Abstract
An outbreak of betacoronavirus SARS-CoV-2 began in Wuhan, China in December 2019. COVID-19, the disease associated with infection, rapidly spread to produce a global pandemic. We report development of a rapid (<40 min), easy-to-implement and accurate CRISPR-Cas12-based lateral flow assay for detection of SARS-CoV-2 from respiratory swab RNA extracts. We validated our method using contrived reference samples and clinical samples from US patients, including 36 patients with COVID-19 infection and 42 patients with other viral respiratory infections. Our CRISPR-based DETECTR assay provides a visual and faster alternative to the US CDC SARS-CoV-2 real-time RT-PCR assay, with 95% positive predictive agreement and 100% negative predictive agreement..
Editor’s summary
SARS-CoV-2 in patient samples is detected in under an hour using a CRISPR-based lateral flow assay.
Over the past 40 years, there have been recurrent large-scale epidemics from emerging viruses such as human immunodeficiency virus (HIV), SARS and MERS coronaviruses, 2009 pandemic influenza H1N1 virus, Ebola virus (EBOV), Zika virus (ZIKV) and most recently SARS-CoV-21,2. All of these epidemics most likely resulted from an initial zoonotic animal-to-human transmission event, with either clinically apparent or occult spread into vulnerable human populations. Each time, a lack of rapid, accessible, and accurate molecular diagnostic testing has hindered the public health response to the emerging viral threat.
In early January 2020, a cluster of cases of pneumonia from a novel coronavirus, SARS-CoV-2 (with the disease referred to as COVID-19), was reported in Wuhan, China1,2. This outbreak has spread rapidly, with over 1.2 million reported cases and 64,500 deaths worldwide as of April 4th, 20203. Person-to-person transmission from infected individuals with no or mild symptoms has been reported4,5. Assays using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) approaches for detection of the virus in 4–6 hours have been developed by several laboratories, including an Emergency Use Authorization (EUA)-approved assay developed by the US CDC6. However, the typical turnaround time for screening and diagnosing patients with suspected SARS-CoV-2 has been >24 hours given the need to ship samples overnight to reference laboratories. Although serology tests are rapid and require minimal equipment, their utility may be limited for diagnosis of acute SARS-CoV-2 infection, because it can take up to 5 days following symptom onset for a patient to mount a detectable antibody response7. To accelerate clinical diagnostic testing for COVID-19 in the United States, on February 28th, 2020, the FDA permitted individual clinically licensed laboratories to report the results of in-house developed SARS-CoV-2 diagnostic assays while awaiting results of an EUA submission for approval8.
Here we report the development and initial validation of a CRISPR -Cas12 based assay9–12 for detection of SARS-CoV-2 from extracted patient sample RNA, called SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR). This assay performs simultaneous reverse transcription and isothermal amplification using loop-mediated amplification (RT-LAMP)13 for RNA extracted from nasopharyngeal or oropharyngeal swabs in universal transport media (UTM), followed by Cas12 detection of predefined coronavirus sequences, after which cleavage of a reporter molecule confirms detection of the virus. We first designed primers targeting the E (envelope) and N (nucleoprotein) genes of SARS-CoV-2 (Fig. 1a). The primers amplify regions that overlap the WHO assay (E gene region) and US CDC assay (N2 region in the N gene)6,14, but are modified to meet design requirements for LAMP. We did not target the N1 and N3 regions used by the US CDC assay, as these regions lacked suitable protospacer adjacent motif (PAM) sites for the Cas12 guide RNAs (gRNAs). Next, we designed Cas12 gRNAs to detect three SARS-like coronaviruses (SARS-CoV-2 accession NC_045512, bat SARS-like coronavirus (bat-SL-CoVZC45, accession MG772933), and SARS-CoV, accession NC_004718) in the E gene and specifically detect SARS-CoV-2 only in the N gene (Supplementary Fig. 1). This design is similar to those used by the WHO and US CDC assays, which use multiple amplicons with probes that are either specific to SARS-CoV-2 or are capable of identifying related SARS-like coronaviruses.
Figure 1.
