Abstract
Purpose
Familial exudative vitreoretinopathy (FEVR) and Norrie disease (ND) are genetic disorders that can be caused by mutations in the NDP gene and affect retinal vasculature growth and development. This study aimed to describe the copy number variations (CNVs) in the NDP gene in Chinese FEVR families and the associated phenotypes.
Methods
This study recruited 651 FEVR families. SeqCNV was used to analyze the CNVs in the families without mutations in known FEVR-associated genes. Multiplex ligation-dependent probe amplification and semiquantitative multiplex PCR were performed to verify the NDP CNVs. The probands and family members underwent complete ocular examinations.
Results
NDP CNVs were identified in four patients from three unrelated families, accounting for 15% of the patients with NDP mutations and 0.46% of the entire FEVR cohort. Exon 2 deletions were detected in two families, and whole gene deletion was identified in one family. The affected individuals were born blind with total retinal detachment.
Conclusions
The findings confirm that CNVs are a common NDP mutation type. The CNV-associated phenotype is congenital blindness with total retinal detachment. Antenatal genetic analyses and fetal ultrasound can facilitate early diagnosis and interventions in patients with NDP mutations.
Introduction
Familial exudative vitreoretinopathy (FEVR; OMIM: 133780) is an inherited disease affecting the growth and development of retinal vessels, with characteristic abnormalities in the retinal vasculature [1]. Its clinical manifestations are avascularity in the peripheral retina [2], retinal folds, vitreous hemorrhage, and retinal detachment [3]. The three inheritance patterns of FEVR are X-linked (XL) recessive, autosomal dominant (AD), and autosomal recessive (AR). The nine following genes have been reported to cause FEVR: LRP5 (OMIM: 603506) [4], TSPAN12 (OMIM: 613138) [5], NDP (OMIM: 300658) [6], FZD4 (OMIM: 604579) [7], ZNF408 (OMIM: 616454) [8], KIF11 (OMIM: 148760) [9], CTNNB1 (OMIM: 116806) [10], JAG1 (OMIM: 601920) [11], and CTNNA1 (OMIM: 116805) [12]. NDP is inherited as an XL trait, whereas LRP5 can be inherited as either AD or AR, and the rest of the genes have been reported to inherit in the AD model.
Norrie disease (ND; OMIM: 310600) is an inherited eye disorder that causes blindness in male infants at birth or soon thereafter [13], combined with neurosensory deafness and central nervous system abnormalities. It is inherited as an XL trait [14] caused by mutations in NDP [15].
NDP, located on chromosome Xp11.4, comprises three exons and encodes a secreted 133-amino-acid protein called Norrie or Norrie disease protein (NDP) [15]. It is a cysteine knot protein that activates the Norrie/β-catenin signaling pathway through a precise connection with FZD4 and LRP5 [16]. Mutations in NDP can progress to X-linked FEVR, ND, and retinopathy of prematurity [17]. Thus far, 185 mutations in NDP have been listed in the Human Gene Mutation Database (HGMD). They are classified as missense or nonsense, splicing, small deletions, small insertions, small indels, gross deletions, and gross insertions.
Copy number variations (CNVs), which occupy many genetic variants, include deletion, insertion, and duplication events longer than 1 kb has been maintained [18]. CNVs in NDP, first reported by Chen et al. [19], are mainly gross deletions. To date, 25 gross deletions have been recorded in the HGMD. However, CNVs in NDP remain poorly understood. The aim of this study was to describe the CNVs in the NDP gene in Chinese families and the associated phenotypes.
Methods
Patients
This study adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Review Board of Zhongshan Ophthalmic Center, Sun Yat-sen University (2014MEKY048). Informed consent forms were obtained from the participants or their guardians.
In total, 651 probands with a diagnosis of FEVR were recruited at Zhongshan Ophthalmic Centre, Sun Yat-sen University, from January 2014 to April 2021. The criteria for clinical diagnosis of FEVR were the same as in previous studies [20,21], and the FEVR stage was determined using the classification system developed by Trese et al. [22]. Patients with a birthweight of less than 1,500 g or a gestational age of less than 32 weeks were excluded, as were patients with obvious systemic conditions. The diagnosis of FEVR was made at the initial visit to the ophthalmic center. Patients with fibrous and vascular changes of the retina at birth or soon after birth, as well as hearing loss and mental retardation or other systemic conditions, were diagnosed as ND.
