatics analysis. It is found that MIAT can promote the expression of STAT3 and PD‐L1 by sponging miR‐411‐5p, providing a novel idea for HCC immunotherapy research targeting PD‐L1.
2. MATERIALS AND METHODS
2.1. Clinical samples and animal models
All human HCC samples were obtained from the Fujian Medical University Union Hospital from August 2019 to October 2021, and the ethical consent was granted from the Ethics Committee of the Fujian Medical University Union Hospital. The clinical characteristics of the patient are shown in Tables S1 and S2, according to the American Joint Committee on Cancer version 8 (AJCC8). 5 × 106 Huh7 cells transfected with sh‐NC and sh‐MIAT were inoculated into the armpits of 6‐ to 8‐week‐old nude mice. When the diameter of the mouse tumour is greater than or equal to 2 cm, the mice were sacrificed under inhalation anaesthesia with isopentobarbital, and the tumour tissues were removed for subsequent experiments. The animal experiments were approved by the Animal Ethics Committee of the Fujian Medical University.
2.2. Cell lines and transfection
Human hepatoblastoma cell line HepG2 (HB‐8065; ATCC) and HCC cell line Huh7 (JCRB0403, Japan) were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 12% foetal bovine serum (FBS; Gibco) at 37°C with 5% CO2. Specific siRNAs against MIAT and NC sequences and miR‐411‐5p mimic/inhibitor or control sequences were synthesized by RiboBio. The sh‐MIAT and sh‐NC plasmids mediated by lentiviral vectors were designed and packaged according to the siRNA sequence. For dual‐luciferase reporter assay, the MIAT and STAT3 3’UTR wild‐type (WT) sequences and their corresponding mutant (Mut) sequences were cloned into the pmirGLO vector by Boshang Biotech (Shanghai, China).
2.3. RNA extraction and qRT‐PCR analysis
Total RNA was extracted from tissue and cells using TRIzol (TianGen). 3 μg total RNA was subjected to reverse transcription reaction using EasyScript One‐Step gDNA Removal and cDNA Synthesis SuperMix (TransGen). qRT‐PCR analyses were performed using EvaGreen 2X qPCR MasterMix‐Low ROX (ABM). For miRNA detection, cDNA was synthesized using miRNA cDNA Synthesis Kit (ABM) and then amplified using miRNA qPCR profiling kits (ABM). GAPDH is used as an internal reference for lncRNA and mRNA, and U6 is used as an internal reference for miRNA. The primer sequences used for qRT‐PCR are shown in Table S3.
2.4. Western blot and antibodies
Total protein was extracted from the cells with RIPA lysis buffer (Beyotime, Shanghai, China). An equal amount of protein was electrophoresed on an SDS–polyacrylamide gel and onto a polyvinylidene fluoride (PVDF) membrane (Merck Millipore). After blocking with 5%–10% milk, the membrane was incubated with the primary antibody at 4°C overnight and then stained with the corresponding horseradish peroxidase‐labelled secondary antibody for 1h. Finally, BeyoECL Plus (Beyotime) was used to observe the immunoreactivity and the Bio‐Rad chemiluminescence imager was used for detection. GAPDH serves as an internal control. The following primary antibodies were used: GAPDH (Servicebio, GB11002), STAT3 (ABclonal, A1192) and PD‐L1 (Affinity, DF6526). A relative protein expression was quantified using the ImageJ software.
2.5. Dual‐luciferase reporter assay
MIAT WT/Mut or STAT3 WT/Mut and NC or miR‐411‐5p mimic were co‐transfected into HepG2 and Huh7 cells. After 48 h of transfection, the cell lysate was collected and the luciferase activity was detected by Dual‐Luciferase® Reporter Assay System (Promega). The data were normalized with Renilla luciferase.
2.6. RNA immunoprecipitation
RNA immunoprecipitation (RIP) was performed according to the instructions of Millipore Magna RIP Kit (Millipore, Darmstadt, Germany). HepG2 cells were lysed in RIP lysis buffer, argonaute RISC catalytic component 2 (AGO2) antibody or IgG antibody was added, and they were incubated at 4°C overnight. Then, the RNA was purified, and the immunoprecipitated RNA was extracted with TRIzol and analysed by qRT‐PCR detection. The primers used in this analysis are described in Table S3.
