Abstract
The organization of chromatin into different types of compact versus open states provides a means to fine tune gene regulation. Recent studies have suggested a role for phase-separation in chromatin compaction, raising new possibilities for regulating chromatin compartments. This perspective discusses some specific molecular mechanisms that could leverage such phase-separation processes to control the functions and organization of chromatin.
Keywords: Histone, nucleosome, heterochromatin, phase-separation, auto-inhibition
The nucleus is a crowded mixture of macromolecules with chromatin comprising a dominant component (Hancock and Jeon 2014). Despite such crowding, distinct sophisticated activities occur within the nucleus with high specificity. Phase-separation, a long-observed property of macromolecules at high concentration, provides one vantage point to explain how intra-nuclear organization might occur in a crowded milieu. In its simplest form, phase-separation refers to the process where a mixture of components in solution de-mix (separate) into two or more distinct phases with different physical and chemical properties (Hyman et al. 2014; Alberti et al. 2019). At one extreme, this process describes the non-functional aggregation of misfolded proteins. At another extreme, this process describes the formation of functional liquid-like compartments formed by the macromolecules that de-mix from solution. This latter form of phase-separation has been termed liquid–liquid phase-separation (LLPS). In LLPS driven states, nucleation and condensation of the component macromolecules are proposed to drive the formation of liquid droplets with differentiated material and chemical micro-environments (Hyman et al. 2014; Alberti et al. 2019). Over the last several decades, chemists, physicists and material scientists have studied the conditions that promote LLPS in model systems (Hancock and Jeon 2014). These studies have identified several key determinants that promote LLPS: (i) the polymeric nature of the component macromolecules; (ii) the ability of molecules to form multi-valent interactions; (iii) intrinsically disordered regions (IDRs) within proteins that can participate in multivalent interactions; and (iv) macromolecular crowding. The eukaryotic nucleus has an abundance of these determinants. For example, (i) chromatinized DNA provides long polymers, (ii) intrinsically disordered histone tails participate in multi-valent interactions between nucleosomes, and (iii) macromolecular concentrations within a eukaryotic nucleus can reach up to 400 mg/ml (Hancock and Jeon 2014) (figure 1). Consistent with these determinants, a series of studies are showing how LLPS processes could drive and regulate chromatin organization within the nucleus (figure 2). Below, I first summarize the findings of these studies and then discuss how these findings suggest ideas for mechanisms that regulate phase-separation based chromatin organization.
