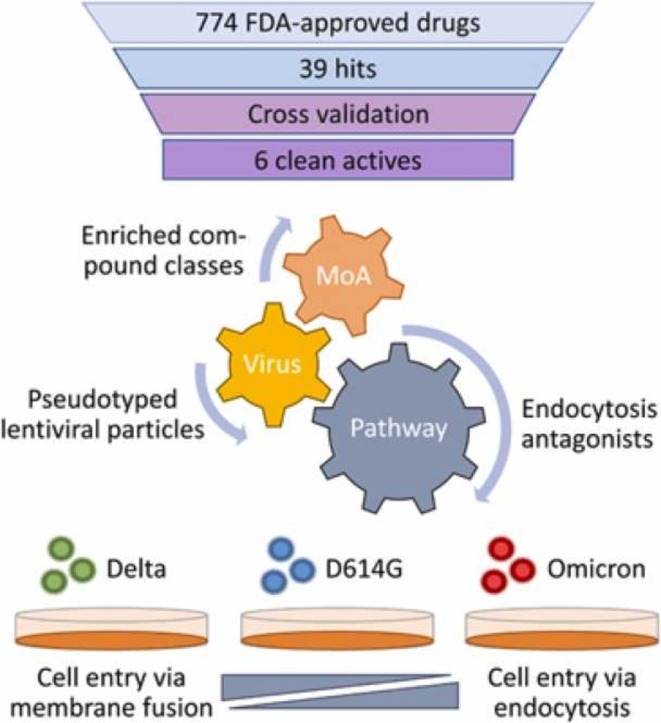
Abstract
The Severe Acute Respiratory Syndrome Coronavirus type 2 (SARS-CoV-2) has continuously evolved, resulting in the emergence of several variants of concern (VOCs). To study mechanisms of viral entry and potentially identify specific inhibitors, we pseudotyped lentiviral vectors with different SARS-CoV-2 VOC spike variants (D614G, Alpha, Beta, Delta, Omicron/BA.1), responsible for receptor binding and membrane fusion. These SARS-CoV-2 lentiviral pseudoviruses were applied to screen 774 FDA-approved drugs. For the assay we decided to use CaCo2 cells, since they equally allow cell entry through both the direct membrane fusion pathway mediated by TMPRSS2 and the endocytosis pathway mediated by cathepsin-L. The active molecules which showed stronger differences in their potency to inhibit certain SARS-CoV-2 VOCs included antagonists of G-protein coupled receptors, like phenothiazine-derived antipsychotic compounds such as Chlorpromazine, with highest activity against the Omicron pseudovirus. In general, our data showed that the various VOCs differ in their preferences for cell entry, and we were able to identify synergistic combinations of inhibitors. Notably, Omicron singled out by relying primarily on the endocytosis pathway while Delta preferred cell entry via membrane fusion. In conclusion, our data provide new insights into different entry preferences of SARS-CoV-2 VOCs, which might help to identify new drug targets.
Keywords: SARS-CoV-2, Pseudovirus, Lentiviral vector, Variant of concern, D614G, Delta, Omicron, BA.1, Drug repurposing, Cellular entry, Infection route, G protein coupled receptor antagonist
Graphical Abstract
1. Introduction
Since its first isolation in late 2019, the Severe Acute Respiratory Syndrome Coronavirus type 2 (SARS-CoV-2) has continuously evolved, resulting in the manifestation of several variants, some of which became a special threat to health systems worldwide as so-called variants of concern (VOCs). According to the WHO there are several criteria a SARS-CoV-2 VOC has to fulfill [1]. These criteria include elevated transmissibility, increased severity of disease, immune escape, a changed clinical manifestation and a decreased effectiveness of standard procedures. The latter comprise public health measures, therapeutic plans and vaccines. Until now five SARS-CoV-2 variants were considered VOCs based on at least a selection of these criteria. The earliest VOCs were Alpha (B.1.1.7) and Beta (B.1.351), designated VOCs on the 18th of December 2020. They were followed by Gamma (P1, January 11, 2021), then Delta (B.1.617.2, May 11, 2021) and the latest Omicron (B.1.1.529), which was declared a VOC on November 26, 2021 [27].
The first detected SARS-CoV-2 mutation that spread in the human population contained the amino acid exchange D614G in the spike protein [2]. Later, the Alpha variant had acquired 23 mutations consisting of six synonymous mutations, four deletions and 13 non-synonymous mutations [3]. Two of the deletions and six of the non-synonymous mutations were located within the spike protein affecting affinity for its cellular receptor and recognition by human proteases. The Beta variant was reported to have 21 mutations with nine of them in the spike domain [4], [5]. In the Gamma variant 34 mutations were found, 21 non-synonymous mutations, three deletions, a 4-nucleotide insertion and 10 synonymous mutations, with 12 of these mutations in the spike region [6], [7]. Of the 27 mutations in the Delta variant seven non-synonymous mutations and two deletions are located in the spike protein [8], [9]. Up until now Omicron is the variant with the highest number of mutations, 72 for Omicron/BA.1, with half of them within the spike protein (30 non-synonymous mutations and six deletions). The spike mutations are responsible for enhanced receptor binding affinity and decreased antibody recognition [9], [10]. In summary, all variants accumulated mutations within the spike protein. Being the first point of interaction with the host cell and immune system, spike protein mutations potentially alter virus transmissibility and infectivity, immune escape and cell tropism.
Infection of a host cell with SARS-CoV-2 starts with binding of the virus to the surface receptor followed by direct fusion with the plasma membrane or endocytosis. The angiotensin-converting enzyme 2 (ACE2) was shown to interact with the spike protein (S protein), thereby acting as a receptor for SARS-CoV and SARS-CoV-2 [11]. The binding results in a conformational change of the S2 domain of the S protein and initiates the fusion of the viral and host membrane. In addition, it was shown that the spike protein has to be activated by a proteolytic cleavage of cellular exogenous or membrane-bound proteases to facilitate the membrane fusion process. The transmembrane serinprotease 2 (TMPRSS2) was reported to be involved in this activation step [28]. If fusion does not occur, the virus will be taken up through the endosomal pathway, which is the preferred pathway for the majority of human coronaviruses. Recently, it was shown that endocytosis of SARS-CoV-2 is a clathrin-mediated process [29]. The acidic pH inside the vesicles and thus the activation of human cathepsin-L results in spike-protein cleavage at the S2’ site followed by the fusion of viral and endosome membrane [30]. Recent studies showed a reduced ability of the Omicron variant for direct cell membrane fusion and an enhanced preference for the endocytosis route, making the virus less dependent on the TMPRSS2 protease but more dependent on the key proteins of endocytosis such as cathepsin-L [16]. Additionally, furin, which is involved in a variety of viral infections such as influenza, was also proposed to interact with the spike protein to facilitate the entering and later the exit process [31]. Alpha and Delta variants both have mutations in the corresponding furin cleavage site at amino acid 681 (Alpha P681H, Delta P681R), shown to be responsible for enhanced cell entry [16].
Here we present a drug-repurposing screen for the identification of compounds interfering with the SARS-CoV-2 cell entry process using SARS-CoV-2 pseudoviruses based on lentiviral vectors. We validated the sensitivity of SARS-CoV-2 variants of concern Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2) and Omicron/BA.1 (B.1.1.529.1) towards a set of defined hits. We found compounds with differing activities against the various SARS-CoV-2 VOCs, which might help to dissect differences in cell entry mechanisms and to identify novel therapeutic targets.
