Abstract
Ferulic acid (FA) is one of the most abundant hydroxycinnamic acids in the plant world, especially in the cell wall of grain bran, in comparison with forage and crop residues. Previous studies noted that FA was mainly linked with arabinoxylans and lignin in plant cell walls in ester and ether covalent forms. After forages were ingested by ruminant animals or encountered rumen microbial fermentation in vitro, these cross-linkages form physical and chemical barriers to protect cell-wall carbohydrates from microbial attack and enzymatic hydrolysis. Additionally, increasing studies noted that FA presented some toxic effect on microbial growth in the rumen. In recent decades, many studies have addressed the relationships of ester and/or ether-linked FA with rumen nutrient digestibility, and there is still some controversy whether these linkages could be used as a predicator of forage digestibility in ruminants. The authors in this review summarized the possible relationships between ester and/or ether-linked FA and fiber digestion in ruminants. Rumen microbes, especially bacteria and fungi, were found capable of breaking down the ester linkages within plant cell walls by secreting feruloyl and p-coumaroyl esterase, resulting in the release of free FA and improvement of cell wall digestibility. The increasing evidence noted that these esterases secreted by rumen microbes presented synergistic effects with xylanase and cellulase to effectively hydrolyze forage cell walls. Some released FA were absorbed through the rumen wall directly and entered into blood circulation and presented antioxidant effects on host animals. The others were partially catabolized into volatile fatty acids by rumen microbes, and the possible catabolic pathways discussed. To better understand plant cell wall degradation in the rumen, the metabolic fate of FA along with lignin decomposition mechanisms are needed to be explored via future microbial isolation and incubation studies with aims to maximize dietary fiber intake and enhance fiber digestion in ruminant animals.
Keywords: Rumen microbes, Ferulic acid, Fiber digestion
1. Introduction
The global human population is projected to reach 9 to 10 billion by 2050, raising the hugely increased demand for foodstuffs and animal products (McNeill et al., 2013). Ruminant animals are important livestock to provide humans with quantity and quality of meat and milk, as well as leather and wool (Li, 2021). Plant cell walls are the most abundant renewable chemical energy on earth which amounts to 1011 tons synthesized annually (Topakas and Paul, 2007). About 70% of the metabolizable energy of feedstuffs is provided to ruminants by rumen microbial fermentation (Bergman, 1990). Due to the shortage of food, timber and fossil carbon, comprehensive utilization of plant cell walls has raised interest. Because of the presence of active and diverse microbial populations, the rumen functions as a large natural fermentation chamber to degrade nutrients from plant cell wall (Buanafina et al., 2008). Therefore, making good use of fiber-rich forage in the rumen can lead to considerable economic benefits. However, the complex structure of plant cell walls, mainly consisted of polysaccharides, phenolic acids and protein, impedes the further utilization of these biomass (Hatfield et al., 2017).
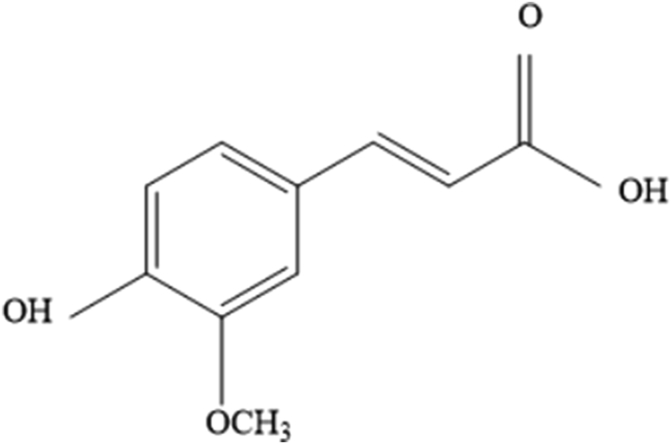
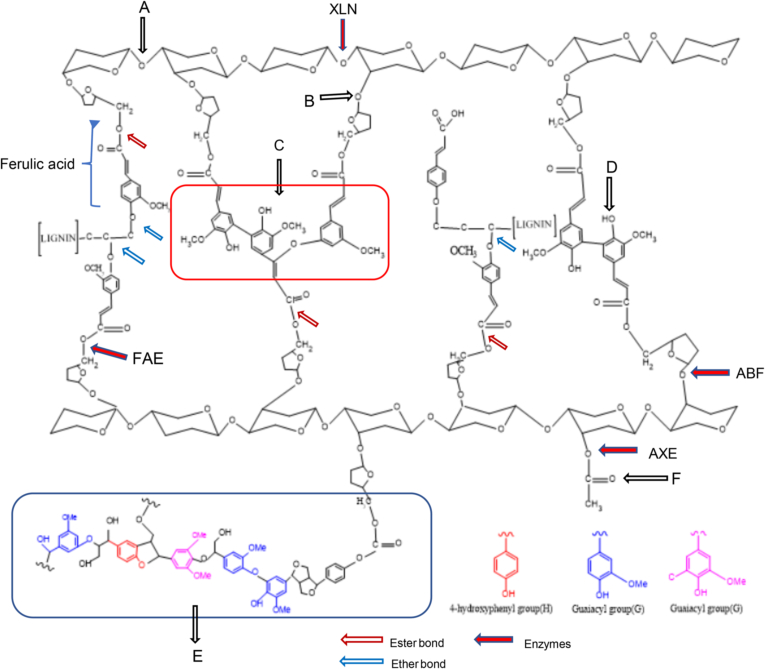
