Abstract
Objectives
Because of the spread of the Omicron variant, many countries have experienced COVID-19 case numbers unseen since the start of the pandemic. We aimed to compare the epidemiological characteristics of Omicron with previous variants and different strains of influenza to provide context for public health responses.
Methods
We developed transmission models for SARS-CoV-2 variants and influenza, in which transmission, death, and vaccination rates were taken to be time-varying. We fit our model based on publicly available data in South Africa, the United States, and Canada. We used this model to evaluate the relative transmissibility and mortality of Omicron compared with previous variants and influenza.
Results
We found that Omicron is more transmissible and less fatal than both seasonal and 2009 H1N1 influenza and the Delta variant; these characteristics make Omicron epidemiologically more similar to influenza than it is to Delta. We estimate that as of February 7, 2022, booster doses have prevented and Omicron infections in the United States and Canada, respectively.
Conclusion
Our findings indicate that the high infectivity of Omicron will keep COVID-19 endemic, similar to influenza. However, because of Omicron's lower fatality rate, our work suggests that human populations living with SARS-CoV-2 are most likely.
Keywords: Death rate, Effective reproduction number, Influenza, Omicron variant, Transmission rate
Introduction
On January 30, 2020, the World Health Organization (WHO) declared COVID-19, which is caused by SARS-CoV-2, to be a public health emergency (World Health Organization, 2021c). The ongoing COVID-19 pandemic presents great threats to public health and significant challenges to global economic development. As of February 20, 2022, the COVID-19 epidemic has caused more than 422 million confirmed cases worldwide and a number of confirmed deaths approaching 5.8 million (World Health Organization, 2021a). The rapid mutation rate of the COVID-19 virus is also an important reason for its great and long-lasting impact (Yu et al., 2022): the successive emergence of SARS-CoV-2 variants has caused a worldwide multi-wave epidemic of COVID-19. In November 2021, the Omicron variant (B.1.1.529) was first discovered in Gauteng, South Africa; its swift spread led to the fourth wave of the COVID-19 pandemic (Maslo et al., 2022; Planas et al., 2022).
In the first 2 months after its discovery, the Omicron variant was identified in 110 countries across all six WHO regions (World Health Organization, 2021b), and its infection rate has been identified as being significantly faster than that of the Delta variant (Wei et al., 2021; World Health Organization, 2021b). As a result of its fast spread, Omicron has come under intense study (Cameroni et al., 2022; Cao et al., 2022; Cele et al., 2022; Liu et al., 2022; Planas et al., 2022). Using high-throughput yeast display screening, Cao et al. (2022) found that mutations present in Omicron allowed it to escape from more than 85% of tested neutralizing antibodies targeting its receptor-binding domain. Furthermore, Liu et al. (2022) found that Omicron is markedly resistant to neutralization by serum not only from convalescent patients but also from individuals vaccinated with one of four widely used COVID-19 vaccines (Pfizer, Moderna, J & J, and AstraZeneca). Planas et al. (2022) found that Omicron often escapes from monoclonal and vaccine-elicited antibodies. However, their results also included that antibodies generated by a recent booster vaccine dose could neutralize Omicron (Planas et al., 2022), albeit less effectively than other SARS-CoV-2 variants.
Mathematical models can be used to understand the dynamics of SARS-CoV-2 variants and help inform effective control strategies, and many modeling studies have hence been performed to make projections about the spread of these variants (Liu and Rocklöv, 2021; Yu et al., 2022). In a literature review, Liu and Rocklöv (2021) found that estimates of Delta's basic reproductive number ranged from 3.2 to 8, with a mean of 5.08, much higher than that of the ancestral strain. Yu et al. (2022) used mathematical models to estimate the relative transmissibility of Omicron using variant proportion data in South Africa, finding that it is more transmissible than Delta by a factor of approximately 3.8. Similarly, Chen et al. (2022) found that Omicron is more transmissible than Delta by a factor of 2.8 using an AI model. However, as the Omicron variant is still new, our understanding of its infectivity and fatality, and how these compare to other variants, is still evolving. Additionally, to the best of our knowledge, the question of how the epidemiological properties of Omicron compare to those of other fast-spreading viruses such as influenza has not yet been addressed. To evaluate the properties of the Omicron variant, we develop a mathematical model in which transmission, death, and vaccination rates are all time-varying.
We fit our model using the numbers of new confirmed cases of the Delta and Omicron variants, and all other variants in aggregate (e.g., Beta, Gamma, Epsilon), in South Africa, the United States, and Canada, and the number of individuals who have been fully vaccinated and who have received booster doses in those countries. After this, we computed the transmission rate, death rate, and effective reproduction number of Delta, Omicron, and the other variants in each study jurisdiction. We also computed these statistics for seasonal influenza and the 2009 strain of H1N1 influenza in the United States and Canada by fitting case and death data to another model (see Supporting Information Appendix). The epidemic curves produced by our model fit the data very well, indicating that our model captures the dynamics of the COVID-19 virus variants.
