Abstract
Two strains of Lactococcus lactis subsp. lactis were used to determine the influence of lactose and arginine on viability and amino acid use during carbohydrate starvation. Lactose provided energy for logarithmic-phase growth, and amino acids such as arginine provided energy after carbohydrate exhaustion. Survival time, cell numbers, and ATP concentrations increased with the addition of arginine to the basal medium. By the onset of lactose exhaustion, the concentrations of glycine-valine and glutamate had decreased by as much as 67% in L. lactis ML3, whereas the serine concentration increased by 97% during the same period. When no lactose was added, the concentrations of these amino acids remained constant. Similar trends were observed for L. lactis 11454. Without lactose or arginine, L. lactis ML3 was nonculturable on agar but was viable after 2 days, as measured by fluorescent viability stains and intracellular ATP levels. However, L. lactis 11454 without lactose or arginine remained culturable for at least 14 days. These data suggest that lactococci become viable but nonculturable in response to carbohydrate depletion. Additionally, these data indicate that amino acids other than arginine facilitate the survival of L. lactis during carbohydrate starvation.
Lactococcus lactis subsp. lactis is used widely in the cheese industry as a starter culture for cheese production. Starter cultures face carbohydrate starvation conditions with less than 0.1% lactose in the cheese curd after pressing (34). Starvation conditions decrease the ability to synthesize ATP, generate proton motive force (PMF), and accumulate nutrients necessary to maintain viability over time (13).
In optimum growth, L. lactis is a homofermentative lactic acid bacterium that ferments lactose to lactic acid and ATP. Lactococci lack a cytochrome system and are unable to produce ATP by oxidative phosphorylation (29). They rely on glycolysis and substrate-level phosphorylation to generate compounds that serve as energy donors for solute transport and growth (31). In lactococci, lactose is transported into the cell by a phosphoenolpyruvate (PEP)-mediated phosphotransferase system (PTS), sugar transport ATPases, ion-linked sugar transport mechanisms, and sugar exchange mechanisms for a net yield of four ATP molecules per lactose molecule via glycolysis (19, 28, 33).
The absence of lactose causes immediate energy starvation because lactococci do not contain carbohydrate storage polymers (18, 22). During starvation, the intracellular levels of the glycolytic intermediates PEP, 3-phosphoglycerate (3-PG), and 2-phosphoglycerate increase and constitute the PEP potential, which provides a link between sugar transport and energy-yielding reactions of glycolysis (28, 29). During starvation, PEP is metabolized slowly to pyruvate and ATP due to the regulation of pyruvate kinase (28). This maintenance of large PEP pools may provide necessary maintenance energy (ATP) for the organism during starvation as well as permit the rapid accumulation of PTS sugars when they become available again (28, 29).
Upon energy starvation of L. lactis, the PMF dissipates, the pH gradient collapses, and ATP levels decrease below 0.1 mM (13, 18). Nevertheless, many organisms have the ability to use alternate carbon sources for energy. In response to carbohydrate starvation, many bacteria become viable but nonculturable (VBNC) rather than die and lyse, as suggested for lactococci (6). This state has been observed for bacteria such as Vibrio, Pseudomonas, Micrococcus, Enterococcus, Brevibacterium, Escherichia coli, and others (16). During the VBNC state, cells continue to transport and metabolize nutrients but do not form colonies on solid agar. Shigella dysenteriae becomes VBNC during starvation, and the cells continue to transport and incorporate methionine into cellular proteins (23).
Transport of amino acids in lactococci occurs via three different mechanisms. The majority of amino acids are transported by the PMF-driven transport that links amino acid uptake to the PMF. The driving force for H+ translocation and the PMF is usually supplied by the free energy of ATP hydrolysis (12). L. lactis catalyzes the translocation of leucine, isoleucine, valine, alanine, glycine, serine, threonine, methionine, histidine, proline, cysteine, lysine, phenylalanine, tyrosine, tryptophan, and arginine together in symport with one proton via separate mechanisms (12, 19). During starvation of lactococci, the proton-linked amino acid transporter and the arginine or ornithine antiporter still function and are only moderately affected during incubation (13).
The driving force for arginine uptake into cells is supplied by the arginine and ornithine concentration gradient (20). This transport mechanism is beneficial because no additional metabolic energy is required for the transport of arginine across the membrane (12). The excess arginine may be metabolized by the arginine deiminase (ADI) pathway to produce energy (ATP) in L. lactis (4). The ADI pathway is widely distributed among bacteria and serves either as the sole or as an additional source of energy, carbon, and nitrogen in lactic acid bacteria, bacilli, Pseudomonas, Aeromonas, clostridia, Mycoplasma, and halobacteria (4).
In this study, the influence of lactose and arginine on the amino acid metabolism and culturability of L. lactis was examined. This study was initiated to determine if L. lactis could become VBNC after the exhaustion of lactose when present with arginine and other amino acids. Amino acids may be utilized for protein synthesis during starvation as well as provide an additional energy source. Our results indicate that lactococci remain viable in the absence of lactose or arginine. The cells were able to use other amino acids to survive, produce ATP, and maintain cellular integrity without being culturable on agar.