2. Results
2.1. Pseudovirus generation and assay development
Lentiviral vectors can efficiently be pseudotyped with envelope (env) proteins from a large variety of other enveloped viruses, including SARS-CoV [12] and SARS-CoV-2 [13]. The resulting (lentiviral) pseudotypes or pseudoviruses mimic receptor binding and membrane fusion of the original virus, including all cellular factors involved. Based thereon, these crucial steps of the viral life cycle can be studied using a well-established and safe vector system. After the membrane fusion, all subsequent steps up to marker gene expression are carried out solely by the mechanisms of the underlying lentiviral vector. Beside its safety, another important benefit of the vector system is the possibility to express easy-to-detect marker genes like fluorescent proteins or luciferase. In addition, the lentiviral vectors used are replication-incompetent, avoiding virus spreading within the culture and making the overall assay a well-defined procedure. Here we used the vector LeGO-Luc2-iG2 from our LeGO vector platform [14], [15] that confers concomitant expression of eGFP and firefly luciferase in transduced cells. The vector was pseudotyped with spike protein from one of the five SARS-CoV-2 variants D614G, Alpha, Gamma, Delta and Omicron/BA.1, respectively. Expression of the fluorescent protein allowed to determine the functional titer of pseudovirus preparations on 293T-ACE2 cells by flow cytometry, whereas luciferase expression was used as high-throughput compatible marker in the assay on CaCo2 cells to screen the drug repurposing library. Titers of non-concentrated pseudoviruses with SARS-CoV-2 spike protein (truncated) were in the range of 0.5–2.9 × 105 /mL, which was about 100 times lower than standard VSV-G pseudotyped vectors produced as control, but still high enough for all planned experiments even without further purification or concentration steps. As reported in the literature [13], deletion of the terminal 18–20 amino acids was essential to achieve efficient packaging. Indeed, pseudotyping with the full-length spike resulted in approx. 500–1000 times lower titers compared to the truncated spike protein (19 amino acids deleted). It has been suggested that this N-terminal amino-acid stretch comprises the endoplasmic reticulum retention signal of the spike protein. Counterintuitively, but also in line with the literature [13], we did not observe increased surface expression of the truncated spike protein (data not shown). This might hint to an alternative explanation for impaired pseudotyping with full-length spike, namely steric interference during packaging.
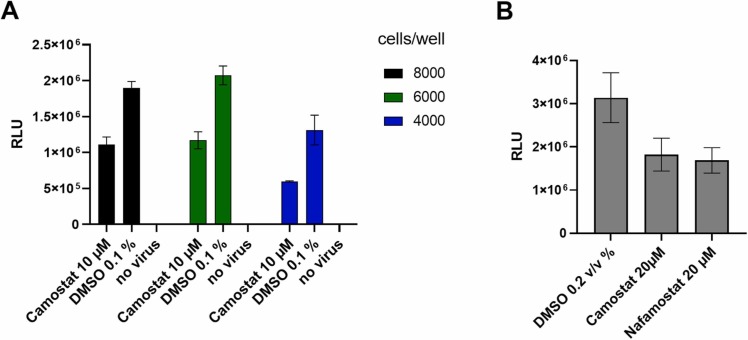
The SARS-CoV-2 pseudoviruses based on lentiviral vectors were utilized to develop a cell-based assay to identify inhibitors of viral entry processes. CaCo2 cells, reported to express both the ACE2 receptor and TMPRSS2, were used [35]. The assay was optimized regarding cell number, pseudovirus concentration and incubation time with compounds prior to pseudovirus addition and post transduction. Best results with signal to background ratio (S/B) of 1.7 were achieved using 8000 cells/well in 20 µL media with 10 µL of lentiviral pseudovirus-containing supernatant. Lower cell numbers resulted in increased variation in the DMSO-solvent control. To evaluate the assay, we used camostat and nafamostat as positive controls for inhibition of viral entry. A maximum response of 50% inhibition could be achieved at non-toxic concentrations ( Fig. 1).
Fig. 1.
: Adaptation and pharmacological validation of SARS-CoV-2 pseudovirus assay for screening, (A). Titration of cell number per well in a 384-well plate, calculation of signal/background ratio using camostat as positive control and DMSO as negative control (B). Validation of assay response to known inhibitors of SARS-CoV-2 entry. Compounds were added 30 min prior to virus addition to 8000 cells per well. Data shown is based on the average and standard deviation of three independent experiments. Detection of luminescence signal was performed after 48 h at 37 °C. RLU: relative light units measured as counts per 100 ms/well.
2.2. Screening of a repurposing library
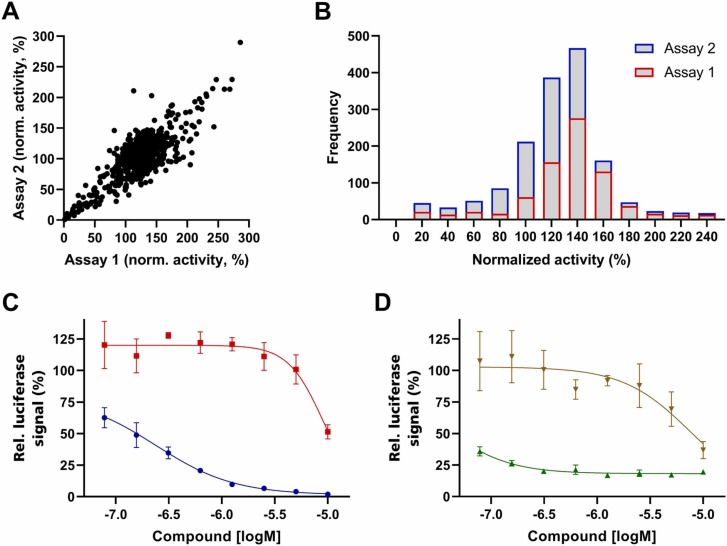
To demonstrate the applicability of the developed assay system we screened the 774 drugs of the SCREEN-WELL FDA approved drug library V2 (BML-2843–0100; Enzo Life Sciences Inc.) for their ability to inhibit SARS-CoV-2 lentiviral pseudovirus entry with spike variant D614G on CaCo2 cells. Camostat was used as a pharmacological control, but failed to inhibit virus entry sufficiently at the given concentration of 10 µM to generate assay robustness with a Z’> 0.5. We therefore normalized the dataset based on vehicle control, DMSO, and repeated the primary screen to assess robustness and predictability. Both screens performed relatively similar ( Fig. 2 A), with a correlation coefficient of 0.72, and showed a slight plate effect resulting in an average signal of 124% of the normalized average signal for the first and 105% for the second assay (Fig. 2 B).
Fig. 2.
SARS-CoV-2 lentiviral pseudovirus entry inhibition assay. Correlation of two independent screens of 774 FDA-approved drugs (A), frequency distribution of the two screens (B), dose response curves of Decitabine (blue) and Trifluoperazine (red) as example for DNA replication inhibitor and dopamine receptor antagonist (C), dose response curves of Docetaxel (green) and Alprostadil (brown) as example for a microtubule polymerization inhibitor and Prostaglandin E1 (D). Data shown in A) and B) are based on singlicates, data shown in C) and D) are based on the average and standard deviation of three independent experiments.
In total, 70 hits were identified based on an average inhibition of the reporter gene of at least 70% (e.g., Fig. 2 C and D). These hits were compared to in-house toxicity data on CaCo2 cells and compounds decreasing viability to below 75% were labeled toxic; thus 10 toxic compounds were removed. To reduce the number of relevant molecules further, the hit population was compared against in-house data sets of HEK293T and MCF7 cell toxicity as well as a dataset of inhibitors of a lentiviral vector reporter gene assay from an unrelated project [17], removing a number of false-positive hits specific to lentiviral vectors. It has to be noted that not all compounds had been screened in this counter assay, therefore two DNA synthesis inhibitors remained in the data set. The additional toxicity data sets eliminated another 26 compounds, and 12 compounds were identified as false-positives due to inhibition of the unrelated pseudovirus assay [17] also based on lentiviral vectors. In summary, mainly compounds interfering with viral nucleic acid production or microtubule function were eliminated. All six compounds with an IC50 below 10 µM, which were also not active in the aforementioned counter assays, are shown in Table 1.