Wang et al. (2012), in our previous laboratory study, collected 13 fibrous feeds including grass hay, alfalfa hay, grain bran, crop straw and/or stalks, and noted that both ferulic acid (FA) and p-coumaric acid (pCA) are the most two abundant phenolic acids, and the FA content much richer than pCA in grain brans comparison with hays crop straw and/or stalks. As shown in Fig. 1, FA is chemically named as 3-(4-hydroxy-3-methoxyphenyl) propenoic acid. Through the shikimic acid pathway, FA participates in the formation of plant cell walls (Seigler, 1998). Ferulate polysaccharide esters are the initiation and nucleation sites for lignification and are then incorporated into the lignin through peroxidase-H2O2-mediated formation of ether linkages between the FA molecules and phenolic monomers in the growing lignin polymer (Delmer and Stone, 1988; Jung and Allen, 1995; Ralph et al., 1995). As shown in Fig. 2, FA in plant cell walls form ether linkages with lignin through the hydroxyl groups in the aromatic ring and is ester-linked via its carboxylic acid group to the C(O)5 position of the arabinofuranosyl side group attached at the C(O)2 to the xylan chains (Barron et al., 2007; Wong et al., 2013). These ester- and ether-linked FA could be measured through mild and harsh alkaline hydrolysis, respectively (Wong, 2006). Lignin and arabinoxylans were connected by ferulate molecules through these linkages (Ralph et al., 1994). Ferulic acid dehydrodimers are ester-linked to plant cell wall polysaccharides and other cell wall components including proteins and lignin (McKinnon and Christensen, 2005; Pedersen et al., 2015). More recently, dehydrotriferulic acids and dehydrotetraferulic acids also have been isolated from plant cell walls (Bunzel et al., 2006). These cross-linkages limit the growth of cell walls, increase the mechanical strength and reduce the utilization of the plant (Iiyama et al., 2001; McKinnon and Christensen, 2005).
Fig. 1.
Chemical structures of ferulic acid.
Fig. 2.
The linkage of ferulic acid in cell wall and the enzymes sites in rumen (Bunzel et al., 2006; Cao et al., 2016a, Cao et al., 2016b; Dilokpimol et al., 2016). XLN = β-1,4-endoxylanase; FAE = feruloyl esterase; ABF = α-arabinofuranosidase; AXE = acetyl xylan esterase. The red arrows present for the ester bond, blue arrows for the ether bond, the red solid arrows for enzymes. A: β-(1-4) linked xylan backbone; B: xylose-arabinose linkage; C: 5-5/8-O-4 dehydrotriferulic acid (TriFA); D: 5-O-diferuloyl group (5-5 linked dimer); E: the example of a lignin structure containing the most frequent bonds and the corresponding monomers; F: 3-O-acetyl group.
In this article, the effect of FA on fiber digestion, the release of bound FA and the possible metabolism pathways of FA in rumen were comprehensively reviewed and discussed.
2. Effect of ferulic acid on fiber digestibility in the rumen
2.1. The relationship between ferulic acid in plant cell wall and rumen digestibility
Forage cell walls are major source of nutritional energy in the rumen. However, less than 50% of these fractions are utilized by the ruminant hosts (Badhan et al., 2014). In an earlier study, ryegrass hay presented a highly positive correlation (r = 0.98) between cell wall digestibility to the ratio of FA to pCA (Hartley, 1972). Then, Casler (2001) highlighted that both lignin and phenolic acids were the main factors limiting the use of energy in rumen. In recent decades, in vitro rumen fiber digestion and gas production and in situ rumen degradation trials have been completed. Ester-linked FA (FAest) and ether-linked FA (FAeth) attracted attention to address their relationships with extent and rate of fermentation and/or digestion.
