Abstract
An alkaliphilic bacterium, Bacillus sp. strain K-1, produces extracellular xylanolytic enzymes such as xylanases, β-xylosidase, arabinofuranosidase, and acetyl esterase when grown in xylan medium. One of the extracellular xylanases that is stable in an alkaline state was purified to homogeneity by affinity adsorption-desorption on insoluble xylan. The enzyme bound to insoluble xylan but not to crystalline cellulose. The molecular mass of the purified xylan-binding xylanase was estimated to be approximately 23 kDa. The enzyme was stable at alkaline pHs up to 12. The optimum temperature and optimum pH of the enzyme activity were 60°C and 5.5, respectively. Metal ions such as Fe2+, Ca2+, and Mg2+ greatly increased the xylanase activity, whereas Mn2+ strongly inhibited it. We also demonstrated that the enzyme could hydrolyze the raw lignocellulosic substances effectively. The enzymatic products of xylan hydrolysis were a series of short-chain xylooligosaccharides, indicating that the enzyme was an endoxylanase.
Xylan, a major component of plant cell wall hemicellulose, is composed of a backbone of β-1,4-linked xylose units which are substituted with arabinose and acetate residues (16). For complete hydrolysis of xylan, many xylanolytic microorganisms often synthesize the multiple groups of xylanolytic enzymes for cooperative actions (10). These enzymes include endo-β-1,4-xylanases (EC 3.2.1.8), β-xylosidase (EC 3.2.1.37), and enzymes which cleave side chain sugars from the xylan backbone, such as α-arabinofuranosidases (EC 3.2.1.55) and acetyl esterases (EC 3.1.1.6). Interest in the application of xylanases in the pulp and paper industry has increased during recent years. Xylanases may be used in pulp-prebleaching process to remove the hemicelluloses which bind to the pulp. The hydrolysis of pulp-bound hemicelluloses releases the lignin in the pulp, reducing the amount of chlorine required for conventional chemical bleaching and minimizing the toxic, chloroorganic waste. Therefore, xylanases from alkaliphilic bacteria and actinomycetes have been studied widely (15, 17, 24). The noncatalytic polysaccharide-binding regions of plant cell wall hydrolases play an important role in the efficient hydrolysis of cellulosic substances (7, 9). Many plant cell wall hydrolases include noncatalytic domains, i.e., cellulose-binding domains (CBDs) (3, 6, 7). However, only a few of these enzymes contain xylan-binding domains (XBDs) together with CBDs. Both CBDs and XBDs occur in xylanases from Thermomonospora fusca (8), Cellulomonas fimi (2), and Streptomyces thermoviolaceus (24) and in arabinofuranosidase from Streptomyces lividans (27). However, there have been no reports of xylanase with only XBDs. As previously reported, our strain, an alkaliphilic bacterium, Bacillus sp. strain K-1, produces alkali-stable xylanases without cellulase and one of them has a strong affinity for insoluble xylan (19). In this study, we describe the purification and properties of the xylan-binding endoxylanase from Bacillus sp. strain K-1.
MATERIALS AND METHODS
Bacterial strain.
Bacillus sp. strain K-1, used in this study, was isolated from a wastewater treatment plant of a pulp and paper manufacturer (19).
Characterization of the bacterium.
The morphological properties and taxonomic characteristics of the bacterium were studied by the methods in Bergey’s Manual of Systematic Bacteriology (21). The isolated bacterium was an aerobic, gram-positive, motile, spore-forming, rod-shaped organism (0.7 μm by 2.9 to 4.6 μm). It showed a positive reaction for the production of catalase. Acid was produced from d-glucose, d-xylose, and d-mannitol. Based on these characteristics, the bacterium was identified as belonging to the genus Bacillus according to Sneath (21). The isolated bacterium was also identified as 93% identical to Bacillus pumilus by the API system (bio-Mérieux, Marcy L’Etoile, France). The culture is available upon request.
Culture medium.
The culture medium used was Berg’s mineral salt medium (1) supplemented with 0.5% xylan. This medium was adjusted to pH 10.5 with 1% Na2CO3 after autoclaving.
Enzyme assays.