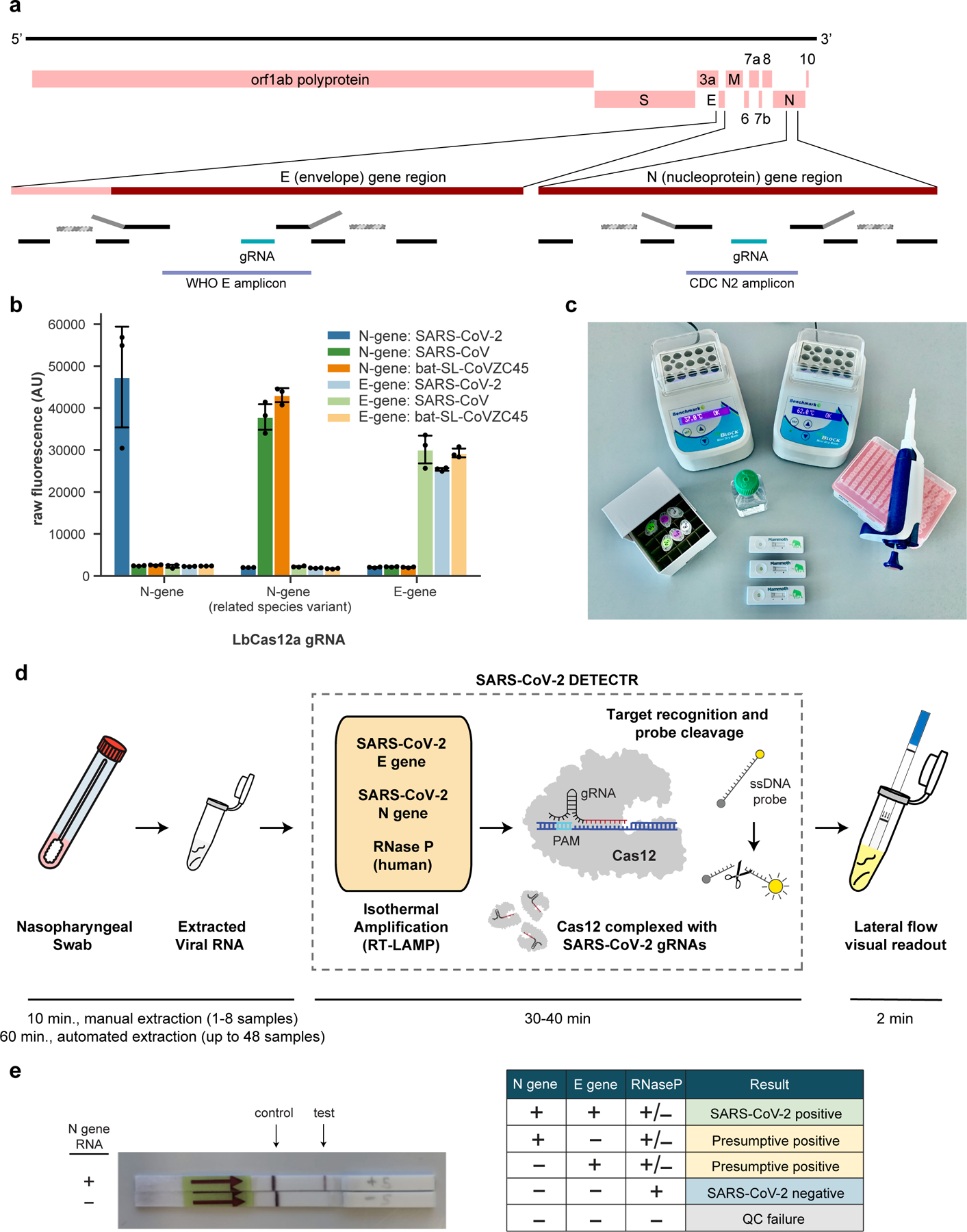
A CRISPR-Cas12 based assay for detection of SARS-CoV-2. (a) Genome map showing primers, probes and gRNAs. Visualization of primers and probes on the SARS-CoV-2 genome. RT-LAMP primers are indicated by black rectangles, the binding position of the F1c and B1c half of the FIP primer (grey) is represented by a striped rectangle with dashed borders. (b) gRNA specificity. Cas12 gRNAs are programmed to specifically target SARS-CoV-2 or broadly detect related coronavirus strains. The N gene gRNA used in the assay (left) is specific for SARS-CoV-2, whereas the E gene gRNA is able to detect 3 SARS-like coronavirus strains (right). A separate N gene gRNA designed to target SARS-CoV and a bat coronavirus fails to detect SARS-CoV-2 (middle). (c) Minimum equipment needed to run protocol. With appropriate biosafety level 2 requirements, the minimum equipment required to run the protocol includes Eppendorf tubes with reagents, heat blocks or water bath (37°C and 62°C), nuclease-free water, pipettes and tips, and lateral flow strips. (d) Schematic of SARS-CoV-2 DETECTR workflow. Conventional RNA extraction can be used as an input to DETECTR (LAMP pre-amplification and Cas12-based detection for E gene, N gene and RNase P), which is visualized by a fluorescent reader or lateral flow strip. (e) Lateral flow strip assay readout. A positive result requires detection of at least one of the two SARS-CoV-2 viral gene targets (N gene or E gene, as indicated in the interpretation matrix).