2.1.1. In vitro
As a rapid method for measuring gas production, the in vitro technique has been an important tool in evaluating ruminant feeds (Menke et al., 1979). Bermuda grass hays with higher FAest content presented higher digestibility of organic matter, neutral detergent fiber (NDF) and acid detergent fiber (ADF) (Jung and Allen, 1995; Mandebvu et al., 1999). A positive correlation between the FAest content on a NDF basis and 24 and 96-h in vitro dry matter digestibility (IVDMD) were observed in brown midrib corn silage (r = 0.78 for 24 h, r = 0.60 for 96 h, P < 0.05) and mature grasses (r = 0.94 for 24 h, r = 0.60 for 96 h, P < 0.05) (Raffrenato et al., 2017). The 48-h IVDMD (r = 0.79) and in vitro neutral detergent fiber digestibility (IVNDFD) (r = 0.82, P < 0.05) of meadow hay were positively correlated to the concentration of FAest (Rodrigues et al., 2007). Different breeds of perennial grasses exhibited different correlations between FAest and IVNDFD. Smooth bromegrass and cocksfoot had a positive correlation between FAest and 96-h IVNDFD while in reed canarygrass the relativity between FAest and 24-h IVNDFD was negative but not notable (Casler and Jung, 2006). In vitro digestion constants such as sharpness of the switching characteristic for the profile of the first phase (r = 0.94, P < 0.01) and asymptotic gas production of the second phase (r = 0.89, P < 0.05) were positively correlated with FAest (Rodrigues et al., 2007). However, a few negative correlations between FAest and IVDMD were reported. The FAest content on an ADF basis of mature grasses was negatively correlated to the 96-h IVDMD (r = −0.75, P < 0.05) (Raffrenato et al., 2017). Jung and Vogel (1992) found an occasional negative relationship between FAeth and 48-h IVNDFD.
The concentration of FAeth of perennial grasses negatively related to the 24 and 96-h IVNDFD (Casler and Jung, 2006), suggesting that the content of ferulated cross-linkages were more pronounced for NDF digestion after a 96-h microbial incubation in comparison with the 24 h incubation (Casler and Jung, 2006). Through an in vitro study, a significant negative effect occurred between 24-h cell wall polysaccharide (r = −0.71, P < 0.01), arabinose (r = −0.42, P < 0.01), glucose (r = −0.33, P < 0.05) and uronic (r = −0.32, P < 0.05) degradability and FAeth in maize stem internodes and maize internodes (Jung et al., 1998; Jung and Buxtono, 1994). A strong negative influence of FAeth on IVNDFD was found in smooth bromegrass, and this influence was independent of the lignin in fiber fraction (Casler and Jung, 1999). According to Raffrenato's (2017) study the Klason lignin had more negative effect on the 24-h extent digestion of bmr corn silage, whereas FAest and acid detergent lignin showed more negatively affected on controversial corn silages. However, Rodrigues et al. (2007) found a positive effect of FAeth fraction (r = 0.93, P < 0.01) on 48-h IVNDFD. 24 and 96-h IVDMD of BMR corn silage and immature grasses were positively correlated to FAeth on both NDF basis (P < 0.05) (Raffrenato et al., 2017).
2.1.2. In situ
The in situ technique was firstly used by Quin et al. (1941) to evaluate rumen degradability of feeds. Through an in situ study, Du and Yu (2011) revealed that FA in barley hull was highly and positively correlated to rumen indigestible dry matter (DM), NDF and ADF at either 12 or 24 h. Effective degradability of NDF (r = −0.98, P < 0.01), cellulose (r = −0.98, P < 0.05) and hemicellulose (r = −0.98, P < 0.05) of crop bran and husks inversely related to the content of FAeth (Cao et al., 2015). Rodrigues et al. (2007) found a high positive relationship between the potential degradation and FAeth (P < 0.05).
In most of the aforementioned studies, the concentration of FAest was positively correlated with the rumen fiber digestion. As mentioned, the deposition of FA during the primary cell wall development accompanies incorporation of other cell wall components (Rodrigues et al., 2007) and the ester-linked ferulic acids mainly exist in the primary cell wall which are more approachable for rumen microbes (Cao et al., 2016a). Thus, the content of FAest only affects the rate instead of the extent of cell wall digestion (Jung and Allen, 1995).
The FAeth was thought to be an indicator of cross-linkages between lignin and arabinoxylans and generally presented a negative effect on cell wall digestibility (Cao et al., 2015; Jung and Casler, 2006; Jung et al., 2011). Due to the lignin polymer, the ether linkage of FA was hard to access by microbes. These linkages could not be cleaved under anaerobic environments (Jung and Allen, 1995), and the negative effect of FAeth on cell-wall digestibility was considered to cause reduced cell wall digestion, but not a reduced digestion rate, in the rumen (Jung and Allen, 1995). Indigestible components of dietary fiber were associated with other food constituents, such as phenolic acid compounds, and this could be ascribed to the ability of polysaccharides to bind and trap phenolic compounds at several sites (Sauracalixto, 2011).
As noted in the above, with the correlation between feed digestion and the content of phenolic acids, the release of hydroxycinnamic acids from the animal fodder indirectly implicated an increase of feed digestibility. Cao et al. (2015) found that the in situ ruminal disappearance of FAest was positively correlated with disappearances of cellulose, hemicellulose and NDF.
The authors in this review summarize that the relationships between bound FA and forage degradation were affected by the plant species, breed, fractions and stage of maturity. Even though the effect on the rate and extent of fiber digestion are not the same, the properties of linkages play an inevitable role in forage digestion (Raffrenato et al., 2017). As stated in the above studies, it is still a somewhat controversial issue whether these ferulated linkages can be used as a predicator of forage digestibility in ruminants.