Clinical examinations
All probands and their family members underwent comprehensive ophthalmic examinations, including best-corrected visual acuity (BCVA), slit-lamp biomicroscopy, ophthalmoscopy, and intraocular pressure. Children underwent either RetCam examination (Clarity Medical Systems, Pleasanton, CA) or scanning laser ophthalmoscopy (Nidek F-10; Nidek, Gamagori, Japan). B-scan ultrasonography (Compact Touch 4.0; Quantel Medical, Cournon-d’Auvergne, France) was performed in patients with cloudy cornea or lens opacity. Fundus fluorescein angiography was performed using a Spectralis HRA (Heidelberg Engineering, Heidelberg, Germany) or RetCam (Clarity Medical Systems).
Targeted gene panel sequencing, whole-exome sequencing, and verification
Peripheral venous blood was collected from the FEVR probands and family members. The DNA extraction method was detailed in our previous study [23]. DNA samples from the probands were analyzed with targeted gene panel (TGP) sequencing (from January 1, 2015, to December 31, 2017) and whole exome sequencing (WES; from January 1, 2018), and variant verification and segregation analysis was performed with Sanger sequencing. The WES data were analyzed using a multistep bioinformatic analysis as described in our previous study [24]. For patients with no detected pathogenic variants, SeqCNV was used for CNV analysis [25].
Semiquantitative multiplex PCR (qPCR) was used to validate the CNVs. Primers of NDP exons 1, 2, and 3 were designed, and exon 6 of SPATA7 and exon 14 of TTLL5 were used as positive controls (Supplementary Table 1). The qPCR process was repeated three times. The detailed qPCR reaction was the same as in a previous study [26]. CNVs of NDP were identified when the relative amount of the female sample was half or the male sample was 0 compared with the control group. Multiplex ligation-dependent probe amplification (MLPA) was also used to determine the CNVs using a single multiplex–based reaction. An MLPA kit (SALSA MLPA Probemix P285-C3 LRP5; FALCO Biosystems–MRC Holland, Amsterdam, Netherlands) was used according to the manufacturer’s instructions. The relevant information on the probes is shown in Supplementary Table 2. The reaction products were run on a genetic analyzer (ABI PRISM 3130 Genetic Analyzer; Applied Biosystems, Foster City, CA) and then analyzed by Coffalyser.Net software (MRC Holland, Amsterdam, Netherlands).
Results
Clinical features of probands with NDP CNVs
Using TGP and WES, NDP mutations were detected in 20 of the 651 FEVR families (3.07%); of these, NDP CNVs were identified in three families (probands XDW1, DX1740, and DX1906), accounting for 15% of the patients with NDP mutations and 0.46% of the entire FEVR cohort. All three probands with NDP CNVs had total retinal detachment (Figure 1).
Figure 1.
Ultrasound of the three probands. Retinal detachment was detected in A,B: the right and left eyes of XDW1, C,D: the right and left eyes of DX1740, and E,F: the right and left eyes of DX1906.
XDW1 was referred to our outpatient clinic 12 days after birth. He was full term and delivered uneventfully. Fetal ultrasound detected vitreous stalk-like changes at 36 weeks of gestation. On the first day after birth, B-scan ultrasonography showed bilateral retinal detachment and choroidal detachment. XDW1 was diagnosed with autism during follow-up, at 4 years old. His uncle had a similar phenotype, with no light perception bilaterally.
In DX1740, bilateral leukocoria was noticed a few days after birth. Ophthalmological examination revealed corneal opacities and shallow anterior chambers. B-scan ultrasonography showed total retinal detachment of bilateral eyes. No systemic abnormalities were noticed at 3 years old.
In DX1906, leukocoria was noticed 3 months after birth. The patient visited our hospital at 6 months of age. B-scan ultrasonography showed bilateral retinal detachment and ophthalmatrophia. Brainstem auditory evoked potential and electroencephalogram were unremarkable. At his last visit at the age of 3 years old, DX1906 still could not walk or speak. All four affected male patients had a similar phenotype: They all had total retinal detachment at an early age of onset, even during the prenatal period.