Figure 1.
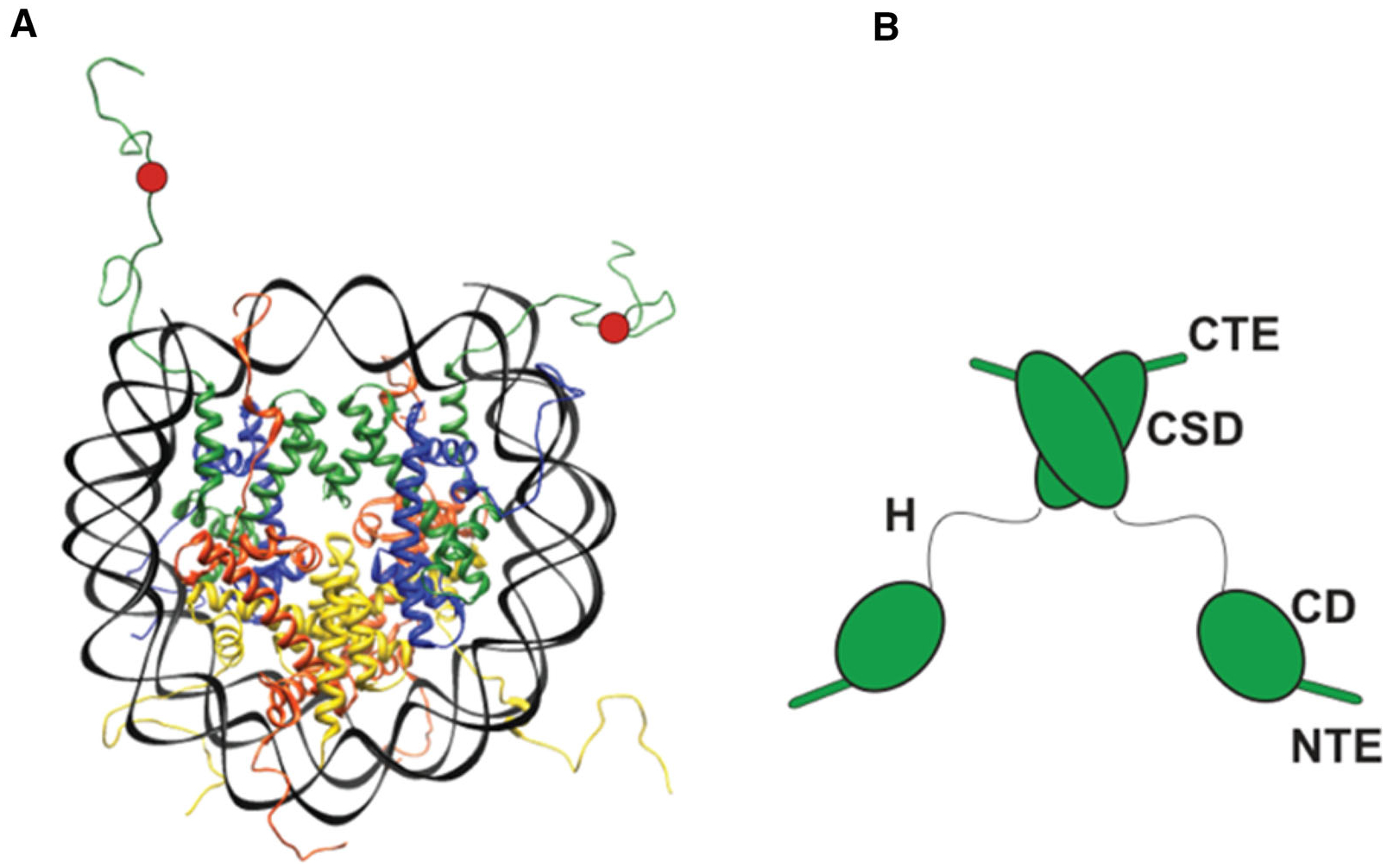
(A) The structure of a nucleosome. DNA is in black. Histone H2A is in red, H2B in yellow, H3 in green and H4 in blue. The H3K9me3 mark is schematically shown as a red circle. (B) Domain diagram of an HP1 dimer. HP1 has two structured domains, the CD, which binds the H3K9me3 mark and the CSD, which forms a dimer that serves as binding interface for various protein ligands. HP1 has three intrinsically disordered regions (IDRs), an N-terminal extension (NTE), a hinge (H) and a C-terminal extension (CTE). Interactions made by the NTE and hinge mediate higher-order HP1 oligomerization.
Figure 2.

The schematic shows multiple LLPS droplets across a stretch of the genome and highlights different mechanisms for regulating LLPS in the context of chromatin organization. The droplets are shown in blue and grey. HP1 proteins are shown in green, and other hypothetical chromatin regulators are shown in purple and maroon. Nucleosomes in blue are shown schematically as adopting different shapes to represent different conformations. The H3K9 methyl mark is depicted in red. Thicker arrows represent droplets with higher material strength compared to thinner arrows.
1. Uncovering how phase separation mechanisms can regulate chromatin organization
A substantial portion of the histones extends out from the folded octamer core in the form of largely unstructured N- or C-terminal tails (Luger et al. 1997) (figure 1A). Previous work has shown that these tails, which are largely positively charged, participate in inter-nucleosomal interactions that drive the compaction of long stretches of nucleosomes. Recent work has shown that the interactions mediated by the histone tails enable chromatin to form phase-separated droplets under physiological salt conditions (Gibson et al. 2019). This phase-separation process further results in substantial condensation of the chromatin. Even more interesting is what the authors find in terms of how chromatin phases are regulated. They find that the nature of the DNA spacing between the nucleosomes plays a key role in regulating the critical concentration of chromatin needed for phase-separation. The commonly found 10n + 5 bp spacing is more favorable for the phase-separation of chromatin compared to the 10n bp spacing. Additionally, the authors find that the viscosity of chromatin phases depends strongly on the presence of the linker histone H1, which binds to the outside of the nucleosomal unit.