Table 1.
Hits of the SARS-CoV-2 lentiviral pseudovirus assay after partial removal of false-positives. The IC50 value is based on a triplicate measurement of the viral reporter gene, and the viability data are means and standard deviations of three measurements at 10 µM.
| Name | IC50 [µM] | Viability [%] | Mode of action |
|---|---|---|---|
| Fluorouracil | 7.01 | 97.0 ± 1.7 | Nucleic acid pathway inhibitor (thymidine synthesis) |
| Cladribine | 7.74 | 107.5 ± 2.7 | Nucleic acid pathway inhibitor (purine analog) |
| Docetaxel | < 0.08 | 80.8 ± 3.0 | Microtubule inhibitor |
| Chlorpromazine | 9.63 | 88.6 ± 4.3 | G-protein receptor antagonist (D2 dopaminergic receptor) |
| Econazole | 8.96 | 88.2 ± 1.8 | Antifungal imidazole (steroid synthesis) |
| Alprostadil | 7.42 | 100.4 ± 2.9 | Prostaglandin E1 |
Next to a number of compounds interfering with different pathways related to viral nucleic acid production, other modes of action putatively relevant for viral entry were identified. All of these compounds were analyzed in more detail in a second screening.
2.3. Variant specificity of entry inhibitors
In a second assay round, we assessed the activity of the hits against different spike protein variants in our SARS-CoV-2 lentiviral pseudovirus assay. In order to get a more general picture and potentially identify more potent compounds, we analyzed hit compound classes rather than the individual hits, only. Particularly, we identified and screened similar compounds for the nucleic acid pathway inhibitors, the microtubule function inhibitors and the G-protein coupled receptor antagonists.
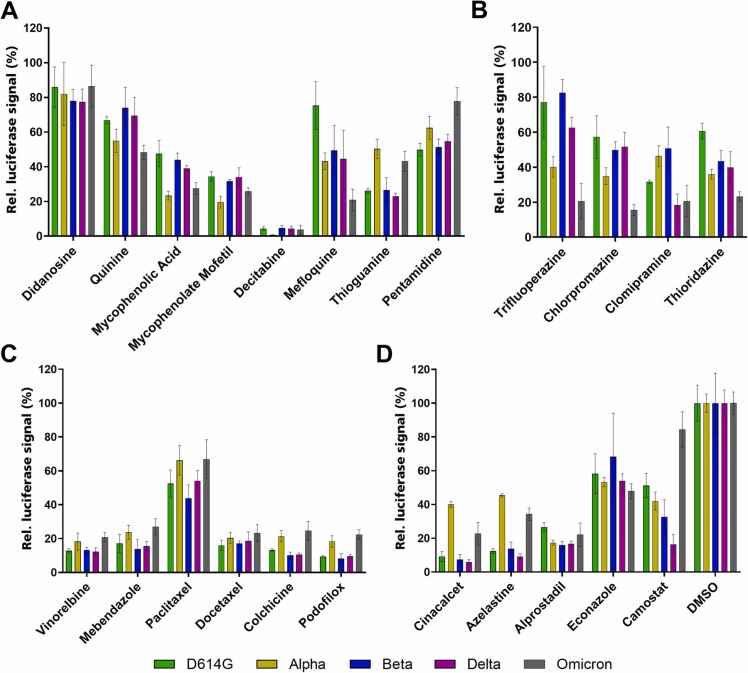
The largest class of active molecules, inhibitors of nucleic acid related pathways, was examined using eight molecules ( Fig. 3 A). Within this class, Mycophenolic acid, an inhibitor of the rate-limiting step of GMP synthesis, was also included alongside its slightly more potent prodrug, Mycophenolate mofetil. Additionally, Mefloquine and Quinine, best known as parasite inhibitors, were tested, as they showed activity in the initial SARS-CoV-2 lentiviral pseudovirus assay and also inhibit purine nucleoside phosphorylase in Plasmodium, pointing to an additional mode of action. In the case of Mycophenolic acid, Alpha and Omicron spike protein variants seem to be affected to a larger degree. Other datasets, like Thioguanine or Pentamidine, from the same set of compounds show an opposing trend. Quite a few compounds and most of the microtubule inhibitors actually followed the trend and were less effective against Alpha and Omicron variants. Nevertheless, they were analyzed, as Docetaxel was a very potent hit in the initial screening, and microtubule inhibitors were previously reported to be virostatic (Fig. 3 C) [18], [19]. In total, six microtubule inhibitors were evaluated and showed very similar effects for all spike variants tested. Next to the aforementioned inhibitors, four antagonists of G-protein coupled receptors, mainly Dopamine receptor antagonists, were selected and evaluated (Fig. 3 B). Interestingly, all four antagonists belong to the group of phenothiazine-derived antipsychotic compounds, and the Omicron spike variant was more sensitive to these inhibitors than the other variants.
Fig. 3.
: Screening of active compound classes against SARS-CoV-2 lentiviral pseudoviruses with different SARS-CoV-2 spike variants. Compounds interfering with nucleic acid synthesis (A), G-protein coupled receptor antagonists (B), inhibitors of microtubule polymerization (C) and compounds with different mode of actions (D). All data were normalized to DMSO (shown in D). SARS-CoV-2 spike D614G variant shown in green, Alpha in yellow, Beta in blue, Delta in pink and Omicron in grey. Data shown is based on the average and standard deviation of three independent experiments. Compound structures are given in Supporting Fig. 1.
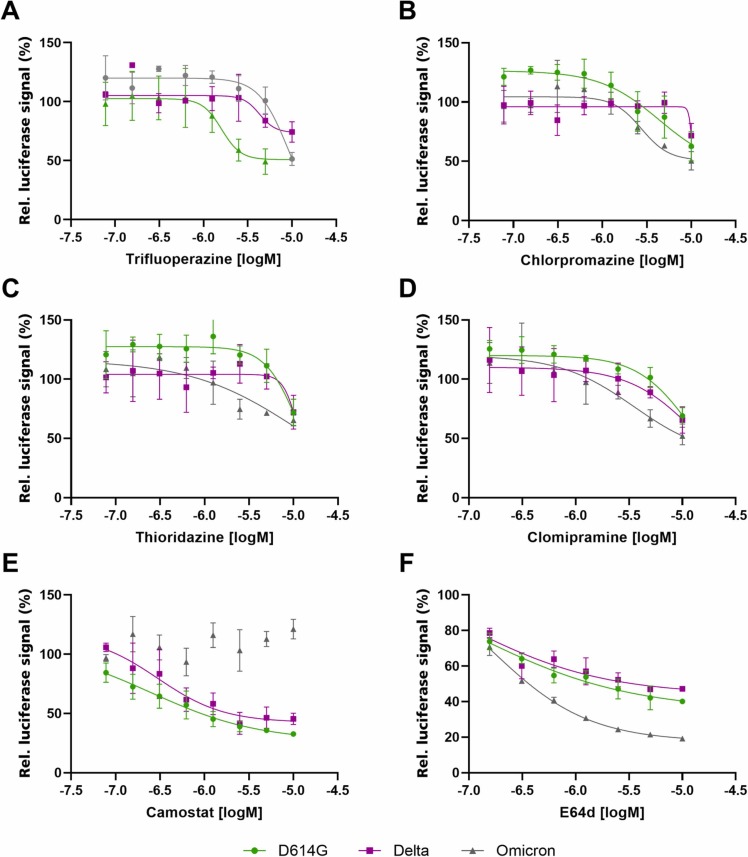
Camostat, the known TMPRSS2 inhibitor used as control, showed an opposite effect, suggesting an alteration in cellular factors required for Omicron uptake (Fig. 3 D). To evaluate this effect in more detail, dose response curves were obtained using the four G-Protein coupled receptor antagonists, with Camostat and E64d as controls ( Fig. 4). All compounds were evaluated against D614G, Delta and Omicron variants in the SARS-CoV-2 lentiviral pseudovirus assay. The four G-protein coupled receptor antagonists were more active against Omicron compared to Delta and D614G variants (Fig. 4 A-D), similarly to the previously reported Cathepsin-L inhibitor E64d involved in endocytosis inhibition. Camostat (Fig. 4 E) and E64d (Fig. 4 F) therefore had opposing effects on Omicron, whereas no change of inhibition potency was observed for D614G and Delta.