2.7. Cytotoxicity assay
Peripheral blood mononuclear cells (PBMCs) were obtained from healthy individuals and cultured in T‐cell culture medium supplemented with IL‐2 (PeproTech) and ImmunoCult™ Human CD3/CD28/CD2 T Cell Activator (Stemcell, Canada) for one week. Correspondingly, the treated tumour cells were co‐cultured with activated T cells for 12 h, in the ratio of cancer cells to activated cells (1:3), and then, the HCC cells were stained with crystal violet.
2.8. Immunohistochemical analysis
The paraffin‐embedded sections were deparaffinized and then incubated with STAT3 (ABclonal, A1192) or PD‐L1 (Abcam, ab213524) antibody overnight at 4°C. After washing three times with PBST, the tissue was incubated with a reaction enhancer (ZSGB‐BIO) for 20 min at room temperature. At last, the tissue was incubated with a secondary antibody labelled with horseradish peroxidase (HRP) and then reacted with DAB to achieve signal amplification and colour development. Finally, the nuclei were counterstained with haematoxylin. The samples were scored according to an H‐score method combining the values of immunoreaction intensity and percentage of staining tumour cell.
2.9. Statistical analysis
The GraphPad Prism 8 software was used for statistical analysis. The data were expressed as mean ±SD. The Pearson linear correlation analysis was used to determine the correlation between the two variables. Student's t test was used to compare two independent samples. p < .05 was considered statistically significant.
3. RESULTS
3.1. MIAT promotes the expression of PD‐L1 and immune escape of HCC cells
PD‐L1 appears to function as a significant biomarker in the poor prognosis of HCC and provided implications to estimate the risk of HCC patients. 17 It is of great significance to explore the molecules related to its changes and their regulatory mechanism. In particular, the relationship between lncRNA and PD‐L1 has not been fully explored. Our previous bioinformatics analysis revealed that the expression of lncRNA MIAT in HCC was positively correlated with immunosuppressive molecules (such as PD‐1, PD‐L1 and CTLA4). 11 In this study, we further used 36 cases of fresh HCC samples collected in recent two years to verify again whether the correlation analysis showed a significant positive correlation between MIAT and PD‐L1 (Figure 1A). We also detected the expression of MIAT and PD‐L1 in another 18 pairs of HCC tissues and adjacent tissues and found that they were significantly higher in cancer tissues than in paired adjacent tissues (Figure 1B). Then, we designed and constructed the sh‐MIAT and sh‐NC plasmids according to the siRNA sequence, 11 which were transfected into HepG2 and Huh7 cells; qRT‐PCR found that the MIAT expression was decreased (Figure 1C). PD‐L1 mRNA expression and protein expression were decreased in HCC cells with MIAT knockdown (Figure 1D, E). In order to study whether MIAT affects the function of PD‐L1, we performed cytotoxicity assay. As shown in Figure 1F, after incubating with PBMC for 24 h, the HCC cells with MIAT knockdown are more sensitive to T‐cell cytotoxicity.