Using synthetic, in vitro transcribed (IVT) SARS-CoV-2 RNA gene targets in nuclease-free water, we demonstrated that CRISPR-Cas12 based detection can distinguish SARS-CoV-2 with no cross-reactivity for related coronavirus strains using the N gene gRNA and with the expected cross-reactivity for the E gene gRNA (Fig. 1b, Supplementary Fig. 2). We then optimized the conditions for the SARS-CoV-2 DETECTR assay on the E gene, N gene and the human RNase P gene as a control, which consists of an RT-LAMP reaction at 62°C for 20–30 min and Cas12 detection reaction at 37°C for 10 min. The DETECTR assay can be run in approximately 30–40 min and visualized on a lateral flow strip (Fig. 1c, d). The SARS-CoV-2 DETECTR assay is considered positive if there is detection of both the E and N genes, or presumptive positive if there is detection of either the E or N gene (Fig. 1e). This interpretation is consistent with that of current FDA EUA guidance and recently approved point-of-care diagnostics under the EUA15.
Visualization of the Cas12 detection reaction is achieved using a FAM-biotin reporter molecule and lateral flow strips designed to capture labeled nucleic acids (Fig. 2a)12. Uncleaved reporter molecules are captured at the first detection line (control line), whereas indiscriminate Cas12 cleavage activity generates a signal at the second detection line (test line). To compare the signal generated by Cas12 when using fluorescence or lateral flow, we carried out RT-LAMP using 5 fM or 0 fM IVT template using N gene primers and monitored the performance of the Cas12 readout on identical amplicons using a fluorescent plate reader and by lateral flow at 0, 2.5, 5, and 10 minutes (Fig. 2b, c). The Cas12 fluorescent signal was detectable in <1 minute, and a visual signal by lateral flow was achieved within 5 minutes.
Figure 2.
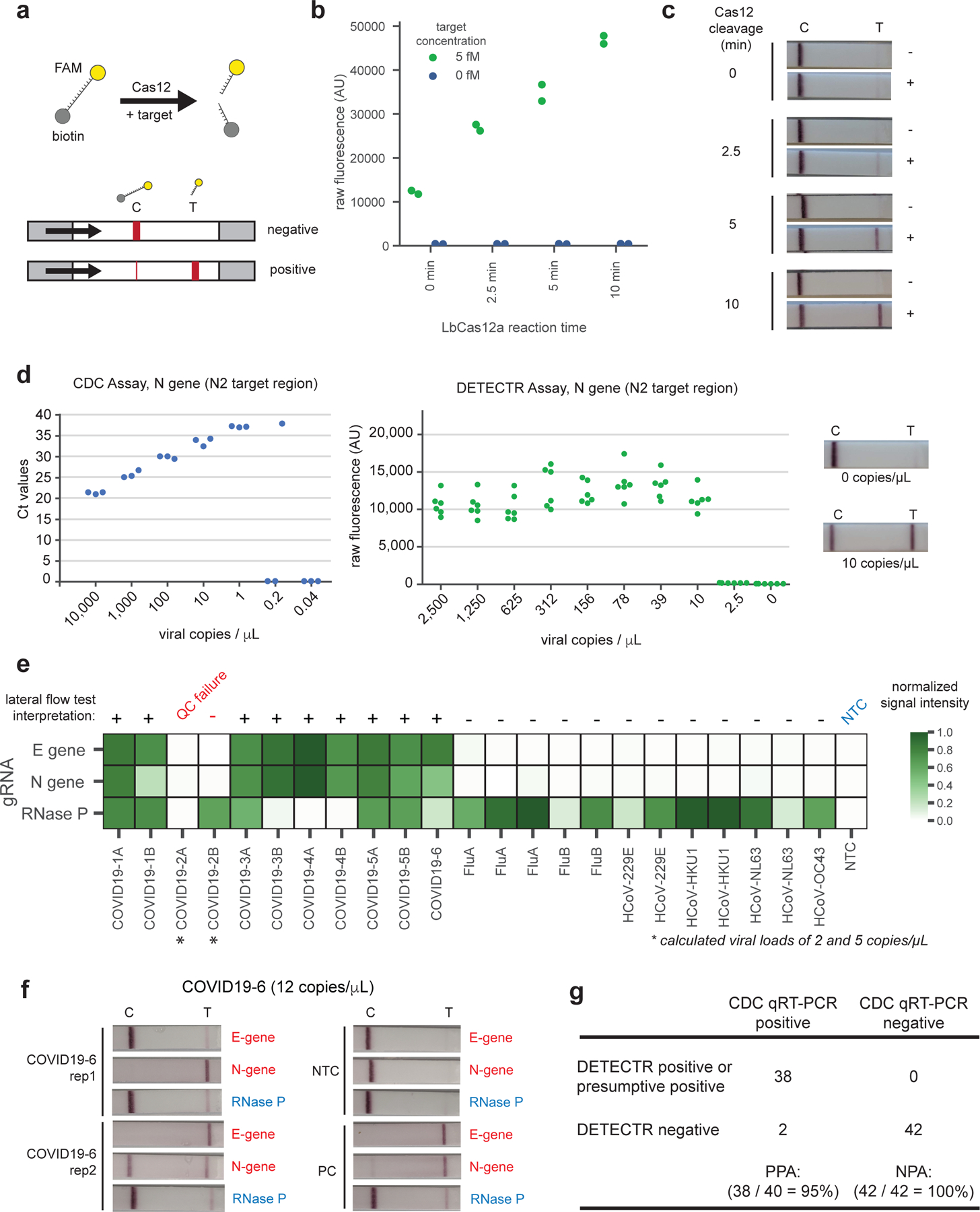
Detection of SARS-CoV-2 in contrived and clinical nasopharyngeal or oropharyngeal swab samples. (a) Schematic of DETECTR coupled with lateral flow readout. The intact FAM-biotinylated reporter molecule flows to the control capture line. Upon recognition of the matching target, the Cas-gRNA complex cleaves the reporter molecule, which flows to the target capture line. (b-c) Comparison of fluorescence to lateral flow. (b) Fluorescence signal of LbCas12a detection assay on RT-LAMP amplicon for SARS-CoV-2 N gene saturates within 10 min. RT-LAMP amplicon generated from 2 µL of 5 fM or 0 fM SARS-CoV-2 N gene IVT RNA by amplifying at 62°C for 20 min. Error bars: mean±SD.