The xylanase activity was measured by determining the amount of reducing sugar released from oat spelt xylan (Sigma Chemical Co., St. Louis, Mo.). The reaction mixture consisted of 1% xylan in 100 mM Tris-HCl buffer (pH 7.0) and enzyme to give a final volume of 0.6 ml (12). After incubation for 15 min at 50°C, the increase in the amount of reducing sugar was determined by the Somogyi-Nelson method (18). Xylanase activity was expressed as micromoles of reducing sugar released per minute per milliliter of enzyme solution.
The cellulase activity was measured under the same conditions as described above using carboxymethyl cellulose (Sigma Chemical Co.) as a substrate.
The β-xylosidase assay mixture consisted of 0.9 mM p-nitrophenyl-β-d-xylopyranoside (Sigma Chemical Co.) in 50 mM phosphate buffer (pH 7.0) and enzyme to give a final volume of 1.1 ml. The reaction mixture was incubated for 30 min, and then 2.0 ml of 0.4 M sodium carbonate was added to terminate the reaction. The amount of p-nitrophenol released was measured by monitoring the optical density at 405 nm (12).
β-Glucosidase and arabinofuranosidase activities were measured under the same conditions as β-xylosidase activity as mentioned above except for the substrates. For the β-glucosidase assay, 1 mM p-nitrophenyl-β-d-glucopyranoside (Sigma Chemical Co.) was used as the substrate, and for the arabinofuranosidase assay, 0.83 mM p-nitrophenyl-α-l-arabinofuranoside (Sigma Chemical Co.) was used. The amount of p-nitrophenol released was measured by monitoring the optical density at 405 nm.
The acetyl esterase assay mixture consisted of 1 mM p-nitrophenylacetate (Sigma Chemical Co.) in 50 mM Tris-HCl buffer (pH 7.0) and enzyme to give a final volume of 2.5 ml. The substrate, dissolved in 50% (vol/vol) methanol, was prepared immediately prior to use. After 10 min of incubation at 30°C, the amount of p-nitrophenol released was measured by monitoring the optical density at 405 nm (15). β-Xylosidase, β-glucosidase, arabinofuranosidase, and acetyl esterase activities were expressed as micromoles of p-nitrophenol released per minute per milliliter of enzyme solution.
Protein determination.
Protein was assayed by the method of Lowry et al. (14) with bovine serum albumin as a standard. The protein content of the eluate of bound xylanase was measured by monitoring the optical density at 280 nm.
Binding assay.
Xylanase-binding assays and preparation of insoluble xylan were performed by the method of Irwin et al. (8). The binding assay was conducted by adding 0.23 mg of protein from the culture supernatant to 50 mg of insoluble xylan (oat spelt), Avicel (Sigma Chemical Co.), α-cellulose (Sigma Chemical Co.) or starch (cassava) in 1.0 ml of 100 mM Tris-HCl buffer (pH 9.0) in 1.5-ml Eppendorf tubes. Samples were shaken at intervals at 4°C for 30 min before being subjected to centrifugation. The amount of enzyme remaining in the supernatant was determined by the standard xylanase assay method. The activity lost from the supernatant was assumed to be the activity bound.
Purification of xylan-binding xylanase.
The bacterial cultures were incubated at 37°C in a rotary shaker, rotating at 200 revolutions/min, for 3 days before centrifugation. The culture supernatant was used as a source of xylan-binding xylanase. For purification of xylan-binding xylanase, 4 mg of protein from the culture supernatant and 500 mg of insoluble xylan in 10 ml of 100 mM Tris-HCl buffer (pH 9.0) were shaken periodically at 4°C for 30 min. The xylan-bound protein complex was washed four times with the same volume of 100 mM Tris-HCl buffer (pH 9.0) and then eluted with 1% triethylamine. The eluate was dialyzed, assayed for xylanase activity, and freeze-dried.
Gel electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (13). Samples were dissolved in a sample application buffer containing 2% (wt/vol) SDS, 5% (vol/vol) 2-mercaptoethanol, 10% (vol/vol) glycerol, and 15 mM Tris-HCl buffer (pH 6.8) and heated in a boiling-water bath for 3 min. The stacking and separating gels consisted of 5 and 10% polyacrylamide, respectively. After electrophoresis, the proteins were stained with Coomassie brilliant blue R-250. The molecular weight standards used were from the low-molecular-weight calibration kit (Bio-Rad, Richmond, Calif.).