NDP CNVs
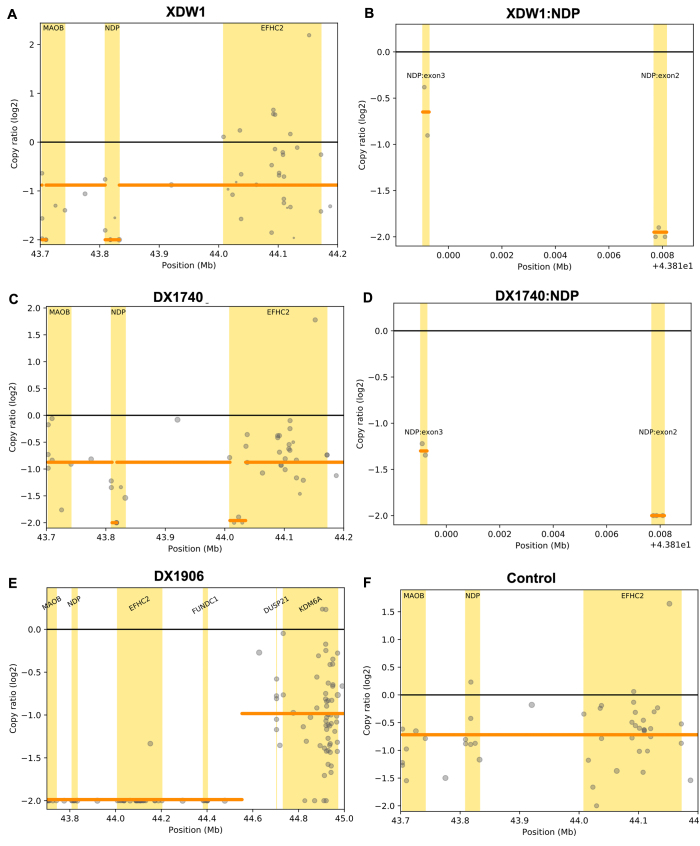
The SeqCNV analysis of the WES data detected NDP CNVs in three families (Figure 2). XDW1 and XDW1uncle had NDP exon 2 deletion, and XDW1M and XDW1aunt were female carriers. DX1740 also had exon 2 deletion, which he inherited from his mother. DX1906 had whole gene deletions of NDP (including exons 1, 2, and 3) and the MAOB, EFHC2, and FUNDC1 genes. These CNVs were confirmed by qPCR (Figure 3) and MLPA (Figure 4).
Figure 2.
NDP copy number variations (CNVs) detected by SeqCNV. A: NDP exon 2 deletion in XDW1. B: The amplified region of NDP with exon 2 deletion in XDW1. C: NDP exon 2 deletion in DX1740. D: The amplified region of NDP with exon 2 deletion in DX1740. E: A 1.73-Mb deletion in DX1906. D: Normal control with no deletion.
Figure 3.
Confirmation of NDP copy number variations (CNVs) using semiquantitative multiplex PCR. Histograms of NDP exons 1, 2, and 3 from A: XDW1, B: DX1740, and C: DX1906 compared with other family members. TTLL5 exon 14 and SPATA7 exon 6 were used as controls. The relative NDP exon 1, 2, and 3 amplicon copy number ratios in the affected males were close to 0 (the normal copy number ratio should be 1 in males and 2 in females), confirming the deletion in a hemizygous state. The copy number ratios of TTLL5 exon 14 and SPATA7 exon 6 were around 2. A: XDW1 and XDW1uncle had NDP exon 2 deletion. B: DX1740 had NDP exon 2 deletion. C: DX1906 had whole NDP gene deletion.
Figure 4.
Confirmation of NDP copy number variations (CNVs) using multiplex ligation-dependent probe amplification (MLPA). A: MLPA analysis of XDW1 (left) and XDW1M (right). The arrow indicates no peak in XDW1 and a half-reduced relative peak height in XDW1M, corresponding to the NDP exon 2 probe. B: MLPA analysis of DX1740 (left) and DX1740M (right). The arrow indicates no peak in DX1740 and a half-reduced relative peak height in DX1740M, corresponding to the NDP exon 2 probe. C: MLPA analysis of DX1906 (left) and DX1906M (right). The arrow indicates no peak in DX1906 and a half-reduced relative peak height in DX1906M, corresponding to the NDP exon 1, 2, and 3 probes.
Discussion
In this study, we identified three families with NDP CNVs and detailed their ocular manifestations. All probands with NDP CNVs had a severe ocular phenotype with total retinal detachment. CNVs accounted for 15% of the families with NDP mutations in our cohort.