Fig. 4.
: The effects of different phenothiazine-derived antipsychotic compounds (A - D) against SARS-CoV-2 lentiviral pseudoviruses with different spike variants. TMPRSS2 inhibitor Camostat (E) and Cathepsin-L-mediated endocytosis inhibitor E64d (F) were used as positive controls. Spike variant D614G is shown in green, Delta in pink and Omicron in gray. Data shown is based on the average and standard deviation of three independent experiments. Compound structures are provided in Supporting Fig. 2.
2.4. Evaluation of endocytosis key factors used by Omicron for cell entry
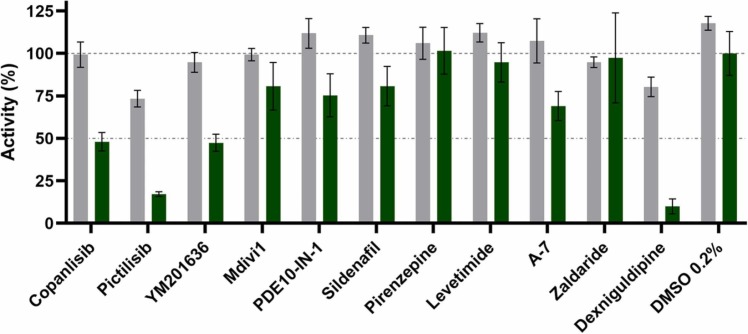
Knowing that SARS-CoV-2 uses clathrin-mediated endocytosis for entry and that the phenothiazine-derived antipsychotic compounds evaluated in Fig. 4 exhibit an inhibitory activity against this form of endocytosis, we evaluated which key proteins might be used by the virus for entry [20]. To do so, we expanded the hit list with representatives of known inhibitors of key proteins of endocytosis such as dynamin I, phosphodiesterases, calmodulin and phosphatidylinositol 3-kinase (PI3K). The same compounds were also tested for cytotoxicity to exclude any unspecific effects in the SARS-CoV-2 lentiviral pseudovirus assay. Tested at 10 µM, none of the compounds reduced the cell viability by more than 30% ( Fig. 5). The obtained data showed that among the selected targets PI3K inhibition had the strongest effect on entry of Omicron SARS-CoV-2 lentiviral pseudovirus (Fig. 5). Also, calmodulin inhibitors showed significant inhibition, especially Dexniguldipine, which is also an inhibitor of the P-gp efflux pump.
Fig. 5.
: Toxicity and virostatic effect of different endocytosis inhibitors. Toxicity (shown as cell viability (%), grey) was tested using the CaCo2 cell line and virostatic activity (green) was analyzed using the Omicron SARS-CoV-2 lentiviral pseudovirus. All compounds were tested at 10 µM. Five different proteins of the endocytosis pathway were analyzed. PI3K using Copanlisib, Pictilisib and YM201636; Dynamin using Mdivi1; PDE using PDE10-IN-1 and Sildenafil; ß-arrestin using Pirenzepine and Levetimidine; Calmodulin using A-7, Zaldaride, Dexniguldipine (also a p-gp efflux pump inhibitor). Data shown is based on the average and standard deviation of three independent experiments.
2.5. Synergy of compound-mediated inhibition of viral entry
As SARS-CoV-2 uses different cellular factors for infection of the host cell, we analyzed how the compound-derived inhibition of viral entry can be enhanced by targeting different key enzymes of these entry pathways.
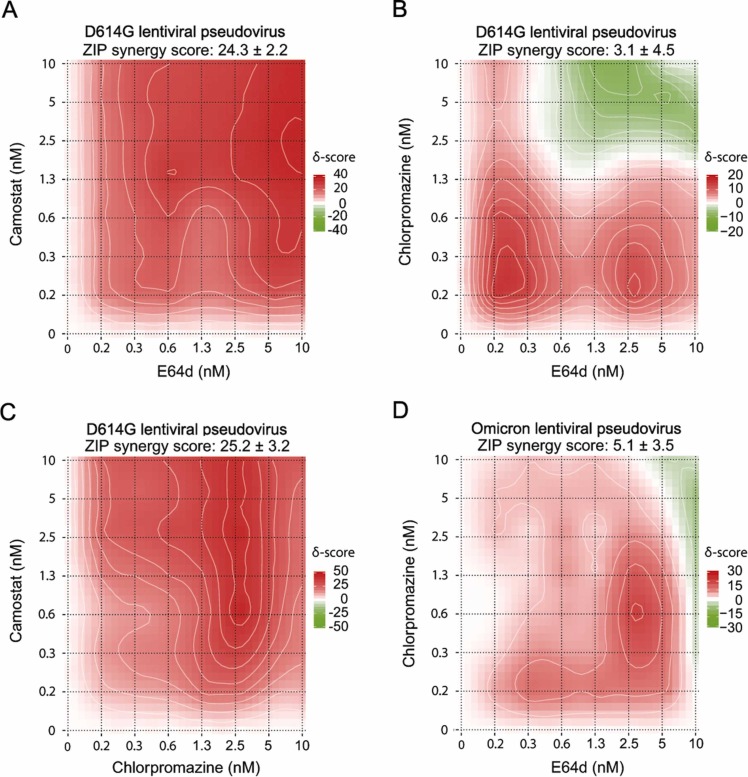
As shown in Fig. 4, the D614G SARS-CoV-2 variant can be inhibited in two ways - by camostat targeting TMPRSS2 and thus direct fusion, and by E64d targeting Cathepsins and thus the endocytosis pathway. We combined the inhibitors in a matrix titration. The results showed a strong synergistic effect reaching a maximum of about 90% inhibition, compared to a maximum of about 50% if compounds were tested alone ( Fig. 6 A). In the next step, we used Chlorpromazine, one of the compounds with higher specificity for Omicron compared to other VOC pseudoviruses, and tested its activity against E64d and Camostat. While the comparison of Chlorpromazine with Camostat showed a clear synergy in D614G (Fig. 6 C), the matrix titration of Chlorpromazine with E64d had only additive effects in D614G and Omicron SARS-CoV-2 lentiviral pseudovirus assays (Fig. 6 B and D). The results showed an inhibition response similar to the addition of the respective single compound IC50 (ZIP score between 0 and 10). This behavior points to the pathway inhibited: While E64d and Camostat as well as Chlorpromazine and Camostat act on different entry routes and are therefore synergistic (ZIP score greater than 10), E64d and Chlorpromazine act additively and therefore most likely target similar routes.
Fig. 6.
: Matrix titration of identified SARS-CoV-2 lentiviral pseudovirus entry inhibitors. Titration of E64d and Camostat (A), E64d and Chlorpromazine (B), Chlorpromazine and Camostat (C) in SARS-CoV-2 D614G lentiviral pseudovirus assay. Titration of E64d and Chlorpromazine (D) in SARS-CoV-2 Omicron lentiviral pseudovirus assay. ZIP score calculated using SynergyFinder 2.0 [21]. Data shown is based on the average and standard deviation of three independent experiments. Compound structures are given in Supporting Fig. 2.