Zymogram analysis.
The zymogram analysis was a modification of the published method (17). The culture supernatant in the sample application buffer was boiled for 3 min and subjected to electrophoresis on an SDS–10% polyacrylamide gel containing 0.1% xylan as described above. After electrophoresis, the gel was soaked in 25% (vol/vol) isopropanol with gentle shaking to remove the SDS and renature the proteins in the gel. The gel was then washed four times for 30 min at 4°C in 0.1 M acetate buffer (pH 5.5). After further incubation for 60 min at 50°C, the gel was soaked in 0.1% Congo red solution for 30 min at room temperature and washed with 1 M NaCl until excess dye was removed from the active band. After the gel was submerged in 0.5% acetic acid, the background turned dark blue and the activity bands were observed as clear colorless areas.
Analysis of xylan hydrolysis products.
The xylan-binding xylanase (1 U) was mixed with 1.6% xylan in 0.1 M Tris-HCl buffer (pH 7) and incubated at 50°C. At the appropriate time, xylan hydrolysis products were removed and qualitatively determined by thin-layer chromatography on silica gel 60 F254 plates (Merck 1.05554; 20 by 20 cm) with a mixture of isopropanol–acetone–0.1 M lactic acid (4:4:2) as a solvent system. The sugar spots were detected on the plates by spraying them with 4 ml of aniline–4 g of α-diphenylamine–200 ml of acetone–30 ml of 80% H3PO4. d-Xylose and xylobiose were used as standards.
Hydrolysis of lignocellulosic substances.
The insoluble lignocellulosic substances such as corn hull, bagasse, and rice straw were ground (40 mesh) and washed several times in hot distilled water to remove the free sugars that remained. Each substance and insoluble xylan (1% [dry weight]) were hydrolyzed with 1 U of the xylan-binding xylanase at pH 5.5 and 50°C. After a 1-min incubation, samples were removed and the amount of reducing sugars produced was determined.
RESULTS
Enzyme production by Bacillus sp. strain K-1.
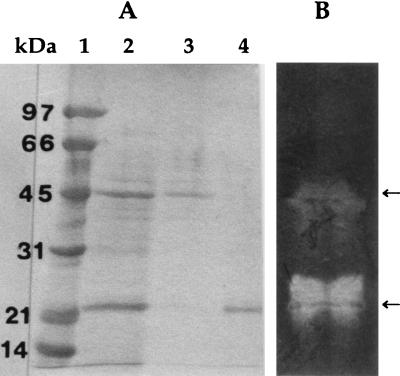
When alkaliphilic Bacillus sp. strain K-1 was grown in an alkaline xylan medium, the extracellular xylanase, β-xylosidase, arabinofuranosidase, and acetyl esterase were detected at 4.8, 0.21, 0.15 and 0.24 U/mg protein, respectively, in the culture supernatant. However, the culture supernatant showed no activity of carboxymethyl cellulase and β-glucosidase. The SDS-PAGE revealed that the culture supernatant contained two major and several minor protein bands (Fig. 1A, lane 2). The major protein bands corresponded to molecular masses of 23 and 45 kDa, respectively. The zymogram analysis (Fig. 1B) showed the two active xylanases, detected by Congo red staining, indicating that the two xylanases were the major extracellular enzymes of the bacterium.
FIG. 1.
SDS-PAGE of purified xylan-binding xylanase by adsorption-desorption on insoluble xylan. (A) Coomassie brilliant blue R-250 staining of the gel. Lanes: 1, molecular mass standards; 2, culture supernatant (40 μg of protein); 3, unbound xylanase (10 μg of protein); 4, bound xylanase (10 μg of protein). (B) Zymogram (40 μg of protein) for detecting xylanase activity.
Binding of xylanase to insoluble substances.
Previously, we reported that the alkaliphilic bacterium Bacillus sp. strain K-1 produced two forms of xylanase and that one of them is a specific xylan-binding xylanase (19). Thus, to evaluate the ability of xylanase to bind to insoluble substances, the culture supernatant was incubated with insoluble xylan, Avicel, α-cellulose, or starch and the unbound fraction was assayed for xylanase activity. Sixty percent of the xylanase activity was bound to insoluble xylan, but with Avicel, α-cellulose, and starch, all the activity remained in the unbound fraction, suggesting that the enzyme was not bound to substances except insoluble xylan.