A total of 185 NDP mutations have been reported in the HGMD. These include 98 (53%) missense mutations, 16 (8.6%) nonsense mutations, 7 (3.8%) splicing, 2 (1.1%) regulatory, 23 (12.4%) small deletions, 25 (13.5%) gross deletions, and 1 (0.5%) gross insertion. CNVs are a common NDP mutation type. These are deletions of exons 1, 2, or 3 or whole-gene deletions along with deletions of the MAOA, MAOB, and EFHC2 genes [19,27-38].
The phenotypes of patients with NDP CNVs in this study were severe, including blindness at birth or soon thereafter. Eisuke Arai et al. described the phenotypes of patients with exon 2 deletions. Leukocoria with total retinal detachment was observed in the patients soon after birth; they were diagnosed with mental retardation between the ages of 8 and 12 years old [39]. To further assess the clinical phenotypes of patients with NDP CNVs, especially their ocular features, we reviewed studies on phenotypes related to NDP CNVs (Table 1) [28,30-33,35,37,39-41]. The review demonstrated that all patients had bilateral blindness—many of them at birth—and ocular abnormalities detected antenatally. All patients had retinal detachment. Other common manifestations were cataract, corneal opacity, and shallow anterior chambers. Despite differences in NDP deletions, the ocular phenotypes were similar.
Table 1. Ocular manifestations of patients with NDP CNVs.
| Patient ID | Mutations (deletions) | Evaluation age | Ocular manifestations | Other problems | Reference |
|---|---|---|---|---|---|
| 1 |
incl. ex. 2 |
2 years |
Born blind |
NA |
[35] |
| 2 |
incl. ex. 2–3 |
5 years |
Born blind |
hearing loss |
[35] |
| 3 |
incl. ex. 3 |
2 years |
Born blind |
mental retardation, microcephaly,
seizures, short stature |
[35] |
| 4 |
incl. ex. 3 |
8 months |
Born blind |
NA |
[35] |
| 5 |
incl. ex. 3 |
4 months |
Born blind |
NA |
[35] |
| 6 |
incl. ex. 3 |
36 years |
Born blind |
mental retardation, seizures |
[35] |
| 7 |
3′ UTR region of ex. Three incl. |
6 years |
blindness with nystagmus, corneal opacities, posterior synechiae, cataract and shallow anterior chamber |
learning difficulty |
[33] |
| 8 |
3′ UTR region of ex. Three incl. |
10 years |
blindness with nystagmus, corneal opacities, posterior synechiae, cataract and shallow anterior chamber |
learning difficulty |
[33] |
| 9 |
ex. 2 |
NA |
blindness |
NA |
[27] |
| 10 |
494.1 kb incl. entire gene+MAOA and MAOB |
At birth |
At birth bilateral leucocoria, exudative vitreoretinopathy |
seizure, developmental delay, language problem and motor development delay |
[37] |
| 11 |
244 bp incl. ex. Three & 3′ UTR region |
NA |
bilateral corneal opacities, shallow anterior chamber, posterior synechiae, retrolenticular fibrovascular plaque, andcomplete congenital retinal detachment. |
NA |
[25] |
| 12 |
e×.1–3 incl. entire gene |
8 years |
blindness since birth, oculodigital sign and wandering eye movement, retinal detachment |
NA |
[39] |
| 13 |
e×.1–3 incl. entire gene |
4 years |
blindness since birth, corneal opacity, total retinal detachment |
NA |
[39] |
| 14 |
494.6 kb incl. entire gene and MOAB and EFHC2 |
At birth |
bilateral retinal detachment, microphthalmia, atrophic irides, disappearance of the anterior chamber, corneal opacification, and cataracts |
No |
[29] |
| 15 |
whole gene deletion |
NA |
FEVR (stage 5/stage 5) |
NA |
[30] |
| 16 |
ex. 2 |
7 months |
Leukocoria with total retinal detachment |
mental retardation |
[38] |
| 17 |
ex. 2 |
5 months |
total retinal detachment |
mental retardation |
[38] |
| 18 |
ex. 2 |
9 days |
total retinal detachment |
mental retardation |
[38] |
| 19 | incl. ex. 2–3 | 4 years | bilateral cataracts, poor vision, retinal detachment | NA | [31] |
incl, include; ex, exon; yrs. years old; UTR, untranslated region; NA, not available.