FIGURE 1.
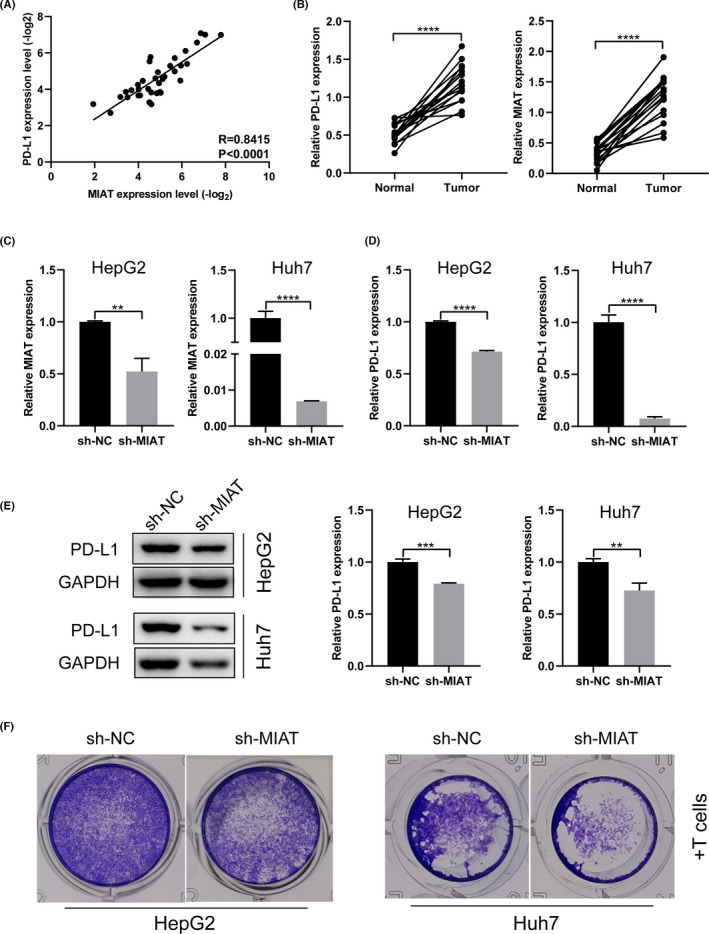
MIAT promotes the expression of PD‐L1 and immune escape of HCC cells. (A) The expression of MIAT and PD‐L1 is positively correlated in HCC tissue. (B) MIAT and PD‐L1 mRNA levels in HCC tissues and paracancerous tissues. (C) sh‐NC and sh‐MIAT plasmids were transfected into HepG2 and Huh7 cells to verify the knockdown effect. (D, E) The expression of PD‐L1 mRNA (D) and protein (E) was detected in HepG2 and Huh7 cells with MIAT knockdown. (F) Cytotoxicity assay was used to detect the cytotoxicity of activated T cells on HepG2 and Huh7 cells with MIAT knockdown. **p <.01, ***p <.001 and ****p <.0001
3.2. MIAT sponges miR‐411‐5p
At present, competitive endogenous RNA (ceRNA), the most widely accepted, is one of the main mechanisms of lncRNA regulation. 18 We speculate that MIAT may regulate the expression of PD‐L1 through the ceRNA mechanism. We used the online database StarBase 3.0 (http://starbase.sysu.edu.cn/index.php) to predict the miRNAs bound by MIAT. It was found that miR‐411‐5p directly binds to MIAT. miR‐411‐5p is negatively correlated with MIAT in HCC tissues (Figure 2A). The level of miR‐411‐5p was significantly increased in HCC cell lines with MIAT knockdown (Figure 2B). miR‐411‐5p mimic was transfected into HCC cells, and the expression of MIAT was found to be decreased (Figure 2C); miR‐411‐5p inhibitor was transfected into HCC cells, and the expression of MIAT was found to be increased (Figure 2D). In order to evaluate the interaction between MIAT and miR‐411‐5p, we constructed the MIAT WT and Mut reporter vectors (Figure 2E). The dual‐luciferase reporter assay showed that the transfection of miR‐411‐5p mimic in HepG2 and Huh7 cells decreased the luciferase activity of the MIAT WT plasmid, but had no effect on that of the MIAT Mut plasmid (Figure 2F).
FIGURE 2.
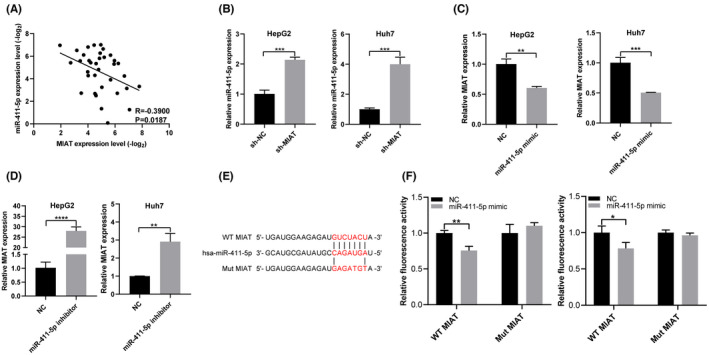
MIAT sponges miR‐411‐5p. (A) The expression of MIAT and miR‐411‐5p is negatively correlated in HCC tissue. (B) miR‐411‐5p mRNA level was detected in HepG2 and Huh7 cells with MIAT knockdown. (C, D) MIAT mRNA levels were detected in HepG2 and Huh7 cells transfected with miR‐411‐5p mimic (C) or miR‐411‐5p inhibitor (D). (E) Interacting sequences on MIAT for miR‐411‐5p were obtained from StarBase 3.0. The putative binding sites of miR‐411‐5p were mutated in MIAT. (F) The dual‐luciferase reporter assay detects the interaction between MIAT and miR‐411‐5p. *p <.05, **p <.01, ***p <.001 and ****p <.0001
3.3. STAT3 is a functional target of miR‐411‐5p