Purification of xylan-binding xylanase.
The purified xylan-binding xylanase protein appeared as single protein band on SDS-PAGE and had a molecular mass of 23 kDa (Fig. 1A, lane 4). The 23-kDa xylanase remained bound to the insoluble xylan after treatment of the culture supernatant with insoluble xylan. The 45-kDa xylanase was found as unbound xylanase (lane 3). The yield of the 23-kDa xylan-binding xylanase was about 60% of the relative xylanase activity of the culture supernatant, and the enzyme had a specific activity of 32.7 U/mg of protein.
Effect of pH on activity and stability.
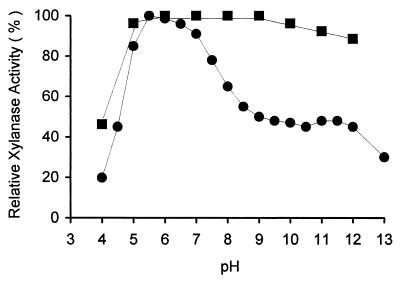
The activity of the xylan-binding xylanase at various pH values was measured by using oat spelt xylan as the substrate. The reaction pHs were adjusted to 4.0 to 13.0 with various buffers at 100 mM. The enzyme showed a broad pH activity profile and relatively high activity under alkaline conditions, with an optimum at pH 5.5 (Fig. 2). The stability of the enzyme was determined by incubating at 30°C for 24 h at different pHs (50 mM), and the residual activity was measured by the standard assay method. The xylan-binding xylanase was stable at pH 5.0 to 9.0 but had 88% activity at pH 12 (Fig. 2).
FIG. 2.
Effect of pH on activity (solid circles) and stability (solid squares) of xylan-binding xylanase. The reaction pH values were adjusted with the following buffer systems: acetate buffer (pH 4.0 to 5.5), phosphate buffer (pH 6.0 to 6.5), Tris-HCl buffer (pH 7.0 to 9.0), Na2CO3-NaHCO3 buffer (pH 9.5 to 11.0), Na2HPO4-NaOH buffer (pH 11.5 to 12.0), or KCl-NaOH buffer (pH 13.0).
Effect of temperature on activity and stability.
The optimum temperature of the purified enzyme was determined by varying the reaction temperature at pH 7.0. The enzyme had an optimum temperature of 60°C. The thermal stability of the xylan-binding xylanase was measured by incubating it at pH 7.0 for 30 min at different temperatures ranging from 30 to 80°C. The residual activity was determined by the standard assay. The enzyme was stable at temperatures up to 50°C. At 60°C, the enzyme showed 90% activity.
Effect of metal ions on the xylanase activity.
As shown in Table 1, the enzyme activity was greatly increased by 1 mM FeCl2, CaCl2, and MgCl2. In contrast, it was strongly inhibited by Mn2+.
TABLE 1.
Effects of metal ions and EDTA on xylanase activitya
| Ion or EDTA (mM) | Relative activity (%) |
|---|---|
| None | 100 |
| EDTA (1) | 42.2 |
| EDTA (10) | 22.2 |
| EDTA (50) | 0 |
| CaCl2 | 130.7 |
| CuCl2 | 51.2 |
| CoCl2 | 54.4 |
| FeCl2 | 261.6 |
| MgCl2 | 172.0 |
| MnCl2 | 16.5 |
| NiCl2 | 60.7 |
| ZnCl2 | 46.1 |
| CaSO4 | 191.6 |
| CuSO4 | 33.3 |
| CoSO4 | 76.3 |
| FeSO4 | 289.0 |
| MgSO4 | 174.0 |
| MnSO4 | 12.0 |
For the case of metal ions, the enzyme solution was mixed with the final concentration of 50 mM EDTA and dialyzed with Tris-HCl buffer (pH 7.0). Then the EDTA-treated enzyme solutions were preincubated in a mixture containing various chemicals (1 mM) at 30°C for 1 h, after which the reactivated activity was measured under the standard assay conditions.
Substrate specificity of the xylan-binding xylanase.