2.2. Reasons for the negative effect of ferulic acid on plant cell wall digestion in the rumen
2.2.1. Barrier of plant cell walls
The cross-linkages form an obstruction through substitution and steric hindrance for the accessing of hydrolytic enzymes to their polysaccharide substrate (Grabber et al., 1995; Várnai et al., 2014). Though a biomimetic model, Grabber et al. (2009) pointed that the reduction of ferulate linkages increased the extent and the rate of cell wall enzymatic hydrolysis and noted that compared with lignin content, ferulate cross-linking had a more profound effect in determining the rate and extent of digestion of hemicellulose. Etherification protects FA from being decarboxylated during oxidative coupling which can impede the access of microbe and enzymes. Wong et al. (2019) noted that diferulates were responsible for the control of plant cell growth, and reduced cell wall digestibility as a biological protection against pathogenic attack. The hydrophobicity ability of phenolic compounds also impedes the access of enzyme to cell wall polysaccharides (Besle et al., 1994). Thus, feruloyl polysaccharides are critical entities in directing cell wall cross-linking and in limiting biodegradability by microorganisms (Ishii, 1997; Jung, 2003).
2.2.2. Toxic effects
Phenolic monomers inhibited digestibility of cellulose and xylan by influencing the attachment of the fibrolytic microorganisms to fiber particles (Hartley and Akin, 1989; Varel and Jung, 1986). The FA can inhibit the growth of microbes (such as bacteria, protozoa and fungi) (Zuhainis Saad et al., 2008; Zuhainis et al., 2007). At concentrations over 5 mmol/L, FA can suppress the growth of the cellulolytic strains Ruminococcus albus, Ruminococcus flavefaciens, and Bacteroides succunigenes (Chesson et al., 1982). The growth rate of R. albus and IVDMD at 24 and 48 h was inhibited by FA at 10 mmol/L (Borneman et al., 1986). The ester-linked feruloyl showed the ability to inhibit the growth of ruminal bacteria R. flavefaciens FD1, Seknomonas ruminantium HD4, and Butyrivibrio fibrisolvens 49 (Akin et al., 1993). Dehydrodimers of FA inhibit the growth of microbes. Boutigny et al. (2010) found that the 8-5′-benzofuran dimer FA showed the same extent in inhibit trichothecene biosynthesis as the monomer of FA. FA inhibits the digestibility of cellulose, not only producing the negative effect on rumen microbes, but also depressing enzyme activity. Free and bound phenolic-rich extracts from shaddock peels show a negative effect on α-amylase, α-glucosidase and angiotensin Ⅰ-converting enzyme activity in a dose-dependent manner (Schmidt et al., 2014). The intestinal sucrases and maltases were inhibited by ferulic and isoferulic acid in a mixed type manner (Adisakwattana et al., 2009). The activities of cellulolytic enzymes (including carboxymethylcellulase, filterpaperase, xylanase and β-glucosidase) and the production of total volatile fatty acid (VFA) of Neocallimastix frontalis B9 were depressed by FA addition (Zuhainis Saad et al., 2008). However, some microbes show ability to detoxicate FA or even use FA as a single carbon source. Zymomonas mobilis ZM4 can detoxify phenolic aldehydes (Gu et al., 2015). Piromyces sp. FNG5 (isolated from feces of wild nil gai) also showed tolerance to phenolic monomers and even had the ability to degrade them (Paul et al., 2003).
With these characteristics, the author in this review speculated that FA could inhibit rumen microbes and their enzymes and lead to low rumen digestibility. But the lowest standard for inhibiting the microbes in rumen still needs further research. It is necessary to take into consideration that the FA amounts consumed under a nutritional intervention may be very different from the intake in the context of a normal diet. Callaway et al. (2009) noticed that carboxylic phenols directly killed Escherichia coli O157:H7 at physiologically unrealistic concentrations, but when phenolics were added to the intestine, the phenolic compounds provided a competitive advantage to the organisms that competed against E. coli O157:H7. Just like the intestine, the concentration of FA may be very low in rumen. Thus, the inhibition of microbes by FA in natural condition seems to be less realistic. An in vitro batch culture carried by Cao et al. (2016b) showed that the free FA was at low ration in culture fluid range from 5.65 to 7.56 μg/75 mL and FAest in culture fluid was 0.90 to 1.85 μg/75 mL. Even though the intake of FA depends on the type and amount of diet, the concentration of free and linked FA in rumen cannot reach the inhibitory level in former studies. Additionally, it is believed that the absorption of FA is quick within the rumen. Also, the released FA can be catabolized by microbes during rumen fermentation. There is still no identification of the toxic dose of FA for rumen microbes. Thus, whether the FA in diets could have toxin effect on rumen microbes and enzymes requires further research.