MATERIALS AND METHODS
Bacterial strains and media.
L. lactis subsp. lactis ML3 was obtained from the Utah State University culture collection. L. lactis subsp. lactis 11454 was obtained from the American Type Culture Collection (Rockville, Md.). Strains were propagated in Elliker broth (Difco Laboratories, Detroit, Mich.) for 16 h at 30°C, harvested by centrifugation (11,750 × g for 5 min at 25°C), washed twice with sterile saline, resuspended in 2 ml of sterile saline, and inoculated (1%) into sterile chemically defined basal medium (CDM) (7, 11). CDM was supplemented with lactose and arginine to make four media: 0.2% lactose and 2% arginine, 0.2% lactose and 0% arginine, 0% lactose and 2% arginine, and 0% lactose and 0% arginine. The media were then adjusted to pH 7.0, buffered with 190 mM sterile 3-[N-morpholino]-propanesulfonic acid (MOPS; Sigma Chemical Co., St. Louis, Mo.), and filter sterilized through a 0.2-μm-pore-size bottle-top filter (Corning Costar, Corning, N.Y.). All cultures were grown in 200 ml of media at 30°C.
Lactose determination.
The lactose concentration was determined by high-pressure liquid chromatography. The sample was prepared by filtration through a sterile 0.2-μm-pore-size syringe filter (Nalge Company, Rochester, N.Y.) to remove cells. The filtered sample (10 μl) was injected with a model 507 chromatograph (Beckman Instruments, Inc., Fullerton, Calif.) autosampler into a Benson carbohydrate BC100 Ca2+ column (0.78 by 30 cm) fitted with a guard cartridge in a cartridge holder (Alltech, Deerfield, Ill.). The column and guard cartridge were heated to 86°C in a Bio-Rad Laboratories (Hercules, Calif.) column heater. Lactose was eluted with water at a flow rate of 0.5 ml/min during a 14-min run with a model 125 pump (Beckman). Detection was done with an LC 30 refractive-index detector (The Perkin-Elmer Corp., Norwalk, Conn.). Peak areas were determined with a model 427 integrator (Beckman) set at an attenuation of 16. A linear standard curve (R2 = 0.99) was observed with 0.125, 0.25, 0.5, 1.0, and 2.5 mg of lactose per ml (data not shown).
Culturable cell estimation.
Samples were taken at various times from the culture suspensions and diluted in sterile saline dilution blanks. From these dilutions, 100 μl was spread on Elliker agar plates with a Spiral System CU (Cincinnati, Ohio). The plates were incubated anaerobically for 48 h at 30°C. The colony count was determined with duplicate plates in accordance with the manufacturer’s instructions.
Viable cell estimation.
Samples were collected from the cell suspensions and stained in accordance with the manufacturer’s instructions by use of the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Inc., Eugene, Oreg.). The kit estimated bacterial viability with two nucleic acid stains that differed in their spectral characteristics and ability to penetrate healthy bacterial cell membranes. The fluorescence emission of each cell suspension was measured with an RF-1501 spectrofluorophotometer (Shimadzu, Pleasanton, Calif.) at an excitation of 480 nm and an emission of 520 nm for the live dye and an excitation of 550 nm and an emission of 580 nm for the dead dye. Results are expressed in relative fluorescence units.
ATP quantitation.
The ATP concentration in cell extracts was quantified by measuring luminescence with an ATP assay kit (Calbiochem-Novabiochem Corporation, San Diego, Calif.) as described by the manufacturer. The assay is based on the luciferase-catalyzed oxidation of d-luciferin in the presence of an ATP-magnesium salt and oxygen to produce light. Luminescence was measured with an LS6500 scintillation counter (Beckman).
Amino acid determination.
Samples were prepared by filtering cell suspensions through a 0.2-μm-pore-size syringe filter (Nalge) and were centrifuged (5,000 × g for 3 h at 4°C) through a 1K Centricon (Pall Filtron, Ann Arbor, Mich.). An aliquot was derivatized with 3-(4-carboxybenzoyl)quinoline-2-carboxaldehyde in accordance with the instructions enclosed in the ATTO-TAG CBQ derivatization kit (Molecular Probes). Norleucine was added as an internal standard to each reaction mixture prior to the addition of the derivatizing agent.
Amino acids were monitored by micellar electrokinetic chromatography with capillary electrophoresis and fluorescence (26, 32). Sample electrophoresis was done by use of a P/ACE2100 automated capillary electrophoresis system with System Gold software (Beckman) and 50 mM sodium borate (pH 9.5) containing 150 mM sodium dodecyl sulfate and 10 mM tetrahydrofuran as the run buffer. Each analysis was run for 60 min at 22.5 kV and 25°C with a 1-s pressure injection into a fused silica capillary (75-μm inner diameter by 57 cm). Each amino acid was detected by laser-induced fluorescence (laser-induced fluorometer [LIF] model 488; Beckman) with a 488-nm argon ion laser as the excitation source and a 560-nm emission filter.