The xylan-binding xylanase was assayed with various substrates to study the substrate specificity. The xylan-binding xylanase had hydrolytic activity toward xylan, but no activity toward p-nitrophenyl-β-d-xyloside, p-nitrophenyl β-d-glucoside, arabinofuranoside, p-nitrophenylacetate, or carboxymethyl cellulose (data not shown).
Hydrolysis of lignocellulosic substances.
The initial hydrolysis of the lignocellulosic substances was compared with insoluble xylan. Corn hull, bagasse, and rice straw were hydrolyzed at 62, 57, and 56 μg of reducing sugars per min, respectively, whereas the corresponding value for insoluble xylan was 98 μg/min. These results indicated that xylan-binding xylanase was capable of hydrolyzing the hemicellulose present in lignocellulosic substances.
Action of xylan-binding xylanase on oat spelt xylan.
The hydrolysis products of oat spelt xylan by xylan-binding xylanase were analyzed by thin-layer chromatography. The hydrolysis products were x2, x3, x4, x5, x6, and larger xylooligosaccharides (data not shown), indicating that the xylan-binding xylanase was an endoxylanase.
DISCUSSION
There have been many reports of cellulose-binding domains located not only in cellulases but also in xylanases and other hemicellulases that remove side chains from the xylan backbone (2, 6, 8, 10). The xylanases from Thermomonospora fusca (8), Cellulomonas fimi (2), and Streptomyces thermoviolaceus (25) contained both noncatalytic domains, XBDs and CBDs, and appeared to have affinity for both cellulose and xylan. The functions of these noncatalytic domains are for binding required for the hydrolysis of cellulosic substances (7, 9). Irwin et al. (8) reported that a xylanase gene was cloned from T. fusca and expressed in Streptomyces lividans TK24, producing xylanase which bound to both cellulose and insoluble xylan. However, after the treatment with a protease, a proteolyzed 24-kDa catalytically active fragment did not bind to cellulose but bound to xylan. Similarly, xylanase from C. fimi, expressed in Escherichia coli JM83, contained a C-terminal CBD and an internal XBD and bound to both cellulose and xylan. Deletion of the C-terminal CBD abolished the capacity of the enzyme to bind to cellulose, but the truncated xylanase retained its xylan-binding properties (2). However, xylanase with an XBD that exhibits no affinity for cellulose has not been found in nature. Therefore, this is the first time that a xylanase that exhibits the absolute affinity for only insoluble xylan was found in Bacillus sp. strain K-1.
The xylan-binding xylanase was purified easily by specific interaction between the xylan-binding region of the enzyme and insoluble xylan. The estimated molecular weight (23,000) of the purified xylanase is lower than other xylanases containing XBDs (2, 8, 25). Some plant cell wall hydrolases have multiple activities such as β-xylosidase/β-glucosidase (26), and β-xylosidase/α-arabinofuranosidase (20) and some enzymes exhibited both XBDs and CBDs (2, 8, 25). However, xylan-binding xylanase from Bacillus sp. strain K-1 is specific and binds only to xylan. The enzyme showed no β-xylosidase, β-glucosidase, arabinofuranosidase, acetyl esterase, or cellulase activity. The yield of the purified xylanase was about 60% of the relative xylanase activity of the culture supernatant. Homogeneity of the purified enzyme was confirmed by its appearing as a single protein band on SDS-PAGE.
The properties of the purified enzyme indicated that the xylanase activity remained considerable in the alkaline pH range. The enzyme was most active at pH 5.5 and quite stable at alkaline pHs. The xylanases from other alkaliphilic microorganisms are also stable at pHs up to 11 (23, 24). The optimum temperature of our purified enzyme was 60°C, and it was quite stable up to 50°C. Thin-layer chromatography of enzymatic hydrolyzate identified the xylan-binding xylanase as an endoxylanase.
The effect of metal ions on xylanase activity is unclear. Xylan-binding xylanase was slightly inhibited by 1 mM EDTA. The xylanase activity decreased as the EDTA concentration increased and was completely inhibited by 50 mM EDTA. The xylan-binding xylanase activity was strongly inhibited by Mn2+. In contrast, the activity was greatly elevated by the addition of Fe2+, Ca2+, and Mg2+. These results were not similar to the other xylanases (11, 24, 28). However, we suggest that metal ions may not affect only the active site of the xylan-binding xylanase but also the noncatalytic xylan-binding region, which is involved in the efficient hydrolysis of the substrate (4, 5, 22).