To explore how miR‐411‐5p regulates the expression of PD‐L1 in HCC cells, we used StarBase 3.0 to predict the targeting genes that miR‐411‐5p might recognize and found that STAT3 is one of the targets of miR‐411‐5p. Some studies have shown that STAT3, as a transcription factor of PD‐L1, could bind to the promoter of PD‐L1 to regulate its expression. 19 , 20 , 21 , 22 The relationship between miR‐411‐5p, STAT3 and PD‐L1 was verified in HCC tissues (Figure 3A–C). The mRNA and protein levels of STAT3 and PD‐L1 were decreased following the transfection of miR‐411‐5p mimics (Figure 3D, E) and increased with the transfection of the miR‐411‐5p inhibitor in HCC cells (Figure 3F, G). Subsequently, the dual‐luciferase reporter assay was used to verify whether miR‐411‐5p can bind to STAT3 mRNA 3’UTR. We constructed the reporter vectors containing WT and Mut 3’UTR of STAT3 (Figure 3H). The dual‐luciferase reporter assay showed that the relative luciferase activity of STAT3 WT plasmid in HCC cells was inhibited after the co‐transfection of miR‐411‐5p mimic, but did not change the activity of the STAT3 Mut plasmid (Figure 3I). The above one indicates that miR‐411‐5p can regulate PD‐L1 expression through STAT3.
FIGURE 3.
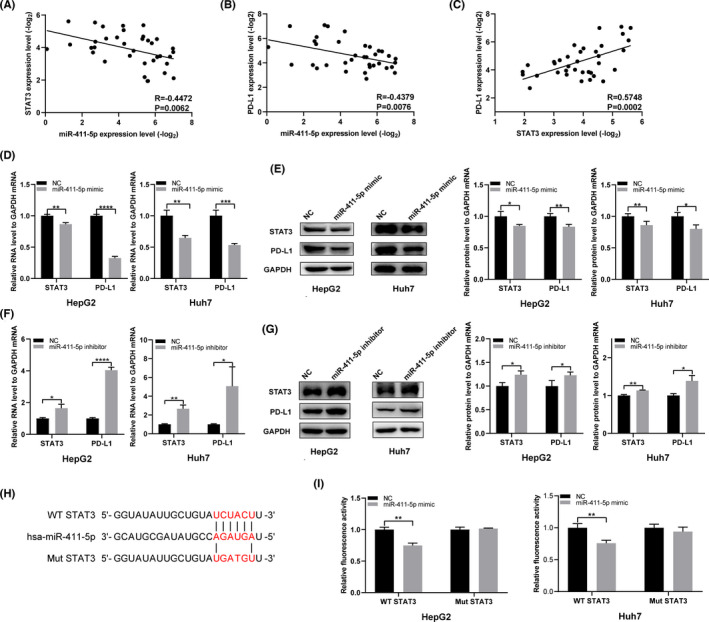
STAT3 is the functional target of miR‐411‐5p. (A‐C) The correlation between miR‐411‐5p, STAT3 and PD‐L1 in HCC tissues. (D, E) STAT3 and PD‐L1 mRNA and protein were detected in HepG2 and Huh7 cells transfected with miR‐411‐5p mimic. (F, G) STAT3 and PD‐L1 mRNA and protein were detected in HepG2 and Huh7 cells transfected with miR‐411‐5p inhibitor. (H) Interacting sequences on STAT3 for miR‐411‐5p were obtained from StarBase 3.0. The putative binding sites of miR‐411‐5p were mutated in STAT3. (I) The dual‐luciferase reporter assay was used to detect the interaction between miR‐411‐5p and STAT3. *p <.05, **p <.01, ***p <.001 and ****p <.0001
3.4. MIAT regulates PD‐L1 expression through the miR‐411‐5p/STAT3 axis
In order to verify whether miR‐411‐5p can bind to MIAT and STAT3, we performed RIP assay with AGO2 antibody in HepG2 cells. MIAT, miR‐411‐5p and STAT3 were all immunoprecipitated by AGO2 (Figure 4A). In HCC tissues, the expression of MIAT and STAT3 was positively correlated (Figure 4B). Since MIAT can sponge miR‐411‐5p, we next determined whether MIAT can affect the expression of STAT3 through competitive binding with miR‐411‐5p. It was found that MIAT knockdown significantly decreased the mRNA and protein levels of STAT3 and PD‐L1 in HepG2 and Huh7 cells (Figure 4C, D). Finally, we examined the expression of STAT3 and PD‐L1 in HCC cells co‐transfected with sh‐MIAT and miR‐411‐5p inhibitors. As expected, the mRNA expression and protein expression of STAT3 and PD‐L1 were partially restored in the co‐transfected cells (Figure 4E, F). These results indicate that MIAT can promote the expression of STAT3 and PD‐L1 through sponging miR‐411‐5p in HCC cells.
