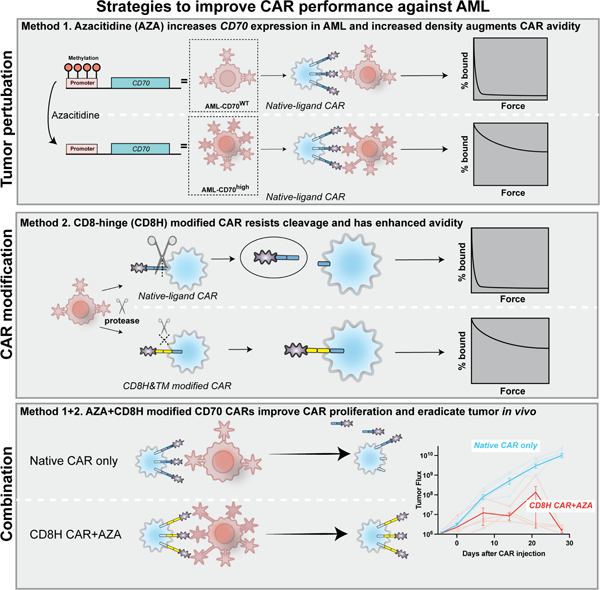
Summary
Chimeric antigen receptor (CAR) T cell therapy is effective in lymphoid malignancies, but there has been limited data in myeloid cancers. Here, we start with a CD27-based CAR to target CD70 (‘native’) in acute myeloid leukemia (AML), and we find modest efficacy in vivo, consistent with prior reports. We then use orthogonal approaches to increase binding on both the tumor and CAR-T cell sides of the immune synapse: a pharmacologic approach (azacitidine) to increase antigen density of CD70 in myeloid tumors, and an engineering approach to stabilize binding of the CAR to CD70. To accomplish the latter, we design a panel of hinge-modified regions to mitigate cleavage of the extracellular portion of CD27. Our CD8 hinge and transmembrane-modified CD70 CAR-T cells are less prone to cleavage, have enhanced binding avidity, and increased expansion, leading to more potent in vivo activity. This enhanced CD70-targeted CAR is a promising candidate for further clinical development.
Keywords: Cell Engineering, Hematologic Neoplasms, Cellular Immunity, Chimeric Antigen Receptors, Combined Modality Therapy
Graphical Abstract
INTRODUCTION
Acute myeloid leukemia (AML) is the most common acute leukemia in adults. While AML was uniformly fatal half a century ago, the advent of intensive induction cytotoxic chemotherapy led to cures in 40% of adults (Döhner et al., 2015). AML treatment changed little over the intervening fifty years, but since 2017, there have been a number of drugs approved by the FDA including inhibitors of hedgehog, BCL-2, FLT3, IDH1/2, a CD33 antibody drug conjugate, and a more potent liposomal formulation of induction chemotherapy (Guerra et al., 2019). While these interventions represent substantial progress, the majority of AML patients still fail to respond, or relapse and die from their disease, with older and relapsed or refractory patients having especially dire outcomes (Döhner et al., 2015).
Immunotherapy with checkpoint blockade has had limited success in AML. One way to bypass the limited natural T cell receptor (TCR) repertoire to neoantigens or tumor-specific antigens is to redirect T cells with chimeric antigen receptors (CARs). CAR-T cell therapy has rapidly revolutionized the treatment of lymphoid malignancies, with six FDA approvals since 2017 (Abramson et al., 2020; Jacobson et al., 2020; Neelapu et al., 2017; Schuster et al., 2017; Wang et al., 2020). These therapies result in destruction of malignant clones, but simultaneously lead to an on-target, off-tumor effect resulting in the elimination of normal B cells or plasma cells, respectively. Fortunately, hypogammaglobulinemia is manageable via routine administration of intravenous immunoglobulin. However, development of CAR-T cell therapy to treat AML has been more difficult, and off tumor effects have been more consequential. The majority of the available surface antigens present on AML blasts are also expressed on many myeloid and stem cell populations, and prolonged ablation of either population is not compatible with survival. CARs targeting various antigens in AML have been described recently (CD123(Luo et al., 2015; Mardiros et al., 2013), CD33(Kim et al., 2018; Wang et al., 2015), FLT3(Jetani et al., 2018)), some of which are currently in phase I clinical trials, though substantial toxicities and deaths have resulted (Hofmann et al., 2019; Melao, 2017).
CD70 is a tumor necrosis factor (TNF) superfamily member that has recently been described as a potential target for AML (Perna et al., 2017; Riether et al., 2020; Sauer et al., 2021). CD70 is the ligand for CD27, which is a T cell costimulatory molecule that provides survival signals and is associated with memory formation. Normal expression of CD70 is restricted to activated immune cells (Bowman et al., 1994; Nolte et al., 2009), though it is also upregulated in a number of cancer types (Flieswasser et al., 2019). Unlike other AML targets, CD70 is not expressed on normal hematopoietic stem cells (Bowman et al., 1994; Junker et al., 2005; Nolte et al., 2009; Perna et al., 2017; Riether et al., 2020; Riether et al., 2017; Ruf et al., 2015; Sauer et al., 2021). An antibody targeting CD70, ARGX-110 (cusatuzumab), has yielded impressive response rates in a phase I trial of newly diagnosed AML patients not fit for traditional intensive induction chemotherapy (Ochsenbein, 2018; Riether et al., 2020). This suggests that CAR-T cells targeting CD70 may also be an attractive option, given that CAR-T cells can have higher potency than traditional antibody-based therapies targeting the same antigen (Slaney et al., 2018).
The first generation CD70-targeted CAR utilized a full length CD27 protein fused to the CD3ζ domain (flCD27- CD3ζ) in a ligand-based targeting approach that was effective at targeting B cell malignancies (Shaffer et al., 2011). Later, a panel of ligand-based CD27 CAR-T cells with different ligand lengths and co-stimulation domains demonstrated that a truncated CD27 protein fused to the 41BB costimulatory domain (trCD27–41BBζ) was the most promising and was brought to a phase I clinical trial for solid tumors (NCT02830724) (Wang et al., 2017). Other investigators have demonstrated efficacy against head and neck cancers and gliomas expressing CD70 using this second-generation trCD27–41BBζ CAR (Jin et al., 2017; Park et al., 2018; Shaffer et al., 2011; Tu et al., 2019; Wang et al., 2017). Most recently, another panel of CD70-targeted CARs was developed for targeting AML, this time utilizing a single chain variable fragment (scFv) with variable spacer regions (Sauer et al., 2021). This panel was compared to the original first generation flCD27-CD3ζ CAR with findings that the panel of scFv-based CARs proved to be inferior to the original flCD27- CD3ζ version.
Despite rapid and intense development, the optimal parameters for CAR-T cell-target cell interactions that will predict in vivo and in-human efficacy are not yet well understood. High affinity (Lynn et al., 2019) interactions and high levels of constitutive CAR expression (Eyquem et al., 2017; Frigault et al., 2015) may paradoxically result in reduced T cell function due to exhaustion. Beyond the binding element, CAR construct design may also directly impact CAR function and potency (Hamieh et al., 2019; Majzner et al., 2020). A recent report of two seemingly very similar CD19-targeted CAR-T cell products, in which the only difference was the length of the linker in the scFv region, showed that the former resulted in inferior immune synapse formation (Singh et al., 2019) and reduced efficacy in pre-clinical models. When a CD19-CD28 CAR used in a phase I clinical trial was modified to include a CD8 hinge rather than the original CD28-derived sequence, in vitro testing revealed substantially reduced cytokine production with the CD8 modification, and a clinical trial confirmed substantial reductions in toxicity with comparable efficacy (Brudno et al., 2020). Thus, the structures of the binding region and the extracellular hinge and transmembrane domains have a role in determining CAR binding, function, and potency. In applying this principle to the CD27-ligand based system, an additional factor is the natural cleavage of CD27 into a soluble (sCD27) form through the action of matrix metalloproteases produced by T cells, AML, and other malignant cells (Burchill et al., 2015; Kato et al., 2007; Pirillo et al., 2020; Zhou et al., 2011). Confirming this issue, high levels of sCD27 were found in the supernatant of CD27-ligand based CARs, but not scFv-based CD70-targeted CARs (Sauer et al., 2021).
CAR-T cells may have a threshold tumor antigen density below which they are unable to kill (Fry et al., 2018), but higher avidity binding may increase T-cell responses to low antigen density tumors (Greenman et al., 2021). Antigen levels have been modulated pharmacologically in leukemia and myeloma, with bryostatin to increase CD22 expression in leukemia (Ramakrishna et al., 2019) and gamma-secretase inhibitors to reduce cleavage of BCMA from the surface of multiple myeloma cells, to augment potency of CAR-T cells to those antigens (Pont et al., 2019; Ramakrishna et al., 2019). Azacitidine (AZA), a hypomethylating agent (HMA) and nucleoside analogue that inhibits DNA methyltransferase (Pubchem) and decreases methylation of the CD70 promoter, resulting in increased CD70 surface expression in solid tumor cell lines as well as in primary AML blasts and leukemic stem cells (LSCs) (Kitajima et al., 2018; Ochsenbein, 2018; Riether et al., 2020). Recent preclinical characterization of the effect of HMAs on the tumor microenvironment of an immunocompetent mouse model demonstrated depletion of immunosuppressive MDSCs and PD-1 expressing T cells, thus generating an overall more immuno-permissive tumor microenvironment, possibly favoring adoptive T cell therapy (Nahas et al., 2019). Importantly, unlike bryostatin or gamma-secretase inhibitors, AZA is already FDA approved for the treatment of hematologic malignancies and is used extensively for the palliative management of patients with AML, especially those who are unfit for intensive, curative therapy (Ossenkoppele and Löwenberg, 2015).
In this study, we sought to optimize CD70-targeted CAR-T-cells for AML. The most potent previously described CAR targeting CD70, the second generation trCD27–41BBζ construct (‘native’) had only modest efficacy in xenograft models of AML. Using genetic and pharmacologic manipulations of AML tumor lines, we demonstrate that CD70-targeted CAR-T cell function correlates with increased antigen density. Next, we rationally designed a panel of hinge-modified CARs designed to abrogate protease-mediated cleavage of the truncated CD27 protein that serves as the extracellular domain of the CAR. We noted that although standard in vitro assays did not reliably distinguish the potency of the CAR variants, the CD8 hinge and transmembrane (CD8H&TM) domain-modified CAR had improved binding avidity, which correlated strongly with superior in vivo activity, both with and without AZA. Our data support further clinical development of this CAR in combination with AZA for AML.
RESULTS
Second generation CD70 CAR-T-cells are effective against AML
We analyzed mRNA expression values for CD70 from 50 AML patients in the Center for Patient Derived Models (CPDM) database at the Dana-Farber Cancer Institute using cBioPortal and found that a subset of patients’ disease overexpresses CD70 in some cases even when other canonical myeloid markers like CD33 and CD123 are reduced (Fig. S1A). Similarly, we examined a panel of AML cell lines and found that most expressed CD70 as measured by flow cytometry (Fig. S1B). We generated a second generation trCD27–41BBζ (native) CAR construct (Fig. 1A) and tested it functionally in vitro against a panel of AML lines for cytotoxicity (Fig. S1C), antigen-specific activation (Fig. S1D), and cytokine production (Fig. S1E). Overall, we found that functional activity largely correlated with target CD70 expression. We next confirmed its activity in an animal model of AML. After injecting 5 ×105 Molm13 cells expressing click-beetle-green (CBG) luciferase intravenously (IV) into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice on day −7, mice were treated on day 0 with 1×106 native CAR-T cells or untransduced T cells (UTD) (Fig. S1F). CAR-T treated mice had reduced tumor burden, prolonged survival, modest peripheral blood expansion, and a reduction in bone marrow blast count, but overall, this CAR-T cell product was only modestly effective against a moderate tumor burden (Fig. S1G-J).
Figure 1. Higher antigen density improves CD70-CAR-T cell avidity and activity.
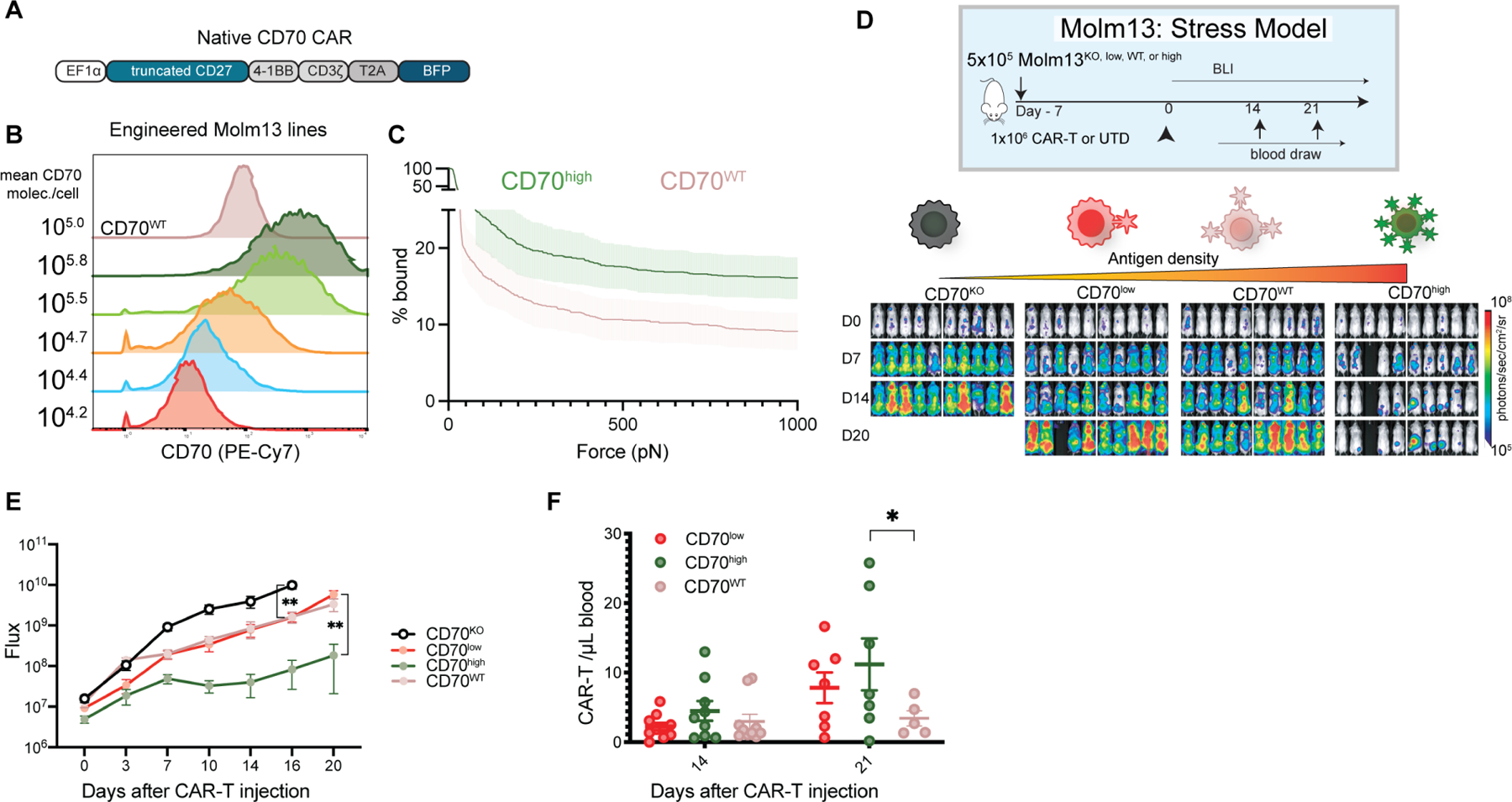
(A) trCD27–41BBζ, ‘native’ CD70-targeted CAR construct. (B) CD70 knockout Molm13 AML cells were transduced at various multiplicities of infection (MOI) with truncated CD70 lacking an intracellular signaling domain. Five populations were selected, and flow sorted for only CD70+ cells, generating five new cell lines, CD70 wild type (CD70WT), CD70 high (CD70high), CD70 high intermediate (CD70high-int), CD70 intermediate (CD70int), CD70 low intermediate (CD70low-int), and CD70 low (CD70low). CD70 expression by flow cytometry and number of molecules per cell are shown. (C) Immune synapse binding avidity to Molm13 CD70high or CD70WT targets assessed via acoustic force microfluidic microscopy. Data represents mean ± SEM and combined experiments from 4 separate microfluidic chips from separate donors. (D) Experimental setup: n=10 NSG mice per group were injected with 5×10⁵ cells from either Molm13 CD70KO, Molm13 CD70low (104.2 mol./cell), Molm13wt (105/mol./cell), or Molm13 CD70high (105.8 mol./cell) and then treated with 1×106 native CAR-T-cells seven days later (day 0) in a stress model of AML. Images represent bioluminescence (BLI) at the indicated time points. (E) Quantification of flux (photons/second). Points represent mean ± SEM. ** p <0.01 by unpaired t test. Comparison between CD70low and CD70WT was 0.17 by unpaired t test. (F) CAR-T cell expansion in the peripheral blood at the indicated time points determined by flow cytometry (CD3+ BFP+). Lines represent mean ± SEM. * p< 0.05 by mixed-effects model using Sidak correction for multiple comparisons. Comparison between CD70low and CD70WT was 0.2318 by the same mixed effects model. Representative of n=1 experiment.