Materials and Methods
Data collection and analysis
To understand the impact of the SARS-CoV-2 variants on the spread of the COVID-19 epidemic, we collected data on the proportions of all SARS-CoV-2 infections that each of the variants accounted for (Our World in Data, 2021). These data were reported every 2 weeks from May 18, 2020, to February 7, 2022, in South Africa, the United States, and Canada (Figure A1). We also collected daily numbers of new confirmed cases and new deaths (Centers for Disease Control and Prevention, 2021; Our World in Data, 2021) and the number of fully vaccinated individuals, broken down into those with and without booster doses (Mathieu et al., 2021). We used the relative prevalence of the SARS-CoV-2 variants to calculate the daily numbers of new confirmed cases infected by Delta, Omicron, and all other variants in our study countries. Similarly, we calculated the daily number of new deaths in the study countries from Delta, Omicron, and all other variants by scaling total COVID-19 deaths by the proportions of each SARS-CoV-2 variant from 2 weeks before.
Model formulation
To mimic the spread of COVID-19, we constructed a disease transmission model with vaccination. The total population (denoted by ) is divided into eight classes, namely , , , , , , , and , representing the numbers of individuals who were (1) susceptible; (2) fully vaccinated, without having received booster doses; (3) fully vaccinated, with booster doses; (4) exposed; (5) infectious (asymptomatic); (6) infectious (symptomatic); (7) recovered; and (8) dead of the infection, respectively. The baseline infection probability among susceptible and vaccinated individuals is defined as
where denotes the transmission rate, , , and are the probabilities of randomly encountering infected individuals in compartments , , and , respectively, and and represent the probabilities of transmission in asymptomatic and exposed individuals, respectively. Susceptible and vaccinated individuals who have encountered SARS-CoV-2 become exposed at the rates , , and , where and denote the reduction in susceptibility to infection conferred by a completed vaccine series and a full vaccine series plus a booster dose, respectively. Susceptible individuals transfer to the fully vaccinated and recovered classes by vaccination at the rates and , respectively, where is the probability of complete protection from COVID-19 after full vaccination, and denotes the rate of vaccine series completion. and are the analogous values for individuals receiving a booster dose. Exposed individuals can become asymptomatic and symptomatic infected individuals at the rates and , respectively, where denotes the proportion of infected individuals who are asymptomatic and denotes the mean length of COVID-19 incubation period. Eventually, all cases in and will either recover or die at the transition rates and , which are the reciprocals of asymptomatic and symptomatic infection periods, respectively. The death rate of asymptomatic infected individuals is extremely low, so we only considered the death rate of symptomatic individuals, , where denotes their probability of death. Our model is given by
| (1) |
The model time step is 1 day. , , , and are all time-varying parameters, which were dynamically fitted using an Markov Chain Monte Carlo (MCMC) method (Haario et al., 2001, 2006). All parameters are listed in Table 1 ; model parametrization is detailed in the Supporting Information Appendix.
Table 1.
The parameter description of Model (1).
| Parameters | Description (units) | Value | Source |
|---|---|---|---|
| Mean duration of COVID-19’s incubation period (day) | 5.2 | Li et al. (2020) | |
| Probability of transmission for asymptomatic infected individuals (dimensionless) | 0.55 |
Li et al. (2020) Hao et al. (2020) |
|
| Probability of transmission for exposed individuals (dimensionless) | 0.55 |
Li et al. (2020) Hao et al. (2020) |
|
| Proportion of infected individuals who are asymptomatic (dimensionless) | 60% | Qiu (2020) | |
| Infectious period of asymptomatic individuals (day) | 8 | Maier and Brockmann (2020) | |
| Infectious period of symptomatic individuals (day) | 14 | Kumar et al. (2021) | |
| Protection against infection generated by a full vaccine series, without booster doses (dimensionless) | 80% or 20% | Ye et al. (2022), AHIR (2021), Ontario (2021) | |
| Protection against infection generated by receiving a booster dose (dimensionless) | 80% or 70% |
Ye et al. (2022) National Health Service (2022) |
|
| Probability that an individual will be completely protected from COVID-19 after two doses of vaccine (dimensionless) | 0 | estimated | |
| Probability that an individual will be completely protected from COVID-19 after booster doses of vaccine (dimensionless) | 0 | Estimated | |
| Probability of deaths among symptomatic individuals (dimensionless) | see Figures 1, B2, B3 | Estimated | |
| Proportion of fully vaccinated individuals who have not received any booster doses () | see Supporting Information | MCMC | |
| Proportion of fully vaccinated individuals who have received booster doses () | see Supporting Information | MCMC | |
| Transmission rate () | see Figures 1, B2, B3 | MCMC | |
| Total population (South Africa) Total population (United States) Total population (Canada) |
55700000 329500000 38000000 |
World Health Organization (2021e) |
According to the next generation matrix approach (van den Driessche and Watmough, 2002), the effective reproduction number, , can be expressed as
| (2) |
incorporating the daily numbers of new cases generated by exposed, symptomatic, and asymptomatic individuals. The basic reproduction number is commonly used to measure transmission potential at the beginning of an epidemic (Diekmann et al., 1990). However, transmission potential varies over the course of an outbreak, and it can be measured at any time by the effective reproduction number.