FIGURE 4.
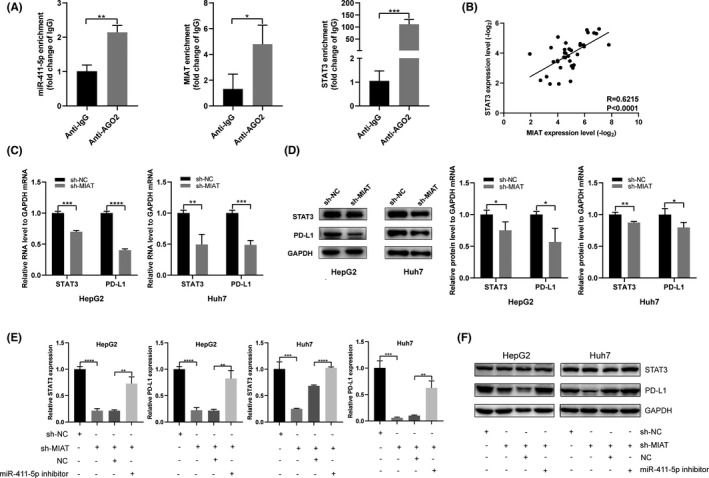
MIAT regulates PD‐L1 expression through the miR‐411‐5p/STAT3 axis. (A) The RIP experiment was performed in HepG2 cells to confirm the interaction between MIAT, miR‐411‐5p and STAT3. (B) The expressions of MIAT and STAT3 in HCC tissues are positively correlated. (C, D) STAT3 and PD‐L1 mRNA (C) and protein (D) levels were detected in HepG2 and Huh7 cells transfected with sh‐NC and sh‐MIAT. (E, F) The sh‐MIAT and miR‐411‐5p inhibitors were co‐transfected into HepG2 and Huh7 cells, and the mRNA (E) and protein (F) levels of STAT3 and PD‐L1 were detected. *P <.05, **p <.01, ***p <.001 and ****p <.0001
3.5. MIAT regulates PD‐L1 through miR‐411‐5p/STAT3 in vivo
In order to further validate the effects of MIAT in vivo, the xenograft tumour model was constructed to examine the role of MIAT. Then, the Huh7 cells transfected with sh‐NC and sh‐MIAT were injected into nude mice. We observed that the expression of MIAT was significantly decreased in the sh‐MIAT group (Figure 5A), and the expression of STAT3 and PD‐L1 mRNA in the MIAT knockdown group was also lower (Figure 5C, D), while the expression of miR‐411‐5p was higher in the sh‐MIAT group (Figure 5B). Next, we used IHC analysis to further evaluate the expression of STAT3 and PD‐L1 proteins in tumour tissues and found that the expression of STAT3 and PD‐L1 in the sh‐MIAT group was lower (Figure 5E, F).
FIGURE 5.