The presence of the noncatalytic substrate-binding domain did not alter the activity of the xylanase against the soluble substrate, but it is advantageous for binding the enzyme to the plant cell wall, where insoluble xylan is present (2). In our study, the xylan-binding xylanase was able to hydrolyze raw agricultural substances without any pretreatment. It is possible that the xylan-binding region plays an important role when the enzyme is presented with the lignocellulosic substances. However, the hydrolysis of these substances occurred at different rates that varied with their xylan content and structural complexity.
The xylan-binding region, which exhibits high affinity to insoluble xylan, appears to be a potential tool to bring the xylanase directly to the surface of the insoluble hemicellulose-containing lignocellulosic substrate. There is currently interest in the use of xylanases for the prebleaching of kraft pulps. The lack of cellulase activity with a wide pH profile, stability under alkaline conditions, and the xylan-binding region of this enzyme make it an attractive candidate for this function. Currently, we are working on an application which uses the culture supernatant and the purified xylan-binding xylanase in the process of prebleaching of kraft pulps.
ACKNOWLEDGMENTS
We acknowledge a support by grant from the National Research Council of Thailand, under the Thai-Japan Cooperative Research Program.
We thank George A. Gale, University of Memphis, Memphis, Tenn., for correcting the English in the manuscript and for making useful comments.
REFERENCES
- 1.Berg B, Hofstan B V, Petterson G. Growth and cellulase formation by Cellvibrio fulvus. J Appl Bacteriol. 1972;35:201–214. doi: 10.1111/j.1365-2672.1972.tb03691.x. [DOI] [PubMed] [Google Scholar]
- 2.Black G W, Hazlewood G P, Millward-Sadler S J, Laurie J I, Gillbert H J. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem J. 1995;307:191–195. doi: 10.1042/bj3070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco J, Coque J J R, Velasco J, Martin J F. Cloning, expressing in Streptomyces lividans and biochemical characterization of a thermostable endo-β-1,4-xylanase of Thermomonospora alba ULJ B1 with cellulose binding ability. Appl Microbiol Biotechnol. 1997;48:208–217. doi: 10.1007/s002530051040. [DOI] [PubMed] [Google Scholar]
- 4.Chauvaux S, Souchon H, Alzari P M, Chariot P, Beguin P. Structural and functional analysis of the metal-binding sites of Clostridium thermocellum endoglucanase CelD. J Biol Chem. 1995;270:9757–9762. doi: 10.1074/jbc.270.17.9757. [DOI] [PubMed] [Google Scholar]
- 5.Choi S K, Ljungdahl L G. Structural role of calcium for the organization of the cellulosome of Clostridium thermocellum. Biochemistry. 1996;35:4906–4910. doi: 10.1021/bi9524631. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira L M A, Wood T M, Williamson G, Faulds C, Hazlewood G P, Black G W. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem J. 1993;294:349–355. doi: 10.1042/bj2940349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall J, Black G W, Ferreira L M A, Millward-Sadler S A, Ali B R S. The non-catalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of avicel. Biochem J. 1995;309:749–756. doi: 10.1042/bj3090749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irwin D, Jung E D, Wilson D B. Characterization and sequence of a Thermomonospora fusca xylanase. Appl Environ Microbiol. 1994;60:763–770. doi: 10.1128/aem.60.3.763-770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karita S, Sakka K, Ohmiya K. Cellulose-binding domains confer an enhanced activity against insoluble cellulose to Ruminococcus albus endoglucanase IV. J Ferment Bioeng. 1996;81:553–556. [Google Scholar]
- 10.Kelett L E, Poole D M, Ferreira L M A, Durrant A J, Hazlewood G P, Gilbert H J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990;272:369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura I, Sasahara H, Tajima S. Purification and characterization of two xylanases and an arabinofuranosidases from Aspergillus sojae. J Ferment Bioeng. 1995;80:334–339. [Google Scholar]