(c) LbCas12a on the same RT-LAMP amplicon produces visible signal through lateral flow assay within 5 min. (d) Limit of detection for CDC qPCR and DETECTR. Ct values using the CDC qPCR assay (n=3) and fluorescence values using SARS-CoV-2 DETECTR (n=6) using SARS-CoV-2 N2 gene IVT-RNA. Representative lateral flow results for the assay shown for 0 copies/µL and 10 copies/µL. (e) Patient sample DETECTR data. Clinical samples from 6 patients with COVID-19 infection (n=11, 5 replicates) and 12 patients infected with influenza or one of the 4 seasonal coronaviruses (HCoV-229E, HCoV-HKU1, HCoV-NL63, HCoV-OC43) (n=12) were analyzed using SARS-CoV-2 DETECTR. Signal intensities from lateral flow strips were quantified using ImageJ and normalized to the highest value within the N gene, E gene or RNase P set, with a positive threshold set at five standard deviations above background. The final determination for SARS-CoV-2 test results was based on the interpretation matrix in Fig. 1e, with results indicated above the heat map. (f) Lateral flow strips showing SARS-CoV-2 DETECTR assay results. Two replicate assays were performed using 2 µL of extracted RNA for each reaction (titer 12 copies/µL). Positive controls used IVT RNA for SARS-CoV-2 targets and total human RNA for RNase P. LbCas12a detection assays were run on lateral flow strips (TwistDx) and imaged after 3 min. (g) Performance characteristics of the fluorescent SARS-CoV-2 DETECTR assay. 83 clinical samples (41 COVID-19 positive, 42 negative) were evaluated using the fluorescent version of the SARS-CoV-2 DETECTR assay (Supp. Fig. 7 a, c, d). One sample (COVID19–3) was omitted due to failing assay quality control. Positive and negative calls are based on criteria described in Fig. 1e. Abbreviations: fM, femtomolar; NTC, no-template control; FLUA, influenza A virus; FLUB, influenza B virus; HCoV, human coronavirus; PPA, positive predictive agreement; NPA, negative predictive agreement.