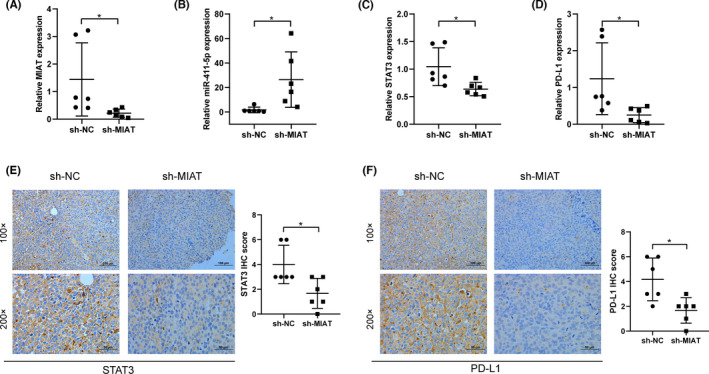
MIAT regulates PD‐L1 through miR‐411‐5p/STAT3 in mice in vivo. (A–D) The expression of MIAT (A), miR‐411‐5p (B), STAT3 (C) and PD‐L1 (D) in mouse tumour tissues was detected by qRT‐PCR. (E, F) IHC comparison of the expression of STAT3 (E) and PD‐L1 (F) in sh‐NC and sh‐MIAT groups. *p <.05
4. DISCUSSION
In this study, we explored the effect of MIAT on HCC and found that MIAT affected the escape of HCC cells from T‐cell cytotoxicity by regulating the expression of PD‐L1. In previous studies of HCC, silencing MIAT could inhibit HCC by inducing senescence and activating P53/P21 and pRb signalling pathways 23 ; MIAT regulates EphA2 by sponging miR‐520d‐3p to promote HCC progress, 24 and MIAT can also promote the proliferation and invasion of HCC cells through sponging miR‐214. However, the immune‐related research of MIAT in HCC remains largely unknown. Xu et al found that HCP5 and MIAT upregulate the expression of PD‐L1 in colon cancer and cervical cancer cells through sponging miR‐150‐5p, 25 which is consistent with our research results.
Emerging evidences show that lncRNA plays an important role in PD‐L1‐mediated immune escape; for example, GATA3‐AS1 promotes the progression of triple‐negative breast cancer by stabilizing PD‐L1 protein and degrading GATA3 protein 26 ; HOTTIP could promote the secretion of IL‐6, upregulate the expression of PD‐L1 and inhibit the activity of T cells in ovarian cancer 27 ; CASC11 recruits EIF4A3 to enhance the stability of E2F1 mRNA. CASC11 and E2F1 impacted the activation of the NF‐κB signalling and PI3K/AKT/mTOR pathway and further regulated the expression of PD‐L1 in HCC 28 ; SNHG14/miR‐5590‐3p/ZEB1 positive feedback loop regulates the PD‐1/PD‐L1 checkpoint in diffuse large B‐cell lymphoma. 29 The potential of lncRNA and its corresponding regulation of immune checkpoints as novel diagnostic biomarkers and therapeutic targets in the treatment of malignant tumours have been emphasized. 30 We conducted cytotoxicity assay and found that the downregulation of PD‐L1 caused by MIAT knockdown made HCC cells to be more sensitive to T‐cell cytotoxicity. The key role of lncRNA in HCC is expected to become a novel drug target for therapeutic intervention.
lncRNA could guide RNA‐induced silencing complex (RISC) to miRNA response element (MRE) by competitively binding miRNAs, thereby inhibiting protein production, which is called the ceRNA hypothesis. 31 lncRNA regulates targeting genes through sponging miRNA and plays an important role in the occurrence and development of HCC. For example, HOXD‐AS1 competitively bound to miR‐130a‐3p, then prevented SOX4 from miRNA‐mediated degradation, and thus activated the expression of EZH2 and MMP2 and facilitated HCC metastasis 32 ; MCM3AP‐AS1 promotes FOXA1 expression by targeting miR‐194‐5p to play a carcinogenic effect in HCC 33 ; SNHG6 targets miR‐204‐5p resulting in an increased E2F1 expression and enhances G1‐S phase transition, thereby promoting HCC 34 ; and tumour‐associated macrophage‐induced H19 promotes the aggressiveness of HCC by triggering and activating the miR‐193b/MAPK1 axis. 35 In view of the mechanism of the lncRNA‐miRNA‐mRNA network, we screened out miR‐411‐5p and the target gene STAT3. To verify whether STAT3 is the functional target of miR‐411‐5p, we first examined the expression and correlation of them. The results indicated that the expression of miR‐411‐5p was negatively correlated with STAT3 and significantly inhibited the expression of STAT3 at the mRNA and protein levels. The luciferase report experiment and RIP results show that MIAT and STAT3 can directly bind to miR‐411‐5p. STAT3 has been shown to bind to the PD‐L1 promoter to regulate its expression in a transcriptional manner, 36 which could induce apoptosis of immune cells, inhibit the lethality of T cells and promote tumour immune escape. 37 In addition, we verified the relationship between MIAT, miR‐411‐5p, STAT3 and PD‐L1 in HCC specimens. In summary, we found that MIAT in HCC cells can promote the expression of STAT3 and PD‐L1 through sponging of miR‐411‐5p.