Results
Infectivity
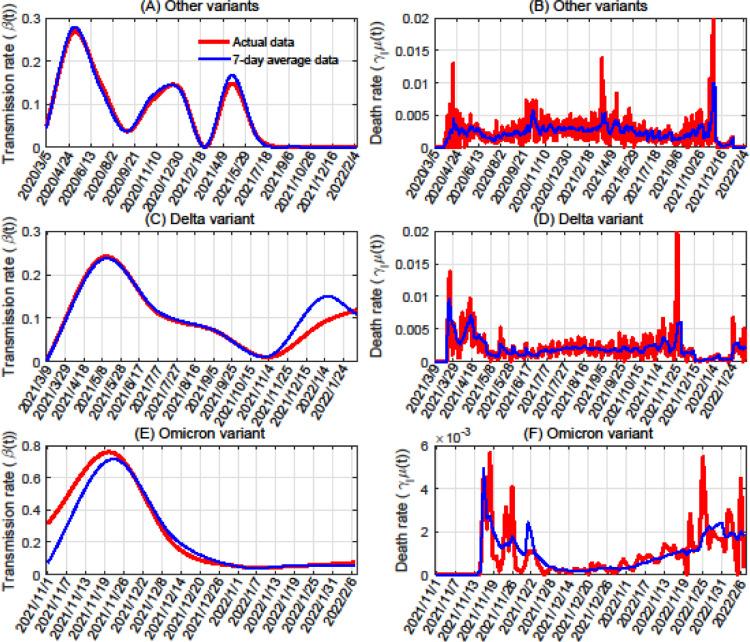
For each SARS-CoV-2 variant, we estimated its transmission rates in South Africa, the United States, and Canada based on data from the time periods in each country during which it first emerged. We performed our analysis with both raw case numbers (Figures 1 , B2, and B3) and 7-day averaged data (Figures E10, E11, and E12). The two cubic splines that we used for transmission rate (based on the raw and smoothed data) were highly similar for all variants and countries (Figures 2 , E13, and E14), showing the robustness of our methods.
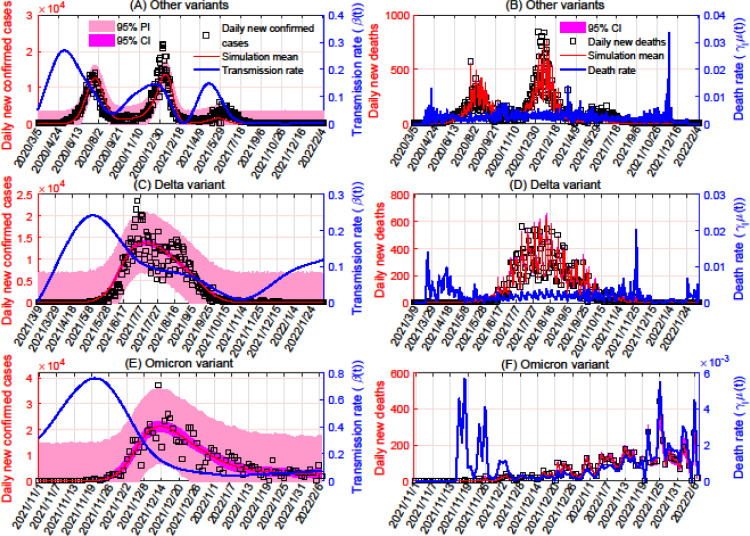
Figure 1.
Model fitting based on data for COVID-19 cases and deaths in South Africa. Panels A, C, and E show the fitting of the transmission rates for variants other than Delta and Omicron, the Delta variant, and the Omicron variant, respectively, based on daily numbers of new confirmed cases of these variants. Panels B, D, and F show the fitting of the variants’ death rates (in the same order), based on daily numbers of new deaths because of each variant. The transmission rates were taken to be cubic spline functions, with numbers of nodes equal to 13 for variants other than Delta and Omicron, seven for Delta, and six for Omicron. In each subplot, the red curve represents the mean simulated number of cases or deaths, and the blue curve represents the transmission or death rate. The 95% PI and CI are plotted in pink and magenta, respectively. CI, confidence interval; PI, prediction interval.