RESULTS AND DISCUSSION
Lactose utilization.
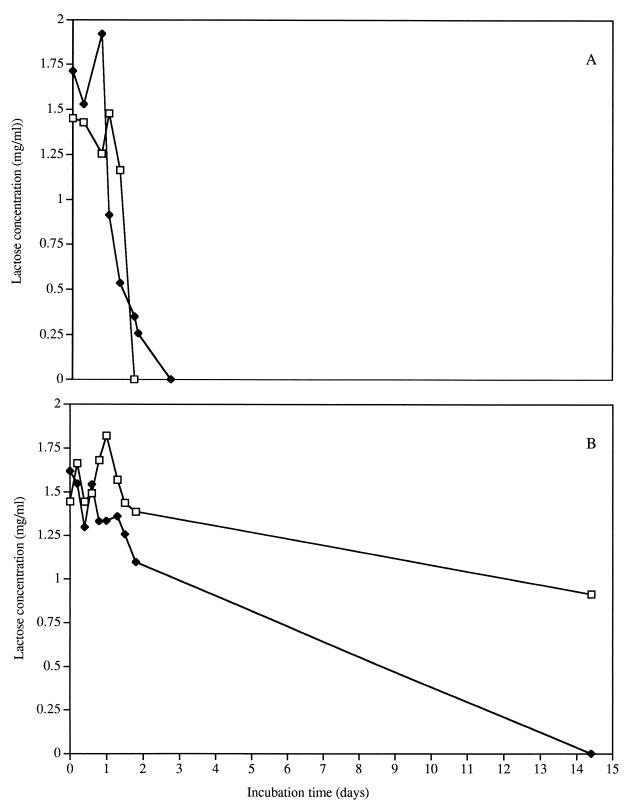
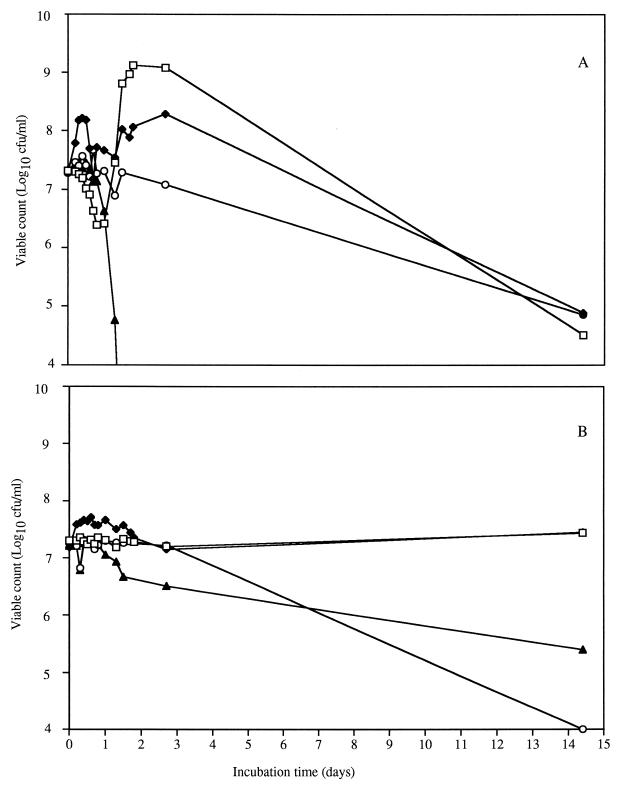
The lactose concentration during the growth of L. lactis subsp. lactis ML3 decreased to nondetectable levels irrespective of arginine content within 3 days (Fig. 1A) and did not change the pH of the media due to the use of MOPS buffer. The culture containing lactose and arginine grew to higher numbers (109 CFU/ml) than did the culture without arginine (108 CFU/ml) after lactose depletion (Fig. 2A). The presence of arginine appeared to play a beneficial role in cell growth and viability when present with lactose.
FIG. 1.
Lactose concentrations in spent media for L. lactis ML3 (A) and L. lactis 11454 (B). Cells were grown in 0.2% lactose and 2% arginine (□) and 0.2% lactose and no arginine (⧫).
FIG. 2.
Culturable cell counts for L. lactis ML3 (A) and L. lactis 11454 (B) grown in various media. Cells were grown in 0.2% lactose and 2% arginine (□), 0.2% lactose and no arginine (⧫), no lactose and 2% arginine (○), and no lactose or arginine (▴).