Treatments have rarely been administered to mitigate the deleterious natural history of ND. Trese et al. [42] reported that early vitrectomy was effective because it could at least preserve light perception and visual acuity in one eye. Clement et al. reported an ND case diagnosed using amniocentesis; in this case, blindness was prevented after laser photocoagulation at birth [43]. Liu et al. reported that antenatal genetic analyses combined with fetal ultrasound could be used for prenatal diagnosis of FEVR and ND [33]. Indeed, our review of the phenotypes of patients with NDP CNVs, all of whom had severe retinal detachment at birth or soon thereafter, suggests that antenatal genetic analyses and fetal ultrasound can facilitate early diagnosis and interventions.
Twenty-five NDP gross deletions or CNVs, detected using different methods, have been reported in the HGMD [19]. Deborah et al. reported six intragenic deletions using Southern blot hybridization experiments with exon-specific PCR fragment probes [37]. Suárez-Merino et al. identified a 1.5-Mb DNA deletion together with the NDP and MAO genes using a method that combines PAC library screening with STS mapping [44]. Rivera-Vega et al. detected a partial deletion in the untranslated 3′ region of exon 3 of the NDP gene using PCR and DNA sequencing [35]. Rodriguez-Revenga et al. detected contiguous deletions of the NDP, MAOA, MAOB, and EFHC2 genes using microarray comparative genomic hybridization and fluorescent in situ hybridization analysis [36]. Staropoli et al. analyzed the microdeletion of NDP in the Xp11.3–p11.4 region using MLPA and comparative genomic hybridization (CGH) [40]. Waroop et al. analyzed the intragenic copy number analysis with exon-level array CGH [27]. CNVs are called from WES data using a relative coverage method, followed by exon CGH, chromosome microarray, MLPA, qPCR, or Sanger sequencing for confirmation [34]. The deletion can also be identified using the integrative genomic viewer directly from the WES data [45]. In this study, we used SeqCNV to analyze the WES data and then MLPA and qPCR to verify the NDP CNVs.
In summary, we identified NDP CNVs in three families, accounting for 15% of patients with NDP mutations and 0.46% of our entire FEVR cohort. The CNV-associated phenotype of NDP was congenital blindness with total retinal detachment. Antenatal genetic analyses and fetal ultrasound may facilitate early diagnosis and early interventions in patients with NDP mutations.
Acknowledgments
The authors are grateful to Qingjiong Zhang and Shiqiang Li for the MLPA analysis. This study was supported by grants from the Fundamental Research Funds of State Key Laboratory of Ophthalmology, research funds of Sun Yat-sen University (15ykjc22d; Guangzhou, Guangdong, China), and Science and Technology Program Guangzhou, China (201803010031; Guangzhou, Guangdong, China), National Natural Science Foundation of China (81700879). The authors declare no potential conflicts of interest. Dr. Xiaoyan Ding (dingxiaoyan@gzzoc.com) and Dr. Zhan Li (13702332216@139.com) are co-corresponding authors for this paper.
Appendix 1. Primers used for qPCR.
To access the data, click or select the words “Appendix 1.”
Appendix 2. MLPA probes of NDP.
To access the data, click or select the words “Appendix 2.”
References
- 1.Criswick VG, Schepens CL. Familial exudative vitreoretinopathy. Am J Ophthalmol. 1969;68:578–94. doi: 10.1016/0002-9394(69)91237-9. [DOI] [PubMed] [Google Scholar]
- 2.Tauqeer Z, Yonekawa Y. Familial Exudative Vitreoretinopathy: Pathophysiology, Diagnosis, and Management. Asia Pac J Ophthalmol (Phila) 2018;7:176–82. doi: 10.22608/APO.201855. [DOI] [PubMed] [Google Scholar]