3. Release of bound ferulic acid in the rumen
To promote the existence of microbes, the rumen is a complex and efficient ecosystem that degrades plant biomass (Kothari et al., 2018). Through mechanical, chemical and enzymatic forces, bound FA are released by microbial action in ruminants (Chesson et al., 1999; Hegde et al., 2006). Three main microbial populations (protozoon, bacteria, fungi) help to degrade the structure of plant cell wall and release FA (Betts and Dart, 1988; Mathew and Abraham, 2006). Ester-linked FA can be extensively disrupted by phenolic acid esterases during the ruminal fermentation, while the ether bonds between FA and lignin cannot be broken in the rumen (Rodrigues et al., 2007). The breakdown of these linkages renders the cell wall more susceptible to enzymatic attack and increases cell wall degradability (Morris et al., 2017).
3.1. Mechanical competence of rumen microbes to release bound ferulic acids
3.1.1. Rumen protozoa
Rumen protozoa (104 to 106 cells/mL) constitute about 50% of the viable biomass in rumen and have long been reported to contribute to plant cell wall degradation (Sirohi et al., 2013). When protozoa are eliminated from rumen liquid, the breakdown of cellulose and hemicellulose is decreased (Williams and Coleman, 1997). Some protozoa could associate with plant cell walls and then penetrate deeply into the broken plant tissues and adhere via a special attachment organelle. Others show primary degradation abilities of plant cell tissues by ingesting small particles (Orpin and Letcher, 1978). Except for ingesting and attachment, enzymes in protozoa may also play an important role in the degradation of plant cell walls. Enzymes such as pectin esterase, amylase, cellulose and polygalactureonase have been found in rumen protozoa (Béra-Maillet et al., 2005; Devillard et al., 2003; Sirohi et al., 2013; Williams, 1979). However, the activity of feruloyl esterase (FAE) has not been detected in rumen protozoa. The degradation of plant tissues by protozoa help fungi and bacteria to approach cell wall more easily which lead to the further release of FA.
3.1.2. Fungi
Fungus (103 to 106 cells/mL) are the most efficient contributors to fiber degradation, even though they only make up 20% of the rumen biomass (Rezaeian et al., 2004). Fungal parasites can penetrate and enter host cell walls. Thereafter, they differentiate specialized intracellular feeding structures, called haustoria, and invaginate the plants' plasma membrane. Meanwhile, a series of highly active hydrolase enzymes (such as cellulase, hemicellulose, esterases and other side chain enzyme) are secreted. The multi-component cellulase system produced by anaerobic fungi is the most important mechanism to degrade lignin and release FA. Rumen fungi shows chemotactic response to phenolic acids (such as ferulic, syringic and coumaric acid) then attacks lignin to release FA from plant cell walls (Wubah and Kim, 1996). Zoospores of fungi also prefer to colonize lignin-rich regions, and upon germination, solubilize these regions (Qi et al., 2011).
3.1.3. Bacteria
Due to their large biomass (1010 to 1011 cells/mL), bacteria play an important role in the release of FA. Rumen bacteria degrade fiber through the adhesion of their fibrolytic enzymes to their substrates (Cai et al., 2010). Cellulosic bacteria are divided into bacteria which adhere to plant fiber and those that do not. When bacteria degrade the plant cell wall, four steps are essential. First, the bacteria access the plant fragments. The second step is the initial non-specific adhesion at appropriate parts of plant debris. The third step is the specific adhesion on binding of the substrate to the surface of the bacteria. Then, adherent bacteria proliferate at specific sites in plant tissues. These mechanical steps of rumen microbes break plants into small fragments, and this step facilitates further enzyme attachment sites.
3.2. Enzymatic competence of rumen microbes to release ferulic acid
After attaching to plant fragments, microbes secrete a series of enzymes. Due to the ability to cleave ester-linkages between FA and the attached sugar, cinnamoyl esterases were regarded to be a “helper” enzyme which facilitate the access of cellulases to the main chain of plant cell wall (Benoit et al., 2008).
Still, rumen microbes lack of the enzyme to degradation the ether bond in plant cell wall (Rodrigues et al., 2007).
3.2.1. Feruloyl esterases
Feruloyl esterases (EC 3.1.1.73, FAE) were first discovered from the culture filtrates of Streptomyces olivochromogenes (Mackenzie et al., 1987). After then, more than 80 kinds of FAE have been discovered and characterized in fungi and bacteria (Oliveira et al., 2020). The FAE are key enzymes to hydrolyze the ester bond arabinoxylan fibers and release FA, esterified diferulates and feruloyl oligosaccharides from agricultural biomass (Andreasen et al., 2001b; Cheng et al., 2012; Uraji et al., 2018). Expect secreted by microbes, FAE were also found in mammalian intestinal epithelial cells and of plant origin (Andreasen et al., 2001a; Kern et al., 2003; Oliveira et al., 2020).