3. Discussion
The SARS-CoV-2 pandemic is a global threat and resulted in countless research projects searching for drugs against viral replication or COVID-19 as the syndrome. Drug repurposing, with its promise for faster and cheaper drug development, is at the forefront of this endeavor [22]. A good example is Remdesivir, a Gilead drug formerly investigated to treat Ebola infection. The drug was tested against COVID-19 in February 2020, was granted an Emergency Use Authorization by the FDA in May and was fully approved in October of the same year. Although Remdesivir was later found to be only moderately effective, the example demonstrates the potential of drug repurposing as a means of fast and safe drug development [23]. Here we describe a repurposing screen aimed at the identification of approved drugs that interfere with the entry process of SARS-CoV-2 in a pseudovirus assay. Moreover, active hits were profiled using pseudoviruses with the spike protein of the VOCs Alpha, Beta, Delta and Omicron/BA.1.
The high-throughput cell-based screen using the D614G SARS-CoV-2 lentiviral pseudovirus showed that the largest group of actives, 15 of 39, were compounds inhibiting nucleic acid synthesis at different steps of the process. Many of these compounds were directly developed against viral infections, like NRTIs and NNRTIs against HIV, whereas others, like Cladribine or Fluorouracil, are marketed cancer drugs. It is important to note that the lentiviral pseudovirus used in our assay is derived from HIV, therefore inhibitors of the HIV reverse transcriptase and integrase enzymes are detected as “false-positives”, as previously analyzed and described by us [17]. These actives were therefore general lentivirus inhibitors and showed no specificity for a VOC spike protein. Another large group of active molecules belonged to the class of microtubule binders. Although many of the compounds showed some signs of toxicity at the tested concentration and incubation time, they still demonstrated specific activity, altogether. Studies showed that this activity is not limited to SARS-CoV-2, but that, for example, Podofilox and Mebendazole inhibited infection with human Cytomegalovirus and Ebola virus, respectively [19], [32]. The specific effects of microtubule inhibitors against SARS-CoV-2 infection have been well documented, but there may also be more general effects reducing the severity of COVID-19 [22]. Colchicine, for example, disrupts different cellular processes including the NLRP3 inflammasome as well as processes leading to cytokine storm and was therefore investigated as a therapeutic option against COVID-19 [33], [34].
The group of active compounds, which showed the largest differences in their potency to inhibit entry of lentiviral pseudoviruses of various SARS-CoV-2 VOCs, included antagonists of G-protein coupled receptors, such as the Dopamine receptor. As Trifluoperazine, Chlorpromazine, Clomipramine and Thioridazine showed enhanced inhibitory effects on Omicron entry, and these compounds have been reported to be involved in inhibition of clathrin-mediated endocytosis, also used by SARS-CoV-2, we evaluated which key proteins were most relevant for the entry of the Omicron SARS-CoV-2 lentiviral pseudovirus. Chlorpromazine was also reported to inhibit calmodulin, which is involved in control of ACE2 ectodomain shedding [20], [24]. The expansion of the hit list with compounds inhibiting targets of endocytosis, which are involved in G-protein controlled pathways or calmodulin interactions, revealed an important role of PI3K for entry of Omicron. In addition, the calmodulin inhibitor Dexniguldipine showed strong inhibition of Omicron SARS-CoV-2 lentiviral pseudovirus entry, whereas other calmodulin inhibitors showed only weak inhibitory activity. Dexniguldipine is not only a calmodulin inhibitor but also a P-gp inhibitor reported to inhibit the efflux pump involved in multi-drug resistance in, e.g., cancer cells, thus increasing its own intracellular concentration as well as the activity of co-administrated drugs [25].
Our data demonstrate that lentiviral vectors pseudotyped with Spike proteins of the various SARS-CoV-2 VOCs exhibit different preferences for direct membrane fusion or entry through endocytosis. Confirming previously reported data [16] we show that the Omicron variant has a preference for the endocytosis pathway, whereas D614G uses both pathways and the Delta variant prefers entry though direct membrane fusion. In general, it is important to note that the effect of a tested compound might strongly be affected by the cellular system used for the assay. As recently reported by Willett et al. [16] and others, the effect of cathepsin-L inhibitor E64d, involved in uptake by endocytosis, might be neglectable in those cell lines which primarily support cell entry through direct membrane fusion regulated by TMPRSS2. Here we used CaCo2 cells that equally allow cell entry through both the direct membrane fusion pathway mediated by TMPRSS2 and the endocytosis pathway mediated by cathepsin-L, as shown by inhibition of TMPRSS2 using Camostat and inhibition of cathepsin-L using E64d. We observed that targeting both, TMPRSS2 and Cathepsins, with a combination of Camostat and E64d resulted in strongly enhanced inhibition of D614G pseudovirus entry compared to the effects when the two compounds were added individually. Most probably, this observation was due to the concomitant inhibition of both entry pathways used by the D614G variant. It seems likely that SARS-CoV-2 variants are able to switch between entry mechanisms as seen for the Delta VOC. In line with this finding, we observed almost complete inhibition (up to 80–90%) of virus entry by endocytosis inhibitors towards the Omicron SARS-CoV-2 lentiviral pseudovirus, which was, on the contrary, not inhibited by TMPRSS2 inhibitors. In parallel, the combination of the identified hits resulted in relatively weak additive inhibitory effects, pointing to a linear inhibition within the same pathway.
In line with previously reported data [16], our study shows that SARS-CoV-2 variants can utilize different routes of cell entry, with the exact proportion of each route depending on cell type and inhibition applied. Omicron appears to be an exception, apparently almost solely relying on the endocytosis mechanism. For variants using both direct membrane fusion and endocytosis, e.g., D614G and Delta, key enzymes of both mechanisms need to be inhibited to achieve efficient suppression of cell entry. Based on our results it is tempting to speculate that targeting calmodulin and PI3K together with other factors may be beneficial for inhibition of SARS-CoV-2 entry. Enhancement of ACE2 ectodomain shedding through inhibition of calmodulin hinders all SARS-CoV-2 variants from using ACE2 as receptor to bind to the cell surface. The additional inhibition of PI3K, directly or through calmodulin inhibition, might be beneficial for inhibition of SARS-CoV-2 variants which prefer the endocytosis pathway, such as Omicron. Identification of further key factors needed for viral entry, such as the specific protease of ACE2 shedding, will reveal more starting points for development of combined therapies. Fig. 7.
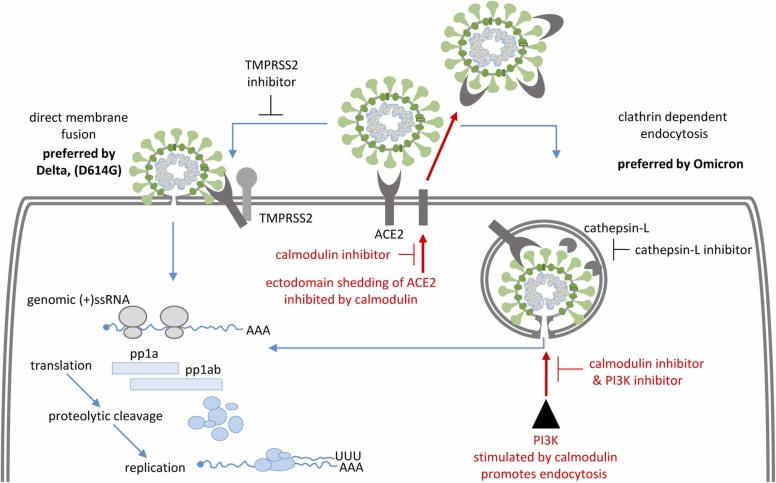
Fig. 7.
Schematic representation of two entry mechanisms of SARS-CoV-2. Whereas Delta VOC prefers direct membrane fusion (left), Omicron VOC prefers uptake by endocytosis and subsequent membrane fusion (right). The specific points of intervention found within this screening campaign are highlighted in red.