See also Figures S1-S2.
Higher tumor antigen density increases CD70 CAR activity
We next sought to explore the effects of antigen density in our system through genetic perturbation of target cells. Starting with the Molm-13 cell line, which was the most reliable and consistent line to use in xenograft mouse models, we generated a panel of isogenic cell lines with variable levels of CD70 expression by first genetically knocking out CD70 (Fig. S1K) and then re-introducing it with viral transduction and selection of single-cell clones with stable CD70 expression (Fig. 1B). CD70 expression levels had no discernable effect on in vitro growth of the Molm-13 isogenic cell lines (Fig. S1L). We then performed our standard panel of in vitro functional assays, including cytotoxicity and activation, confirming a correlation between antigen density and CAR activation (Fig. S1M,N). Notably, low level CAR activation due to tonic CD3ζ signaling can lead to antigen independent clustering and activation (Long et al., 2015). Additionally, we evaluated binding avidity via acoustic force microscopy finding that native CAR-T cells demonstrated higher binding avidity to the CD70high tumor than CD70WT (Fig. 1C, S2). To determine how antigen density affected in vivo activity, we injected NSG mice intravenously with Molm13 CD70KO, CD70low, CD70WT, or CD70high AML, followed by a non-curative dose (1×106/mouse) of native CAR-T cells in a stress model to stratify responses (“stress models” being low-dose CAR T cells.) Mice bearing CD70high tumors had improved tumor control (Fig. 1D, E), and enhanced CAR proliferation over CD70WT bearing mice (Fig. 1F). Interestingly, CD70low and CD70WT expressing Molm13 were not differentially controlled by, and did not induce differential engraftment, in mice treated with the native CD70 CAR in vivo. Recent work examining the relationship between CAR activation and antigen density suggests that our results may be explained by the sigmoidal relationship between antigen density and CAR responsiveness. Our CD70low and CD70WT tumors appear to lay within a ‘flat’ portion of the sigmoidal antigen density-responsiveness curve, while increase to our CD70high levels result in substantial increases in CAR potency (Hernandez-Lopez et al., 2021; Majzner et al., 2020).
CD70 antigen density can be increased pharmacologically with azacitidine in AML
Use of agents to augment tumor antigen density have shown synergy with CAR-T cells (Ramakrishna et al., 2019). Azacitidine (AZA), a drug routinely used in the treatment of AML, has been shown to increase CD70 expression by hypomethylating the CD70 promoter (Riether et al., 2020). We hypothesized that AZA would potentiate native CAR-T cells by increasing antigen density (Fig. 2A). We confirmed increased CD70 antigen in multiple AML cell lines (Fig. 2B, Fig. S3A) after exposure to physiologically relevant concentrations of AZA. Increases in CD70 antigen expression were achievable in vivo through systemic administration of AZA. NSG mice were engrafted with Molm13 cells and then treated with 5 days of intraperitoneal (IP) AZA or vehicle (PBS); bone marrow evaluation indicated higher expression of CD70, but not the myeloid marker CD33 (Fig. S3B). We next asked whether AZA could enhance the potency of native CAR-T cells in a high tumor burden model of AML. NSG mice were injected with Molm13 cells IV on day −22, followed by 5 days of AZA or vehicle starting on day −4, and CD70 CAR-T cells on day 0 (Fig. 2C). In all AZA in vivo experiments, mice were pre-treated with AZA and at least 5 pharmacokinetic half-lives (based on human data) passed prior to administration of CAR-T cells on the same day with the goal of interrogating AZA’s effect CD70 modulatory effect on tumor (rather than CAR-T cells). Mice treated with AZA and native CAR-T cells had improved tumor control (Fig. 2D, 2E) and prolonged survival compared to the control group (Fig. 2F). CAR-T cells expanded in blood (Fig. 2G), and the AZA+CAR group had lower tumor burden in the bone marrow at the time of euthanasia or death (Fig. 2H).
Figure 2. Azacitidine augments CD70-CAR potency by increasing antigen expression on AML blasts in a high tumor burden model.
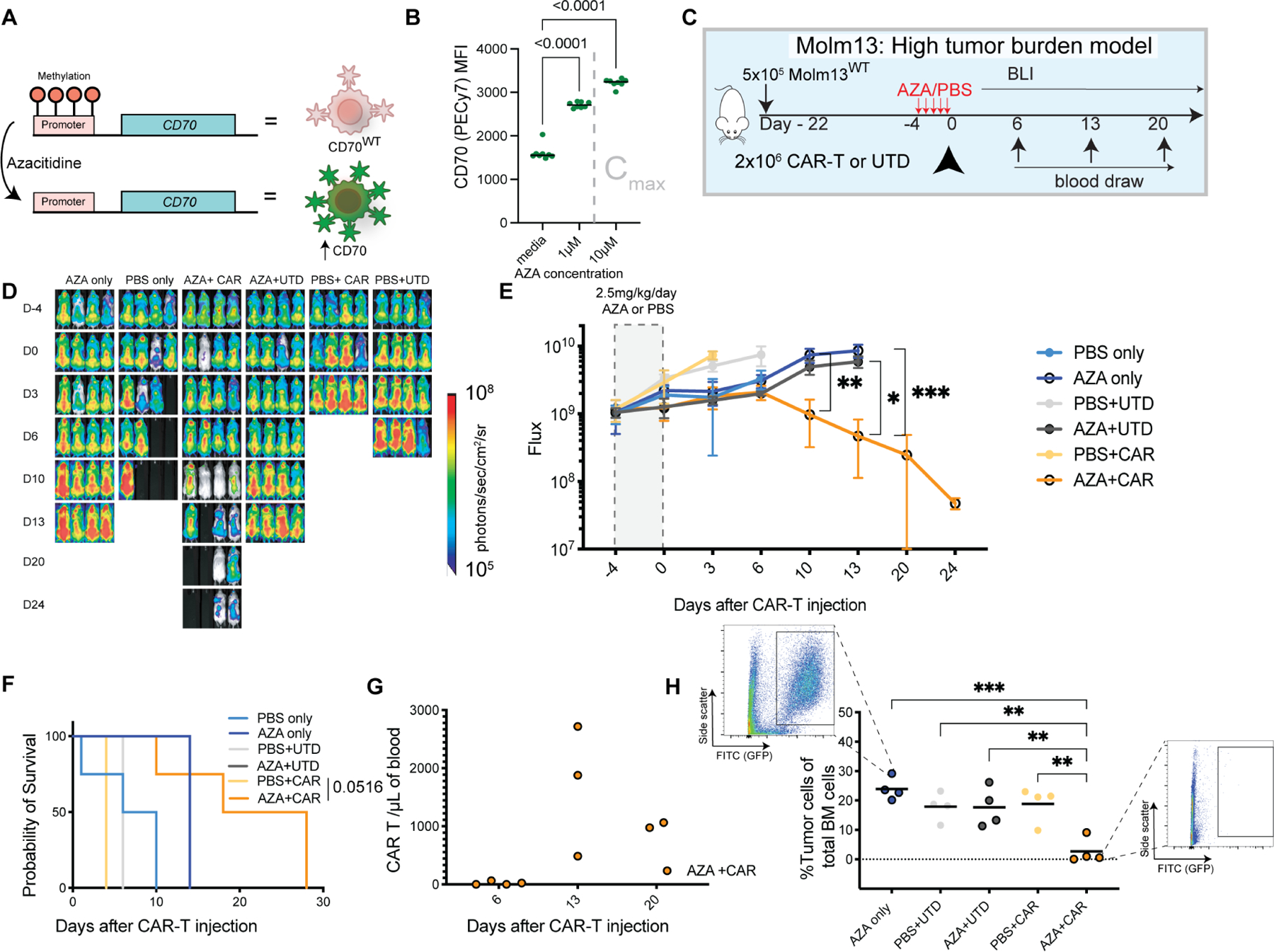
(A) Mechanism of CD70 antigen increase after CD70 expression (Riether et al., 2020). (B) CD70 expression levels by flow cytometry after Molm13 exposure to the indicated concentrations of azacitidine for 72 hours. Gated on live (DAPI-) cells. Cmax represents the maximal concentration in humans after a single dose of 75mg/m2 (Actavis_Pharma). Representative of n=3 independent experiments. Horizontal lines represent the median. (C) Experimental setup: NSG mice were injected with 5×10⁵ Molm13 on day −22 (n=4 mice per group). After prolonged tumor engraftment and randomization, mice received intraperitoneal injections of 2.5mg/kg/day azacitidine or vehicle (PBS) starting on day −4. On day 0 they were treated with either: no intervention, a single dose of 2 ×106 native CD70-targeted CAR-T-cells, or the equivalent number of untransduced T cells (UTD). (D) BLI of AML xenografts over time in the indicated groups. (E) Quantification of flux [photons/second] in the experimental groups at the indicated time points. Points represent mean ± SEM. * p< 0.05, ** p<0.01, *** p<0.001 by mixed effect model and Dunnett’s correction for multiple hypotheses. (F) Kaplan-Meier survival curves of the treatment groups. P-value as shown for AZA+CAR comparison to PBS+CAR control p by Log-rank (Mantel-Cox) test with Bonferroni correction. (G) Quantification of CAR-T-cells (CD3+ BFP+) measured in the peripheral blood by flow cytometry at the indicated time points. (H) Percentage of GFP positive cells in the femur at the time of death or euthanasia as assessed by flow cytometry gated as shown which were representative of the identified conditions. Bars represent the mean. ** p< 0.01, *** p< 0.001 by one-way ANOVA with Dunnett’s correction for multiple hypotheses to the AZA+CAR condition. Representative of n=2 independent experiments.
See also Figure S3.
CD70 CAR hinge modifications produce less cytokines despite comparable in vitro cytolytic capacity and expansion to the native CAR.
In order to further improve upon the in vivo potency of the native CD70 CAR, we systematically mapped out the structural elements of the CD27 protein, including disulfide bonds, glycosylation sites, and other elements thought to be crucial for secondary and tertiary structure based on the solved crystal structure (Teplyakov et al., 2017). We then utilized an in silico prediction algorithm to identify putative matrix metalloprotease cleavage sites within the CD27 protein that could produce a peptide product comparable to the known size of sCD27 (Cieplak and Strongin, 2017). To reduce the size of the construct and improve its binding to CD70, we generated four variants: “truncated” in which the stalk was partially deleted, “deleted” in which the entirety of the stalk was deleted, “flexible” in which the endogenous stalk was replaced with an equivalent length flexible linker, and CD8 hinge and transmembrane (CD8H&TM) in which the stalk was replaced with versions from the human CD8 protein (Fig. 3A). Structural modeling and difference distance mapping of these variants revealed a more rigid conformation of the truncated and deleted variants, while the flexible variant was predicted to maintain a similar conformation to the native structure, with the CD8H&TM CAR adopting an intermediate state (Fig. 3B). While CD70 is expressed transiently on activated T cells and could theoretically lead to fratricide, we saw no deficit in expansion with any of the variants relative to UTD (Fig. 3C). In vitro cytotoxicity revealed that the deleted variant was not functional, but the others were essentially indistinguishable against Molm13 targets (Fig. S4A). The CD8H&TM CAR-T cells slightly outperformed other variants against the OCI-AML3 line (Fig. 3D), but all the variants were comparable against the Monomac1 line (Fig. S4B). Similarly, levels of activation, as determined by CD69 expression upon co-culture with AML cells, were comparable across the variant constructs (Fig S4C). In vitro cytokine production among all variants on co-culture with Molm13 was substantially lower than the native CAR-T cells (Fig. 3E). To determine the effects of long-term antigen stimulation, we stimulated the CAR-T cell variants with an equimolar ratio of irradiated CD70-expressing K562 weekly for five weeks and interrogated their memory phenotype and expression of exhaustion markers, finding that they were similar among all the variant constructs (Fig. 3F). Finally, we took these recursively stimulated CAR-T cells and assessed their functional capacity in a real-time cytotoxicity assay. We found that the flexible and CD8H&TM variants had increased cytotoxic capacity relative to the truncated version (Fig. 3G).
Figure 3. Most hinge variants have comparable in vitro cytolytic capacity and expansion relative to the native CAR.