Figure 2.
Estimation of transmission and death rates in South Africa using actual data versus a 7-day rolling average. Panels A, C, and E show transmission rates for variants besides Delta and Omicron, the Delta variant, and the Omicron variant, respectively. Panels B, D, and F show the variants’ death rates, in the same order.
In South Africa, the average transmission rate of the Omicron variant was 0.4201 from November 1, 2021, to December 26, 2021 (Figure 1A). For Delta, this was 0.1264 from March 9, 2021, to May 2, 2021 (Figure 1C), and for all other variants, it was 0.1629 from March 5, 2020, to April 28, 2020 (Figure 1E). We estimate that Omicron's transmission rate is 3.3 times that of Delta and 2.6 times that of the other variants in South Africa; these values are consistent with previous studies (Chen et al., 2022; Yu et al., 2022).
In the United States, Omicron's average transmission rate was 0.6794 from November 21, 2021, to December 26, 2021 (Figure B2A). This was 0.1981 for Delta from May 2, 2021, to June 6, 2021 (Figure B2C), and 0.3299 for other variants from January 23, 2020, to February 27, 2020 (Figure B2E). In the United States, the transmission rate of Omicron is higher than that of Delta and other variants by factors of 3.4 and 2.1, respectively.
In Canada, Omicron had an average transmission rate of 0.5735 from November 15, 2021, to December 26, 2021 (Figure B3A), whereas that of Delta was 0.0373 from February 23, 2021, to April 5, 2021 (Figure B3C), and the rate for the other variants was 0.1693 from January 26, 2020, to March 7, 2020 (Figure B3E). Omicron's transmission rate in Canada is 15.4 times and 3.4 times that of Delta and other variants, respectively.
Fatality
We estimated the lethality of the different variants by using their average death rates. For Delta, Omicron, and other variants in aggregate, these are 0.0020, , and 0.0022 in South Africa (Figures 1B, D, F), respectively. There, Delta's death rate is 2.3 times that of Omicron, whereas the death rate of the variants other than Delta and Omicron is 1.1 times and 2.5 times those of Delta and Omicron, respectively. In the United States, we found that the death rates of Delta, Omicron, and the other variants are, in order, , , and 0.0019 (Figures B2B, D, F). There, the death rate of Delta is 4.7 times that of Omicron, and the death rate of the other variants is 10.8 times that of Omicron. In Canada, the death rate of Delta is , whereas this is for Omicron and 0.0014 for the other variants (Figures B3B, D, F). Delta's death rate in Canada is higher than that of Omicron by a factor of 3.1; for the other variants, this factor is 7.4. The previously mentioned analysis shows that Delta and especially Omicron have lower death rates than previous variants.
Effective reproduction number
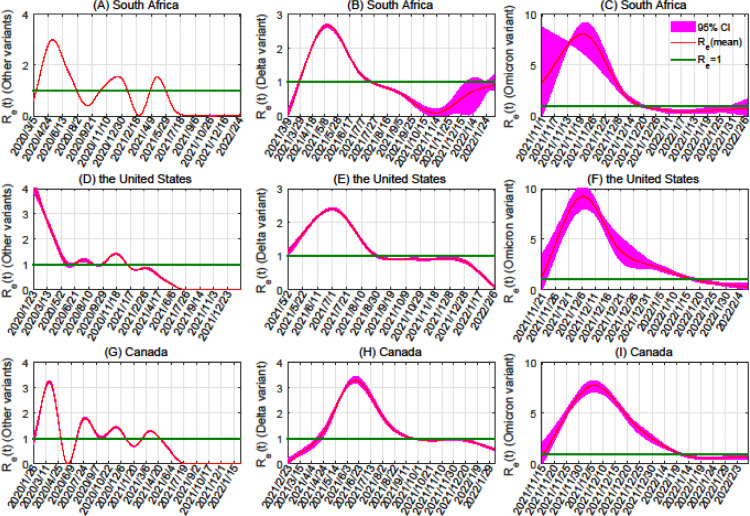
To calculate the effective reproduction numbers for each SARS-CoV-2 variant, we substituted the estimated parameters into Eq. (2) (Figure 3 ). We subsequently found the average effective reproduction numbers for the considered regions, considering the time from the date the first case was reported until 1 month later. We found that for Omicron, the average effective reproduction numbers in South Africa, the United States, and Canada are 6.39, 6.34, and 5.44, respectively. For Delta, these are 0.78, 1.50, and 0.31, and for variants other than Omicron and Delta, these are 1.24, 3.74, and 1.57. Thus, Omicron's average effective reproduction number is 8.2 times that of Delta in South Africa, 4.2 times that of Delta in the United States, and 17.5 times that of Delta in Canada. Likewise, the average effective reproduction number for Omicron was 5.2, 1.7, and 3.5 times that of variants other than Omicron and Delta in those three countries.