4. Methods
4.1. Cloning of SARS-CoV-2 spike expressing plasmids
Standard molecular cloning techniques were used to generate spike protein expressing plasmids needed for the production of lentiviral pseudoviruses. New spike-encoding plasmids were deposited at Addgene as indicated. A human codon-optimized version of the full-length cDNA of SARS-CoV-2 Spike protein (wild type, Wuhan-Hu-1) was synthesized (Twist Biosciences, San Francisco, CA) and cloned into the mammalian expression plasmid pEF1a-Puro under control of the human elongation factor 1α promoter resulting in pEF1a-Puro-SARS-CoV-2-Spike-wt (Addgene #183413). The mutation D614G was introduced by PCR, resulting in pEF1a-Puro-SARS-CoV-2-Spike-D614G (Addgene #183415). To remove the putative N-terminal endoplasmic reticulum retention signal, the part encoding the last 19 amino acids was removed by PCR, resulting in pEF1a-Puro-SARS-CoV-2-Spike-wt-d19 (Addgene #183414) and pEF1a-Puro-SARS-CoV-2-Spike-D614G-d19 (Addgene #183416). The cDNA of Spike Alpha (B.1.1.7) was synthesized based on the same codon-usage (GeneArt / ThermoFisher, Darmstadt, Germany), resulting in pEF1a-Puro-SARS-CoV-2-Spike-Alpha-d19 (Addgene #183417).
The following three plasmids [26] expressing SARS-CoV-2 spike variants under control of a CMV promoter were a gift from David Nemazee, ordered through Addgene.org (Watertown, MA), pcDNA3.3-SARS-CoV-2-Spike-Beta-d18 (B.1.351, Addgene #170449), pcDNA3.3-SARS-CoV-2-Spike-Delta-d18 (B.1.617.2, Addgene #172320). The cDNA of Spike Omicron/BA.1 (B.1.1.529.1) was synthesized (GeneArt/ThermoFisher, Darmstadt, Germany) with a codon-usage based on the Beta and Delta cDNA, resulting in pcDNA3.3-SARS-CoV-2-Spike-Omicron-BA.1-d18 (Addgene #180843).
4.2. Production of lentiviral pseudoviruses with SARS-CoV-2 spike protein
Cell-free viral particles containing supernatants were generated by transient transfection of 293T cells with four plasmids using the calcium phosphate transfection method as described previously [14], [15]. Protocols are available at the LeGO website (http://www.LentiGO-Vectors.de). The four plasmids used for transfection were the 3rd generation HIV1-derived lentiviral SIN vector LeGO-Luc2-iG2 (Addgene #183418) (15 µg), expressing eGFP and firefly luciferase (Luc2, Promega), the 3rd-generation packaging plasmids pMDLg/pRRE (Addgene #12251) (10 µg) and pRSV-Rev (Addgene #12253) (5 µg) and one of the SARS-CoV-2 spike protein expressing plasmids (4 µg) per 5 × 106 293 T cells plated the day before on a 10 cm cell culture dish in 10 mL DMEM (4.5 g/L glucose, GlutaMAX, Gibco #31966–021) with 10% fetal bovine serum (Sigma F7524), penicillin/streptomycin (100 U/mL, 100 μg/mL Gibco #15140–122), and 25 mM HEPES (Gibco #15630–056). Per plate, three supernatants were collected (8 mL overnight, 6 mL over day, 8 mL overnight), filtrated though 0.45-µm syringe filters (Whatman #10462100), kept at 4 °C until all three were pooled, then aliquoted into 2 mL tubes and frozen at −80 °C.
Supernatants containing the pseudovirus particles were titrated on 293T-ACE2-Puro-2G7 cells, a single cell derived clone stably overexpressing human ACE2, plated on 24-well plates with 50.000 cells per well in 500 µL DMEM. For the titration, 8 µg/mL polybrene was added to the medium and after addition of the pseudovirus, plates were centrifuged for 1 h at 1000 g and 25 °C (spin-inoculation). Gene transfer rates were analyzed 2–3 days after transduction by flow cytometry based on eGFP expression. Titers of 0.5–2.9 × 105 /mL SARS-CoV-2 spike pseudotyped vector particles were obtained non-concentrated, whereas VSV-G pseudotyped vectors as control typically reach 1–5 × 107 /mL, for comparison.
4.3. High-throughput screening and hit profiling
We used the SCREEN-WELL FDA approved drug library V2 (BML-2843–0100; Enzo Life Sciences Inc.) for identification of compounds interfering with the entry of the lentiviral pseudovirus SARS-CoV-2 D614G variant.
CaCo2 cells were obtained from Cell Lines Service (CLS, #300137) and used between passage 5 and 25. Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM High Glucose (4.5 g/L), without L-Glutamine, without Phenol Red, Capricorn, #DMEM-HXRXA) with 10% fetal bovine serum (Capricorn, #FBS-12A), L-Glutamine (Capricorn, #GLN-B), streptomycin (100 μg/mL), and 100 U/mL penicillin (Capricorn, #PS-B). For plating, cells were washed with Dulbecco’s PBS, w/o Ca2+ & Mg2+, w/o Phenol Red (Capricorn, #PBS-1A), trypsinized (Trypsin-EDTA Capricorn, #Try-1B), resuspended, and seeded into white 384-well microtiter plates (Greiner Bio-One, #781073) at 8000 cells/20 µL/well. The cells were then incubated at 37 °C in the presence of 5% CO2 for 24 h. Compounds (20 nL/well of 10 mM stock concentration in 100% v/v DMSO) and controls (Camostat 20 nL/well of 10 mM stock concentration in 100% v/v DMSO, and 20 nL/well of 100% v/v DMSO (Carl Roth, #HN47.1)) were added to cells using the Echo 550 Liquid Handler and incubated for 30 min at 37 °C in the presence of 5% CO2. 10 µL/well of lentiviral pseudovirus with SARS-CoV-2 spike D614G were added to the cells and incubated for 48 h at 37 °C in the presence of 5% CO2. 10 µL of Steady-Glo Luciferase Detection Reagent (Promega, #E2520) were added per well, the microplate was incubated at the dark for 15 min. Luminescence was measured using the EnSight multimode plate reader with 100 ms detection time/well.
If not otherwise mentioned, hit profiling was performed using the conditions of high-throughput screening but with selected hits in dose response, starting with 10 µM end concentration in 1:2 dilution/ 8 steps using lentiviral pseudovirus with SARS-CoV-2 spike D614G and of the variants of concern Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2) and Omicron/BA.1 (B.1.1.529.1).
Data analysis was performed using Microsoft Excel and GraphPad Prism 8. Test compound results were normalized relative to the DMSO control that represents 0% inhibition of lentiviral pseudovirus entry. Dose response curves were fitted to 4-parameter logistic functions in GraphPad Prism 8 with constrains to minimal response of 0% inhibition.
4.4. Cytotoxicity assay
The general cytotoxic and cytostatic assessment was performed using the measurement of the intracellular ATP level with Promega CellTiter-Glo (#G7570) assay kit. CaCo2 cells, were obtained from Cell Lines Service (CLS, #300137) and used between passage 5 and 25. CaCo2 cells grown in Dulbecco’s Modified Eagle Medium (DMEM High Glucose (4.5 g/L), without L-Glutamine, without Phenol Red, Capricorn, #DMEM-HXRXA) with 10% fetal bovine serum (Capricorn, #FBS-12A), L-Glutamine (Capricorn, #GLN-B), streptomycin (100 μg/mL), and 100 U/mL penicillin (Capricorn, #PS-B) were washed in Dulbecco’s PBS, w/o Ca2+ & Mg2+, w/o Phenol Red (Capricorn, #PBS-1A), trypsinized (Trypsin-EDTA Capricorn, #Try-1B), resuspended, and seeded into white 384-well microtiter plates (Greiner Bio-One, #781073) at 8000 cells/20 µL/well and incubated at 37 °C in the presence of 5% CO2 for 24 h. One microplate is used as a reference signal plate for untreated cells on the day of compound addition. Therefore, 10 µL of the CellTiter-Glo Detection Reagent were added to each well, the microplate was centrifuged (1 min, 400 g) and incubated at the dark for 15 min. The generated luminescence signal was detected using the EnVision Multilabel 2103 Reader with 100 ms reading time/well. In other microplates compound (20 nL of 10-mM stock concentration in 100% v/v DMSO) controls (Natrium Selenite 20 nL/well of 2.5 mM stock concentration in 100% v/v DMSO, and 20 nL/well of 100% v/v DMSO (Carl Roth, #HN47.1)) were added to cells using the Echo 550 Liquid Handler and incubated for 48 h at 37 °C in the presence of 5% CO2. Luminescence signal was measured according to the procedure of the reference plate.