Generally, based on the substrate utilization and amino acid sequences, FAE are classified into four types (Type A, B, C and D) (Crepin et al., 2004). Recently, new types of FAE have been identified (Uraji et al., 2018). In this system, FAE were divided into 12 sub-families. All of these FAE possess a similar three-dimensional structure with α/β-hydrolase having a serine, histidine, and aspartic acid catalytic triad (Goldstone et al., 2010; Li et al., 2011). Kühnel et al. (2012) summarized the activity of different types of FAE. Type A FAE prefer methyl ferulate, methyl p-coumarate and methyl sinapinate and can release 5,5′-diferulic acid. Until now, the production of Type A FAE was only observed in fungi (Mogodiniyai Kasmaei and Sundh, 2019). Type B FAE toward to hydroxycinnamic acid ester with free hydroxyl substituents advance. Type C FAE can hydrolyze all hydroxycinnamic acid methyl esters. Type D esterases are unspecific esterases to hydrolyze all hydroxycinnamic acid methyl esters (Wong, 2006). When pretreated or co-incubated with endo-xylanases, Type A and type D are able to hydrolyze synthetic diferulate (Araf-FA-FA-Araf) while Type B and Type C FAE could not release free diferulates (Wong et al., 2019). A full range of FAE contribute to the high-efficiency fiber digestion by rumen microbes (Wong et al., 2019). Most A, B and C were found from aerobic fungi and worked in concert to hydrolyze s the ester linkages between polysaccharide main chain with phenolic acids (Li et al., 2011; Qi et al., 2011).
Because no less than 1% of microbes in rumen can be cultured, only a few of FAE produced by rumen microbes have been identified (Cheng et al., 2012). Presently, Aspergillus, Fusarium, Neocallimastix, Fusarium Orpinomyces, Piromyces, Penicillium and Talaromyces are reported to secrete FAE in the rumen or gastrointestinal tract (Benoit et al., 2008; Cao et al., 2013; Li, 2021; Paul et al., 2003). Compared with fungi, only a few bacteria are able to secrete FAE. Butyrivibrio, Prevotella, Clostridium, Cellulosilyticum, Butyrivibrio, Streptomyces, Lactobacillus and Dickeya have been reported to produce FAE (Li et al., 2011; Stewart et al., 1988; Wang et al., 2005). More recently, Mogodiniyai Kasmaei and Sundh (2019) used whole-genome shotgun metagenomics and genome binning to explore novel prokaryotic FAE in cow, horse, sediment, and soil datasets and nominated Candidatus Rhabdochlamydia genus as a novel FAE to produce taxonomic unit. In this study, four genomes: B. fibrisolvens, Butyrivibrio proteoclasticus, R. albus, and R. flavefaciens belonging to Clostridiales, were reported to produce FAE. Limited research has been conducted on the structure of FAE in ruminants. A promiscuous feruloyl esterase with a broad activity to release FA, coumaric acid, coumarin-3-carboxylic acid, and cinnamic acid was isolated from the rumen bacterium B. proteoclasticus (Goldstone et al., 2010). The structure of this FAE is identified in two different space groups, in both the apo-form, and the ligand bound form with FA located in the active site. The presence of three different conformations adopted by different molecules in the crystals shows the flexibility of the lid domain.
FAE are accessory enzymes which assist cellulose and hemicellulose to be more available to xylanolytic and pectinolytic enzymes (Dilokpimol et al., 2016; Gopalan et al., 2015). These enzymes break down the polysaccharides into low molecular weight fragments, which are more suitable as esterase substrate (Mathew and Abraham, 2004). Thus, the ability was enhanced with the addition of these cell wall degrading enzymes such as xylanase, cellulases, pectinases, hemicellulases and arabinofuranosidas (Mathew and Abraham, 2004; Shin et al., 2006). After pretreating wheat bran with xylanase, the diFA released by human faecal bacteria increased from 40% to 80% (Vardakou et al., 2007). Borneman et al. (1990) found that the activity of FAE may be facilitated by β-xylosidase.
3.2.2. p-Coumaroyl esterase
Compared with FAE, only a few studies have focus on p-coumaroyl esterase (3.1.1.B10, pCAE). Trans-p-coumaroyl esterase have been found in anaerobic rumen fungi (Borneman et al., 1990). Borneman et al. (1991) noted that p-coumaroyl esterase secreted by Neocallimastix can release p-coumaroyl groups from O-[5-O-((E)-p-coumaroyl)-α-L-arabinofuranosyl]-(l→3)-O--D-xylopyranosyl-(1→4)-D-xylopyranose. A feruloyl/p-coumaroyl esterase purified from Penicillium pinophilum released 100% of the alkali-extractable FA from water-soluble wheat straw xylan (Castanares and Wood, 1992). This enzyme has a remarkably wide substrate specificity: hydrolyzing phenolic acids from model substrate and releasing acetic acid from p-nitrophenyl acetate and from an acetylated xylan. A recombinant purified enzyme hydrolyzing methyl MpCA and chlorogenic acid resulted in classification as a pCAE, with a distinct chlorogenic acid esterase side activity (Nieter et al., 2017). Both of FAE and pCAE can release FA and pCA from xylan polysaccharides, there exist differences in their specificity for hydrolysis of methyl ferulate (MFA) and MpCA, respectively (Nieter et al., 2017). Still, Aspergillus awamori, P. pinophilum, Neocallimastix and Rhizoctonia solani have been reported to secrete pCAE (Castanares and Wood, 1992; Borneman et al., 1991; Nieter et al., 2017).