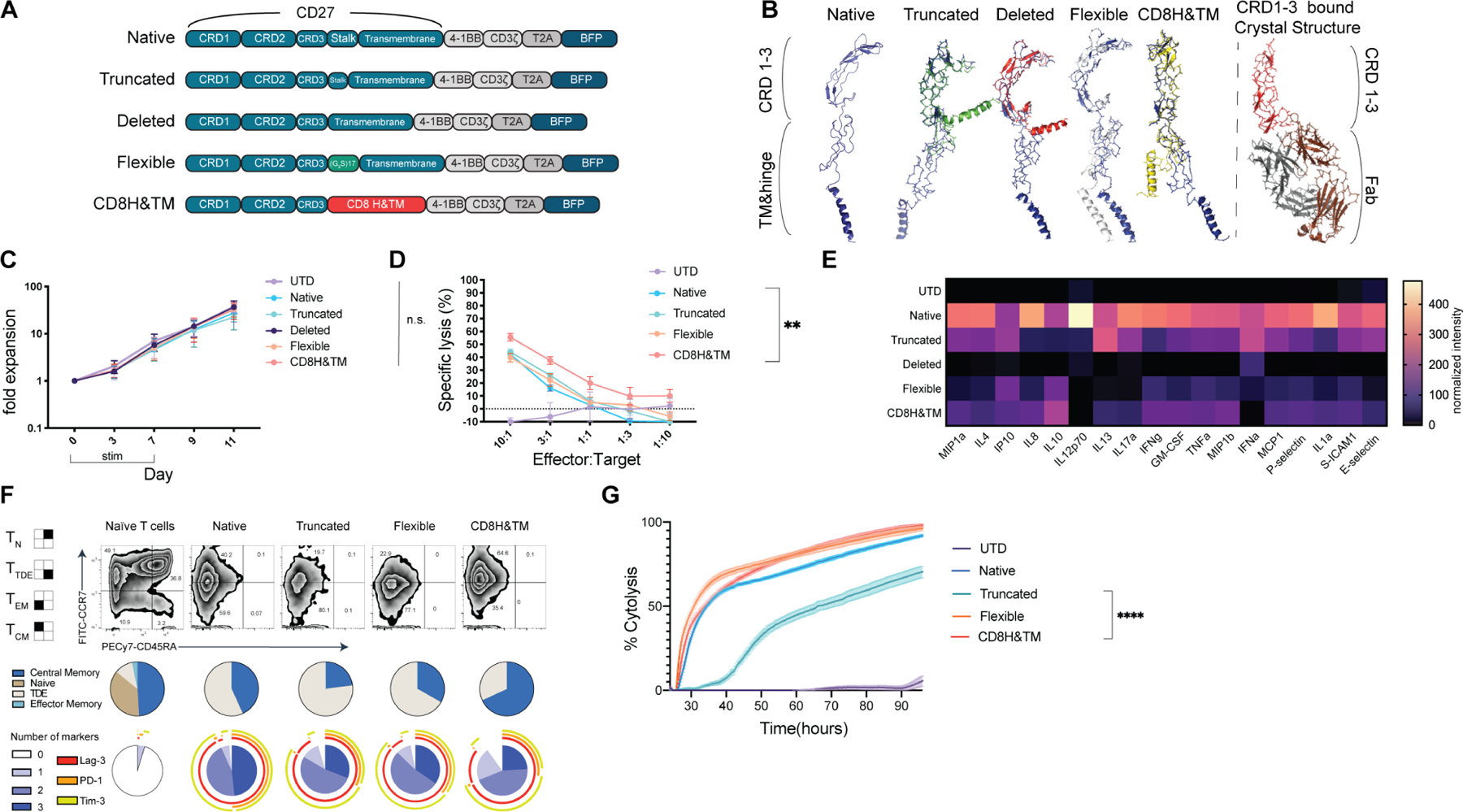
(A) Construct designs for CD70-targted CAR hinge variants. (B) Predicted structure of the CD70-targeted CAR cysteine rich domains (CRDs) as well as the hinge and transmembrane domains using the Phyre2 (Kelley et al., 2015) predictive engine with each aligned in Pymol3 with the MUSCLE plugin to the Native CRD (blue structure). Published crystal structure for CRD 1,2, & 3 is shown for comparison binding to a Fab. (C) CD70 hinge variant CAR expansion after lentiviral transduction. All differences are nonsignificant (ns) by 2-way ANOVA with Dunnett’s correction for multiple hypotheses. Points represent mean ± SEM of T-cells from 3 healthy donors. (D) Cytotoxicity as assessed in a luciferase-based killing assay for 16hrs with the indicated CAR and OCI-AML3 target. Data points indicate mean ± SEM of technical triplicates also performed in biological triplicate with T-cells derived from 3 healthy donors. * p< 0.05, **** p< 0.0001 by one-way ANOVA with Dunnett’s multiple hypotheses correction at the 10:1 E:T ratio compared to the native CAR. (E) Heatmap of normalized cytokines in the supernatants of CD70-CAR hinge variants after overnight co-culture with Molm13 at a 1:1 ratio. Cytokines were measured by Luminex assay in technical duplicates and biological triplicate with T cells derived from 3 healthy donors. Representative of n=1 independent experiment. (F) CD70-CAR hinge variants were stimulated with 5 doses of weekly CD70-expressing irradiated K562s at a 1:1 ratio and assessed at day 28 for phenotype and exhaustion markers. T cells from the corresponding unstimulated healthy donor are shown for comparison. Exhaustion markers from naïve T cells from the same unstimulated healthy donor are shown for reference. Performed in 3 independent donors, data is representative of a single donor. (G) The same CARs from day 28 were used in a real-time cytotoxicity assay against Molm13 targets at a 1:1 effector: target ratio (relative to Day 0 tumor seeding). Assay performed in quadruplicate. Means are plotted ± SEM. **** p <0.0001 by one-way ANOVA assessed at the terminal time point.
See also Figure S4.
To further delineate the nature of the binding between the CD70-CARs and AML tumor cells, we measured the binding avidity of CAR-T cells transduced with our variants using acoustic force microfluidic microscopy (Fig. 4A, B, Movie S1). We found a higher median level of force required to detach CD8H&TM CAR-T cells than the native CAR-T cells and other variants. Because our variants were in the membrane-proximal portions of the extracellular domain of the CAR, we hypothesized that the observed differences were more likely to be due to decreased cleavage of the extracellular portion (as opposed to changes in binding affinity, which would occur in the membrane-distal portion of the CAR). To assess for cleavage of the CARs, we performed an ELISA for soluble CD27 (sCD27) in the supernatant of the CAR variants in the presence or absence of tumor. We found significant increases in sCD27 in the supernatant of the native and truncated CARs, but not the UTD, or those in which the entirety of the hinge had been replaced (flexible and CD8H&TM) suggesting abrogation of proteolytic cleavage (Fig. 4C-D). The observation that the flexible CAR did not shed but still had lower avidity than the CD8H&TM CAR suggests that the structure of hinge itself plays a role in binding.
Figure 4. Decreased proteolytic cleavage of the CD8H&TM CD70-CAR results in enhanced binding avidity.
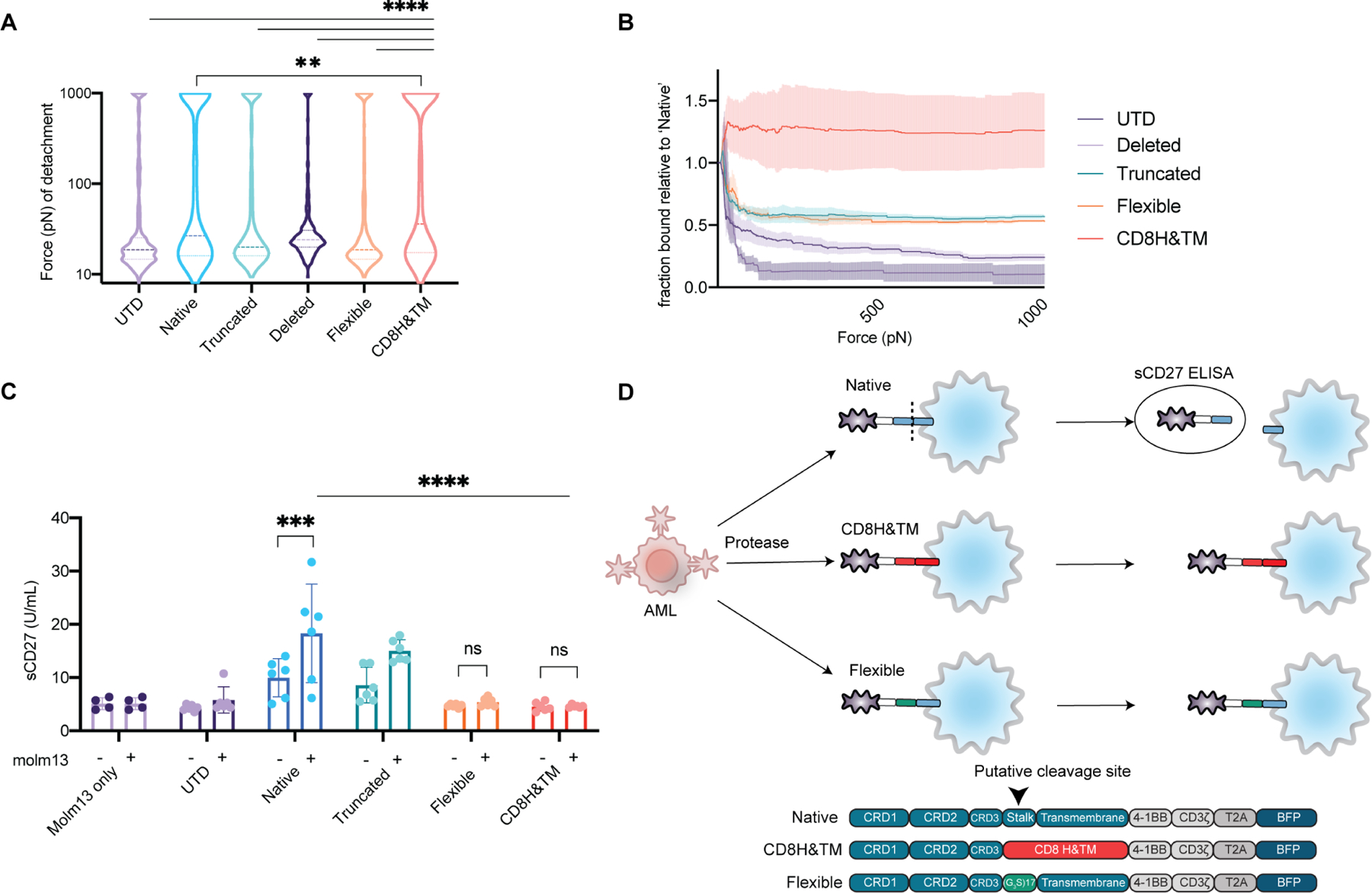
(A) CD70-targeted CAR hinge variants immune synapse binding affinity to Molm13 targets assessed via acoustic force microfluidic microscopy. Solid line represents median with dotted lines representing quartiles. Data represents combined experiments from at least 3 separate microfluidic chips over separate days comprising at least 330 effector-target binding events. **p <0.01, **** p <0.001 by Mixed-effects analysis with Dunnett’s multiple comparisons test to CD8H&TM. (B) Fraction of cells bound relative to the native CD70 CAR from (A) as a function of applied acoustic force. Curves represent mean ± SEM. (C) Supernatant from CD70-CAR hinge variants or UTDs in the presence or absence of Molm13 was obtained at a 1:1 ratio from overnight co-culture and assayed for soluble CD27 in an ELISA. Assay performed technical duplicates and biological triplicate with T cells derived from 3 healthy donors. Bars represent mean ± SEM. *** p<0.001, **** p<0.0001 by separate 2-way ANOVA tests (+/− Molm13 for each CAR, and between CARs) with Dunnette’s and Sidak’s multiple hypotheses correction respectively. Differences between CARs for the Molm13 absent conditions were non-significant. (D) Schematic of the assay and proposed mechanism from (C).
See also Movie S1.
High-avidity CD8H&TM CD70 CAR has enhanced in vivo anti-AML activity
To compare the in vivo anti-leukemic capacity of our CAR variants, we injected AML-bearing NSG mice with each of the functional CAR variants with a low CAR dose (1×106/mouse) in our non-curative stress AML model to best stratify responses (Fig. 5A). The CD8H&TM variant demonstrated significantly improved tumor control (Fig. 5A,B) and survival (Fig. 5C). CD8H&TM CAR-T cell expansion in the blood was also several orders of magnitude higher by day 21 than the other variants, including native (Fig. 5D), and contracted after day +35 (Fig. S5A. Peripheral blood CD8H&TM CAR-T cell characterization on day 35 revealed that the majority of CAR-T cells displayed a central memory phenotype (Fig. 5D). CD8H&TM CAR treated mice were the only group with a bone marrow effectively sterilized of tumor (Fig. 5E) and with high levels of CAR-T cells in the marrow (Fig. 5F). Immunohistochemistry for T cells (CD3), and tumor (CD33) revealed high levels of CD3 staining only in the CD8H&TM group and absent CD33 staining also only in this group (Fig. 5G). To date, analyses to determine the most potent CAR-T cell constructs have relied on pre-clinical mouse models, since in vitro assays often fail to distinguish among variants. We compared three in vitro measures of CAR-T cell potency (cytotoxicity, IFNγ secretion, and avidity) for their association with anti-tumor effect in the in vivo model. Goodness of fit-R2 value was highest (0.906) for the association with binding avidity, and low in the comparisons to in vitro cytolysis and IFN-γ production (0.5982, and 0.548 respectively), indicating that among the preclinical assessments, avidity was the most predictive of the in vivo results (Fig. 5H). We further explored this in a different ligand-based CAR where there was no clear winning construct in standard in vitro assays of CAR-T cell effector function. In this model, we used a monomeric APRIL binder or a trimeric APRIL (TriPRIL) binder to target BCMA and TACI in multiple myeloma cell lines. Our in vivo data demonstrated that TriPRIL CAR-T cells were significantly more effective than APRIL CAR-T cells (Schmidts et al., 2019). Here, we tested the original APRIL and TriPRIL CARs using acoustic force microscopy, and found a similar correlation between binding avidity and in vivo outcomes (Fig. S5B,C).
Figure 5. Avidity-enhanced CD8H&TM CD70 CARs have increased tumor control and proliferation in a stress model of AML.