L. lactis subsp. lactis 11454 used lactose at a slower rate than did L. lactis ML3 (Fig. 1B). Lactose was present after 14 days of incubation of L. lactis 11454 in CDM/L+/A+ (CDM containing lactose [L+] and arginine [A+]). L. lactis 11454 did not increase in cell density with lactose present but maintained higher cell numbers than L. lactis ML3 during incubation. With arginine present, the cells did not enter carbohydrate starvation even after 14 days of incubation. These data indicate that lactose use is different between L. lactis ML3 and L. lactis 11454 and is linked to the arginine content of the medium. Differences in lactose fermentation in lactococci have been attributed to the activity of the Lac-PTS transport system (5, 12) and altered phospho-β-galactosidase activity (3). The mechanism of lactose use was not investigated in this study. One possible explanation for these data may be the transport mechanism, as noted for L. lactis 11454 strain T-1. This strain lacks at least one of the proteins necessary for the active transport of lactose into the cell (5). Alternatively, the slow metabolism of L. lactis 11454 may have been due to a deficiency of a nutrient in CDM or a lack of phospho-β-galactosidase. Irrespective of the mechanism for the difference in lactose metabolism, it is important to note that the carbohydrate content was higher in 11454 than in ML3 in the same time frame and had an impact on cellular metabolism during incubation.
Viability and culturability.
The culturability of L. lactis was estimated by determining the cell population from the plate counts, while viability was determined with the use of fluorescence as a result of healthy cell membranes and the amount of ATP present. L. lactis ML3 grown in CDM/L+/A+ reached a cell density of 109 CFU/ml, which decreased to 105 CFU/ml after 14 days (Fig. 2A). Conversely, in CDM/L−/A− (CDM lacking lactose [L−] and arginine [A−]), strain ML3 was culturable for 2 days and then became nonculturable on solid agar. The addition of either lactose or arginine increased cell density and culturability time for ML3.
The cell density of L. lactis 11454 in CDM/L+/A+ remained constant during incubation at about 107 CFU/ml (Fig. 2B). In CDM/L−/A−, the cell numbers decreased to 105 CFU/ml in 14 days, but the cells remained culturable. Data from cells grown in CDM/L−/A− suggested that L. lactis 11454 was using amino acids as a source of energy to maintain culturability, whereas L. lactis ML3 lost the ability to form colonies on solid agar.
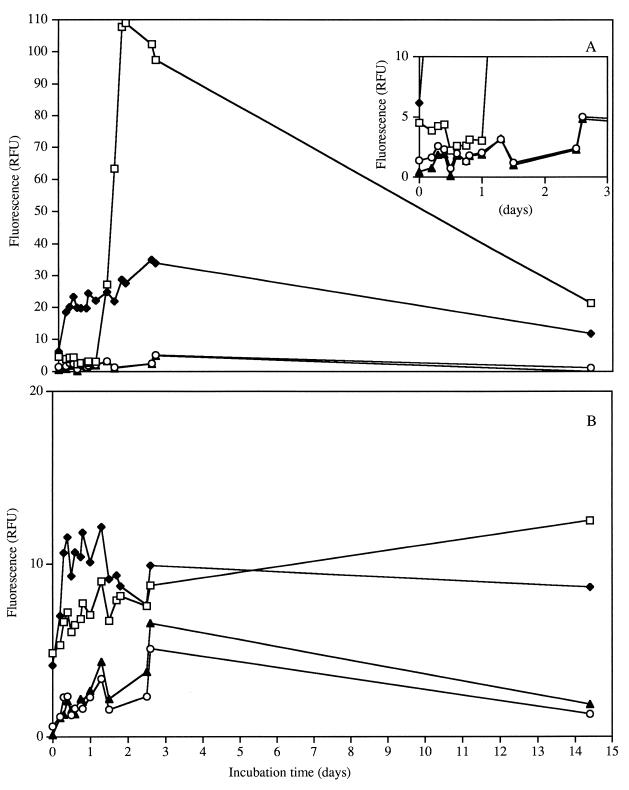
As an independent measure of viability, cellular fluorescence was estimated with the LIVE/DEAD BacLight kit. The relative fluorescence of the live population was similar to the growth curve and estimated the viable population without further subculturing (Fig. 3). L. lactis ML3 grown in CDM/L+/A+ showed increased fluorescence, which correlated with the plate count data, while fluorescence for L. lactis 11454 remained constant. L. lactis ML3 grown in CDM/L−/A− had fluorescent viable cells, even though the cells were not culturable on solid agar after 2 days. These data suggested that lactococcal viability can be estimated without plate counts and supported the use of this technique to estimate bacterial viability and the VBNC state (10, 15, 17).
FIG. 3.
Estimation of viability with fluorescence for L. lactis ML3 (A) and L. lactis 11454 (B) grown in various media. Cells were grown in 0.2% lactose and 2% arginine (□), 0.2% lactose and no arginine (⧫), no lactose and 2% arginine (○), and no lactose or arginine (▴). The inset in panel A depicts data from days 0 to 3 for ML3. RFU, relative fluorescence units.
ATP concentration.