We next compared the analytic limits of detection (LoD) of the RT-LAMP/Cas12 DETECTR fluorescent assay relative to the US FDA Emergency Use Authorization (EUA)-approved CDC assay for detection of SARS-CoV-2 (Table 1; Fig. 2d). A standard curve for quantitation was constructed using 7 dilutions of a control IVT viral nucleoprotein RNA (“CDC VTC nCoV Transcript”)6, with 3 replicates at each dilution (Fig. 2d, left; Extended Data 1). Ten two-fold serial dilutions of the same control nucleoprotein RNA were then used to run the fluorescent DETECTR assay, with 6 replicates at each dilution (Fig. 2d, center; Supplementary Fig. 3). We confirmed the ability of the assay to generate a visual signal by lateral flow at the limit of detection (Fig. 2d, right). In comparison to the quantitative CDC RT-PCR assay, our DETECTR lateral flow assay generates an easy-to-interpret qualitative readout for the presence or absence of the virus. The estimated LoD for the CDC assay tested by California Department of Public Health is 1 copy/µL reaction, consistent with the analytic performance in the FDA package insert, versus 10 copies/µL reaction for the DETECTR assay.
Table 1.
Comparison of the DETECTR (RT-LAMP/Cas12) assay with the CDC qRT-PCR assay for detection of SARS-CoV-2
| SARS-CoV-2 DETECTR, RT-LAMP/Cas12 | CDC SARS-CoV-2 qRT-PCR | |
|---|---|---|
| Target | E gene & N gene* | N gene (3 amplicons, N1, N2, and N3) |
| Sample control | RNase P | RNase P |
| Limit of Detection | 10 copies/µL input | 3.2 copies/µL input |
| Assay reaction time (approximate) | 30–40 min | 120 min |
| Assay sample-to-result time (approximate) | 45 min (with manual RNA extraction) | 4 hr (including RNA extraction) |
| Assay results | Qualitative | Quantitative |
| Assay components | RT-LAMP (62°C, 20–30 min) Cas12 (37°C, 10 min) Lateral flow strip (RT, 2 min; no additional time if using fluorescence readout) |
UDG digestion (25°C, 2 min), reverse transcription (50°C, 15 min), denature (95°C, 2 min) amplification, (95°C, 3 sec; 55°C 30 sec; 45 cycles) |
| Bulky instrumentation required | No | Yes |
| FDA EUA approval | Pending clinical validation | Yes |
E gene primers target same amplicon region as in the WHO protocol; N gene primers target same N2 amplicon region as in the CDC protocol
We then assessed the capability of the RT-LAMP assay to amplify SARS-CoV-2 nucleic acid directly from raw sample matrix consisting of nasopharyngeal swabs from asymptomatic donors placed in universal transport medium (UTM) or phosphate buffered saline (PBS) and spiked with SARS-CoV-2 IVT target RNA. Assay performance was diminished at reaction concentrations of ≥10% UTM and ≥10% PBS by volume, with estimated limits of detection decreasing to 15,000 and 500 copies/µL, respectively (Supplementary Fig. 4).
We next tested extracted RNA from 11 respiratory swab samples collected from 6 PCR-positive COVID-19 patients (COVID19–1A/B to COVID19–5A/B, where A refers to a nasopharyngeal swab and B refers to a oropharyngeal swab, and COVID19–6, a single nasopharyngeal swab) and 12 nasopharyngeal swab samples from patients with influenza (n=5), and common human seasonal coronavirus infections (n=7, representing OC43, HKU1, 229E and NL63) using SARS-CoV-2 DETECTR assay with fluorescence-based and lateral flow strip readouts (Fig. 2e, f; Supplementary Fig. 5, 6, 7). SARS-CoV-2 was detected in 9 of the 11 patient swabs and did not cross react with the other respiratory viruses. The two negative swabs from COVID-19 patients were confirmed to be below the established limit of detection.
Given the high concordance between lateral flow and fluorescence-based readouts (23 of 24 tests, or 95.8%) (Supplementary Fig. 7, 8), we used a fluorescence-based readout to blindly test an additional 60 nasopharyngeal swab samples from patients with acute respiratory infection for SARS-CoV2 using our DETECTR assay. Of the 60 samples, 30 were positive for COVID-19 infection by qRT-PCR testing and 30 were negative for COVID-19 infection but either positive for another viral respiratory infection by respiratory virus panel (RVP) multiplex PCR testing or negative by all testing. The positive predictive agreement (PPA) and negative predictive agreement (NPA) of SARS-CoV-2 DETECTR relative to the CDC qRT-PCR assay were 95% and 100%, respectively, for detection of the coronavirus in 83 total respiratory swab samples.