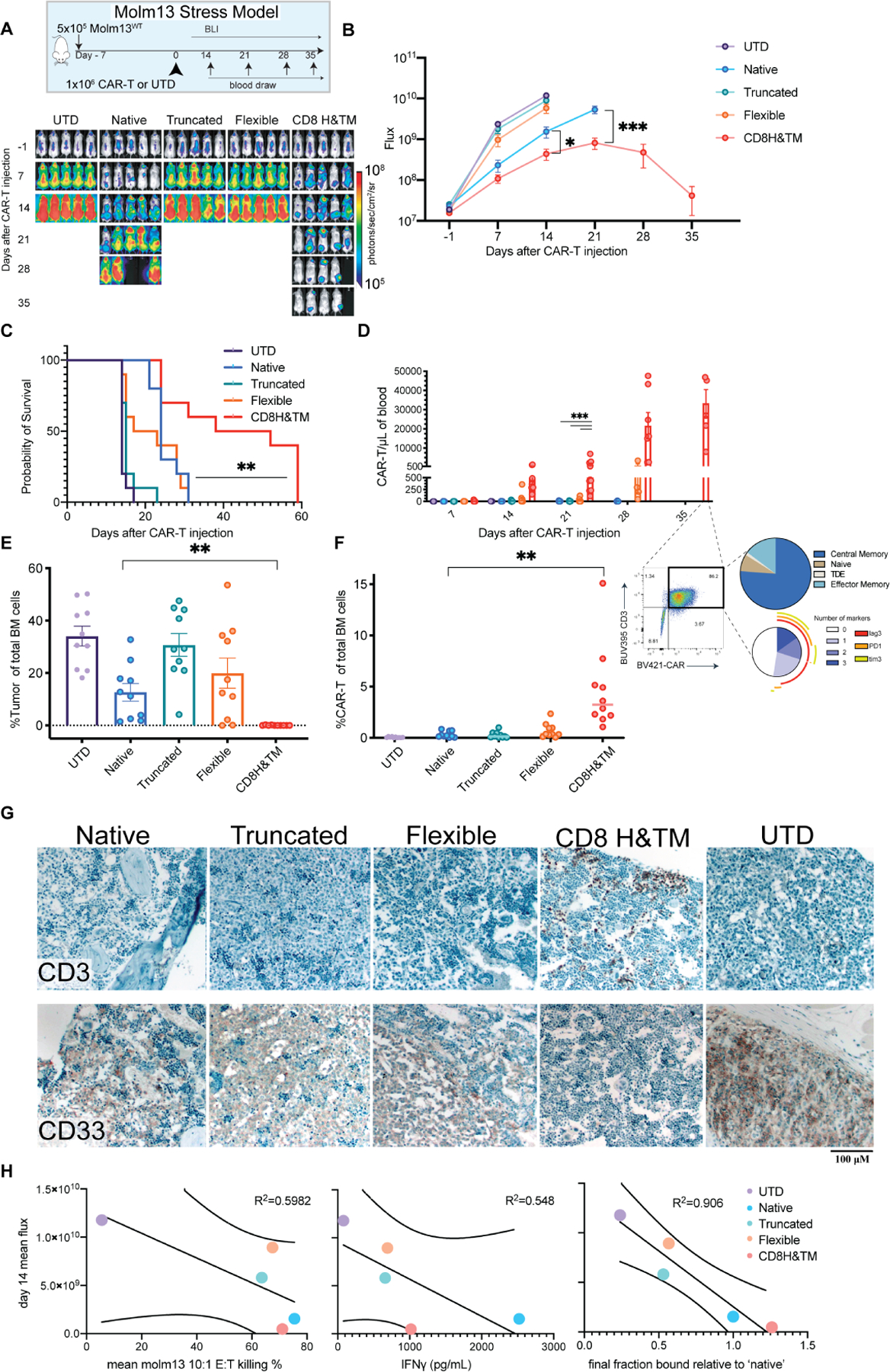
(A) Mice were engrafted with 5×10⁵ Molm13 cells on day −7. On day 0, ten mice per group received 1×106 CAR-T cells from the indicated CD70-CAR hinge variant or UTD. Experiment completed with two separate biological donors. Representative of n=1 experiment. (B) Quantitative flux measurements * p< 0.05, *** p <0.001 by unpaired T-test. Points represent mean ± SEM. (C) Survival ** p<0.01 Native compared to CD8H&TM by log-rank (Mantel-Cox) with Bonferroni correction. (D) Weekly CAR-T cell expansion assessed by flow cytometry. Bars represent mean ± SEM. *** p<0.001 by 2-way ANOVA with Dunnett’s multiple hypotheses correction. Representative T cell subsets and exhaustion markers are shown at day +35. (E) Bone marrow tumor burden assessed by flow cytometry as a percentage of total single cells in the marrow. Bars represent mean ± SEM. ** p <0.01 by unpaired T-test (F) CAR-T-cells as fraction of total bone marrow cells. Lines represent the median. ** p <0.01 by unpaired T-test. (G) Representative immunohistochemistry for the indicated markers is shown. Images underwent individual white balancing correction using the white balance correction_1.0 plugin for ImageJ. Identical brightness and contrast adjustment applied to all images to improve clarity. (H) Correlations between mean peak flux for each in vivo experimental group for which all groups were still evaluable (day +14) and remaining bound cells relative to the native CAR-T cells after maximal force application in the avidity experiments from figure 3, mean in vitro Molm13 killing at a 10:1 ratio, and mean IFNγ production after Molm13 co-culture are shown. R2 value represents a linear regression goodness of fit, lines represent 95% confidence bands of the best fit line.
See also Figures S5.
We then compared our leading candidate CD8H&TM CAR-T cells to the native CAR-T cells in a patient-derived xenograft (PDX) model of AML (Fig. 6A). CD70 expression as assessed by flow cytometry was moderate, but lower than our other xenograft models (Fig. 6B). 5 × 106 fresh PDX cells were injected on day −4 followed by 2 × 106 CAR-T cells of either native or CD8H&TM on day 0. Serial blood monitoring by flow cytometry revealed steady tumor growth in the untreated and native CAR-T cell treated mice with low-level delayed CAR-T cell expansion, while the CD8H&TM mice did not have detectable circulating tumor, they had high level CAR-T cell expansion by day 14 and 21, and continued survival (Fig. 6C-E). To demonstrate that the increase in sCD27 shedding was a result of cleavage of the native CAR and not due to cleavage of endogenous CD27, we first identified that Jurkat T cells lacked endogenous CD27 expression (Fig. 6F). We then transduced Jurkat T cells with either the native or CD8H&TM constructs and achieved equivalent transduction efficiencies. CAR-transduced Jurkat cells were incubated overnight with Molm13 AML cells, and sCD27 was measured by ELISA. We found substantially reduced sCD27 in the supernatant of the CD8H&TM construct relative to the native (Fig. 6G), demonstrating that sCD27 originates from cleavage of the CAR itself.
Figure 6. CD8H&TM but not native CD70 CAR-T cells eradicate AML in a PDX model.
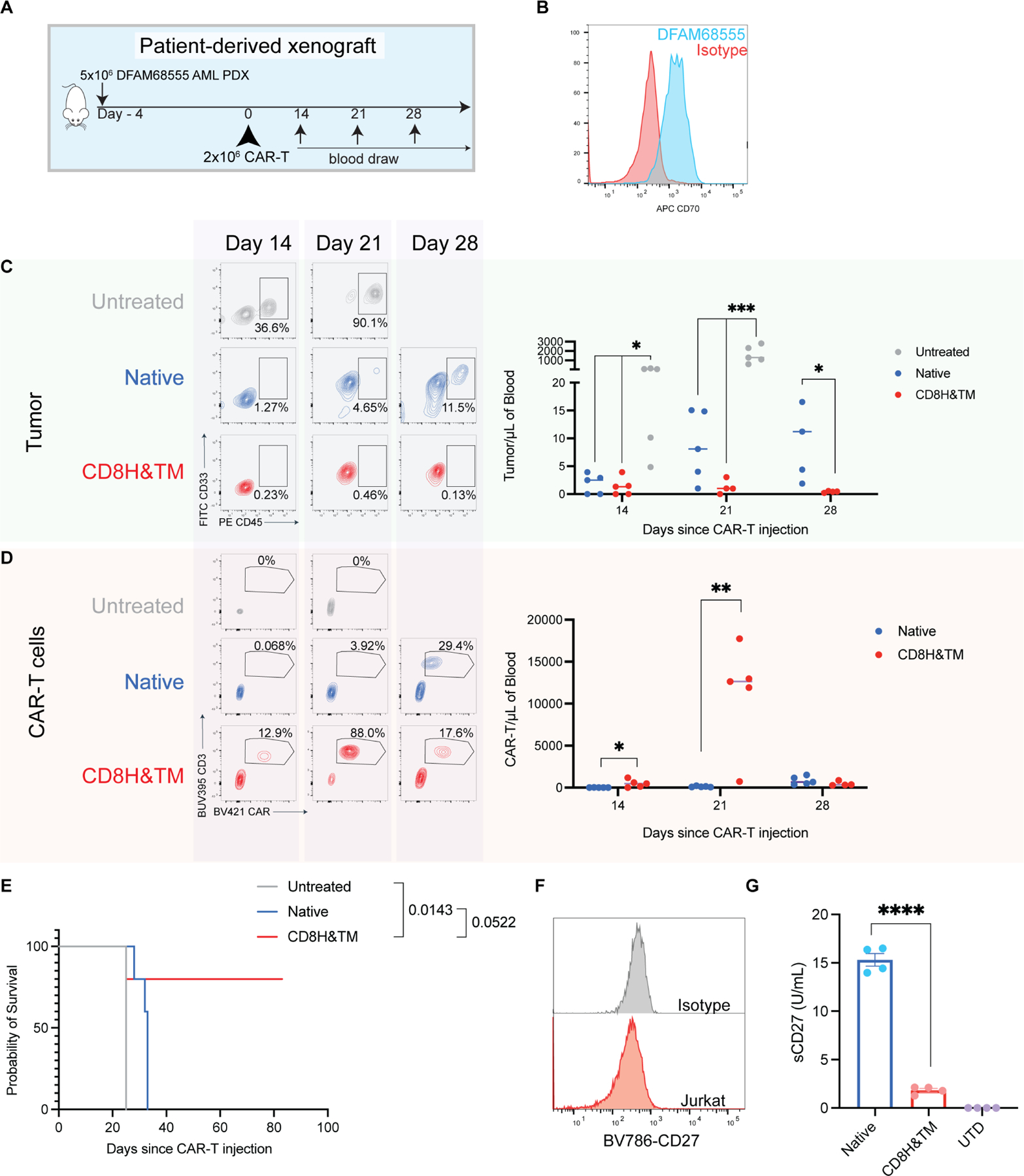
(A) Experimental schema: Mice were engrafted with 5×106 DFAM68555 AML PDX cells from an adult AML patient with monocytic, therapy-related AML on day −4. On day 0 they received no treatment or 2×106 of either native or CD8H&TM CAR-T cells and underwent weekly blood draws to monitor tumor (CD33+CD45+) and CAR-T (CD3+BFP+) expansion in the blood. (B) DFAM68555 CD70 expression relative to isotype control. (C,D) Representative tumor (C) and CAR (D) expansion at the indicated time points * p <0.05 by one-way ANOVA with Dunnett’s correction for multiple comparisons, *** p<0.001 by one-way ANOVA with Dunnett’s correction for multiple comparisons, * p<0.05 and ** p<0.01 both by unpaired T-test. Horizontal lines represent the median. (E) Kaplan-Meier survival curve. P values by Mantel-Cox test shown for the indicated comparisons without multiple hypotheses correction. (F) Jurkat T cells were assessed via flow cytometry for CD27 expression. (G) Jurkat T cells were transduced with the indicated CAR-T cells and co-cultured overnight at a 1:1 ratio with Molm13. Supernatant was assayed for sCD27 via ELISA. **** p <0.0001 by unpaired t test. Representative of n=1 independent experiment. Bars represent mean ± SEM.
Finally, we employed our two orthogonal strategies to increase potency of CD70-directed CAR-T cells for AML, aiming to increase binding by AZA-mediated enhancement of antigen density and use of the high-avidity, cleavage-resistant CD8H&TM variant CAR. We injected NSG mice with 5×105 OCI-AML3 on day −7, treated with 5 days of intraperitoneal AZA, and then administered 2×106 of either native or CD8H&TM CAR-T cells on day 0 (Fig. 7A). By day 20 neither AZA alone nor CAR alone were sufficient to provide tumor control, however, combination of the two agents eliminated tumor by day 30. On day 30 the mice were sacrificed, and bone marrow was analyzed. The CD8H&TM CAR-T cells either with or without AZA were superior to the native CAR-T cells, but only the addition of AZA resulted in eradication of tumor (Fig. 7A, B, C). While the CD8H&TM CAR with or without azacitidine was able to sterilize the bone marrow, only the combination of the two was sufficient to eradicate the extramedullary disease that OCI-AML3 is known to cause due to its strong proclivity for migration and invasion (Faderl et al., 2011; Xu et al., 2016; Yang et al., 2019). Expansion of CD8H&TM CAR-T cells were again orders of magnitude higher than native CAR-T cells and greater in the AZA than vehicle treated mice (Fig. 7D). Mice treated with AZA and the CD8H&TM CAR-T cells had substantially higher levels of sentinel CAR-T cells in the marrow than all other conditions (Fig 7. E). In a similar experiment using our other xenograft model, Molm13, we found that pretreatment with AZA, followed by either native or CD8H&TM CAR-T cells resulted in substantially improved tumor control, survival, and CAR-T cell expansion (Fig. S6 A-F). Thus, the combination of pharmacologically mediated antigen density with a cleavage-resistant, ligand-binder CAR-T, resulted in increased in vitro avidity and enhanced in vivo potency in AML.
Figure 7. Avidity enhanced CD8H&TM CARs outperform native CD70 CARs and eradicate tumor when combined with azacitidine.
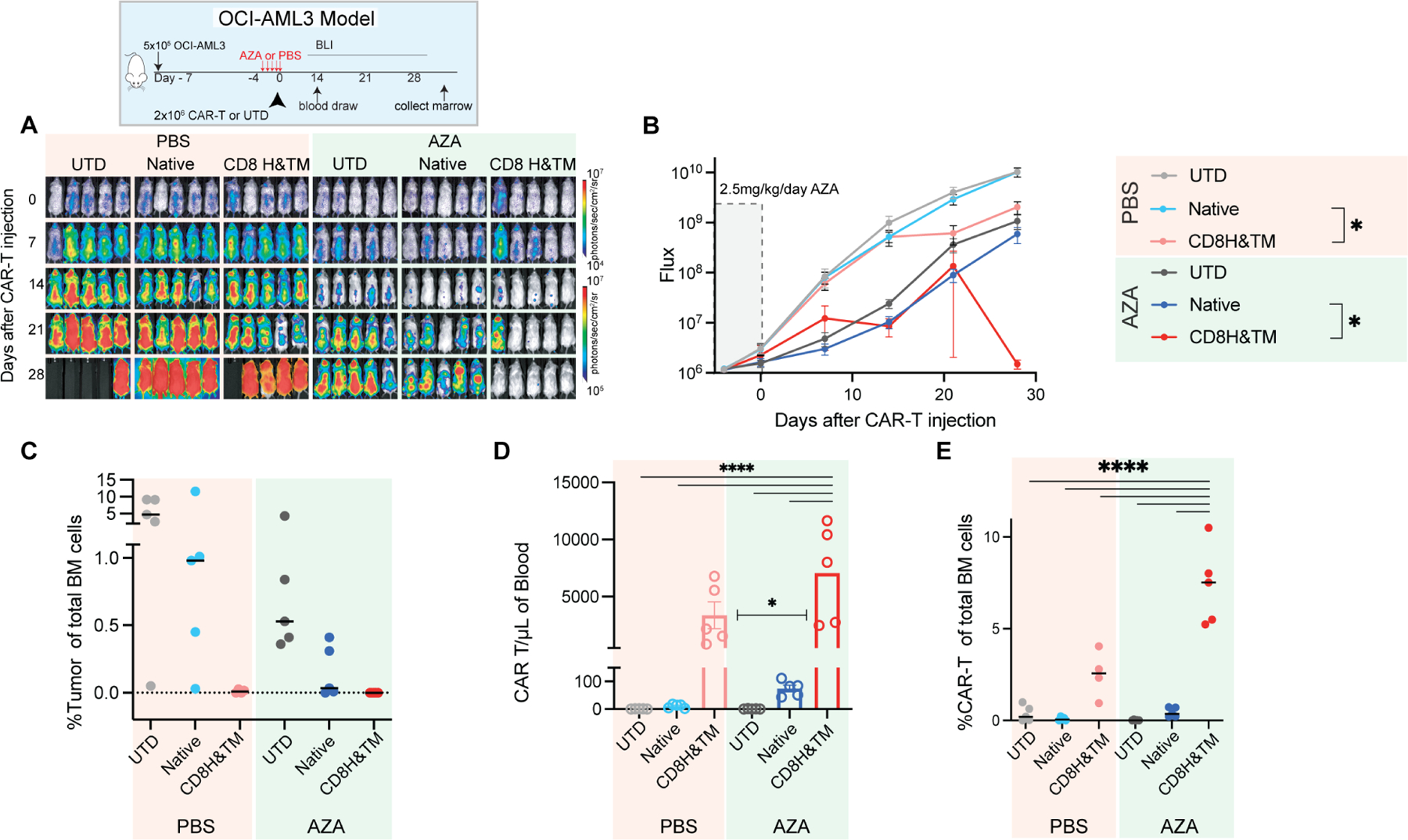
(A) NSG mice were injected with 5×10⁵ OCI-AML3 AML cells and tumor burden was monitored by BLI. After tumor engraftment and randomization, mice received IP injections of 2.5mg/kg/day AZA starting on day −4 for a duration of 5 days. On day 0 they were treated with the indicated effector cell at a dose of 2×106. On day 30 all mice were sacrificed, and bone marrow was evaluated via flow cytometry for tumor and CAR-T cells. (B) Quantification of flux [photons/second] in the experimental groups at the indicated time points. Mean ± SEM is shown. * p< 0.05 by unpaired T-test. (C) Bone marrow tumor burden assessed by flow cytometry as a percentage of total single cells in the marrow. Lines represent the median. (D) Quantification of CAR-T cells (CD3+ BFP+) measured in the peripheral blood by flow cytometry at day 14. Bars represent mean ± SEM. * p<0.05, **** p<0.0001 by one-way ANOVA with Dunnett’s multiple comparisons test. (E) CAR-T cells as percentage of total bone marrow cells. Lines represent median. **** p <0.0001 by one-way ANOVA with Dunnett’s multiple comparisons test. Representative of n=1 independent experiment.