- 3.Poulter JA, Ali M, Gilmour DF, Rice A, Kondo H, Hayashi K, Mackey DA, Kearns LS, Ruddle JB, Craig JE, Pierce EA, Downey LM, Mohamed MD, Markham AF, Inglehearn CF, Toomes C. Mutations in TSPAN12 cause autosomal-dominant familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:248–53. doi: 10.1016/j.ajhg.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao X, Ventruto V, Trese MT, Shastry BS, Hejtmancik JF. Autosomal recessive familial exudative vitreoretinopathy is associated with mutations in LRP5. Am J Hum Genet. 2004;75:878–84. doi: 10.1086/425080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikopoulos K, Gilissen C, Hoischen A, van Nouhuys CE, Boonstra FN, Blokland EA, Arts P, Wieskamp N, Strom TM, Ayuso C, Tilanus MA, Bouwhuis S, Mukhopadhyay A, Scheffer H, Hoefsloot LH, Veltman JA, Cremers FP, Collin RW. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:240–7. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ZY, Battinelli EM, Fielder A, Bundey S, Sims K, Breakefield XO, Craig IW. A mutation in the Norrie disease gene (NDP) associated with X-linked familial exudative vitreoretinopathy. Nat Genet. 1993;5:180–3. doi: 10.1038/ng1093-180. [DOI] [PubMed] [Google Scholar]
- 7.Robitaille J, MacDonald ML, Kaykas A, Sheldahl LC, Zeisler J, Dube MP, Zhang LH, Singaraja RR, Guernsey DL, Zheng B, Siebert LF, Hoskin-Mott A, Trese MT, Pimstone SN, Shastry BS, Moon RT, Hayden MR, Goldberg YP, Samuels ME. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat Genet. 2002;32:326–30. doi: 10.1038/ng957. [DOI] [PubMed] [Google Scholar]
- 8.Collin RW, Nikopoulos K, Dona M, Gilissen C, Hoischen A, Boonstra FN, Poulter JA, Kondo H, Berger W, Toomes C, Tahira T, Mohn LR, Blokland EA, Hetterschijt L, Ali M, Groothuismink JM, Duijkers L, Inglehearn CF, Sollfrank L, Strom TM, Uchio E, van Nouhuys CE, Kremer H, Veltman JA, van Wijk E, Cremers FP. ZNF408 is mutated in familial exudative vitreoretinopathy and is crucial for the development of zebrafish retinal vasculature. Proc Natl Acad Sci USA. 2013;110:9856–61. doi: 10.1073/pnas.1220864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat. 2016;37:235–41. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun W, Xiao X, Li S, Jia X, Wang P, Zhang Q. Germline Mutations in CTNNB1 Associated With Syndromic FEVR or Norrie Disease. Invest Ophthalmol Vis Sci. 2019;60:93–7. doi: 10.1167/iovs.18-25142. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Zhang X, Xu H, Huang L, Zhang S, Liu W, Yang Y, Fei P, Li S, Yang M, Zhao P, Zhu X, Yang Z. Exome sequencing revealed Notch ligand JAG1 as a novel candidate gene for familial exudative vitreoretinopathy. Genet Med. 2020;22:77–84. doi: 10.1038/s41436-019-0571-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhu X, Yang M, Zhao P, Li S, Zhang L, Huang L, Huang Y, Fei P, Yang Y, Zhang S, Xu H, Yuan Y, Zhang X, Zhu X, Ma S, Hao F, Sundaresan P, Zhu W, Yang Z. Catenin alpha 1 mutations cause familial exudative vitreoretinopathy by overactivating Norrin/beta-catenin signaling. J Clin Invest. 2021;131:e139869. doi: 10.1172/JCI139869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburg M. Norie’s disease (atrofia bulborum hereditaria). Acta Ophthalmol (Copenh) 1963;41:134–46. doi: 10.1111/j.1755-3768.1963.tb03533.x. [DOI] [PubMed] [Google Scholar]
- 14.Berger W, Meindl A, van de Pol TJ, Cremers FP, Ropers HH, Doerner C, Monaco A, Bergen AA, Lebo R, Warburg M. Isolation of a candidate gene for Norrie disease by positional cloning. Nat Genet. 1992;1:199–203. doi: 10.1038/ng0692-199. [DOI] [PubMed] [Google Scholar]
- 15.Berger W, van de Pol D, Warburg M, Gal A, Bleeker-Wagemakers L, de Silva H, Meindl A, Meitinger T, Cremers F, Ropers HH. Mutations in the candidate gene for Norrie disease. Hum Mol Genet. 1992;1:461–5. doi: 10.1093/hmg/1.7.461. [DOI] [PubMed] [Google Scholar]