Data analysis was performed using Microsoft Excel and GraphPad Prism 8. Test compound results were normalized relative to respective controls with DMSO control representing 0% inhibition of cell viability and Na-Selenite control representing 100% inhibition of cell viability. Control well outliers were eliminated according to the three-sigma method.
4.5. Chemical compounds
Chemical compounds studied in this article: Camostat (PubChem CID: 5284360), Nafamostat (PubChem CID: 5311180), Decitabine (PubChem CID: 451668), Trifluoperazine (PubChem CID: 5566), Docetaxel (PubChem CID: 148124), Alprostadil (PubChem CID: 5280723), Fluorouracil (PubChem CID: 3385), Cladribine (PubChem CID: 20279), Chlorpromazine (PubChem CID: 2726), Econazole (PubChem CID: 3198), Didanosine (PubChem CID: 135398739), Quinine (PubChem CID: 3034034), Mycophenolic acid (PubChem CID: 446541), Mycophenolate Mofetil (PubChem CID: 5281078), Mefloquine (PubChem CID: 40692), Thioguanine (PubChem CID: 2723601), Pentamidine (PubChem CID: 8813), Clomipramine (PubChem CID: 68539), Thioridazine (PubChem CID: 5452), Vinorelbine (PubChem CID: 5311497), Mebendazole (PubChem CID: 4030), Paclitaxel (PubChem CID: 36314), Colchicine (PubChem CID: 6167), Podofilox (PubChem CID: 10607), Cinacalcet (PubChem CID: 156418), Azelastine (PubChem CID: 2267), E64d (PubChem CID: 65663), Copanlisib (PubChem CID: 135565596), Pictilisib (PubChem CID: 17755052), YM201636 (PubChem CID: 9956222), Mdivi1 (PubChem CID: 3825829), PDE10-IN-1 (PubChem CID: 72709059), Sildenafil (PubChem CID: 135398744), Pirenzepine (PubChem CID: 71405), Levetimide (PubChem CID: 30844), A-7 (PubChem CID: 54601194), Zaldaride (PubChem CID: 65909), Dexniguldipine (PubChem CID: 6918097).
CRediT authorship contribution statement
BE and KR designed the study; KR, TE and JH designed and cloned the spike expression plasmids; KR and JW developed the SARS-CoV-2 lentiviral pseudoviruses and performed flow cytometry analyses; MK and BE developed and executed the high throughput assay and performed data acquisition; MK and AZ performed data and medicinal chemistry analysis; BF, BE and KR supervised the experiments. All authors wrote and approved the final version of the manuscript.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgements
The authors appreciate the help of the UKE Cytometry and Cell Sorting Core Unit. This work has been supported by the Deutsche Forschungsgemeinschaft (DFG), Germany, within SFB841 (SP2 to BF) and the Friedrich-Ebert-Stiftung, Germany (to JW). This work is part of the doctoral theses of MK and JW.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2022.113104.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
Data will be made available on reasonable request.
References
- 1.World Health Organisation, Variants of Concern, 2022. 〈https://www.who.int/emergencies/diseases/novel-coronavirus-2019/coronavirus-disease-answers?query=variants+of+concern&referrerPageUrl=https%3A%2F%2Fwww.who.int%2Femergencies%2Fdiseases%2Fnovel-coronavirus-2019%2Fcoronavirus-disease-answers〉 (accessed Feb 2022).
- 2.Zhan Xiao-Yong, Zhang Ying, Zhou Xuefu, Huang Ke, Qian Yichao, Leng Yang, Yan Leping, Huang Bihui, He Yulong. Molecular evolution of SARS-CoV-2 structural genes: evidence of positive selection in spike glycoprotein. bioRxiv. 2020 doi: 10.1101/2020.06.25.170688. 06.25.170688. [DOI] [Google Scholar]
- 3.M. Chand, S. Hopkins, G. Dabrera, C. Achison, W. Barclay, N. Ferguson, E. Volz, N. Loman, A. Rambaut, J. Barrett, Investigation of novel SARS-COV-2 variant, 2020. 〈https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdf〉 (accessed 22 February 2022).
- 4.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N.-Y., Korsman S., Davies M.-A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Kosakovsky Pond S.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 5.Yadav P.D., Sarkale P., Razdan A., Gupta N., Nyayanit D.A., Sahay R.R., Potdar V., Patil D.Y., Baradkar S., Kumar A., Aggarwal N., Shete A.M., Kaur H., Mohandas S. Isolation and characterization of SARS-CoV-2 Beta variant from UAE travelers. J. Infect. Public Health. 2022;15:182–186. doi: 10.1016/j.jiph.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido Dd.S., Mishra S., Crispim M.A.E., Sales F.C., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., de Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., Kraemer M.U., Gaburo N., Camilo Cd.C., Hoeltgebaum H., Souza W.M., Rocha E.C., de Souza L.M., de Pinho M.C., Araujo L.J.T., Malta F.S.V., de Lima A.B., Silva Jd.P., Zauli D.A.G., de S Ferreira A.C., Schnekenberg R.P., Laydon D.J., Walker P.G.T., Schlüter H.M., Dos Santos A.L.P., Vidal M.S., Del Caro V.S., Filho R.M.F., Dos Santos H.M., Aguiar R.S., Modena J.L.P., Nelson B., Hay J.A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Suchard M.A., Bowden T.A., Pond S.L.K., Wu C.-H., Ratmann O., Ferguson N.M., Dye C., Loman N.J., Lemey P., Rambaut A., Fraiji N.A., Carvalho Md.P.S.S., Pybus O.G., Flaxman S., Bhatt S., Sabino E.C. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 doi: 10.1101/2021.02.26.21252554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.E. Hodcroft, Variant: 20J (Gamma, V3): also known as S.501Y.V3, 2022. 〈https://covariants.org/variants/20J.Gamma.V3〉 (accessed 22 February 2022).
- 8.E. Hodcroft, Variant: 21A (Delta): also known as 21A/S:478K, 2022. 〈https://covariants.org/variants/21A.Delta〉 (accessed 22 February 2022).
- 9.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.E. Hodcroft, Variant: 21K (Omicron): also known as BA.1, 2022. 〈https://covariants.org/variants/21K.Omicron〉 (accessed 22 February 2022).