Lots of factors affect the activity of cinnamoyl esterases in ruminants. Yu et al. (2002) noticed that FAE (produced by Aspergillus) could not release FA from oat hull ground through 1 mm screen but when using 250 μm screen, a small amount of FA was released. Thus, the substrate size affects the activity of FAE (Borneman et al., 1991; Yu et al., 2002). McKinnon and Christensen (2005) explained that the large substrate may restrict the access of FAE to feruloyl groups and lead to an undetectable release of FA. Wang et al. (2005) isolated a FAE from human intestinal bacterium Lactobacillus acidophilus and suggested that with the addition of FA, the concentration of FAE was increased, then reached a peak and declined. Thus, the concentration of FA affects the activity of FAE. Even under the same conditions, the FA released from barley spent grain, wheat bran and maize bran by the same FAE were different. The complex physical and steric factors of plant cell wall could affect the FAE to release FA (Bartolome et al., 1997). The activities of FAE are influenced by the substrate of forage and complex cell wall materials (Oliveira et al., 2020). Benoit et al. (2008) found that the expression of faeA and faeB (genes encoded FAE) were induced by FA and other aromatic compounds. The activity of FAE were stability under pH 8.0–9.0 and it was inhibited by Ca2+, Co2+, Mg2+, Zn2+, Mn2+, K+, Fe2+ Cu2+ (Cao et al., 2013; Yang et al., 2009).
In recent years, cinnamoyl esterases are widely used in agriculture waste utilization, food industries, pharmaceutical and biofuel industries. Making good use of FAE leads to the comprehensive utilization of forage and favorable economic benefits.
4. Catabolism of ferulic acid in the rumen
After release from plant cell walls, FA and diferulic acids are absorbed in the rumen or further catabolized by microbes (Soberon et al., 2012). Microbes catabolize FA mainly from addition or deletion of side groups, production of other organic molecules and/or incorporation of carbon from other phenolic acids into microbial biomass, and polymerization (Martin and Haider, 1976; Singh et al., 2001). Using natural substrates through microbial fermentation to produce high-value products seems to be a commercial and environmentally-friendly renewable energy source (Wong, 2006). Some industries use microbes to convert FA to high value products such as vanillin.
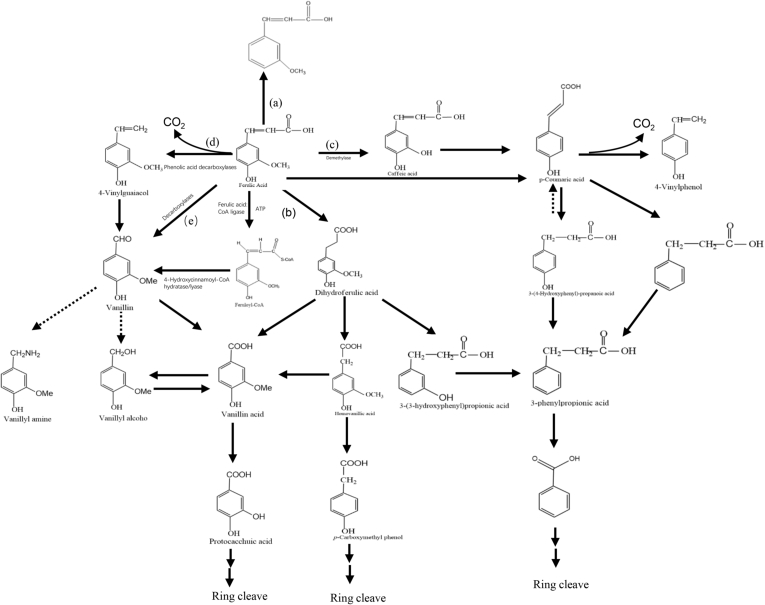
The FA is catabolized by microbes through non oxidative decarboxylation (Mishra et al., 2014), β-oxidation (Delneri et al., 1995), demethylation (Grbić-Galić and Pat-Polasko, 1985), side chain reduction (Falconnier et al., 1994), coenzyme A independent deacetylation and direct deacetylation (Plaggenborg et al., 2001). Theoretically, the rumen microbes are able to fully metabolize phenolic acids under anaerobic conditions. But, for the distribution of π electrons over its ring structure, the benzene nucleus become very stable (Besle et al., 2010). Thus, many microbes only hydrogenate the alkyl side chains but not transform further. The author summarized the possible pathways to catabolize FA in rumen as shown in Fig. 3.
Fig. 3.
The possible pathways to catabolize ferulic acid in the rumen (Besle et al., 2010; Ohmiya et al., 1986; Sheng et al., 2015; Tripathi et al., 2002). (a) Dihydroxylation; (b) reduction; (c) demethylation; (d) decarboxylation; (e) deacetylation.