Figure 3.
Effective reproduction number for variants of SARS-CoV-2. Each row of graphs shows different variants within a given country (from top to bottom: South Africa, the United States, and Canada). The columns show results for particular variants (from left to right: variants other than Delta and Omicron, the Delta variant, the Omicron variant). The 95% CIs are plotted in magenta. CI, confidence interval; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Booster dose effectiveness
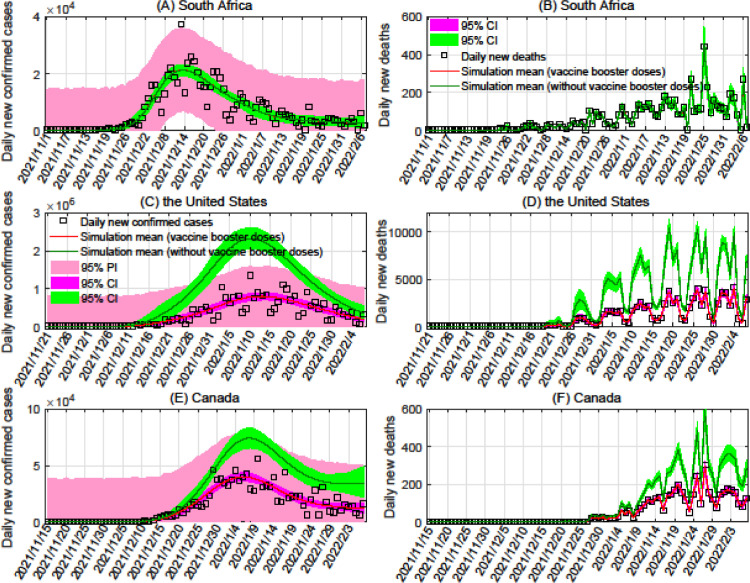
We additionally determined the protection rate of booster doses against infection by Omicron in the United States and Canada, as shown in Figure 4 (Because the distribution of booster doses in South Africa began on January 17, 2022, we did not consider the effectiveness of booster doses on Omicron there.). As of February 7, 2022, we found that vaccination has reduced the number of infected people by and in the United States and Canada, respectively, and reduced the number of deaths by and in those countries. These results imply that with 70% protection against Omicron (see Table 1), booster doses of currently available vaccines can significantly reduce mortality.
Figure 4.
Effectiveness of vaccines against the Omicron variant in South Africa, the United States, and Canada. The black boxes, red curves, and green curves represent the number of reported cases or deaths, simulated mean under the scenario where booster doses are administered at rates fit to current data, and simulated mean under the scenario where no booster doses are administered, respectively.
COVID-19 and seasonal influenza
To estimate the transmission and death rates of seasonal influenza, we collected weekly numbers of new confirmed cases and deaths from August 6, 2017, to December 22, 2019, in the United States and Canada. We used these to fit a seasonal influenza transmission model (see Supporting Information Appendix). Our parameter fitting, using the same methods as for model (1), is shown in Figures D6 and D7. We then compared these rates with the corresponding ones for Omicron over the full course of its outbreak to capture transmission dynamics in all outbreak phases. We chose a longer interval to model dynamics over because seasonal influenza is not an emerging disease. In the United States, the average transmission rates of seasonal influenza and Omicron were 0.120 and 0.407, respectively; the rate for seasonal influenza ranged from 0.041 to 0.202, whereas that of Omicron ranged from 0.0418 to 1.064. The average death rate of seasonal influenza in the United States was 0.0064 (ranging from 0.0013 to 0.0202), whereas that of Omicron was (ranging from 0 to ). In Canada, seasonal influenza and Omicron had average transmission rates of 0.158 (ranging from 0.0183 to 0.266) and 0.360 (ranging from 0.0817 to 0.838), respectively. The average death rates of seasonal influenza and Omicron were 0.0019 and , respectively, with ranges from 0 to 0.0816 for seasonal influenza and from 0 to 0.0018 for Omicron. These results indicate that Omicron's profile (high transmissibility, low mortality) is a more exaggerated version of seasonal influenza, suggesting that Omicron outbreaks may be more like those of seasonal influenza than those of Delta.