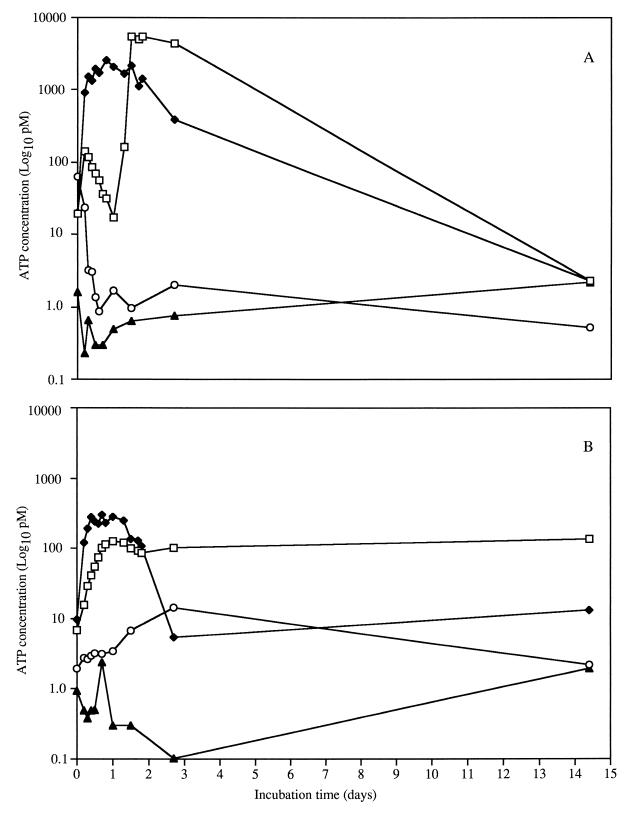
As a third measure of viability, the ATP concentration was determined during growth. The ATP concentration during the growth of L. lactis ML3 followed the same pattern as plate counts and viable fluorescent population estimations (compare Fig. 2A, 3A, and 4A). However, in CDM/L−/A−, the ATP concentration decreased initially and then increased to the original level. The presence of ATP with the plate counts and fluorescence viability estimations showed viable cells present at a low concentration, but no cells could be cultured on solid agar. These observations suggested that L. lactis ML3 cells were viable but nonculturable, since ATP was still present in the cells.
FIG. 4.
ATP concentrations for L. lactis ML3 (A) and L. lactis 11454 (B). Cells were grown in 0.2% lactose and 2% arginine (□), 0.2% lactose and no arginine (⧫), no lactose and 2% arginine (○), and no lactose or arginine (▴).
When grown in CDM/L+/A+, the L. lactis 11454 cultures produced ATP levels that increased during the first 3 days and then remained constant (Fig. 4B), as did the plate counts and estimates of viability with fluorescence. In CDM/L+/A−, the ATP concentration decreased after carbohydrate exhaustion and then leveled off (Fig. 4B). For both strains grown in CDM/L−/A−, the ATP concentration increased after 1 to 3 days, suggesting that energy sources other than lactose and arginine were available to the cells (Fig. 4).
Arginine was below femtomole levels after 2 weeks in CDM/L+/A+, presumably due to the activity of the ADI pathway. This energy source is important because arginine is converted to ornithine, with the production of 1 mol of ATP per mol of arginine consumed (4). Analysis of arginine and ornithine exchange activities in membrane vesicles isolated from growing and starving cells of L. lactis ML3 indicated that the transport system was not inactivated during starvation and that activity was maintained (13). Arginine metabolism allowed for a longer period of growth during carbohydrate starvation by providing an additional source of ATP. This result is in agreement with that of Thomas and Batt (27), who found that the survival time of L. lactis ML3 increased in the presence of arginine.
Without lactose present initially, arginine concentrations were not depleted during the 14-day incubation in each strain (data not shown). Observations from both strains suggested that the cells must metabolically deplete lactose, thereby inducing the ADI pathway (2, 4). When cells were grown in a lactose-containing medium and then inoculated into a lactose-deficient medium, they did not grow or use arginine, even though it was present (data not shown). The data suggested that an unknown induction factor produced during lactose metabolism played a role in regulating the ADI pathway.
Amino acid profile.