Here we combined isothermal amplification with CRISPR-Cas12 DETECTR technology to develop a rapid (30–40 min) test for detection of SARS-CoV-2 in clinical samples. The use of existing qRT-PCR based assays is hindered by the need for expensive lab instrumentation, and availability is currently restricted to public health laboratories. Importantly, the DETECTR assay developed here has comparable accuracy to qRT-PCR and use routine protocols and commercially available, “off-the-shelf” reagents. As the DETECTR assay uses similar sample collection and RNA extraction methods as the CDC assay and other qRT-PCR assays, it is subject to the same potential limitations with regards to the availability of personal protective equipment (PPE)16 and extraction kits and reagents17. However, some key advantages of our approach over qRT-PCR include isothermal signal amplification obviating the need for thermocycling, rapid turn-around time, single nucleotide target specificity, integration with accessible and easy-to-use reporting formats such as lateral flow strips, and no requirement for complex laboratory infrastructure. The time taken to develop and validate this SARS-CoV-2 DETECTR assay (<2 weeks for SARS-CoV-2, Supplementary Fig. 9). shows that this technology can be quickly mobilized to diagnose infections from emerging zoonotic viruses.
Although most cases of COVID-19 during the first month of the epidemic were traced back to the city of Wuhan in Hubei province in China, the ongoing increase in cases around the world seems now to be driven by local community transmission18,19. Therefore, there is an urgent public health need for rapid diagnostic tests for SARS-CoV-2 infection. The documented cases of asymptomatic infection and transmission in COVID-19 patients4,5 greatly increase the pool of individuals who need to be screened. Viral titers in hospitalized patients can fluctuate day-to-day with no correlation to disease severity20–22, and a single negative qRT-PCR test for SARS-CoV-2 does not exclude infection. SARS-CoV-2 is shed in stool2=3, raising the possibility of environmental contamination that might contribute to local disease outbreaks. Tests such as the DETECTR assay reported here are amenable to periodic repeat testing of patient samples. Clinical validation of this assay in response to recent draft guidance from the US FDA8 is ongoing in a CLIA (Clinical Laboratory Improvement Amendments)-certified microbiology laboratory.
The major pandemics and large-scale epidemics of the past half century have all been caused by zoonotic viruses. A diagnostic method that can be readily adapted to detect infection from emergent viruses is urgently needed. We report that our CRISPR-based DETECTR technology can be reconfigured within days to detect SARS-CoV-2 (Supplementary Fig. 9). The future development of portable microfluidic-based cartridges and lyophilized reagents to run the assay could enable point-of-care testing outside of the clinical diagnostic laboratory, such as airports, local emergency departments and clinics, and other locations.
ONLINE METHODS
Nucleic acid preparation
SARS-CoV-2 target sequences were designed using all available genomes available from GISAID24 as of January 27, 2020. Viral genomes were aligned using Clustal Omega. Next, LbCas12a target sites on the SARS-CoV-2 genome were filtered against SARS-CoV, two bat-SARS-like-CoV genomes and common human coronavirus genomes. Compatible target sites were compared to those used in current protocols from the CDC and WHO. LAMP primers for SARS-CoV-2 were designed against regions of the N gene and E gene using PrimerExplorer v5 (https://primerexplorer.jp/e/) with compatible gRNAs. As a sample control, a compatible gRNA was designed to previously published RNase P POP7 RT-LAMP primers25.
Target RNAs were generated from synthetic gene fragments of the viral genes of interest. First a PCR step was performed on the synthetic gene fragment with a forward primer that contained a T7 promoter. Next, the PCR product was used as the template for an in vitro transcription (IVT) reaction at 37°C for 2 hours. The IVT reaction was then treated with TURBO DNase (Thermo) for 30 min at 37°C, followed by a heat-denaturation step at 75°C for 15 min. RNA was purified using RNA Clean and Concentrator 5 columns (Zymo Research). RNA was quantified by Nanodrop and Qubit and diluted in nuclease-free water to working concentrations.
DETECTR assays
DETECTR assays were performed using RT-LAMP for pre-amplification of viral or control RNA targets and LbCas12a for the trans-cleavage assay. RT-LAMP was prepared as suggested by New England Biolabs (https://www.neb.com/protocols/2014/10/09/typical-rt-lamp-protocol) with a MgSO4 concentration of 6.5 mM and a final volume of 10 µL. LAMP primers were added at a final concentration of 0.2 µM for F3 and B3, 1.6 µM for FIP and BIP, and 0.8 µM for LF and LB. Reactions were performed independently for N gene, E gene, and RNase P using 2 µL of input RNA at 62°C for 20–30 min.