COVID-19 and 2009 H1N1 influenza
To compare the transmission and death rates of the 2009 strain of H1N1 influenza with those of Omicron, we collected daily numbers of cumulative confirmed cases and deaths of H1N1 from April 23, 2009, to July 6, 2009, in the United States and Canada. We used the same model for H1N1 as for seasonal influenza; the parameter fitting is shown in Figures D8 and D9 (see Supporting Information Appendix). We subsequently found the average transmission and death rates for the considered regions, calculated using the rates from the date the first case was reported until 2 months later. In the United States, the average transmission rate of H1N1 was 0.347 (ranging from 0.0783 to 0.590), and that of Omicron over a similar time period of approximately 2 months from the first detection was 0.509 (ranging from 0.142 to 1.064). The average death rates of H1N1 and Omicron were 0.0020 and , respectively, with ranges from to 0.0125 for H1N1 and from 0 to for Omicron. In Canada, the average transmission rates of H1N1 and Omicron were 0.320 and 0.470, respectively; the rate for H1N1 ranged from 0.0903 to 0.751, whereas that for Omicron ranged from 0.0893 to 0.8383. The average death rate of H1N1 in Canada is (ranging from to 0.0024); for Omicron, this is (ranging from 0 to ). Hence, compared with those of H1N1, Omicron's average transmission rate was 46% higher in the United States and 47% higher in Canada, whereas Omicron's average death rate was 97% lower in the United States and 90% lower in Canada, results analogous to how Omicron is more transmissible and less deadly than seasonal influenza.
Discussion
The successive emergence of COVID-19 virus variants has caused multiple COVID-19 outbreak waves across the world. In October 2020, the Delta variant (B.1.617.2) was discovered in Maharashtra, India (del Rio et al., 2021), which was a driving factor in the second wave of COVID-19 in that country. In November 2021, the Omicron variant (B.1.1.529) was first discovered in Gauteng, South Africa, and quickly spread to other countries (Maslo et al., 2022; Planas et al., 2022). This led to the fourth wave of the COVID-19 pandemic: Omicron replaced Delta as the dominant strain after 8 weeks in South Africa and later accounted for more than 90% of all cases after 8 and 10 weeks of circulation in the United States and Canada, respectively.
To understand the SARS-CoV-2 variants’ epidemiological properties, we developed a model describing their dynamics, featuring time-varying rates of transmission, death, and vaccination. We created an inverse method to estimate the time-varying death rate of the COVID-19 virus variants, which greatly simplifies the complexity of parameter estimation. Using this method, we found that the transmission rate of Omicron is 3.3 times that of Delta in South Africa, and the death rate of Delta is 2.3 times that of Omicron there. Correspondingly, these numbers are 3.4 and 4.7 in the United States and 15.4 and 3.1 in Canada. This makes it clear that Omicron is more infective but less lethal than Delta. We also found that with a complete vaccine series plus a booster dose providing 70% protection against Omicron, vaccination has reduced the number of infected people by and in the United States and Canada, respectively.
During the COVID-19 pandemic, comparisons with seasonal influenza have been frequently made by public officials (Faust and del Rio, 2020), and influenza has been used as a point of reference for clinical studies of patients with COVID-19 (Brehm et al., 2021; Xie et al., 2020). Previous variants of SARS-CoV-2 were characterized by higher mortality than seasonal influenza, even after accounting for the underreporting of influenza deaths (Faust and del Rio, 2020). However, our results indicate that the Omicron variant has a lower death rate than seasonal influenza, the opposite of other SARS-CoV-2 variants. Similarly, the COVID-19 pandemic was compared with H1N1 in 2009, only a few months after the beginning of the former (Jhaveri, 2020). Clinically, the two diseases result in similar immune responses (Morris et al., 2021), and many computed tomography imaging features are common to both (Yin et al., 2020). In contrast to early variants of SARS-CoV-2, which were observed to have higher mortality rates than 2009 H1N1 influenza (da Costa et al., 2020), we found Omicron to be less deadly than that strain. We also found Omicron to be more transmissible than the tested varieties of influenza. Our results indicate that although Omicron must be taken seriously because of its high infectivity, its low fatality suggests that it can serve as a less dangerous replacement for other SARS-CoV-2 strains, outcompeting them but causing less damage.