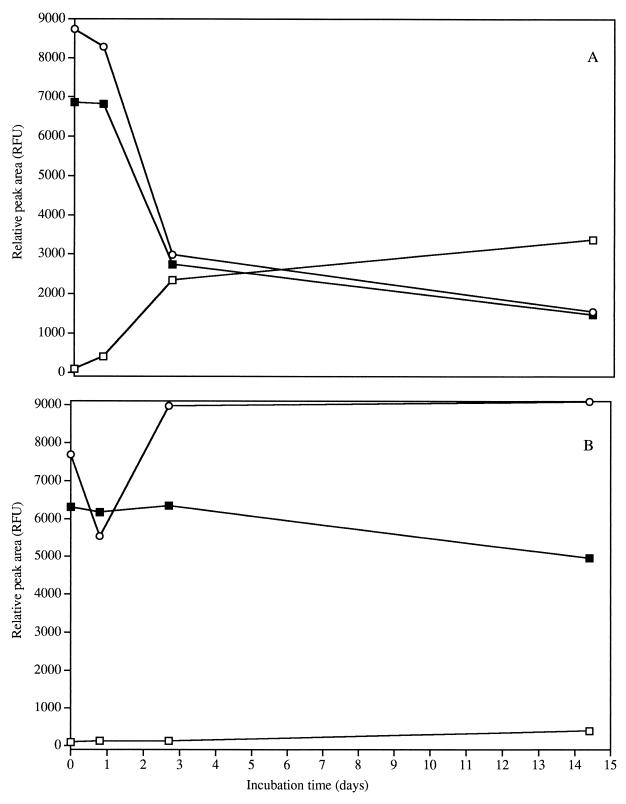
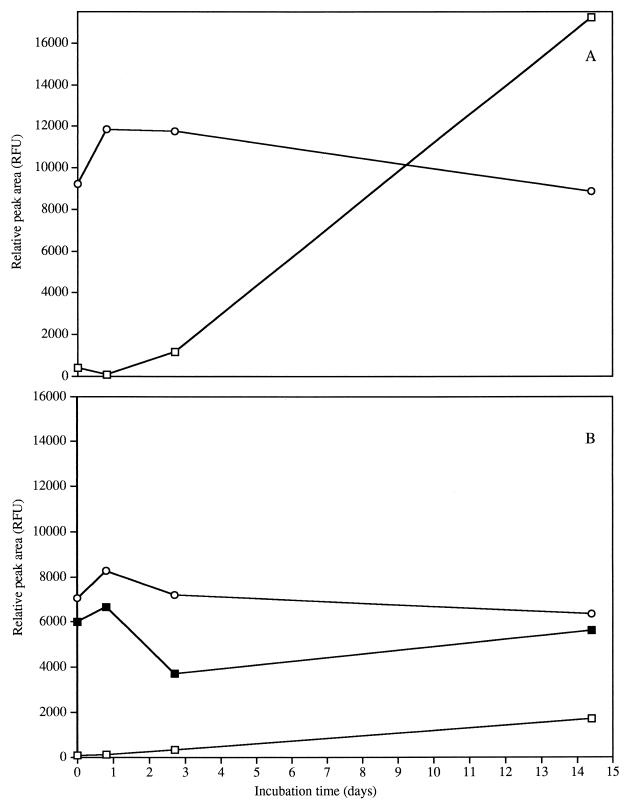
Extracellular amino acid concentrations were estimated by capillary electrophoresis and laser-induced fluorometry. Concentrations of histidine, methionine, glutamine, asparagine, threonine, tyrosine, alanine, leucine, phenylalanine, proline, and isoleucine were unchanged during 14 days of incubation with all media and strains tested (data not shown). In L. lactis ML3 cultures, glycine-valine and glutamate concentrations decreased by as much as 67% in CDM/L+/A+ until the lactose was depleted, after which they decreased only slightly (Fig. 5A). The decrease in extracellular glutamate and glycine or valine concentrations is in agreement with the observation of an increase in the intracellular concentrations of glutamate and glycine at the onset of starvation (22). Rapid but variable use of glutamate by lactococci was also found with L. lactis 133 and is associated with osmotic stress and ATP content for transport (12, 19, 21, 31). The rate of glutamate uptake by L. lactis increases more than 30-fold when the intracellular pH is raised from 6.0 to 7.4 due to osmotic stress (21). These observations have also been made for E. coli and Salmonella typhimurium in connection with the intracellular accumulation of potassium and glutamate, which enhances survival by regulating osmotic pressure and intracellular pH (1, 8). Hence, observations with ML3 suggest that glutamate utilization may signal stress during the incubation period (Fig. 5 and 6).
FIG. 5.
Extracellular amino acid profiles for L. lactis ML3 with 0.2% lactose and 2% arginine (A) and without lactose and 2% arginine (B). Amino acids shown are serine (□), glycine or valine (○), and glutamate (■). RFU, relative fluorescence units.
FIG. 6.
Extracellular amino acid profiles for L. lactis 11454 with 0.2% lactose and 2% arginine (A) and without lactose and 2% arginine (B). Amino acids shown are serine (□), glycine or valine (○), and glutamate (■). RFU, relative fluorescence units.
The serine concentration with L. lactis ML3 increased by 97% in CDM/L+/A+; however, in CDM/L−/A+, the serine concentration remained constant (Fig. 5). In CDM/L+ and CDM/L−, serine was produced with L. lactis 11454 (Fig. 6); however, higher production of serine was observed in CDM/L+. The production of serine was unexpected but possible because one mechanism of serine production is from 3-PG (9). Dehydration of 3-PG with subsequent transamination and dephosphorylation yields serine and Pi. 3-PG is a product of glycolysis and a constituent of the PEP potential that increases along with the Pi level during carbohydrate starvation (30). Therefore, the serine production observed under these conditions could have been a result of excess 3-PG which accumulated during carbohydrate starvation.