LbCas12a trans-cleavage assays were performed similar to those previously described9. 50 nM LbCas12a (available from NEB) was pre-incubated with 62.5 nM gRNA in 1X NEBuffer 2.1 for 10 min at 37°C. After formation of the RNA-protein complex, the lateral flow cleavage reporter (/56-FAM/TTATTATT/3Bio/, IDT) was added to the reaction at a final concentration of 500 nM. RNA-protein complexes were used immediately or stored at 4°C for up to 24 hours before use.
Lateral flow readout
After completion of the pre-amplification step, 2 µL of amplicon was combined with 18 µL of LbCas12a-gRNA complex and 80 µL of 1X NEBuffer 2.1. The 100 µL LbCas12a trans-cleavage assay was allowed to proceed for 10 min at 37°C.
A lateral flow strip (Milenia HybriDetect 1, TwistDx) was then added to the reaction tube and a result was visualized after approximately 2 min. A single band, close to the sample application pad indicated a negative result, whereas a single band close to the top of the strip or two bands indicated a positive result, see Supplementary Fig. 6 for additional details in the interpretation of the lateral flow strips. Lateral flow strips can be interpreted visually or were quantified using ImageJ’s gel analyzer tool and signal was normalized to the max signal intensity.
Optimized DETECTR method for patient samples
The patient optimized DETECTR assays were performed using RT-LAMP method as described above with the following modifications: A DNA binding dye, SYTO9 (Thermo Fisher), was included in the reaction to monitor the amplification reaction and the incubation time was extended to 30 min to capture data from lower titre samples.
The fluorescence based patient optimized LbCas12a trans-cleavage assays were performed as described above with modifications; 40nM LbCas12a was pre-incubated with 40nM gRNA, after which 100nM of a fluorescent reporter molecule compatible with detection in the presence of the SYTO9 dye (/5Alex594N/TTATTATT/3IAbRQSp/) was added to the complex. 2 µL of amplicon was combined with 18 µL of LbCas12a-gRNA complex in a black 384-well assay plate and monitored for fluorescence using a Tecan plate reader.
Contrived sample preparation.
In-vitro transcribed RNA (gift from California Department of Public Health (CDPH)), with a titer of 10,000 copies/µL (Ct value of 21) was diluted into 2,500 copies/µL first, then serially diluted in water to concentration of 1250, 625, 312, 156, 78, 39, 10 and 2.5 copies per microliter.
Human clinical sample collection and preparation
Negative nasopharyngeal swabs were acquired from healthy donors in Chiu lab with the approval of the University of California, San Francisco (UCSF) IRB. Clinical nasopharyngeal and oropharyngeal swab samples from patients infected with SARS-CoV-2, influenza, or other coronaviruses were collected in UTM and transported to the CDPH or UCSF lab. Sample RNA of SARS-CoV-2 was extracted following instructions as described in the CDC EUA-approved protocol6 (input 120 µL, elution of 120 µL) using Qiagen DSP Viral RNA Mini Kit (Qiagen) at CDPH and the MagNA Pure 24 instrument (Roche Life Science) at UCSF. Nasopharyngeal swab samples of influenza and common coronavirus were extracted at UCSF using the MagNA Pure 24 instrument.
CDC real-time qRT-PCR assay
The CDC assay was performed using the ABI 7500 Fast DX (Applied Biosystems) or Roche Lightcycler 480 (Roche) instruments according to the CDC EUA-approved protocol6.
Bioinformatic analyses
Multiple sequence alignments to generate viral consensus sequences were performed using Clustal Omega v.1.2.426 and Jalview v.2.1127 software. RT-LAMP primer design was performed using PrimerExplorer v.5 software (Eiken Chemical Co., Ltd).
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by NIH grants R33-AI129455 (CYC) from the National Institute of Allergy and Infectious Diseases and R01-HL105704 (CYC) from the National Heart, Lung, and Blood Institute and the Charles and Helen Schwab Foundation. We thank Jill Hakim and Dustin Glasner for assisting with sample collection, and Vikram Joshi and Maria-Nefeli Tsaloglou for helpful discussions in the preparation of this manuscript.
Footnotes
COMPETING INTERESTS
CYC is the director of the UCSF-Abbott Viral Diagnostics and Discovery Center (VDDC), receives research support funding from Abbott Laboratories, and is on the Scientific Advisory Board of Mammoth Biosciences, Inc. JSC is a co-founder of Mammoth Biosciences, Inc. JPB, CLF, JS and XM are employees of Mammoth Biosciences, Inc. CYC, JPB, XD, CLF, JS, XM and JSC are co-inventors of CRISPR-related technologies.