- 16.Ohlmann A, Tamm ER. Norrin: molecular and functional properties of an angiogenic and neuroprotective growth factor. Prog Retin Eye Res. 2012;31:243–57. doi: 10.1016/j.preteyeres.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Bao Y, Yang J, Chen L, Chen M, Zhao P, Qiu S, Zhang L, Zhang G. A Novel Mutation in the NDP Gene is Associated with Familial Exudative Vitreoretinopathy in a Southern Chinese Family. Genet Test Mol Biomarkers. 2019;23:850–6. doi: 10.1089/gtmb.2019.0099. [DOI] [PubMed] [Google Scholar]
- 18.Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics. 2013;14(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen ZY, Battinelli EM, Hendriks RW, Powell JF, Middleton-Price H, Sims KB, Breakefield XO, Craig IW. Norrie disease gene: characterization of deletions and possible function. Genomics. 1993;16:533–5. doi: 10.1006/geno.1993.1224. [DOI] [PubMed] [Google Scholar]
- 20.Kashani AH, Brown KT, Chang E, Drenser KA, Capone A, Trese MT. Diversity of retinal vascular anomalies in patients with familial exudative vitreoretinopathy. Ophthalmology. 2014;121:2220–7. doi: 10.1016/j.ophtha.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 21.Ranchod TM, Ho LY, Drenser KA, Capone A, Jr, Trese MT. Clinical presentation of familial exudative vitreoretinopathy. Ophthalmology. 2011;118:2070–5. doi: 10.1016/j.ophtha.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Pendergast SD, Trese MT. Familial exudative vitreoretinopathy. Results of surgical management. Ophthalmology. 1998;105:1015–23. doi: 10.1016/S0161-6420(98)96002-X. [DOI] [PubMed] [Google Scholar]
- 23.Tang M, Ding X, Li J, Hu A, Yuan M, Yang Y, Zhan Z, Li Z, Lu L. Novel mutations in FZD4 and phenotype-genotype correlation in Chinese patients with familial exudative vitreoretinopathy. Mol Vis. 2016;22:917–32. [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Sun L, Li S, Huang L, Zhang T, Wang Z, Yu B, Luo X, Ding X. Novel variants in familial exudative vitreoretinopathy patients with KIF11 mutations and the Genotype-Phenotype correlation. Exp Eye Res. 2020;199:108165. doi: 10.1016/j.exer.2020.108165. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Zhao L, Wang Y, Cao M, Gelowani V, Xu M, Agrawal SA, Li Y, Daiger SP, Gibbs R, Wang F, Chen R. SeqCNV: a novel method for identification of copy number variations in targeted next-generation sequencing data. BMC Bioinformatics. 2017;18:147. doi: 10.1186/s12859-017-1566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang L, Lu J, Zhang L, Zhang Z, Sun L, Li S, Zhang T, Chen L, Cao L, Ding X. Whole-Gene Deletions of FZD4 Cause Familial Exudative Vitreoretinopathy. Genes (Basel) 2021;12:980. doi: 10.3390/genes12070980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aradhya S, Lewis R, Bonaga T, Nwokekeh N, Stafford A, Boggs B, Hruska K, Smaoui N, Compton JG, Richard G, Suchy S. Exon-level array CGH in a large clinical cohort demonstrates increased sensitivity of diagnostic testing for Mendelian disorders. Genet Med. 2012;14:594–603. doi: 10.1038/gim.2011.65. [DOI] [PubMed] [Google Scholar]
- 28.Halpin C, Sims K. Twenty years of audiology in a patient with Norrie disease. Int J Pediatr Otorhinolaryngol. 2008;72:1705–10. doi: 10.1016/j.ijporl.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Hong S, Wang L, Zhao D, Zhang Y, Chen Y, Tan J, Liang L, Zhu T. Clinical utility in infants with suspected monogenic conditions through next-generation sequencing. Mol Genet Genomic Med. 2019;7:e684. doi: 10.1002/mgg3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia B, Huang L, Chen Y, Liu S, Chen C, Xiong K, Song L, Zhou Y, Yang X, Zhong M. A novel contiguous deletion involving NDP, MAOB and EFHC2 gene in a patient with familial Norrie disease: bilateral blindness and leucocoria without other deficits. J Genet. 2017;96:1015–20. doi: 10.1007/s12041-017-0869-5. [DOI] [PubMed] [Google Scholar]