See also Figure S6.
DISCUSSION
In this study we investigated orthogonal methods to improve cellular avidity of a CD70-CAR construct in models of AML. We show enhanced avidity and in vivo efficacy of the native CD70-CAR against high antigen density targets and that increases in antigen density can be achieved via treatment with AZA. To improve CD70 CAR-T avidity, we screened a panel of hinge variants designed to mitigate the unique property of CD27-ligand-based CARs to undergo cleavage at the hinge. We identified that replacement of the stalk region of CD27 with either a flexible linker or the CD8H&TM abrogated hinge proteolysis, but only the CD8H&TM CAR had enhanced binding avidity. Interestingly, of the pre-clinical assays used to compare CAR constructs, only binding avidity correlated with in vivo performance.
Pre-clinical selection of optimal candidate CARs for clinical translation has proven challenging. Typical in vitro assays include assessment of cytolysis, cytokine production, and short- and long-term proliferation. Ideal performance for a future clinical product on these assays is unclear and may be paradoxical. For example, cytokine production in vitro may or may not predict anti-tumor efficacy or cytokine release syndrome toxicity, despite its use as a potency assay in commercial CAR-T products targeting CD19. A recent report of a hinge-modified CD19-targeted CAR-T cells demonstrated substantially reduced in vitro cytokine production compared to a nearly identical older version, but the modified CAR resulted in substantially reduced toxicity despite comparable efficacy (Brudno et al., 2020). In our study, cytotoxicity assays and cytokine production in response to antigen stimulation quickly identified our deleted CAR as ineffective, but was not predictive of the differences in potency in vivo among the variants.
Attempts to improve CAR-T cell binding have generally focused on the affinity of the targeting moiety. However, increased affinity can increase bystander damage and toxicity (Castellarin et al., 2020; Liu et al., 2015). Additionally, inordinately high CAR surface expression can lead to exhaustion (Lynn et al., 2019). Similarly, poor clustering of CARs on the surface can result in early exhaustion and poor functionality (Long et al., 2015). With TNF superfamily-based CARs, higher order surface clustering and the hinge region has been implicated as playing a critical role in binding and successful signal transduction (Richter et al., 2012; Vanamee É and Faustman, 2018; Wyzgol et al., 2009). Indeed, we have previously shown that a trimeric ligand-based CAR using APRIL, another TNF family member, required higher-order clustering for adequate binding (Schmidts et al., 2019). Due to the size of the extracellular domain of CD27, we did not find that it was feasible to improve binding or function by multimerizing the extracellular domain of CD27. Instead, we focused on improving binding by reducing cleavage of the extracellular domain with a series of rationally constructed variants. Our data demonstrate that both the CD8H&TM and flexible CARs reduced cleavage, but only the CD8H&TM modification improved avidity and in vivo efficacy. In addition, the CD8H&TM appeared to confer more favorable long term signaling characteristics, as after repeated antigen stimulation, our CD8H&TM modified CARs maintained a high level of expansion with the majority exhibiting a central memory phenotype (Fraietta et al., 2018a). As with most CAR-T cells, our CD70-targeted CAR-T cells exhibit some degree of basal signaling which may be the consequence of prior activation from the manufacturing process, low-level allogenic activation upon incubation with non-MHC matched targets, or tonic signaling due to high levels of CAR expression (Boroughs et al., 2020; Long et al., 2015).
Recent careful modeling of a CD19-CAR system demonstrated that hinge modification alone was sufficient to drive the formation of an improved immunological synapse via more efficient receptor clustering (Majzner et al., 2020). Given the number of factors impacting cellular avidity, the improvement seen with our CD8H&TM CAR may be multifactorial--including decreased hinge proteolysis and more favorable immune synapse formation. While the flexible CAR was also able to abrogate cleavage, the suboptimal avidity and in vivo performance suggests that unfavorable immune synapse formation may be a separate factor. Indeed, molecular modeling of glycine-serine linkers suggests that longer polymeric structures like the flexible construct may not have the requisite rigidity for stable synapse formation which is an important parameter for signaling in TNF family members (Li et al., 2016; Richter et al., 2012). Together these data support the hypothesis that reducing cleavage is important, and the hinge itself plays additional roles (Alabanza et al., 2017; Majzner et al., 2020).
Whereas affinity refers to the specific binding strength between an scFv and its target, cellular avidity engenders the entirety of the cellular synapse including sustained expression of the CAR binding domain, co-receptor binding, cell adhesion, target antigen density, cytoskeleton, and other interactions, thus integrating the net effect of a complex web of attachments (Greenman et al., 2021). Recently, a strong correlation between avidity and CAR receptor expression levels reflected the dominant contribution of the receptor to avidity, and increased avidity resulted in increased sensitivity to low antigen concentrations (Greenman et al., 2021). Increased binding could also be achieved with dual-antigen targeted bicistronic CARs (Fernández de Larrea et al., 2020). We leveraged a relatively new tool to measure avidity between cells by applying acoustic force in a microfluidic device. Similar to Greenman 2021, we use the term cellular avidity to encompass the strength of the entirety of the cellular synapse, which includes sustained expression of the CAR binding domain, co-receptor binding, cell adhesion, target antigen density, cytoskeleton, and other interactions (Greenman et al., 2021). Although we did not measure co-receptor binding or other interactions specifically, we would expect these to be the same among the different conditions tested. The instrument we uses exerts ramped up force in the z-plane between the tumor cell layer and the T cells; thus, the quantification of bound cells as the force increases is an integrated measure of the web of interactions between the T cells and their targets. As the use of this measurements increases, it may be that another term, besides “avidity,” enters the mainstream use. In our system, despite inhibition of proteolysis by both the flexible and CD8H&TM CAR, only the latter demonstrated superior avidity, suggesting a role for the hinge domain on avidity as well. It remains to be seen if the correlation we observed between avidity and in vivo efficacy will be generalizable to all tumor-target systems. Preclinical and clinical studies have found that the inclusion of a CD8H&TM rather than a CD28 hinge and transmembrane results in lower levels of activation induced cell death, lower cytokine production in vitro, and dramatically lower systemic toxicity in patients despite comparable efficacy (Alabanza et al., 2017; Brudno et al., 2020; Majzner et al., 2020). Though these findings were in the context of CD19-targeted CARs with CD28-costimulation, we also observed lower cytokine production despite comparable cytotoxicity with the CD8H&TM, suggesting that this might be an intrinsic feature of the CD8H&TM module across CAR systems.
In AML, the first clinical trial utilizing a CD70-targeting agent was a monoclonal antibody in combination with AZA. This trial demonstrated favorable responses in individuals with AML unfit for intensive therapy and is being developed in additional clinical trials (NCT04023526, NCT04241549, NCT04150887) (Riether et al., 2020). CD70 represents a unique CAR target, unlike other conventional CAR antigens like CD19, CD22, and BCMA in that it confers a signaling advantage to AML blasts and LSCs, which is desirable in a single target setting (Riether et al., 2020; Riether et al., 2017). Additionally, while CD70 appears to be transiently expressed on a limited number of activated immune cells, it is not expressed in other normal cells, including hematopoietic stem cells. This expression profile makes CD70 a highly attractive target when compared to other myeloid targets (Bowman et al., 1994; Nolte et al., 2009; Perna et al., 2017; Sauer et al., 2021). Since CD70-CD27 signaling provides a survival stimulus to LSCs, it is theoretically possible that strategies like AZA to increase its expression might paradoxically augment tumor growth, but our study demonstrates a clear net effect of anti-leukemia. Importantly, while AZA treatment globally reduces methylation across AML samples, a comprehensive multi-omics analysis revealed that effects on individual surface protein levels may be heterogeneous between cell lines, mirroring clinical practice where patients can have dramatically different responses to therapy (Leung et al., 2019). Reassuringly, the AZA combination with an anti CD70-targeted antibody appeared to be synergistic in a small study compared to historical controls (Riether et al., 2020). Finally, while CD70 expression levels appear important for CD70 CAR-T cell function, other variables including effector to target ratio, immune synapse formation, antigen density, tumor growth rate, and intrinsic ability of the tumor to evade killing (e.g. downregulation or loss of apoptotic family members (Singh et al., 2020) ) certainly cooperate as well.
A limitation of our study is the focus on pre-treatment (rather than concurrent) use of AZA prior to CAR-T cell treatment. While mice were pre-treated with AZA and at least 5 half-lives elapsed prior to CAR-T cell injection for animal models, it is possible that some residual AZA might have been bioavailable and modulated CAR-T cell performance, though we anticipate this would be very limited, if present. Further exploration will be necessary to characterize potential effects of AZA on CAR-T cells. Encouragingly, recent work has shown that hypomethylating agents may further potentiate CAR-T cells directly; indeed, a single low dose of decitabine during CAR-T expansion enhanced anti-tumor potential in one pre-clinical study (Wang et al., 2021). A case report describing the effect of biallelic loss of the epigenetic regulator TET2 in a CD19 CAR-T cell for chronic lymphocytic leukemia raises the idea that pharmacologic epigenetic reprogramming may generally augment CAR function and remains to be explored in greater detail (Fraietta et al., 2018b; Jain et al., 2020). A potential alternative hypothesis is that AZA selectively depletes CD70-low blasts (rather than directly mediating CD70 increases on a per-cell basis), but this alternative hypothesis would also lead to augmented anti-tumor responses with the combination of AZA with CAR-T cells. One strength of our work is the use of xenografts with substantially higher tumor burdens than typically used in CAR-T cell studies, coupled with treatment doses in the low range for CAR-T cells (Bonifant et al., 2016; Duong et al., 2019; Petrov et al., 2018). Like most CAR-T cell studies, our work relied on the use of T cells obtained from healthy donors which may not recapitulate intrinsic disease-specific deficits that might be present in leukemia patients.
The most important first step in any clinical translation is a demonstration of safety. A CD70-targeted antibody has been successfully utilized with AZA in a clinical trial without significant toxicity, and a CD70-targeted second-generation CAR-T trial for solid tumors (NCT02830724) has not reported toxicity since enrollment began in 2016 (Riether et al., 2020). Further efforts to quantify tumor CD70 expression and/or promoter methylation status would be very beneficial for clinical translation. AZA is currently the de facto standard of care in patients unfit for intensive chemotherapy and could be leveraged into a combination clinical trial without great difficulty in this patient population. In summary, our work proposes a combination strategy of pharmacologic therapy that potentiates CAR-T cellular immunotherapy and warrants further study in AML with a high unmet clinical need.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Marcela Maus (MVMAUS@mgh.harvard.edu)
Materials availability
Plasmids generated in this study will be made available upon request and completion of a Material Transfer Agreement.
Data and code availability
Novel datasets were not generated with this study. Bulk-RNA data was obtained from the CPDM Pan-Cancer Patient Derived Models (https://www.dana-farber.org/research/departments-centers-andlabs/integrative-research-centers/center-for-patient-derived-models/ordering-patient-derived-models/).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines, cell culture, and PDX tumors
Jurkat, Molm13, MM.1S, THP-1, SupT1, HEK293T, and K562 cell lines were obtained from the American Type Culture Collection (ATCC) and OCI-AML3, MonoMac-1 were obtained from DSMZ-German Collection of Microorganisms and Cell Cultures. Nomo1 was a gift from Dr. Christopher Ott and Molm13 was a gift from Dr. David Weinstock. DFAM68555 PDX cells were obtained from the ProXe (https://www.proxe.org/) database (Townsend et al., 2016). Human T cells were purified (Stem Cell Technologies, 15061) from healthy donor leukopaks purchased from the Massachusetts General Hospital blood bank under an institutional review board-approved protocol.
Mice
6–10 week old male or female in-house bred NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) (The Jackson Laboratory) mice were housed in cages of up to 5 mice under pathogen-free conditions. Animals were housed at temperatures of 21.1–24.5C (70–76F), 30–70% humidity, and 12:12 light dark cycles. Mice were maintained at the MGH Center for Cancer Research and all care and conducted experiments were carried out using protocols approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee.
METHOD DETAILS
Study design
The objective of the study was to improve the potency of CD70-directed CAR-T-cells for AML via pharmacologic and genetic avidity enhancements. An existing CD70-targeted second generation trCD27–4-1BB CAR was characterized after insertion into primary human T-cells through assays for cytotoxicity, activation, and cytokine production. Genetic or pharmacologic methods to increase avidity via tumor antigen density augmentation were explored, followed by trCD27–4-1BB CAR hinge structural modifications to mitigate proteolysis and augment avidity. In vitro replicates were variable and are indicated in the figure legends. The effect of these parameters on our system were investigated using similar in vitro assays in addition to direct avidity measurement through acoustic force application. Mouse models of leukemia were evaluated by bioluminescence, bone marrow aspirate via histology and flow cytometry; human T-cells were enumerated in the bone marrow and blood. For in vivo experiments, mice were randomly assigned to treatment groups based on tumor burden.
CAR design
Transgenes were designed in Geneious (version 11.1.5), then synthesized and cloned into third generation lentiviral vectors under the regulation of a human EF-1α promoter. The native trCD27–4-1BB-CD3ζ CAR was engineered based on a previously published design (Wang et al., 2017) utilizing a blue fluorescent protein (BFP) reporter after a self-cleaving peptide sequence. Hinge variants were designed using annotations from clinvar (https://www.ncbi.nlm.nih.gov/clinvar/), sequence motifs from the published crystal structure,(Teplyakov et al., 2017) and an in silico matrix metalloprotease cleavage predictive algorithm (Cieplak and Strongin, 2017).