- 11.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. 271-280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giroglou T., Cinatl J., Rabenau H., Drosten C., Schwalbe H., Doerr H.W., von Laer D. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 2004;78:9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson M.C., Lyddon T.D., Suarez R., Salcedo B., LePique M., Graham M., Ricana C., Robinson C., Ritter D.G. Optimized pseudotyping conditions for the SARS-COV-2 spike glycoprotein. J. Virol. 2020;94 doi: 10.1128/JVI.01062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber K., Mock U., Petrowitz B., Bartsch U., Fehse B. Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther. 2010;17:511–520. doi: 10.1038/gt.2009.149. [DOI] [PubMed] [Google Scholar]
- 15.Weber K., Bartsch U., Stocking C., Fehse B. A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol. Ther. 2008;16:698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- 16.Willett B.J., Grove J., MacLean O.A., Wilkie C., Logan N., De Lorenzo G., Furnon W., Scott S., Manali M., Szemiel A., Ashraf S., Vink E., Harvey W.T., Davis C., Orton R., Hughes J., Holland P., Silva V., Pascall D., Puxty K., da Silva Filipe A., Yebra G., Shaaban S., Holden M.T.G., Pinto R.M., Gunson R., Templeton K., Murcia P.R., Patel A.H., The COVID-19 Genomics UK (COG-UK) Consortium, Haughney J., Robertson D.L., Palmarini M., Ray S., Thomson E.C. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv. 2022 doi: 10.1101/2022.01.03.21268111. (medRxiv) (medRxiv) [DOI] [Google Scholar]
- 17.Ellinger B., Pohlmann D., Woens J., Jäkel F.M., Reinshagen J., Stocking C., Prassolov V.S., Fehse B., Riecken K. A high-throughput HIV-1 drug screening platform, based on lentiviral vectors and compatible with biosafety level-1. Viruses. 2020;12 doi: 10.3390/v12050580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouznetsova J., Sun W., Martínez-Romero C., Tawa G., Shinn P., Chen C.Z., Schimmer A., Sanderson P., McKew J.C., Zheng W., García-Sastre A. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg. Microbes Infect. 2014;3 doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen T., Schwarz T.M., Vigant F., Gardner T.J., Hernandez R.E., Lee B., Tortorella D. The microtubule inhibitor podofilox inhibits an early entry step of human cytomegalovirus. Viruses. 2016;8 doi: 10.3390/v8100295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniel J.A., Chau N., Abdel-Hamid M.K., Hu L., von Kleist L., Whiting A., Krishnan S., Maamary P., Joseph S.R., Simpson F., Haucke V., McCluskey A., Robinson P.J. Phenothiazine-derived antipsychotic drugs inhibit dynamin and clathrin-mediated endocytosis. Traffic. 2015;16:635–654. doi: 10.1111/tra.12272. [DOI] [PubMed] [Google Scholar]
- 21.Ianevski A., Giri A.K., Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yousefi H., Mashouri L., Okpechi S.C., Alahari N., Alahari S.K. Repurposing existing drugs for the treatment of COVID-19/SARS-CoV-2 infection: a review describing drug mechanisms of action. Biochem. Pharmacol. 2021;183 doi: 10.1016/j.bcp.2020.114296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali K., Azher T., Baqi M., Binnie A., Borgia S., Carrier F.M., Cavayas Y.A., Chagnon N., Cheng M.P., Conly J., Costiniuk C., Daley P., Daneman N., Douglas J., Downey C., Duan E., Duceppe E., Durand M., English S., Farjou G., Fera E., Fontela P., Fowler R., Fralick M., Geagea A., Grant J., Harrison L.B., Havey T., Hoang H., Kelly L.E., Keynan Y., Khwaja K., Klein G., Klein M., Kolan C., Kronfli N., Lamontagne F., Lau R., Lee T.C., Lee N., Lim R., Longo S., Lostun A., MacIntyre E., Malhamé I., Mangof K., McGuinty M., Mergler S., Munan M.P., Murthy S., O'Neil C., Ovakim D., Papenburg J., Parhar K., Parvathy S.N., Patel C., Perez-Patrigeon S., Pinto R., Rajakumaran S., Rishu A., Roba-Oshin M., Rushton M., Saleem M., Salvadori M., Scherr K., Schwartz K., Semret M., Silverman M., Singh A., Sligl W., Smith S., Somayaji R., Tan D.H.S., Tobin S., Todd M., Tran T.-V., Tremblay A., Tsang J., Turgeon A., Vakil E., Weatherald J., Yansouni C., Zarychanski R. Remdesivir for the treatment of patients in hospital with COVID-19 in Canada: a randomized controlled trial. CMAJ. 2022 doi: 10.1503/cmaj.211698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai Z.W., Lew R.A., Yarski M.A., Mu F.-T., Andrews R.K., Smith A.I. The identification of a calmodulin-binding domain within the cytoplasmic tail of angiotensin-converting enzyme-2. Endocrinology. 2009;150:2376–2381. doi: 10.1210/en.2008-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmeira A., Sousa E., Vasconcelos M.H., Pinto M.M. Three decades of P-gp inhibitors: skimming through several generations and scaffolds. Curr. Med. Chem. 2012;19:1946–2025. doi: 10.2174/092986712800167392. [DOI] [PubMed] [Google Scholar]
- 26.Cho H., Gonzales-Wartz K.K., Huang D., Yuan M., Peterson M., Liang J., Beutler N., Torres J.L., Cong Y., Postnikova E., Bangaru S., Talana C.A., Shi W., Yang E.S., Zhang Y., Leung K., Wang L., Peng L., Skinner J., Li S., Wu N.C., Liu H., Dacon C., Moyer T., Cohen M., Zhao M., Lee F.E., Weinberg R.S., Douagi I., Gross R., Schmaljohn C., Pegu A., Mascola J.R., Holbrook M., Nemazee D., Rogers T.F., Ward A.B., Wilson I.A., Crompton P.D., Tan J. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci. Transl Med. 2021;13(616) doi: 10.1126/scitranslmed.abj5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 28.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. National Academy Sc. United States Am. 2020;117(13) doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayati A., Kumar R., Francis V., McPherson Peter S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biological Chem. 2021;296 doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomes C.P., Fernandes D.E., Casimiro F., da Mata G.F., Passos M.T., Varela P., Mastroianni-Kirsztajn G., Pesquero J.B. Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics. Frontiers Cell. Infection Microbiol. 2020;10 doi: 10.3389/fcimb.2020.589505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C., Zheng M., Yang Y., Gu X., Yang K., Li M., Liu Y., Zhang Q., Zhang P., Wang Y., Wang Q., Xu Y., Zhou Y., Zhang Y., Chen L., Li H. Furin: A Potential Therapeutic Target for COVID-19. iSci. 2020;23(10) doi: 10.1016/j.isci.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fanunza E., Frau A., Corona A., Tramontano E. Antiviral Agents Against Ebola Virus Infection: Repositioning Old Drugs and Finding Novel Small Molecules. Annu. Rep. Med. Chem. 2018;51:135–173. doi: 10.1016/bs.armc.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitiello A., Ferrara F., Pelliccia C., Granata G., La Porta R. Cytokine storm and colchicine potential role in fighting SARS-CoV-2 pneumonia. Italian J. Med. 2020;14(2) doi: 10.4081/itjm.2020.1284. [DOI] [Google Scholar]
- 34.Deftereos S.G., Siasos G., Giannopoulos G., Vrachatis D.A., Angelidis C., Giotaki S.G., Gargalianos P., Giamarellou H., Gogos C., Daikos G., Lazanas M., Lagiou P., Saroglou G., Sipsas N., Tsiodras S., Chatzigeorgiou D., Moussas N., Kotanidou A., Koulouris N., Oikonomou E., Kaoukis A., Kossyvakis C., Raisakis K., Fountoulaki K., Comis M., Tsiachris D., Sarri E., Theodorakis A., Martinez-Dolz L., Sanz-Sánchez J., Reimers B., Stefanini G.G., Cleman M., Filippou D., Olympios C.D., Pyrgakis V.N., Goudevenos J., Hahalis G., Kolettis T.M., Iliodromitis E., Tousoulis D., Stefanadis C. The Greek study in the effects of colchicine in COVID-19 complications prevention (GRECCO-19 study): Rationale and study design. Hellenic J. Cardiol. 2020;61(1):42–45. doi: 10.1016/j.hjc.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S., Yoon G.Y., Myoung J., Kim S.J., Ahn D.G. Robust and persistent SARS-CoV-2 infection in the human intestinal brush border expressing cells. Emerg Microbes Infect. 2020;9(1):2169–2179. doi: 10.1080/22221751.2020.1827985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on reasonable request.