The ability of lactococci to survive periods of carbohydrate starvation may depend on their ability to transport and metabolize certain amino acids. During starvation, the proton-linked transport of carriers of the branched-chain amino acids lysine, methionine, phenylalanine, serine, and alanine are affected only moderately (13). The free-amino-acid pool created by amino acid transport and protein degradation is used for protein synthesis during starvation. The synthesis of new proteins during starvation was observed for L. lactis ML3 and E. coli (13, 14) and allowed cells to remain viable longer during periods of starvation (24). Survival for up to 60 h for L. lactis ML3 in growth medium without lactose, vitamins, and bicarbonate is attributable to the accumulation of Mg2+ and amino acids (27). Amino acids may prolong the survival of L. lactis by providing an energy source, supplying amino acids for cell turnover, and minimizing the breakdown of essential proteins (27).
In addition to arginine, other amino acids, such as serine, might provide an additional energy source. When arginine, serine, and tryptophan were added together, the growth rate of L. lactis increased notably (11). Serine produced during starvation could be used as an energy reservoir for the production of energy during starvation. Anaerobically grown Staphylococcus epidermidis is able to ferment serine when glucose is limited (25). Fermentation of serine yields pyruvate, acetate, and NH3. About 90% of the pyruvate formed after the initial deamination of serine is converted to lactate, acetyl coenzyme A, and CO2. Acetyl coenzyme A yields 0.52 mol of ATP via phosphate acetyltransferase and acetate kinase (25). These results indicate a possible role of serine and other amino acids as starvation energy sources that can extend the time that lactococci can survive carbohydrate starvation and remain viable.
From these results, it can be concluded that lactose and arginine provided metabolic products and energy that allowed cells to survive for long periods of carbohydrate starvation. Lactose was used for greater logarithmic-phase growth until depleted, and amino acids were used for energy after carbohydrate exhaustion. The rate of lactose metabolism was strain dependent and showed that the Lac-PTS system in L. lactis 11454 functions differently from that in ML3. Arginine influenced the growth and viability of L. lactis subsp. lactis and provided energy for larger cell numbers and survival times compared to those in cultures without arginine. In L. lactis ML3, the ADI pathway allowed arginine to be metabolized when lactose was depleted. Arginine was not depleted with L. lactis 11454 or L. lactis ML3 grown in CDM/L−. This result is presumed to be related to an unknown connection between lactose and the ADI pathway.
L. lactis was viable after 2 weeks in the absence of known energy sources, such as fermentable carbohydrate and arginine. Lactococci metabolized other endogenous energy sources, such as amino acids, to remain VBNC. L. lactis ML3 could enter the VBNC state and maintain ATP levels and cellular integrity for at least 14 days. Further research needs to be done to deduce the possible role of serine, as well as that of other amino acids, that could be energy sources for cells during carbohydrate starvation. These alternate sources could allow cells to remain metabolically active, yet unculturable, and cells could survive the periods of carbohydrate starvation that occur in harsh environments such as cheese.
ACKNOWLEDGMENTS
This project was supported by the Utah State University Agricultural Experiment Station, Utah Mineral Lease, and Dairy Management, Inc.
We thank Marie Strickland for technical assistance with lactose analysis and Madhavi Ummadi for expertise with the capillary electrophoresis and the amino acid analysis.
Footnotes
Contribution 7050 of the Utah Agricultural Experiment Station.
REFERENCES
- 1.Botsford J L, Alvarez M, Hernandez R, Nichols R. Accumulation of glutamate by Salmonella typhimurium in response to osmotic stress. Appl Environ Microbiol. 1994;60:2568–2574. doi: 10.1128/aem.60.7.2568-2574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crow V L, Thomas T D. Arginine metabolism in lactic streptococci. J Bacteriol. 1982;150:1024–1032. doi: 10.1128/jb.150.3.1024-1032.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crow V L, Thomas T D. Properties of a Streptococcus lactis strain that ferments lactose slowly. J Bacteriol. 1984;157:28–34. doi: 10.1128/jb.157.1.28-34.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demko G M, Blanton S J B, Benoit R E. Heterofermentative carbohydrate metabolism of lactose-impaired mutants of Streptococcus lactis. J Bacteriol. 1972;112:1335–1345. doi: 10.1128/jb.112.3.1335-1345.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox P F, Law J, McSweeney P L H, Wallace J. Biochemistry of cheese ripening. In: Fox P F, editor. Cheese: chemistry, physics, and microbiology. New York, N.Y: Chapman & Hall; 1993. pp. 389–439. [Google Scholar]
- 7.Gao S, Oh D H, Broadbent J R, Johnson M E, Weimer B C, Steele J L. Aromatic amino acid catabolism by lactococci. Lait. 1997;77:371–381. [Google Scholar]