Molecular modeling
Molecular models of hinge variants were generated using the Protein Homology/analogy Recognition Engine V2.0 (Phyre2) (Kelley et al., 2015). These were then visualized in Pymol (version 2.4.1) with the use of the PyMod (version 3.0.2) plugin and ClustalW tool for alignment the native structure.
Cell maintenance and generation
Cells in culture were maintained in RPMI+Glutamax+HEPES (ThermoFisher Scientific, Cat# 72400047), supplemented with 10% FBS (R10), MM.1S was supplemented with 20% FBS (R20) at 37C. Cell lines were transduced to express click beetle green (CBG) luciferase and enhanced GFP (eGFP) and sorted on a BD FACSAria to obtain a clonal population of transduced cells. CD70 KO Molm13 cells were generated via use of the following CD70 CRISPR guide from the Brunello library “GAGCTGCAGCTGAATCACAC” (Doench et al., 2016). DNA guides were purchased from integrated DNA technologies (IDT) and converted to RNA via the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs, Cat# E2050S). After guide RNA and Cas9 protein electroporation into Molm13 cells, single cell clones were established via sorting on a BD FACSAria and monitored for similar proliferative capacity to parental lines. Finally, one clone was chosen to engineer increasing levels of truncated CD70 protein by lentiviral transduction. These Molm13 null, trCD70 transduced lines were then sorted via BD FACSAria for only CD70 expressing cells to establish lines of variable expression. CD70-expressing K562s were established in a similar fashion after transduction with a truncated version of the protein.
Lentiviral production
HEK293T cells were expanded in RPMI 1640 media with GlutaMAX and HEPES (ThermoFisher Scientific 72400–120) with 10% FBS (R10). Constructs were synthesized and then cloned into a third-generation lentiviral backbone (GenScript). Replication deficient lentivirus was produced by transfecting plasmids into HEK293T cells. Supernatant was collected at 24 and 48 hours after transfection. Virus was filtered and then concentrated by ultracentrifugation followed by storage at −80C.
Transfection, T cell culture, activation, and transduction
Genes were synthesized and cloned into a lentiviral plasmid backbone, which was transfected with lipofectamine into HEK293 cells as previously described (Scarfò et al., 2018; Schmidts et al., 2019). Human T cells were activated using CD3/CD28 Dynabeads (ThermoFisher Scientific, 40203D) on Day 0, followed by transduction with a lentiviral vector encoding the CAR on day 1 (24 hours later). T cells were cultured in R10 media with 20 IU/mL of recombinant human IL-2, penicillin, and streptomycin. T cells were debeaded on day 7 and cryopreserved on day 14. Transduced T cells were normalized to the same level of transduction for individual experiments utilizing untransduced but cultured and activated T cells from the same donor and expansion run.
Cellular cytotoxicity, cytokines, and ELISA assay
Cytotoxicity was assessed via co-culture of CAR-T cells with click beetle green luciferase-expressing tumor targets at the indicated ratios for approximately 16 hours. Luciferase activity was measured using a Synergy Neo2 microplate reader from Biotek. Specific lysis percentage for cytotoxicity assays was calculated using the following equation: %specific lysis = ((target cell only luminescence – luminescence of CAR+target)/target cell only luminescence) x100. For cytokine assays, cell-free supernatants were analyzed for cytokine expression using a Luminex array (Luminex Corp, FLEXMAP 3D) according to the manufacturer’s directions. All samples were measured in technical duplicates for cytokine assessment. sCD27 ELISA was performed using the supernatant from overnight co-cultures using an Invitrogen kit (ThermoFisher Scientific BMS286INST) according to the manufacturer’s instructions.
Real-time cytotoxicity assay
Cell index was recorded as a measure of impedance using the xCELLigence RTCA SP instrument (ACEA biosciences). After confirming robust Molm13 CD71 protein expression, target cells were mobilized on the plate bottom by pre-coating the wells with CD71 antibody (BioLegend, 334102). 150,000 Molm13 cells per well were then plated for 24 hours followed by administration of the indicated number of effector cells relative to the initial Molm13 cells. % Cytotoxicity was calculated using the provided software and untreated reference wells.
Azacitidine exposure assays
Azacitidine (Sigma-Aldrich A1287) was dissolved into R10 media to generate a stock solution. Serial dilutions were then performed to generate the indicated concentrations. All stocks and dilutions were performed immediately prior to experiments due to known breakdown of azacitidine in solution.
In vivo experiments
Mice were engrafted with cell lines as described for the individual experiments and sorted prior to treatment allocation to achieve similar mean flux values across treatment groups. Due to instability in solution, azacitidine stock solutions were made fresh daily and administered via intraperitoneal injection. Cryopreserved CAR-T or untransduced T cells were injected intravenously via tail vein at the indicated time points. Tumor burden was monitored via bioluminescence following intraperitoneal injection of D-luciferin substrate solution using an IVIS Spectrum in vivo imaging system. Animals were euthanized per the experimental protocol or when they met a priori defined endpoints by IACUC. Bioluminescence was quantified using Aura version 3.2 from spectral instruments imaging.
Flow cytometry
The following antibody clones targeting their respective antigens were used for flow cytometric analysis where indicated: CD70 (113–16, Biolegend), PeCy7 isotype (MOPC-21, Biolegend), CD69 (FN50, Biolegend), mouse TER-119 (TER-119, Biolegend), mouse NK-1.1 (PK136, Biolegend), mouse Ly-6G/Ly-6C (Gr-1, Biolegend), mouse CD11b (Biolegend), CD366/Tim3 (7D3, BD), CD223/Lag3 (T47–530, BD), CD279/PD-1 (EH12.1, BD), CD197/CCR7 (150503, BD), CD45RA (Hl100, BioLegend), CD3 (SK7, BD), CD33 (67.6, BioLegend), CD27 (L128, BD), CD45 (HI30, BioLegend). In general, cells were stained for fifteen minutes in the dark at 4 degrees Celsius and washed in PBS with 2% FBS. DAPI or 7-AAD were added to establish live versus dead separation. Trucount tubes (BD Biosciences, 340334) were used for murine blood CAR and tumor quantification according to kit instructions. Quantum Simply Cellular beads (Bangs Laboratories, 815) were used to quantify CD70 surface expression.
Analysis was performed in FlowJo software version 10.6.2 with SPICE 6 (Roederer et al., 2011) (https://niaid.github.io/spice/) for visualization of exhaustion markers and T-cell subsets.
Immunohistochemistry
For IHC, murine femurs were washed in PBS and then incubated overnight in 4% paraformaldehyde (PF Thermo-Fisher Scientific AAJ19943K2), followed by an overnight incubation in Cal-ex decalcifier (Fisher Scientific, CS510–1D) and then storage in 70% ethanol until staining. Staining was performed using the services of the MGH specialized histopathology services core facility. Images were analyzed in Fiji (ImageJ software package, which is continuously updated, downloaded 2021) (Schindelin et al., 2012).
Avidity assessment
Microfluidic chips were coated with poly-L-lysine followed by dehydration in a dry incubator with repeated aspiration of any residual liquid. After rehydration with warm R10 media, Molm13 cells were then seeded at a density of 100e6 cells/mL with continuous microscopic visualization to ensure no bubble formation and adequate density. Cells were then incubated for two hours, given fresh media, then incubated for another hour. MM.1S cells were seeded at a density of 150e6 cells/mL, incubated for 1 hour, given fresh media, then incubated for another hour prior to use. Effectors were stained with Celltrace Far Red Cell Proliferation Kit (Thermo Scientific catalogue C34564) according to the kit instructions. They were then seeded at a density of 10e6/mL with an incubation of 5 minutes prior to force application using the z-Movi cell avidity analyzer to visualize effector detachment. The order of effectors was randomized between chips and up to six different effectors were evaluated per microfluidic chip. Analysis was performed using Ocean software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
In general data were presented as mean ± standard error of the mean, unless otherwise noted in the figure legend, with statistically significant differences determined by tests as indicated in figure legends. Significance was considered at a p<0.05. All statistical tests were two-sided. P values are denoted with asterisks as follows which are also stated within each figure legend (**** = p<0.0001, *** = p<0.001, ** = p<0.01, * = p<0.05), ns = p > 0.05). Number of repeated technical, biological, and experimental replicates are described within the relevant figure legend. Data was analyzed using GraphPad Prism (Version 9.3.1). Specific lysis was calculated via the following equation %specific lysis = (total luciferase/ target cells only luciferase) x 100%. Luminex (Luminex Corp, FLEXMAP 3D) was used to analyze cell-free supernatants for cytokine production according to the manufacturer’s recommendation in technical duplicates.
Supplementary Material
Movie S1. Comparison of avidity binding experiments between hinge CAR variants. Related to Figure 4. Raw microscopic view of fluorescence, bright field, and force graph from representative experiments involving each CD70 CAR variant. Molm13 cells were seeded on a microfluidic chip and exposed to the indicated effector cells for 5 minutes prior to the gradual application of up to 1000 pN of force. Orange circles represent analysis-software identified bound effectors, while green circles represent an area where an effector was bound at the start of the force application but was subsequently dislodged. Note, multiple experiments can be run in succession and fluorescent signals without a green or orange circle represent remaining attached effectors from a prior experimental run. The video was sped up for brevity with the first 1.5 seconds of the video relatively slowed to emphasize the period of time in which most effectors are dislodged.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human CD3-BUV395 (Clone SK7) | BD Biosciences | Cat# 564000; RRID: AB_2744382 |
| Human CD45-PE (Clone HI30) | BioLegend | Cat# 304008; RRID:AB_314396 |
| Human Tim3/CD366-BV711 (clone 7D3) | BD Biosciences | Cat# 565566, RRID: AB_2744370 |
| Human Lag3/CD223-AF647 (Clone T47–530) | BD Biosciences | Cat# 565716; RRID: AB_2744328 |
| Human PD1/CD279-BV786 (Clone EH12.1) | BD Biosciences | Cat# 563789; RRID: AB_2738425 |
| Human CCR7/CD197-FITC (Clone 150503) | BD Biosciences | Cat# 561271; RRID: AB_10561679 |
| Human CD45RA- PE-Cy7 (Clone Hl100) | Biolegend | Cat# 304126; RRID: AB_10708879 |
| Human CD33-FITC (Clone 67.6) | Biolegend | Cat# 366620; RRID: AB_2566422 |
| Human CD70-PE-Cy7 (Clone 113–16) | Biolegend | Cat# 355112; RRID: AB_2687254 |
| Human CD27-BV786 (Clone L128) | BD Biosciences | Cat# 563327; RRID: AB_2744353 |
| Human CD71-FITC (CY1G4) | BioLegend | Cat# 334104; RRID: AB_2201482 |
| Human CD71-unconjugated (CY1G4) | BioLegend | Cat# 334102; RRID: AB_1134247 |
| Human CD69-APC (Clone FN50) | Biolegend | Cat# 310910; RRID: AB_314845 |
| Mouse TER-119-APC (Clone TER-119) | Biolegend | Cat# 116212; RRID: AB_313713 |
| Mouse NK-1.1-APC (Clone PK136) | Biolegend | Cat# 108710; RRID: AB_313397 |
| Mouse CD11b-APC (Clone M1/70) | Biolegend | Cat# 101212; RRID: AB_312795 |
| Mouse Ly-6G/Ly-6C-APC (Clone RB6–8C5) | Biolegend | Cat# 108412; RRID: AB_313377 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Donor Leukopaks | Massachusetts General Hospital Blood Bank | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RosetteSep Human T cell Enrichment Kit | Stem Cell Technologies | Cat# 15061 |
| Cell Therapy Systems CD3/CD28 Dynabeads | ThermoFisher Scientific | Cat# 40203D |
| Recombinant Human IL-2 | Peprotech | Cat# 200–02 |
| 5-Azacytidine | Sigma-Aldrich | Cat# A1287 |
| Paraformaldehyde Solution, 4% in PBS | ThermoFisher Scientific | Cat# AAJ19943K2 |
| Cal-Ex Decalcifier | ThermoFisher Scientific | Cat# CS510–1D |
| Poly-L-lysine | Sigma-Aldrich | Cat# P4707 |
| Celltrace Far Red Proliferation Kit | Thermo Scientific | Cat# C34564 |
| Lipofectamine 3000 | Invitrogen | Cat# L3000075 |
| D-Luciferin | ThermoFisher Scientific | Cat# PI88294 |
| Critical commercial assays | ||
| CD27 ELISA | ThermoFisher Scientific | Cat# BMS286INST |
| Luciferase Assay System | Promega | Cat# E1501 |
| Deposited data | ||
| Cbioportal AML expression data | CPDM Pan-Cancer Derived Models | https://www.cbioportal.org/ |
| Experimental models: Cell lines | ||
| SupT1 (male) | ATCC | Cat# CRL-1942 |
| Molm13 (male) | Dr. David Weinstock (Dana Farber Cancer Institute) | N/A |
| Molm13 CD70- (male) | This manuscript | N/A |
| Molm13 CD70low (male) | This manuscript | N/A |
| Molm13 CD70low-int (male) | This manuscript | N/A |
| Molm13 CD70int (male) | This manuscript | N/A |
| Molm13 CD70high-int (male) | This manuscript | N/A |
| Molm13 CD70high (male) | This manuscript | N/A |
| Nomo1 (female) | Dr. Christopher Ott (Massachusetts General Hospital) | N/A |
| Monomac1 (male) | DSMZ | Cat# ACC 252 |
| THP-1 (male) | ATCC | Cat# TIB-202 |
| OCI-AML3 (male) | DSMZ | Cat# ACC 582 |
| HEK293 (female) | ATCC | Cat# CRL-3216 |
| Jurkat (male) | ATCC | Cat# TIB-152 |
| MM1.S (female) | ATCC | Cat# CRL-2974 |
| K562-CD70 (female) | This manuscript | N/A |
| K562 (female) | ATCC | Cat# CCL-243 |
| Experimental models: Organisms/strains | ||
| Mice: NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) | The Jackson Laboratory | Cat# JAX:005557; RRID:IMSR_JAX:005557 |
| DFAM68555 AML patient derived xenograft (female) | PRoXe database | https://www.proxe.org/ |
| Oligonucleotides | ||
| GAGCTGCAGCTGAATCACAC -CD70 CRISPR guide | Brunello library, Doench et al. 2016 | N/A |
| Recombinant DNA | ||
| trCD27–4-1BB- CD3ζ (native) | Designed per specifications from Wang et. al. 2017 | N/A |
| trCD27–4-1BB- CD3ζ (truncated stalk) | This manuscript | N/A |
| trCD27–4-1BB- CD3ζ (deleted stalk) | This manuscript | N/A |
| trCD27–4-1BB- CD3ζ (flexible linker stalk) | This manuscript | N/A |
| trCD27–4-1BB- CD3ζ (CD8H&TM) | This manuscript | N/A |
| trCD27 protein | This manuscript | N/A |
| Software and algorithms | ||
| Flowjo | FlowJo, LLC | https://www.flowjo.com/ |
| GraphPad Prism 9.3.1 | GraphPad Software Inc. | https://www.graphpad.com/ |
| Aura | Spectral Instruments Imaging | https://spectralinvivo.com/software/ |
| Geneious (11.1.5) | Geneious | https://www.geneious.com/ |
| Phyre2 | Kelley, et al. 2015 | http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index |
| Pymol (2.4.1) with Pymod (3.0.2) and ClustalW tool | Schrödinger | https://pymol.org/2/; https://github.com/pymodproject/pymod |
| SPICE 6 | Roederer et al. 2011 | https://niaid.github.io/spice/ |
| Fiji (Continuously updated, downloaded 2021) with white balance correction_1.0 plugin | Schindelin et al. 2012 | https://imagej.net/software/fiji/; https://github.com/pmascalchi/ImageJ_Auto-white-balance-correction |
| CleavPredict | Cieplak et. al. 2017 | http://cleavpredict.sanfordburnham.org |
| Other | ||
| Quantum Simply Cellular beads | Bangs Laboratories | Cat# 815 |
| HiScribe T7 Quick high yield RNA synthesis Kit | New England Biolabs | Cat# E2050S |
| Trucount tubes | BD Biosciences | Cat# 340334 |
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and virus strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, peptides, and recombinant proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350–13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433–59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124–87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14–486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical commercial assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | PerkinElmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202–1012 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental models: Cell lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09–0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental models: Organisms/strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207–1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106–5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
| Chemicals, peptides, and recombinant proteins | ||
| QD605 streptavidin conjugated quantum dot | Thermo Fisher Scientific | Cat#Q10101MP |
| Platinum black | Sigma-Aldrich | Cat#205915 |
| Sodium formate BioUltra, ≥99.0% (NT) | Sigma-Aldrich | Cat#71359 |
| Chloramphenicol | Sigma-Aldrich | Cat#C0378 |
| Carbon dioxide (13C, 99%) (<2% 18O) | Cambridge Isotope Laboratories | CLM-185–5 |
| Poly(vinylidene fluoride-co-hexafluoropropylene) | Sigma-Aldrich | 427179 |
| PTFE Hydrophilic Membrane Filters, 0.22 μm, 90 mm | Scientificfilters.com/Tisch Scientific | SF13842 |
| Critical commercial assays | ||
| Folic Acid (FA) ELISA kit | Alpha Diagnostic International | Cat# 0365–0B9 |
| TMT10plex Isobaric Label Reagent Set | Thermo Fisher | A37725 |
| Surface Plasmon Resonance CM5 kit | GE Healthcare | Cat#29104988 |
| NanoBRET Target Engagement K-5 kit | Promega | Cat#N2500 |
| Deposited data | ||
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Structure of compound 5 | This paper; Cambridge Crystallographic Data Center | CCDC: 2016466 |
| Code for constraints-based modeling and analysis of autotrophic E. coli | This paper | https://gitlab.com/elad.noor/sloppy/tree/master/rubisco |
| Software and algorithms | ||
| Gaussian09 | Frish et al., 2013 | https://gaussian.com |
| Python version 2.7 | Python Software Foundation | https://www.python.org |
| ChemDraw Professional 18.0 | PerkinElmer | https://www.perkinelmer.com/category/chemdraw |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| Other | ||
| DASGIP MX4/4 Gas Mixing Module for 4 Vessels with a Mass Flow Controller | Eppendorf | Cat#76DGMX44 |
| Agilent 1200 series HPLC | Agilent Technologies | https://www.agilent.com/en/products/liquid-chromatography |
| PHI Quantera II XPS | ULVAC-PHI, Inc. | https://www.ulvac-phi.com/en/products/xps/phi-quantera-ii/ |
Acknowledgments:
We thank Piotr Cieplak for his kind assistance with the matrix-metalloprotease cleavage site algorithm he designed (Cieplak and Strongin, 2017). We also thank the following core facilities of the MGH Cancer Center: Blood Bank, Flow Cytometry, and Histopathology.