We found that the Omicron variant is epidemiologically more similar to influenza than previous SARS-CoV-2 variants; this is evident by its unique mutations, which confer upon it a different evolutionary strategy (Du et al., 2022). Hence, we predict that methods for combating Omicron based on previous public health responses to seasonal influenza will be effective. Earlier in the COVID-19 pandemic, it was found that nonpharmaceutical interventions to suppress COVID-19 in China also had the effect of suppressing seasonal influenza cases because the two diseases share similar transmission methods (Lei et al., 2020). Seasonal influenza is characterized by wintertime outbreak peaks and yearly variability in epidemiological characteristics (Chowell et al., 2008). So far, the COVID-19 pandemic has also exhibited these features, with new SARS-CoV-2 variants emerging at least once per year. Further variants in Omicron's lineage have been observed (Desingu et al., 2022), and because of Omicron's ubiquity, the next dominant variant may be one of its descendants. Therefore, as the future of the COVID-19 pandemic may revolve around managing viruses with similar characteristics as Omicron, applying strategies originally designed for seasonal influenza will prove useful.
Our study still has several limitations. First, because the numbers of recovered and asymptomatic infected individuals are not publicly available yet, our simulations only used incidence data, death cases, and the number of fully vaccinated individuals. Second, the numbers of new confirmed cases and deaths for each variant are intertwined with the reported data and hard to disentangle from it. We used the proportions of cases caused by each SARS-CoV-2 variant to calculate these numbers. Third, we used model (1) to fit three SARS-CoV-2 variants under the assumption that an individual can be simultaneously infected with different variants. Fourth, we did not consider human mobility and imported cases from overseas or the role of environmental factors; these will be studied when such data become available.
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Acknowledgments
Funding source
LX is funded by the National Natural Science Foundation of China 12171116 and Fundamental Research Funds for the Central Universities of China 3072020CFT2402. HW is partially supported by NSERC Individual Discovery Grant RGPIN-2020-03911 and NSERC Discovery Accelerator Supplement Award RGPAS-2020-00090.
Ethical approval statement
This article does not contain any studies involving animals or humans performed by any authors.
Author contributions
HW designed research; all authors conceived the work; SJ, KZ, and HW performed research; all authors analyzed data; LX, SJ, and KZ wrote the initial draft; all authors edited the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.05.031.
Appendix. Supplementary materials
References
- Africa Health Research Institute, 2021. https://www.santheafrica.org/. [Accessed 29 December 2021].
- Brehm TT, van der Meirschen M, Hennigs A, et al. Comparison of clinical characteristics and disease outcome of COVID-19 and seasonal influenza. Sci Rep. 2021;11:5803. doi: 10.1038/s41598-021-85081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E, Bowen JE, Rosen LE, et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, Trends in number of COVID-19 cases and deaths in the US reported to CDC, by state/territory. https://covid.cdc.gov/covid-data-tracker/#trends_dailycases, 2021 (accessed 27 December 2021).
- Chen J, Wang R, Gilby NB, Wei GW. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62:412–422. doi: 10.1021/acs.jcim.1c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell G, Miller MA, Viboud C. Seasonal influenza in the United States, France, and Australia: transmission and prospects for control. Epidemiol Infect. 2008;136:852–864. doi: 10.1017/S0950268807009144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa VG, Saivish MV, Santos DER, de Lima Silva RF, Moreli ML. Comparative epidemiology between the 2009 H1N1 influenza and COVID-19 pandemics. J Infect Public Health. 2020;13:1797–1804. doi: 10.1016/j.jiph.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio C, Malani PN, Omer SB. Confronting the Delta variant of SARS-CoV-2, summer 2021. JAMA. 2021;326:1001–1002. doi: 10.1001/jama.2021.14811. [DOI] [PubMed] [Google Scholar]
- Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol. 2022;94:1808–1810. doi: 10.1002/jmv.27601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann O, Heesterbeek JA, Metz JA. On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. J Math Biol. 1990;28:365–382. doi: 10.1007/BF00178324. [DOI] [PubMed] [Google Scholar]
- Du X, Tang H, Gao L, et al. Omicron adopts a different strategy from Delta and other variants to adapt to host. Signal Transduct Target Ther. 2022;7:45. doi: 10.1038/s41392-022-00903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust JS, Del Rio C. Assessment of deaths from COVID-19 and from seasonal influenza. JAMA Intern Med. 2020;180:1045–1046. doi: 10.1001/jamainternmed.2020.2306. [DOI] [PubMed] [Google Scholar]