Beyond the genome packaging mediated by chromatin formation, additional levels of packaging enable formation of repressive chromatin states called heterochromatin. A conserved mechanism for heterochromatin formation involves the use of proteins, termed HP1 proteins, that assemble and oligomerize across chromatin to enable its further compaction (Grewal and Elgin 2002; Canzio et al. 2014; Eissenberg and Elgin 2014). HP1 proteins further recognize chromatin that is methylated on histone H3 at lysine 9 (figure 1A). On their own HP1 proteins can form long oligomers through interactions mediated by IDRs present within HP1 proteins (figure 1B) (Larson et al. 2017). The presence of IDR-mediated interactions and the ability to form multi-valent interactions in the form of oligomers makes HP1 proteins candidates for phase-separation. Indeed, work with the human HP1 protein HP1α, showed this protein can phase-separate on its own, but only under conditions that relieved an auto-inhibited conformation of the HP1α protein (Larson et al. 2017). Complementary findings with Drosophila HP1 proteins indicated that HP1-mediated heterochromatin could adopt phase-separated states in vivo and in vitro (Strom et al. 2017). Based on these behaviors identified for HP1 proteins, a simple model can be proposed wherein the assembly of HP1 proteins on chromatin promotes condensation of chromatin into liquid droplets by adding an extra layer of polymeric organization and multi-valent interactions. Yet, recent work with the S. pombe HP1 protein, Swi6, shows that HP1 proteins do not just add to the multi-valency of chromatin. Instead, Swi6 proteins were found to deform individual nucleosomal units to expose buried histone interfaces that promoted LLPS of chromatin (Sanulli et al. 2019). These results have suggested that the buried core of the histone octamer can directly participate in the biochemical interactions that drive nucleation and condensation of chromatin.
Some insight into the material properties of chromatin-based phases formed in cells was provided by a study that used CRISPR-based approaches to target the processes of nucleation and condensation by various phase-separating protein fragments to telomeric DNA sequences (Shin et al. 2018). This study found that phase-separated droplets nucleated at spatially distinct regions of the genome could result in the coalescence of these regions. The authors propose a mechanism that involves the fusion of the phases nucleated at different genomic sites. These synthetically engineered phases thus highlight the potential for phase-separation mechanisms to organize genome packaging. The study further found that the synthetically generated phases could mechanically exclude surrounding regions of chromatin. Interestingly, the type of chromatin most resistant to mechanical disruption by the engineered phase-separating droplets was heterochromatin (Shin et al. 2018). These results raise the possibility that HP1 and other heterochromatin proteins impart specialized physico-chemical and mechanical properties to this chromatin state.
2. Emerging regulatory mechanisms that control phase-separation of chromatin
The recent work highlighted above describes how phase-separation-based mechanisms can play a role in genome organization (figure 2). In the context of these findings it becomes relevant to ask the following question: if the conditions in the nucleus are highly conducive to phase-separation, how is this fundamental physico-chemical process controlled to carry out the diverse processes within the nucleus? The same studies that have provided experimental evidence for chromatin-based phase-separation have also uncovered some potential regulatory mechanisms as discussed below.
2.1. Control through autoinhibition
Work with the human HP1α protein showed that in the absence of any bound ligand or post-translational modification, this protein exists in a compact autoinhibited dimer form (Larson et al. 2017). Autoinhibition is thought to be mediated by interactions between the hinge region of HP1 proteins and the C-terminal extension (figure 1). Two types of cues are suggested to relieve this autoinhibition: phosphorylation of the N-terminal extension (NTE) or binding of the hinge by DNA. It is proposed that by binding the positively charged hinge, the negatively charged phosphorylated NTE displaces the CTE-hinge contacts and promotes contacts between HP1α dimers. These interactions are then proposed to drive multivalent contacts and phase-separation. In the context of DNA binding it is proposed that hinge-DNA interactions displace the inhibitory CTE contacts and allow the opened up HP1α dimer to form interactions with other HP1 dimers eventually leading to the multivalency required for phase-separation. Relief of auto-inhibition could provide an important control point in the biological regulation of HP1α phase-separation. For example, specific non-chromatin HP1 ligands could either promote or relieve the auto-inhibited state based on developmental cues. Analogously, changes in nuclear localization of the NTE kinase in response to extra-cellular signals could switch HP1 between soluble and phase-separated states.