In conclusion, this study reveals a novel molecular regulation mechanism between HCC and immunity—MIAT/miR‐411‐5p/STAT3/PD‐L1, which may serve as a new target for HCC immunotherapy.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
XZ and NT conceived and designed the study. BP and JQ developed methodology. XK, SS and XW performed data curation. XZ and BP wrote, reviewed and revised the manuscript. NT supervised the study.
Supporting information
Table S1‐S3
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (82173167), the Startup Fund for Scientific Research of the Fujian Medical University (2019XQ2020), the Joint Funds for the Innovation of Science and Technology, Fujian Province (2017Y9100), and the Natural Science Foundation of Fujian Province (2018J01296).
Zhang X, Pan B, Qiu J, et al. lncRNA MIAT targets miR‐411‐5p/STAT3/PD‐L1 axis mediating hepatocellular carcinoma immune response. Int J Exp Path. 2022;103:102–111. doi: 10.1111/iep.12440
REFERENCES
- 1. Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability‐adjusted life‐years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3(4):524‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. El‐Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open‐label, non‐comparative, phase 1/2 dose escalation and expansion trial. Lancet (London, England). 2017;389(10088):2492‐2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma LJ, Feng FL, Dong LQ, et al. Clinical significance of PD‐1/PD‐Ls gene amplification and overexpression in patients with hepatocellular carcinoma. Theranostics. 2018;8(20):5690‐5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Itoh S, Yoshizumi T, Yugawa K, et al. Impact of immune response on outcomes in hepatocellular carcinoma: association with vascular formation. Hepatology (Baltimore, MD). 2020;72(6):1987‐1999. [DOI] [PubMed] [Google Scholar]
- 6. Wang JY, Yang Y, Ma Y, et al. Potential regulatory role of lncRNA‐miRNA‐mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:109627. [DOI] [PubMed] [Google Scholar]
- 7. Xu QR, Tang J, Liao HY, et al. Long non‐coding RNA MEG3 mediates the miR‐149‐3p/FOXP3 axis by reducing p53 ubiquitination to exert a suppressive effect on regulatory T cell differentiation and immune escape in esophageal cancer. J Transl Med. 2021;19(1):264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qian M, Ling W, Ruan Z. Long non‐coding RNA SNHG12 promotes immune escape of ovarian cancer cells through their crosstalk with M2 macrophages. Aging. 2020;12(17):17122‐17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu Z, Zhao H, Feng X, et al. Long non‐coding RNA FENDRR acts as a miR‐423‐5p sponge to suppress the treg‐mediated immune escape of hepatocellular carcinoma cells. Mol Ther Nucleic Acids. 2019;17:516‐529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang CJ, Zhu CC, Xu J, et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti‐tumor miRNAs. Mol Cancer. 2019;18(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng L, Chen Y, Ou Q, Wang X, Tang N. LncRNA MIAT correlates with immune infiltrates and drug reactions in hepatocellular carcinoma. Int Immunopharmacol. 2020;89(Pt A):107071. [DOI] [PubMed] [Google Scholar]
- 12. Liao J, He Q, Li M, Chen Y, Liu Y, Wang J. LncRNA MIAT: myocardial infarction associated and more. Gene. 2016;578(2):158‐161. [DOI] [PubMed] [Google Scholar]
- 13. Wang R, Zhao L, Ji L, Bai L, Wen Q. Myocardial infarction associated transcript (MIAT) promotes papillary thyroid cancer progression via sponging miR‐212. Biomed Pharmacother. 2019;118:109298. [DOI] [PubMed] [Google Scholar]
- 14. Huang X, Gao Y, Qin J, Lu S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR‐214. Am J Physiol Gastrointest Liver Physiol. 2018;314(5):G559‐g565. [DOI] [PubMed] [Google Scholar]
- 15. Chang W, Wang Y, Li W, Geng Z. Long non‐coding RNA myocardial infarction associated transcript promotes the proliferation of cholangiocarcinoma cells by targeting miR‐551b‐3p/CCND1 axis. Clin Exp Pharmacol Physiol. 2020;47(6):1067‐1075. [DOI] [PubMed] [Google Scholar]