DATA AVAILABILITY
All reagents used in this manuscript are available from commercial sources.
CODE AVAILABILITY
Generation of viral consensus sequences and RT-LAMP primer design were performed using publicly available software, including Clustal Omega v.1.2.4, Jalview v.2.11, and Primer Explorer v.5 (online methods).
SUPPLEMENTARY FIGURES
AU reformat SI per the AIP letter guidance. No figures in the main text file please.
Any legends should be with the Supplementary or Extended data figures.
REFERENCES
- 1.Wang C, Horby PW, Hayden FG & Gao GF A novel coronavirus outbreak of global health concern. Lancet 395, 470–473, doi: 10.1016/S0140-6736(20)30185-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727–733, doi: 10.1056/NEJMoa2001017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report - 44 8 (World Health Organization, Geneva, Switzerland, 2020). [Google Scholar]
- 4.Bai Y et al. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA, doi: 10.1001/jama.2020.2565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothe C et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med 382, 970–971, doi: 10.1056/NEJMc2001468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.US Centers for Disease Control and Prevention. Real-Time RT-PCR Panel for Detection 2019-nCoV, <https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-detection-instructions.html> (2020).
- 7.Zhang W et al. Molecular and serological investigation of 2019-nCoV infected patients: implications of multiple shedding routes. Emerging Microbes and Infections 9, 386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.US Food and Drug Administration. Policy for Diagnostics Testing in Laboratories Certified to Perform High Complexity Testing under CLIA prior to Emergency Use Authorization for Coronavirus Disease-2019 during the Public Health Emergency (US Food and Drug Administration, 2020). [Google Scholar]
- 9.Chen JS et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439, doi: 10.1126/science.aar6245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu C Cutting-Edge Infectious Disease Diagnostics with CRISPR. Cell Host Microbe 23, 702–704, doi: 10.1016/j.chom.2018.05.016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gootenberg JS et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442, doi: 10.1126/science.aam9321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myhrvold C et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448, doi: 10.1126/science.aas8836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notomi T et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28, E63, doi: 10.1093/nar/28.12.e63 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR, < https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf>. (World Health Organization, Geneva, Switzerland, 2020). [Google Scholar]
- 15.Cepheid. Package Insert - Xpress - SARS-CoV-2, <http://www.cepheid.com/Package%20Insert%20Files/Xpress-SARS-CoV-2-PI/302-3750-rev-A-PACKAGE-INSERT-EUA-XPRESS-SARS-COV2.pdf> (2020).
- 16.Bauchner B Fontanarosa PW, & Livingston EH Conserving supply of personal protective equipment – a call for ideas. JAMA, doi: 10.1001/jama.2020.4770 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Herper M, and Branswell H Shortage of crucial chemicals creates new obstacle to U.S. coronavirus testing. STATNews, <https://www.statnews.com/2020/03/10/shortage-crucial-chemicals-us-coronavirus-testing/>. (Accessed April 4th, 2020).
- 18.Liu J et al. Community Transmission of Severe Acute Respiratory Syndrome Coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis 26, doi: 10.3201/eid2606.200239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.California Department of Public Health. CDC confirms possible first instance of COVID-19 community transmission in California, <https://www.cdph.ca.gov/Programs/OPA/Pages/NR20-006.aspx> (2020).
- 20.Holshue ML et al. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med 382, 929–936, doi: 10.1056/NEJMoa2001191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan Y, Zhang D, Yang P, Poon LLM & Wang Q Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis, doi: 10.1016/S1473-3099(20)30113-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou L et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med, doi: 10.1056/NEJMc2001737 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young BE et al. Epidemiologic Features and Clinical Course of Patients Infected With SARS-CoV-2 in Singapore. JAMA, doi: 10.1001/jama.2020.3204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 24.Elbe S & Buckland-Merrett G Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 1, 33–46, doi: 10.1002/gch2.1018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis KA et al. A multiplexed RT-LAMP assay for detection of group M HIV-1 in plasma or whole blood. J. Virol. Methods 255, 91–97, doi: 10.1016/j.jviromet.2018.02.012 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Waterhouse AM, et al. Jalview Version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191, doi: 10.1093/bioinformatics/btp033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McWilliam H, et al. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Research 41, doi: 10.1093/nar/gkt376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