2.2. Chemical and conformational modifications of chromatin
The work with Swi6 indicates that deformation of the histone core can be a control point for regulating phase-separation of chromatin (Sanulli et al. 2019). If molecules other than Swi6 also deform the nucleosome core, these could regulate chromatin folding by enabling nucleosome conformations that either promote or inhibit chromatin compaction. Additionally, qualitative differences in the type of nucleosome deformation could result in different types of packing architectures within the compacted chromatin. Similar to proteins that bind nucleosomes, one can imagine how chemical modifications on the core histone residues could directly regulate the conformational state of the nucleosome and have effects that propagate across scales to impact the folding of chromatin domains.
2.3. Control through nucleosome arrangement
The finding that the 10n + 5 bp spacing between nucleosomes promotes phase-separation of chromatin indicates that the specific molecular arrangement of the repeating nucleosomal units can regulate phase-separation driven chromatin compaction (Gibson et al. 2019). Thus, one can imagine that ATP-dependent chromatin remodeling complexes that rearrange nucleosome spacing could either promote or disrupt chromatin compaction by altering the spacing between nucleosomes (Zhou et al. 2016).
2.4. Control through regulating material properties
The work in Gibson et al.(2019) has suggested that the presence of linker histones affects the material properties of chromatin, while the work from Shin et al. (2018) raises the possibility that the specific material properties of heterochromatin may make it more resistant to mechanical disruption. In this context, it has been suggested that heterochromatin contributes to conferring mechanical rigidity to the nucleus (Stephens et al. 2019). Thus, it can be imagined that macromolecules that alter the material properties of chromatin-based phases by affecting the types of multi-valent interactions made within the phases would play a major role in controlling the functions of these phase-separated domains.
3. Open questions
The accumulating findings linking phase-separation to chromatin organization have opened up new questions with new regulatory implications. For example, it is unclear if there is a defined structural organization within chromatin-based phases. Understanding the architecture of molecular arrangements within chromatin phases would help better understand how these phases are formed and regulated. How phase-separation affects biochemical reactions is also poorly understood. One way in which phase-separated states can regulate biochemical reactions is by increasing the local concentrations of reacting molecules. However, it is possible that the chemical microenvironments created within chromatin phases also play a role in regulating biochemical processes. Therefore, it is important to study the types of chemical micro-environments that can arise within phase-separated chromatin states. Finally, understanding the dynamic range of material properties that are possible in naturally occurring chromatin phases would help understand the extent to which meso-scale properties of phases play regulatory roles in biology. Future experimental studies that address these questions and that go hand-in-hand with new technology development are sure to reveal a rich landscape of regulatory possibilities enabled by phase-separation.
Acknowledgements
I thank Kalpana Narlikar and Serena Sanulli for assistance in figure preparation. This work was supported by a grant from the NIH to GJN (R35GM127010).
References
- Alberti S, Gladfelter A and Mittag T 2019. Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates. Cell 176 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Larson A and Narlikar GJ 2014. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 24 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC and Elgin SC 2014. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 30 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MW, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S and Rosen MK 2019. Organization of chromatin by intrinsic and regulated phase separation. Cell 179 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI and Elgin SC 2002. Heterochromatin new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev 12 178–187 [DOI] [PubMed] [Google Scholar]
- Hancock R and Jeon KW 2014. Preface. New models of the cell nucleus crowding, entropic forces, phase separation, and fractals. Int. Rev. Cel. Mol. Biol 307 xiii. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA and Julicher F 2014. Liquid–liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30 39–58 [DOI] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S and Narlikar GJ 2017. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF and Richmond TJ 1997. Crystal structure of the nucleosome core particle at 28Å resolution. Nature 389 251–60 [DOI] [PubMed] [Google Scholar]
- Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD and Narlikar GJ 2019. HP1 reshapes the nucleosome core to promote phase separation of heterochromatin. Nature 10.1038/s41586-019-1669-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M and Brangwynne CP 2018. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175 1481–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, Herman C, Backman V, O’Halloran T, Adam SA, Goldman RD, et al. 2019. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30 2320–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X and Karpen GH 2017. Phase separation drives heterochromatin domain formation. Nature 547 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CY, Johnson SL, Gamarra NI and Narlikar GJ 2016. Mechanisms of ATP-dependent chromatin remodeling Motors. Annual Rev. Biophys 45 153–181 [DOI] [PMC free article] [PubMed] [Google Scholar]