Funding:
Training Program in Cancer Biology and Therapeutics, National Institutes of Health Grant : 2T32CA071345–21A1, ASCO Young Investigator Award (ML)
Lymphoma Research Foundation Grantee. (IS)
DKMS John Hansen Research Grant awardee (DKMS-SLS-JHRG-2020–04) (AS)
National Institutes of Health T32 grant (NIH-5T32CA009216–39) (SRB)
Swiss National Science Foundation, Postdoc.Mobility fellowship (P400PM 186739) (MW)
National Institutes of Health R01CA238268, R01CA252940, and R01CA249062 (MVM)
Gabrielle’s Angel Foundation (MVM)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests:
MVM is an inventor on patents related to adoptive cell therapies, held by Massachusetts General Hospital (some licensed to Promab) and University of Pennsylvania (some licensed to Novartis). MVM holds Equity in 2SeventyBio, Century Therapeutics, Genocea, Neximmune, Oncternal, and TCR2 and has served as a consultant for multiple companies involved in cell therapies. MVM is on the Board of Directors of 2Seventy Bio. MVM has Grant/Research support from CRISPR therapeutics, Kite Pharma, Servier, Novartis. MVM has served as a consultant for multiple companies developing cell therapies. MJF has served as a consultant for several cell therapy companies. No other authors report conflicts of interest. MBL and MVM have filed patents on the constructs described herein.
References
- Abramson JS, Palomba ML, Gordon LI, Lunning MA, Wang M, Arnason J, Mehta A, Purev E, Maloney DG, Andreadis C, et al. (2020). Lisocabtagene maraleucel for patients with relapsed or refractory large B-cell lymphomas (TRANSCEND NHL 001): a multicentre seamless design study. The Lancet 396, 839–852. 10.1016/S0140-6736(20)31366-0. [DOI] [PubMed] [Google Scholar]
- Actavis_Pharma, S.P.S. Azacitidine [package insert] U.S. Food and Drug Administration [Google Scholar]
- Alabanza L, Pegues M, Geldres C, Shi V, Wiltzius JJW, Sievers SA, Yang S, and Kochenderfer JN (2017). Function of Novel Anti-CD19 Chimeric Antigen Receptors with Human Variable Regions Is Affected by Hinge and Transmembrane Domains. Mol Ther 25, 2452–2465. 10.1016/j.ymthe.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifant CL, Szoor A, Torres D, Joseph N, Velasquez MP, Iwahori K, Gaikwad A, Nguyen P, Arber C, Song X-T, et al. (2016). CD123-Engager T Cells as a Novel Immunotherapeutic for Acute Myeloid Leukemia. Molecular Therapy 24, 1615–1626. 10.1038/mt.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroughs AC, Larson RC, Marjanovic ND, Gosik K, Castano AP, Porter CBM, Lorrey SJ, Ashenberg O, Jerby L, Hofree M, et al. (2020). A Distinct Transcriptional Program in Human CAR T Cells Bearing the 4–1BB Signaling Domain Revealed by scRNA-Seq. Molecular therapy : the journal of the American Society of Gene Therapy 28, 2577–2592. 10.1016/j.ymthe.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman MR, Crimmins MA, Yetz-Aldape J, Kriz R, Kelleher K, and Herrmann S (1994). The cloning of CD70 and its identification as the ligand for CD27. The Journal of Immunology 152, 1756–1761. [PubMed] [Google Scholar]
- Brudno JN, Lam N, Vanasse D, Shen Y. w., Rose JJ, Rossi J, Xue A, Bot A, Scholler N, Mikkilineni L, et al. (2020). Safety and feasibility of anti-CD19 CAR T cells with fully human binding domains in patients with B-cell lymphoma. Nature Medicine 26, 270–280. 10.1038/s41591-019-0737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill MA, Tamburini BA, and Kedl RM (2015). T cells compete by cleaving cell surface CD27 and blocking access to CD70-bearing APCs. Eur J Immunol 45, 3140–3149. 10.1002/eji.201545749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin M, Sands C, Da T, Scholler J, Graham K, Buza E, Fraietta JA, Zhao Y, and June CH (2020). A rational mouse model to detect on-target, off-tumor CAR T cell toxicity. JCI Insight 5. 10.1172/jci.insight.136012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieplak P, and Strongin AY (2017). Matrix metalloproteinases - From the cleavage data to the prediction tools and beyond. Biochim Biophys Acta Mol Cell Res 1864, 1952–1963. 10.1016/j.bbamcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191. 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H, Weisdorf DJ, and Bloomfield CD (2015). Acute Myeloid Leukemia. New England Journal of Medicine 373, 1136–1152. 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- Duong MT, Collinson-Pautz MR, Morschl E, Lu A, Szymanski SP, Zhang M, Brandt ME, Chang W-C, Sharp KL, Toler SM, et al. (2019). Two-Dimensional Regulation of CAR-T Cell Therapy with Orthogonal Switches. Molecular Therapy - Oncolytics 12, 124–137. 10.1016/j.omto.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJC, Hamieh M, Cunanan KM, Odak A, Gönen M, and Sadelain M (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faderl S, Bueso-Ramos C, Liu Z, Pal A, Bornmann W, Ciurea DV, Harris D, Hazan-Halevy I, Kantarjian HM, and Estrov Z (2011). Kit inhibitor APcK110 extends survival in an AML xenograft mouse model. Invest New Drugs 29, 1094–1097. 10.1007/s10637-010-9459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández de Larrea C, Staehr M, Lopez AV, Ng KY, Chen Y, Godfrey WD, Purdon TJ, Ponomarev V, Wendel H-G, Brentjens RJ, and Smith EL (2020). Defining an Optimal Dual-Targeted CAR T-cell Therapy Approach Simultaneously Targeting BCMA and GPRC5D to Prevent BCMA Escape–Driven Relapse in Multiple Myeloma. Blood Cancer Discovery 1, 146–154. 10.1158/2643-3230.Bcd-20-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flieswasser T, Camara-Clayette V, Danu A, Bosq J, Ribrag V, Zabrocki P, Van Rompaey L, de Haard H, Zwaenepoel K, Smits E, et al. (2019). Screening a Broad Range of Solid and Haematological Tumour Types for CD70 Expression Using a Uniform IHC Methodology as Potential Patient Stratification Method. Cancers 11. 10.3390/cancers11101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, Boesteanu AC, Wang Y, O’Connor RS, Hwang W-T, et al. (2018a). Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nature Medicine 24, 563–571. 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraietta JA, Nobles CL, Sammons MA, Lundh S, Carty SA, Reich TJ, Cogdill AP, Morrissette JJD, DeNizio JE, Reddy S, et al. (2018b). Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312. 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigault MJ, Lee J, Basil MC, Carpenito C, Motohashi S, Scholler J, Kawalekar OU, Guedan S, McGettigan SE, Posey AD Jr., et al. (2015). Identification of chimeric antigen receptors that mediate constitutive or inducible proliferation of T cells. Cancer immunology research 3, 356–367. 10.1158/2326-6066.Cir14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, Wolters P, Martin S, Delbrook C, Yates B, et al. (2018). CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat Med 24, 20–28. 10.1038/nm.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman R, Pizam Y, Haus-Cohen M, Goor A, Horev G, Denkberg G, Sinik K, Elbaz Y, Bronner V, Globerson Levin A, et al. (2021). Shaping Functional Avidity of CAR-T Cells: Affinity, Avidity and Antigen Density that Regulate Response. Molecular Cancer Therapeutics, molcanther.MCT-19–1109-A.2019 10.1158/1535-7163.Mct-19-1109. [DOI] [PubMed]
- Guerra VA, DiNardo C, and Konopleva M (2019). Venetoclax-based therapies for acute myeloid leukemia. Best practice & research. Clinical haematology 32, 145–153. 10.1016/j.beha.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamieh M, Dobrin A, Cabriolu A, van der Stegen SJC, Giavridis T, Mansilla-Soto J, Eyquem J, Zhao Z, Whitlock BM, Miele MM, et al. (2019). CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature 568, 112–116. 10.1038/s41586-019-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Lopez RA, Yu W, Cabral KA, Creasey OA, Lopez Pazmino MDP, Tonai Y, De Guzman A, Mäkelä A, Saksela K, Gartner ZJ, and Lim WA (2021). T cell circuits that sense antigen density with an ultrasensitive threshold. Science 371, 1166–1171. 10.1126/science.abc1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Schubert ML, Wang L, He B, Neuber B, Dreger P, Muller-Tidow C, and Schmitt M (2019). Chimeric Antigen Receptor (CAR) T Cell Therapy in Acute Myeloid Leukemia (AML). Journal of clinical medicine 8. 10.3390/jcm8020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson C, Chavez JC, Sehgal AR, William BM, Munoz J, Salles G, Munshi PN, Casulo C, Maloney D, de Vos S, et al. (2020). Primary Analysis of Zuma-5: A Phase 2 Study of Axicabtagene Ciloleucel (Axi-Cel) in Patients with Relapsed/Refractory (R/R) Indolent Non-Hodgkin Lymphoma (iNHL). Blood 136, 40–41. 10.1182/blood-2020-136834. [DOI] [Google Scholar]
- Jain N, Zhao Z, Iyer AS, Lopez M, Feucht J, Koche RP, Zhan Y, and Sadelain M (2020). Loss of TET2 Uncouples Proliferative and Effector Functions in CAR T Cells. Blood 136, 1–1. 10.1182/blood-2020-142957.32430499 [DOI] [Google Scholar]
- Jetani H, Garcia-Cadenas I, Nerreter T, Thomas S, Rydzek J, Meijide JB, Bonig H, Herr W, Sierra J, Einsele H, and Hudecek M (2018). CAR T-cells targeting FLT3 have potent activity against FLT3−ITD+ AML and act synergistically with the FLT3-inhibitor crenolanib. Leukemia 32, 1168–1179. 10.1038/s41375-018-0009-0. [DOI] [PubMed] [Google Scholar]
- Jin L, Ge H, Long Y, Yang C, Chang Y, Mu L, Sayour EJ, De Leon G, Wang QJ, Yang JC, et al. (2017). CD70, a novel target of CAR T-cell therapy for gliomas. Neuro-Oncology 20, 55–65. 10.1093/neuonc/nox116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker K, Von Eggeling F, Hindermann W, Diegmann J, and Schubert J (2005). CD70- a new tumor specific biomarker for renal cell carcinoma. Cancer Research 65, 742–742. [DOI] [PubMed] [Google Scholar]
- Kato K, Chu P, Takahashi S, Hamada H, and Kipps TJ (2007). Metalloprotease inhibitors block release of soluble CD27 and enhance the immune stimulatory activity of chronic lymphocytic leukemia cells. Experimental Hematology 35, 434–442. 10.1016/j.exphem.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, 845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Yu K-R, Kenderian SS, Ruella M, Chen S, Shin T-H, Aljanahi AA, Schreeder D, Klichinsky M, Shestova O, et al. (2018). Genetic Inactivation of CD33 in Hematopoietic Stem Cells to Enable CAR T Cell Immunotherapy for Acute Myeloid Leukemia. Cell 173, 1439–1453.e1419. 10.1016/j.cell.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Lee KL, Fujioka M, Sun W, You J, Chia GS, Wanibuchi H, Tomita S, Araki M, Kato H, and Poellinger L (2018). Hypoxia-inducible factor-2 alpha up-regulates CD70 under hypoxia and enhances anchorage-independent growth and aggressiveness in cancer cells. Oncotarget 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Nguyen A, Shi T, Tang L, Ni X, Escoubet L, MacBeth KJ, DiMartino J, and Wells JA (2019). Multiomics of azacitidine-treated AML cells reveals variable and convergent targets that remodel the cell-surface proteome. Proc Natl Acad Sci U S A 116, 695–700. 10.1073/pnas.1813666116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Huang Z, Zhang C, Dong BJ, Guo RH, Yue HW, Yan LT, and Xing XH (2016). Construction of a linker library with widely controllable flexibility for fusion protein design. Appl Microbiol Biotechnol 100, 215–225. 10.1007/s00253-015-6985-3. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang S, Fang C, Yang S, Olalere D, Pequignot EC, Cogdill AP, Li N, Ramones M, Granda B, et al. (2015). Affinity-Tuned ErbB2 or EGFR Chimeric Antigen Receptor T Cells Exhibit an Increased Therapeutic Index against Tumors in Mice. Cancer research 75, 3596–3607. 10.1158/0008-5472.CAN-15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, Smith JP, Walker AJ, Kohler ME, Venkateshwara VR, et al. (2015). 4–1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med 21, 581–590. 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Chang L-J, Hu Y, Dong L, Wei G, and Huang H (2015). First-in-Man CD123-Specific Chimeric Antigen Receptor-Modified T Cells for the Treatment of Refractory Acute Myeloid Leukemia. Blood 126, 3778–3778. 10.1182/blood.V126.23.3778.3778. [DOI] [Google Scholar]
- Lynn RC, Weber EW, Sotillo E, Gennert D, Xu P, Good Z, Anbunathan H, Lattin J, Jones R, Tieu V, et al. (2019). c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300. 10.1038/s41586-019-1805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majzner RG, Rietberg SP, Sotillo E, Dong R, Vachharajani VT, Labanieh L, Myklebust JH, Kadapakkam M, Weber EW, Tousley AM, et al. (2020). Tuning the Antigen Density Requirement for CAR T-cell Activity. Cancer Discovery 10, 702–723. 10.1158/2159-8290.Cd-19-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardiros A, Dos Santos C, McDonald T, Brown CE, Wang X, Budde LE, Hoffman L, Aguilar B, Chang W-C, Bretzlaff W, et al. (2013). T cells expressing CD123-specific chimeric antigen receptors exhibit specific cytolytic effector functions and antitumor effects against human acute myeloid leukemia. Blood 122, 3138–3148. 10.1182/blood-2012-12-474056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melao A (2017). FDA Suspends UCART123 Trials After Patient Death. Immuno-Oncology News