- Haario H, Laine M, Mira A, Saksman E. DRAM: efficient adaptive MCMC. Stat Comput. 2006;16:339–354. [Google Scholar]
- Haario H, Saksman E, Tamminen J. An adaptive Metropolis algorithm. Bernoulli. 2001;7:223–242. [Google Scholar]
- Hao X, Cheng S, Wu D, et al. Reconstruction of the full transmission dynamics of COVID-19 in Wuhan. Nature. 2020;584:420–424. doi: 10.1038/s41586-020-2554-8. [DOI] [PubMed] [Google Scholar]
- Jhaveri R. Echoes of 2009 H1N1 influenza pandemic in the COVID pandemic. Clin Ther. 2020;42:736–740. doi: 10.1016/j.clinthera.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, AbdulRahman A, AlAli S, et al. Time Till Viral Clearance of Severe Acute Respiratory Syndrome Coronavirus 2 Is Similar for Asymptomatic and Non-critically Symptomatic Individuals. Front Med. 2021;8:616927. doi: 10.3389/fmed.2021.616927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Xu M, Wang X, et al. Nonpharmaceutical interventions used to control COVID-19 reduced seasonal influenza transmission in China. J Infect Dis. 2020;222:1780–1783. doi: 10.1093/infdis/jiaa570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Rocklöv J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J Travel Med. 2021;28:taab124. doi: 10.1093/jtm/taab124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier BF, Brockmann D. Effective containment explains subexponential growth in recent confirmed COVID-19 cases in China. Science. 2020;368:742–746. doi: 10.1126/science.abb4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 Omicron wave compared with previous waves. JAMA. 2022;327:583–584. doi: 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu E, Ritchie H, Ortiz-Ospina E, et al. A global database of COVID-19 vaccinations. Nat Hum Behav. 2021;5:947–953. doi: 10.1038/s41562-021-01122-8. [DOI] [PubMed] [Google Scholar]
- Morris G, Bortolasci CC, Puri BK, et al. The cytokine storms of COVID-19, H1N1 influenza, CRS and MAS compared. Can one sized treatment fit all? Cytokine. 2021;144 doi: 10.1016/j.cyto.2021.155593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Health Service, Coronavirus (COVID-19). 2022. https://www.nhs.uk/service-search/find-a-walk-incoronavirus-covid-19-vaccination-site. [Accessed 2 February 2022].
- Ontario Dashboard, Current COVID-19 Risk in Ontario by Vaccination Status. 2021. https://covid19-sciencetable.ca/ontario-dashboard/. [Accessed 29 December 2021].
- Our World in Data, SARS-CoV-2 variants in analyzed sequences in South Africa. https://ourworldindata.org/grapher/covid-variants-area?country=∼ZAF, 2021 (accessed 19 December 2021).
- Planas D, Saunders N, Maes P, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Qiu J. Covert coronavirus infections could be seeding new outbreaks. Nature. 2020 doi: 10.1038/d41586-020-00822-x. [DOI] [PubMed] [Google Scholar]
- van den Driessche P, Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math Biosci. 2002;180:29–48. doi: 10.1016/s0025-5564(02)00108-6. [DOI] [PubMed] [Google Scholar]
- Wei C, Shan KJ, Wang W, Zhang S, Huan Q, Qian W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J Genet Genomics. 2021;48:1111–1121. doi: 10.1016/j.jgg.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, Coronavirus disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Update. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/, 2021a (accessed 29 December 2021).
- World Health Organization, Enhancing readiness for Omicron (B.1.1.529): technical brief and priority actions for Member States. https://www.who.int/docs/default-source/coronaviruse/2021-12-23-global-technical-brief-and-priority-action-on-omicron.pdf?sfvrsn=d0e9fb6c_8, 2021b (accessed 29 December 2021).
- World Health Organization, WHO Director-General's statement on IHR Emergency Committee on Novel Coronavirus. https://www.who.int/director-general/speeches/detail/who-director-general-s-statement-on-ihr-emergency-committee-on-novel-coronavirus, 2021c (accessed 29 December 2021).
- World Health Organization, Countries and Centers. 2021e. https://www.paho.org/en/countries-and-centers. [Accessed 29 December 2021].
- Xie Y, Bowe B, Maddukuri G, Al-Aly Z. Comparative evaluation of clinical manifestations and risk of death in patients admitted to hospital with COVID-19 and seasonal influenza: cohort study. BMJ. 2020;371:m4677. doi: 10.1136/bmj.m4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Zhang Q, Wei X, et al. Equitable access to COVID-19 vaccines makes a life-saving difference to all countries. Nat Hum Behav. 2022;6:207–216. doi: 10.1038/s41562-022-01289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Kang Z, Yang D, Ding S, Luo H, Xiao E. A comparison of clinical and chest CT findings in patients with influenza A (H1N1) virus infection and coronavirus disease (COVID-19) AJR Am J Roentgenol. 2020;215:1065–1071. doi: 10.2214/AJR.20.23214. [DOI] [PubMed] [Google Scholar]
- Yu Y, Liu Y, Zhao S, et al. A simple model to estimate the transmissibility of SARS-CoV-2 Beta, Delta and Omicron variants in South Africa. Social Science Research Network. 2022 doi: 10.3934/mbe.2022485. 5 January https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3989919 (accessed 22 December 2021) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.