- 12.Kyu K L, Ratanakhanokchai K, Uttapap D, Tanticharoen M. Induction of xylanase in Bacillus circulans B6. Bioresource Technol. 1994;48:163–167. [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 15.Mackenzie C R, Bilous D, Schneider H, Johnson K G. Induction of cellulolytic and xylanolytic enzyme systems in Streptomyces spp. Appl Environ Microbiol. 1987;53:2835–2839. doi: 10.1128/aem.53.12.2835-2839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millward-Sadler S J, Poole D M, Henrissat B, Hazelwood G P, Clarke J H, Gilbert H J. Evidence for a general role for high-affinity non-catalytic cellulose binding domains in microbial plant cell wall hydrolases. Mol Microbiol. 1994;11:375–382. doi: 10.1111/j.1365-2958.1994.tb00317.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura S, Wakabayashi K, Nakai R, Aono R, Horikoshi K. Purification and some properties of an alkaline xylanase from alkaliphilic Bacillus sp. strain 41 M-1. Appl Environ Microbiol. 1993;59:2311–2316. doi: 10.1128/aem.59.7.2311-2316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson N. A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem. 1944;153:375–380. [Google Scholar]
- 19.Ratanakhanokchai K, Kyu K L. Abstract of the Seminar of JSPS-NRCT/DOST/LIPI/VCC Large-Scale Cooperative Research in the Field of Biotechnology. 1997. Cellulosome structure of thermophilic cellulolytic and alkaliphilic xylanolytic microorganisms. 1. Isolation of an alkaliphilic xylanolytic bacterium which produced specific xylan-binding xylanase, abstr. JSPS-I-A-4; p. 10. [Google Scholar]
- 20.Sakka K, Yoshikawa K, Kojima Y, Karita S, Ohmiya K, Shimada K. Nucleotide sequence of the Clostridium stercorarium xylA gene encoding a bifunctional protein with β-d-xylosidase and α-l-arabinofuranosidase activities, and properties of the translated product. Biosci Biotechnol Biochem. 1993;57:268–272. doi: 10.1271/bbb.57.268. [DOI] [PubMed] [Google Scholar]
- 21.Sneath P H A. Endospore-forming Gram-positive rods and cocci. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins, Co.; 1986. pp. 1104–1139. [Google Scholar]
- 22.Spurway T D, Morland C, Cooper A, Sumner I, Hazlewood G P, O’Donnell A G, Pickersgill R W, Gilbert H J. Calcium protects a mesophilic xylanase from proteinase inactivation and thermal unfolding. J Biol Chem. 1997;272:17523–17530. doi: 10.1074/jbc.272.28.17523. [DOI] [PubMed] [Google Scholar]
- 23.Tabernero C, Sanchez-Torres J, Perez P, Santamaria R I. Cloning and DNA sequencing of xyaA, a gene encoding an endo-β-1,4-xylanase from an alkalophilic Bacillus strain (N137) Appl Environ Microbiol. 1995;61:2420–2424. doi: 10.1128/aem.61.6.2420-2424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsujibo H, Sakamoto T, Nishino N, Hasegawa T, Inamori Y. Purification and properties of three types of xylanases produced by an alkalophilic actinomycete. J Appl Bacteriol. 1990;69:398–405. doi: 10.1111/j.1365-2672.1990.tb01544.x. [DOI] [PubMed] [Google Scholar]
- 25.Tsujibo H, Ohtsuki T, Ilo T, Yamazaki I, Miyamoto K, Sugiyama M, Inamori Y. Cloning and sequence analysis of genes encoding xylanases and acetyl xylan esterase from Streptomyces thermoviolaceus OPC-520. Appl Environ Microbiol. 1997;63:661–664. doi: 10.1128/aem.63.2.661-664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uziie M, Matsuo M, Yasui T. Possible identity of β-xylosidase and β-glucosidase of Chaetomium trilaterale. Agric Biol Chem. 1985;49:1167–1173. [Google Scholar]
- 27.Vincent P, Shareck F, Dupont C, Morosoli R, Kluepfel D. New α-l-arabinofuranosidase produced by Streptomyces lividans: cloning and DNA sequence of the abfB gene and characterization of the enzyme. Biochem J. 1997;322:845–852. doi: 10.1042/bj3220845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaura I, Koga T, Matsumoto T, Kato T. Purification and some properties of endo-1,4-β-d-xylanase from a fresh-water mollusc, Pomacea insularus (de Ordigny) Biosci Biotechnol Biochem. 1997;61:615–620. doi: 10.1271/bbb.61.615. [DOI] [PubMed] [Google Scholar]