- Nahas MR, Stroopinsky D, Rosenblatt J, Cole L, Pyzer AR, Anastasiadou E, Sergeeva A, Ephraim A, Washington A, Orr S, et al. (2019). Hypomethylating agent alters the immune microenvironment in acute myeloid leukaemia (AML) and enhances the immunogenicity of a dendritic cell/AML vaccine. Br J Haematol 185, 679–690. 10.1111/bjh.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, Braunschweig I, Oluwole OO, Siddiqi T, Lin Y, et al. (2017). Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. New England Journal of Medicine 377, 2531–2544. 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte MA, van Olffen RW, van Gisbergen KP, and van Lier RA (2009). Timing and tuning of CD27-CD70 interactions: the impact of signal strength in setting the balance between adaptive responses and immunopathology. Immunol Rev 229, 216–231. 10.1111/j.1600-065X.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- Ochsenbein A (2018). Argx-110 Targeting CD70, in Combination with Azacitidine, Shows Favorable Safety Profile and Promising Anti-Leukemia Activity in Newly Diagnosed AML Patients in an Ongoing Phase 1/2 Clinical Trial Abstract 2680. held in San Diego, CA. [Google Scholar]
- Ossenkoppele G, and Löwenberg B (2015). How I treat the older patient with acute myeloid leukemia. Blood 125, 767–774. 10.1182/blood-2014-08-551499. [DOI] [PubMed] [Google Scholar]
- Park YP, Jin L, Bennett KB, Wang D, Fredenburg KM, Tseng JE, Chang L-J, Huang J, and Chan EKL (2018). CD70 as a target for chimeric antigen receptor T cells in head and neck squamous cell carcinoma. Oral Oncology 78, 145–150. 10.1016/j.oraloncology.2018.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna F, Berman SH, Soni RK, Mansilla-Soto J, Eyquem J, Hamieh M, Hendrickson RC, Brennan CW, and Sadelain M (2017). Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell 32, 506–519.e505. 10.1016/j.ccell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov JC, Wada M, Pinz KG, Yan LE, Chen KH, Shuai X, Liu H, Chen X, Leung L-H, Salman H, et al. (2018). Compound CAR T-cells as a double-pronged approach for treating acute myeloid leukemia. Leukemia 32, 1317–1326. 10.1038/s41375-018-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirillo C, Haltalli M, Anton SG, Tini V, Kong I, Hawkins E, Falini B, Marra A, Duarte D, and Celso CL (2020). The metalloproteinase inhibitor Prinomastat reduces AML growth, prevents stem cell loss and improves chemotherapy effectiveness. bioRxiv, 2020.2012.2001.393157. 10.1101/2020.12.01.393157. [DOI] [PMC free article] [PubMed]
- Pont MJ, Hill T, Cole GO, Abbott JJ, Kelliher J, Salter AI, Hudecek M, Comstock ML, Rajan A, Patel BKR, et al. (2019). gamma-secretase inhibition increases efficacy of BCMA-specific chimeric antigen receptor T cells in multiple myeloma. Blood 10.1182/blood.2019000050. [DOI] [PMC free article] [PubMed]
- Pubchem. PubChem Compound Summary for CID 9444, Azacitidine https://pubchem.ncbi.nlm.nih.gov/compound/Azacitidine.
- Ramakrishna S, Highfill SL, Walsh Z, Nguyen SM, Lei H, Shern JF, Qin H, Kraft IL, Stetler-Stevenson M, Yuan CM, et al. (2019). Modulation of Target Antigen Density Improves CAR T-cell Functionality and Persistence. Clin Cancer Res 25, 5329–5341. 10.1158/1078-0432.Ccr-18-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Messerschmidt S, Holeiter G, Tepperink J, Osswald S, Zappe A, Branschädel M, Boschert V, Mann DA, Scheurich P, and Krippner-Heidenreich A (2012). The tumor necrosis factor receptor stalk regions define responsiveness to soluble versus membrane-bound ligand. Mol Cell Biol 32, 2515–2529. 10.1128/mcb.06458-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riether C, Pabst T, Höpner S, Bacher U, Hinterbrandner M, Banz Y, Müller R, Manz MG, Gharib WH, Francisco D, et al. (2020). Targeting CD70 with cusatuzumab eliminates acute myeloid leukemia stem cells in patients treated with hypomethylating agents. Nature Medicine 10.1038/s41591-020-0910-8. [DOI] [PubMed]
- Riether C, Schürch CM, Bührer ED, Hinterbrandner M, Huguenin AL, Hoepner S, Zlobec I, Pabst T, Radpour R, and Ochsenbein AF (2017). CD70/CD27 signaling promotes blast stemness and is a viable therapeutic target in acute myeloid leukemia. J Exp Med 214, 359–380. 10.1084/jem.20152008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M, Nozzi JL, and Nason MC (2011). SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry Part A 79A, 167–174. 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf M, Mittmann C, Nowicka AM, Hartmann A, Hermanns T, Poyet C, van den Broek M, Sulser T, Moch H, and Schraml P (2015). pVHL/HIF-regulated CD70 expression is associated with infiltration of CD27+ lymphocytes and increased serum levels of soluble CD27 in clear cell renal cell carcinoma. Clin Cancer Res 21, 889–898. 10.1158/1078-0432.Ccr-14-1425. [DOI] [PubMed] [Google Scholar]
- Sauer T, Parikh K, Sharma S, Omer B, Sedloev DN, Chen Q, Angenendt L, Schliemann C, Schmitt M, Müller-Tidow C, et al. (2021). CD70-specific CAR T-cells have potent activity against Acute Myeloid Leukemia (AML) without HSC toxicity. Blood 10.1182/blood.2020008221. [DOI] [PMC free article] [PubMed]
- Scarfò I, Ormhøj M, Frigault MJ, Castano AP, Lorrey S, Bouffard AA, van Scoyk A, Rodig SJ, Shay AJ, Aster JC, et al. (2018). Anti-CD37 chimeric antigen receptor T cells are active against B- and T-cell lymphomas. Blood 132, 1495–1506. 10.1182/blood-2018-04-842708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts A, Ormhøj M, Choi BD, Taylor AO, Bouffard AA, Scarfò I, Larson RC, Frigault MJ, Gallagher K, Castano AP, et al. (2019). Rational design of a trimeric APRIL-based CAR-binding domain enables efficient targeting of multiple myeloma. Blood Advances 3, 3248–3260. 10.1182/bloodadvances.2019000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SJ, Svoboda J, Chong EA, Nasta SD, Mato AR, Anak Ö, Brogdon JL, Pruteanu-Malinici I, Bhoj V, Landsburg D, et al. (2017). Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. New England Journal of Medicine 377, 2545–2554. 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer DR, Savoldo B, Yi Z, Chow KKH, Kakarla S, Spencer DM, Dotti G, Wu M-F, Liu H, Kenney S, and Gottschalk S (2011). T cells redirected against CD70 for the immunotherapy of CD70-positive malignancies. Blood 117, 4304–4314. 10.1182/blood-2010-04-278218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Frey NV, Engels B, Barrett DM, Shestova O, Ravikumar P, Shyu A, Highfill S, Zhao L, Peng L, et al. (2019). Single Chain Variable Fragment Linker Length Regulates CAR Biology and T Cell Efficacy. Blood 134, 247–247. 10.1182/blood-2019-131024. [DOI] [Google Scholar]
- Singh N, Lee YG, Shestova O, Ravikumar P, Hayer KE, Hong SJ, Lu XM, Pajarillo R, Agarwal S, Kuramitsu S, et al. (2020). Impaired Death Receptor Signaling in Leukemia Causes Antigen-Independent Resistance by Inducing CAR T-cell Dysfunction. Cancer discovery 10, 552–567. 10.1158/2159-8290.Cd-19-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaney CY, Wang P, Darcy PK, and Kershaw MH (2018). CARs versus BiTEs: A Comparison between T Cell-Redirection Strategies for Cancer Treatment. Cancer Discov 8, 924–934. 10.1158/2159-8290.Cd-18-0297. [DOI] [PubMed] [Google Scholar]
- Teplyakov A, Obmolova G, Malia TJ, and Gilliland GL (2017). Crystal structure of CD27 in complex with a neutralizing noncompeting antibody. Acta Crystallogr F Struct Biol Commun 73, 294–299. 10.1107/S2053230X17005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend EC, Murakami MA, Christodoulou A, Christie AL, Köster J, DeSouza TA, Morgan EA, Kallgren SP, Liu H, Wu S-C, et al. (2016). The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer cell 29, 574–586. 10.1016/j.ccell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Zhou X, Guo Z, Huang R, Yue C, He Y, Li M, Chen Y, Liu Y, Chang L. j., and Li Y (2019). CD19 and CD70 Dual-Target Chimeric Antigen Receptor T-Cell Therapy for the Treatment of Relapsed and Refractory Primary Central Nervous System Diffuse Large B-Cell Lymphoma. Frontiers in Oncology 9. 10.3389/fonc.2019.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanamee É S, and Faustman DL (2018). Structural principles of tumor necrosis factor superfamily signaling. Sci Signal 11. 10.1126/scisignal.aao4910. [DOI] [PubMed] [Google Scholar]
- Wang M, Munoz J, Goy A, Locke FL, Jacobson CA, Hill BT, Timmerman JM, Holmes H, Jaglowski S, Flinn IW, et al. (2020). KTE-X19 CAR T-Cell Therapy in Relapsed or Refractory Mantle-Cell Lymphoma. New England Journal of Medicine 382, 1331–1342. 10.1056/NEJMoa1914347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. s., Wang Y, Lv H. y., Han Q. w., Fan H, Guo B, Wang L. l., and Han W. d. (2015). Treatment of CD33-directed Chimeric Antigen Receptor-modified T Cells in One Patient With Relapsed and Refractory Acute Myeloid Leukemia. Molecular Therapy 23, 184–191. 10.1038/mt.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QJ, Yu Z, Hanada KI, Patel K, Kleiner D, Restifo NP, and Yang JC (2017). Preclinical Evaluation of Chimeric Antigen Receptors Targeting CD70-Expressing Cancers. Clin Cancer Res 23, 2267–2276. 10.1158/1078-0432.Ccr-16-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tong C, Dai H, Wu Z, Han X, Guo Y, Chen D, Wei J, Ti D, Liu Z, et al. (2021). Low-dose decitabine priming endows CAR T cells with enhanced and persistent antitumour potential via epigenetic reprogramming. Nature Communications 12, 409. 10.1038/s41467-020-20696-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyzgol A, Müller N, Fick A, Munkel S, Grigoleit GU, Pfizenmaier K, and Wajant H (2009). Trimer stabilization, oligomerization, and antibody-mediated cell surface immobilization improve the activity of soluble trimers of CD27L, CD40L, 41BBL, and glucocorticoid-induced TNF receptor ligand. J Immunol 183, 1851–1861. 10.4049/jimmunol.0802597. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang W, Yan XJ, Lin XQ, Li W, Mi JQ, Li JM, Zhu J, Chen Z, and Chen SJ (2016). DNMT3A mutation leads to leukemic extramedullary infiltration mediated by TWIST1. J Hematol Oncol 9, 106. 10.1186/s13045-016-0337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang L, Yang Z, Jin H, Zou Q, Zhan Q, Tang Y, Tao Y, Lei L, Jing Y, et al. (2019). Up-regulation of EMT-related gene VCAN by NPM1 mutant-driven TGF-β/cPML signalling promotes leukemia cell invasion. J Cancer 10, 6570–6583. 10.7150/jca.30223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Xu L, Tseng H, Cao Y, Jiang J, Ciccarelli BT, Yang G, Patterson CJ, Hunter ZR, and Treon SP (2011). Matrix Metalloproteinase-8 Is Overexpressed in Waldenström’s Macroglobulinemia Cells, and Specific Inhibition of this Metalloproteinase Blocks Release of Soluble CD27. Clinical Lymphoma Myeloma and Leukemia 11, 172–175. 10.3816/CLML.2011.n.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Comparison of avidity binding experiments between hinge CAR variants. Related to Figure 4. Raw microscopic view of fluorescence, bright field, and force graph from representative experiments involving each CD70 CAR variant. Molm13 cells were seeded on a microfluidic chip and exposed to the indicated effector cells for 5 minutes prior to the gradual application of up to 1000 pN of force. Orange circles represent analysis-software identified bound effectors, while green circles represent an area where an effector was bound at the start of the force application but was subsequently dislodged. Note, multiple experiments can be run in succession and fluorescent signals without a green or orange circle represent remaining attached effectors from a prior experimental run. The video was sped up for brevity with the first 1.5 seconds of the video relatively slowed to emphasize the period of time in which most effectors are dislodged.
Data Availability Statement
Novel datasets were not generated with this study. Bulk-RNA data was obtained from the CPDM Pan-Cancer Patient Derived Models (https://www.dana-farber.org/research/departments-centers-andlabs/integrative-research-centers/center-for-patient-derived-models/ordering-patient-derived-models/).