- 16. Wu L, Liu C, Zhang Z. Knockdown of lncRNA MIAT inhibits proliferation and cisplatin resistance in non‐small cell lung cancer cells by increasing miR‐184 expression. Oncol Lett. 2020;19(1):533‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li JH, Ma WJ, Wang GG, et al. Clinicopathologic significance and prognostic value of programmed cell death ligand 1 (PD‐L1) in patients with hepatocellular carcinoma: a meta‐analysis. Front Immunol. 2018;9:2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marzec M, Zhang Q, Goradia A, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD‐L1, B7–H1). Proc Natl Acad Sci USA. 2008;105(52):20852‐20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding L, Chen X, Xu X, et al. PARP1 suppresses the transcription of PD‐L1 by Poly(ADP‐Ribosyl)ating STAT3. Cancer Immunol Res. 2019;7(1):136‐149. [DOI] [PubMed] [Google Scholar]
- 21. Tong L, Li J, Li Q, et al. ACT001 reduces the expression of PD‐L1 by inhibiting the phosphorylation of STAT3 in glioblastoma. Theranostics. 2020;10(13):5943‐5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bu LL, Yu GT, Wu L, et al. STAT3 induces immunosuppression by upregulating PD‐1/PD‐L1 in HNSCC. J Dent Res. 2017;96(9):1027‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao L, Hu K, Cao J, et al. lncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR‐22‐3p in HCC cellular senescence. Aging. 2019;11(17):7098‐7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiang Y, Huang Y, Sun H, Pan Y, Wu M, Zhang J. Deregulation of miR‐520d‐3p promotes hepatocellular carcinoma development via lncRNA MIAT regulation and EPHA2 signaling activation. Biomed Pharmacother. 2019;109:1630‐1639. [DOI] [PubMed] [Google Scholar]
- 25. Xu S, Wang Q, Kang Y, et al. Long noncoding RNAs control the modulation of immune checkpoint molecules in cancer. Cancer Immunol Res. 2020;8(7):937‐951. [DOI] [PubMed] [Google Scholar]
- 26. Zhang M, Wang N, Song P, et al. LncRNA GATA3‐AS1 facilitates tumour progression and immune escape in triple‐negative breast cancer through destabilization of GATA3 but stabilization of PD‐L1. Cell Prolif. 2020;53(9):e12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shang A, Wang W, Gu C, et al. Long non‐coding RNA HOTTIP enhances IL‐6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD‐L1 in neutrophils. J Exp Clin Cancer Res. 2019;38(1):411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song H, Liu Y, Li X, et al. Long noncoding RNA CASC11 promotes hepatocarcinogenesis and HCC progression through EIF4A3‐mediated E2F1 activation. Clin Transl Med. 2020;10(7):e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. LncRNA SNHG14/miR‐5590‐3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD‐1/PD‐L1 checkpoint. Cell Death Dis. 2019;10(10):731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang L, Li J, Ding J, Lian Y, Huangfu C, Wang K. Roles of long noncoding RNAs on tumor immune escape by regulating immune cells differentiation and function. Am J Cancer Res. 2021;11(6):2369‐2385. [PMC free article] [PubMed] [Google Scholar]
- 31. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272‐283. [DOI] [PubMed] [Google Scholar]
- 32. Wang H, Huo X, Yang XR, et al. STAT3‐mediated upregulation of lncRNA HOXD‐AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP‐AS1 promotes the growth of hepatocellular carcinoma by targeting miR‐194‐5p/FOXA1 axis. Mol Cancer. 2019;18(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen K, Hou Y, Liao R, Li Y, Yang H, Gong J. LncRNA SNHG6 promotes G1/S‐phase transition in hepatocellular carcinoma by impairing miR‐204‐5p‐mediated inhibition of E2F1. Oncogene. 2021;40(18):3217‐3230. [DOI] [PubMed] [Google Scholar]
- 35. Ye Y, Guo J, Xiao P, et al. Macrophages‐induced long noncoding RNA H19 up‐regulation triggers and activates the miR‐193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469:310‐322. [DOI] [PubMed] [Google Scholar]
- 36. Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD‐L1: a novel role of pro‐survival signalling in cancer. Ann Oncol. 2016;27(3):409‐416. [DOI] [PubMed] [Google Scholar]
- 37. Zhang L, Kuca K, You L, et al. Signal transducer and activator of transcription 3 signaling in tumor immune evasion. Pharmacol Ther. 2022;230:107969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S3