- 8.Gauthier M J, Flatau G N, Le Rudulier D, Clement R L, Combarro M P. Intracellular accumulation of potassium and glutamate specifically enhances survival of Escherichia coli in seawater. Appl Environ Microbiol. 1991;57:272–276. doi: 10.1128/aem.57.1.272-276.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottschalk G. Bacterial metabolism. 2nd ed. New York, N.Y: Springer-Verlag; 1986. pp. 49–50. [Google Scholar]
- 10.Jacobsen C N, Rasmussen J, Jakobsen M. Viability staining and flow cytometric detection of Listeria monocytogenes. J Microbiol Methods. 1997;28:35–43. [Google Scholar]
- 11.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konings W N, Poolman B, Driessen A J M. Bioenergetics and solute transport in lactococci. Crit Rev Microbiol. 1989;16:419–476. doi: 10.3109/10408418909104474. [DOI] [PubMed] [Google Scholar]
- 13.Kunji E R, Ubbink T, Matin A, Poolman B, Konings W N. Physiological responses of Lactococcus-lactis ML3 to alternating conditions of growth and starvation. Arch Microbiol. 1993;159:372–379. [Google Scholar]
- 14.Mandelstam J. Turnover of protein in growing and non-growing populations of Escherichia coli. Biochem J. 1958;69:110–119. doi: 10.1042/bj0690110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molecular Probes, Inc. LIVE/DEAD Baclight bacteria viability kit (L-7012) instruction manual with appendix. Eugene, Oreg: Molecular Probes Inc.; 1995. [Google Scholar]
- 16.Oliver J D. Formation of viable but nonculturable cells. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 239–272. [Google Scholar]
- 17.Olsen P E, Rice W A. Rapid evaluation of peat-base legume inoculant using immunomagnetic beads for cell retrieval and fluorescent nucleic acid probes for viability analysis. Plant Soil. 1996;186:75–79. [Google Scholar]
- 18.Otto R, Vije J, Brink T, Klont B, Konings W N. Energy metabolism in Streptococcus cremoris during lactose starvation. Arch Microbiol. 1985;141:348–352. [Google Scholar]
- 19.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–148. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 20.Poolman B, Driessen A J M, Konings W N. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J Bacteriol. 1987;169:5597–5604. doi: 10.1128/jb.169.12.5597-5604.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poolman B, Hellingwerf K J, Konings W N. Regulation of the glutamate-glutamine transport system by intracellular pH in Streptococcus lactis. J Bacteriol. 1987;169:2272–2276. doi: 10.1128/jb.169.5.2272-2276.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poolman B, Smid E J, Veldkamp H, Konings W N. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J Bacteriol. 1987;169:1460–1468. doi: 10.1128/jb.169.4.1460-1468.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman I, Shahamat M, Kirchman P A, Russek-Cohen E, Colwell R R. Methionine uptake and cytopathogenicity of viable but nonculturable Shigella dysenteriae type 1. Appl Environ Microbiol. 1994;60:3573–3578. doi: 10.1128/aem.60.10.3573-3578.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeve C A, Bockman A T, Matin A. Role of protein degradation in the survival of carbon-starved Escherichia coli and Salmonella typhimurium. J Bacteriol. 1984;157:758–763. doi: 10.1128/jb.157.3.758-763.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sivakanesan R, Dawes E A. Anaerobic glucose and serine metabolism in Staphylococcus epidermidis. J Gen Microbiol. 1980;118:143–157. doi: 10.1099/00221287-118-1-143. [DOI] [PubMed] [Google Scholar]
- 26.Strickland M, Weimer B C, Broadbent J R. Capillary electrophoresis of cheddar cheese. J Chromatogr. 1996;731:305–310. [Google Scholar]
- 27.Thomas T D, Batt R D. Survival of Streptococcus lactis in starvation conditions. J Gen Microbiol. 1968;50:367–382. doi: 10.1099/00221287-50-3-367. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J. Regulation of sugar transport and metabolism in lactic acid bacteria. FEMS Microbiol Rev. 1987;46:221–231. [Google Scholar]
- 29.Thompson J, Thomas T D. Phosphoenolpyruvate and 2-phosphoglycerate: endogenous energy source(s) for sugar accumulation by starved cells of Streptococcus lactis. J Bacteriol. 1977;130:583–595. doi: 10.1128/jb.130.2.583-595.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson J, Torchia D A. Use of 31P nuclear magnetic resonance spectroscopy and 14C fluorography in studies of glycolysis and regulation of pyruvate kinase in Streptococcus lactis. J Bacteriol. 1984;158:791–800. doi: 10.1128/jb.158.3.791-800.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J, Curtis M A, Miller S P F. N5-(1-Carboxyethyl)-ornithine, a new amino acid from the intracellular pool of Streptococcus lactis. J Bacteriol. 1986;167:527. doi: 10.1128/jb.167.2.522-529.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ummadi S, Weimer B C. Poster presented at the Ninth International Symposium on High Performance Capillary Electrophoresis. 1997. Use of capillary electrophoresis-laser induced fluorescence detection to monitor the utilization of amino acids during bacterial growth. [Google Scholar]
- 33.Varnam A H, Sutherland J P. Milk and milk products. London, England: Chapman & Hall, Ltd.; 1994. pp. 215–332. [Google Scholar]
- 34.Weimer B, Dias B, Ummadi M, Broadbent J, Brennand C, Jaegi J, Johnson M, Milani F, Steele J, Sisson D V. Influence of NaCl and pH on intracellular enzymes that influence cheddar cheese ripening. Lait. 1997;77:383–398. [Google Scholar]