- 31.Keser V, Khan A, Siddiqui S, Lopez I, Ren H, Qamar R, Nadaf J, Majewski J, Chen R, Koenekoop RK. The Genetic Causes of Nonsyndromic Congenital Retinal Detachment: A Genetic and Phenotypic Study of Pakistani Families. Invest Ophthalmol Vis Sci. 2017;58:1028–36. doi: 10.1167/iovs.16-20281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li JK, Li Y, Zhang X, Chen CL, Rao YQ, Fei P, Zhang Q, Zhao P, Li J. Spectrum of Variants in 389 Chinese Probands With Familial Exudative Vitreoretinopathy. Invest Ophthalmol Vis Sci. 2018;59:5368–81. doi: 10.1167/iovs.17-23541. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Zhu J, Yang J, Zhang X, Zhang Q, Zhao P. Prenatal diagnosis of familial exudative vitreoretinopathy and Norrie disease. Mol Genet Genomic Med. 2019;7:e00503. doi: 10.1002/mgg3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, Vertino-Bell A, Smaoui N, Neidich J, Monaghan KG, McKnight D, Bai R, Suchy S, Friedman B, Tahiliani J, Pineda-Alvarez D, Richard G, Brandt T, Haverfield E, Chung WK, Bale S. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 35.Rivera-Vega MR, Chinas-Lopez S, Vaca AL, Arenas-Sordo ML, Kofman-Alfaro S, Messina-Baas O, Cuevas-Covarrubias SA. Molecular analysis of the NDP gene in two families with Norrie disease. Acta Ophthalmol Scand. 2005;83:210–4. doi: 10.1111/j.1600-0420.2005.00398.x. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Revenga L, Madrigal I, Alkhalidi LS, Armengol L, Gonzalez E, Badenas C, Estivill X, Mila M. Contiguous deletion of the NDP, MAOA, MAOB, and EFHC2 genes in a patient with Norrie disease, severe psychomotor retardation and myoclonic epilepsy. Am J Med Genet A. 2007;143A:916–20. doi: 10.1002/ajmg.a.31521. [DOI] [PubMed] [Google Scholar]
- 37.Schuback DE, Chen ZY, Craig IW, Breakefield XO, Sims KB. Mutations in the Norrie disease gene. Hum Mutat. 1995;5:285–92. doi: 10.1002/humu.1380050403. [DOI] [PubMed] [Google Scholar]
- 38.Smith SE, Mullen TE, Graham D, Sims KB, Rehm HL. Norrie disease: extraocular clinical manifestations in 56 patients. Am J Med Genet A. 2012;158A:1909–17. doi: 10.1002/ajmg.a.35469. [DOI] [PubMed] [Google Scholar]
- 39.Arai E, Fujimaki T, Yanagawa A, Fujiki K, Yokoyama T, Okumura A, Shimizu T, Murakami A. Familial cases of Norrie disease detected by copy number analysis. Jpn J Ophthalmol. 2014;58:448–54. doi: 10.1007/s10384-014-0334-4. [DOI] [PubMed] [Google Scholar]
- 40.Staropoli JF, Xin W, Sims KB. Co-segregation of Norrie disease and idiopathic pulmonary hypertension in a family with a microdeletion of the NDP region at Xp11.3-p11.4. J Med Genet. 2010;47:786–90. doi: 10.1136/jmg.2010.079301. [DOI] [PubMed] [Google Scholar]
- 41.Sudha D, Ganapathy A, Mohan P, Mannan AU, Krishna S, Neriyanuri S, Swaminathan M, Rishi P, Chidambaram S, Arunachalam JP. Clinical and genetic analysis of Indian patients with NDP-related retinopathies. Int Ophthalmol. 2018;38:1251–60. doi: 10.1007/s10792-017-0589-0. [DOI] [PubMed] [Google Scholar]
- 42.Walsh MK, Drenser KA, Capone A, Jr, Trese MT. Early vitrectomy effective for Norrie disease. Arch Ophthalmol. 2010;128:456–60. doi: 10.1001/archophthalmol.2009.403. [DOI] [PubMed] [Google Scholar]
- 43.Chow CC, Kiernan DF, Chau FY, Blair MP, Ticho BH, Galasso JM, Shapiro MJ. Laser photocoagulation at birth prevents blindness in Norrie’s disease diagnosed using amniocentesis. Ophthalmology. 2010;117:2402–6. doi: 10.1016/j.ophtha.2010.03.057. [DOI] [PubMed] [Google Scholar]
- 44.Suarez-Merino B, Bye J, McDowall J, Ross M, Craig IW. Sequence analysis and transcript identification within 1.5 MB of DNA deleted together with the NDP and MAO genes in atypical Norrie disease patients presenting with a profound phenotype. Hum Mutat. 2001;17:523. doi: 10.1002/humu.1140. [DOI] [PubMed] [Google Scholar]
- 45.Kim JH, Hu HJ, Chung YJ. Web-based database and viewer of East asian copy number variations. Genomics Inform. 2012;10:65–7. doi: 10.5808/GI.2012.10.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]