4.1. Non-oxidative decarboxylation
Ferulic acid is catabolized to 4-hydroxy-3-methoxystyrene (4-vinyl guaiacol) through non-oxidative decarboxylation by phenolic acid decarboxylases (encoded by blpad) (Curiel et al., 2010). The decarboxylases are divided into ferulic acid decarboxylase (encoded by fad) and p-coumaric acid decarboxylases (encoded by pdc) (Hu et al., 2015; Rodríguez et al., 2010; Sheng et al., 2015; Wen et al., 2011). The non-oxidative decarboxylation of FA is a reversible reaction (Li et al., 2008; Hu et al., 2015) and has been found in both bacteria and fungi (Tinikul et al., 2018). The first step of decarboxylation is the deprotonation of the p-hydroxyl group which ensures the electron flow through ferulate. The ortho-carbon atom of the carboxyl group of FA forms a nucleophilic center. Then a proton transfers from a general acid at the active site to nucleophilic C2 carbon, forming a quinone methide intermediate. The carboxyl of the quinoid intermediate starts a second electron flow, and then the C–C bond of the intermediate is cleaved and generates p-vinyl phenol and CO2. For the high price of 4-vinyl guaiacol (25-30 times more expensive than FA), the decarboxylation of FA is thought to be a commercial way to transfer FA into 4-vinyl guaiacol. In rumen, the 4-vinyl guaiacol can be further catabolized (Li et al., 2008; Mavinkurve and Nazareth, 1986). Mathew and Abraham (2006) found that vinyl guaiacol was transformed to acetovanillone and ethyl guaiacol through biocatalytic routes. The position of the ring cleavage of protocatechuic acid and catechol determines the final products of the pathway.
4.2. Demethoxylation
Haematococcus pluvialis demethoxylates FA at the meta position and forms into pCA which can be further oxidated into p-hydroxybenzoic acid (Tripathi et al., 2002). Two different mechanisms of demethxylation have been expounded. One degradation pathway was followed by the production of acetate from methoxyl groups. Another pathway is to degrade the acidic side-chain of FA without the formation of acetate (Bache and Pfennig, 1981; Grbić-Galić, 1986). Enterobacter cloacae O-demethylates FA into caffeic acid as an intermediary or as an end product in both aerobic and anaerobic conditions (Micard et al., 2002).
4.3. Reduction
During catabolism, the reduction of FA to non-toxic compounds is always the first step (Jugal et al., 1981; Besle et al., 2010). The effect of the reductive anaerobic transformation is to dispose redox equivalents generated in oxidative reactions (Heider and Fuchs, 1997). Compared with demethylation and dihydroxylation, reduction is relatively rapider and faster (Chesson et al., 1999). The FA can be reduced into coniferyl aldehyde then transformed into coniferyl alcohol by white-rot fungus Pycnoporus cinnabarinus 1-937 (Falconnier et al., 1994). Reductases take part in this reduction pathway. Another kind of reduction happens in the double-bond of the side chain. Clostridium strains can reduce cinnamic acid, o-, m- and p-methoxycinnamic acid, caffeic acid, FA to their corresponding 3-phenylpropionic acid derivatives. For example, pCA can be reduced to p-hydroxyhydrocinnamic acid (3- (4-hydroxyphenyl)) propionic acid (Chamkha et al., 2001). Wolinella succinogenes isolated from rumen reduced FA to dihydroferulic acid in the absence of hydrogen acceptors (Ohmiya et al., 1986; Poquet et al., 2008).
4.4. Dehydroxylation
The dehydroxylation of FA can reduce the double in the side-chain (Grbić-Galić, 1986). Enterobacter and Escherichia transformed FA through O-demethylation, dehydroxylation, reduction and decarboxylation under strictly anaerobic conditions (Grbić-Galić, 1986). The dehydroxylation of FA occurs at C4 and has been detected in human feces (Duncan et al., 2016). Kern et al. (2003) summarized that diferulic acid can be metabolized by colonic bacteria and open the hydrofuran rings. Dehydroxylation at C3 is more rarely encountered (Chesson et al., 1999).
4.5. Deacetylation
The deacetylation of FA has been noticed in several strains such as Bacillus subtilis, Pseudomonas acidovorans and Streptomyces setonii (Plaggenborg et al., 2001; Toms and Wood, 1970). In the direct deacetylation, the hydroxylated dihydroferulic acid is formed as an intermediate. After C–C bond cleaved, acetate and vanillic acid are formed (Toms and Wood, 1970). The ferulic-acid-converting enzymes produced by P. acidovorans only can convert phenolic substrates with the hydroxy group in the para position; while in S. setonii, non-phenolic aromatic compounds also can be converted (Muheim and Lerch, 1999). A new coenzyme A-dependent, non-β-oxidation pathway and not direct deacetylation of FA was revealed in Pseudomonas fluorescens AN103, Delftia acidovorans and Amycolatopsis sp. strain HR167 (Overhage et al., 1999; Plaggenborg et al., 2001). The FA is activated by feruloyl-CoA synthetase (encoded by fcs) to feruloyl-CoA. Then the feruloyl-CoA is hydrated to 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl- CoA as a transient intermediate product by enoyl-CoA hydratase (encoded by ech) and subsequently cleaved to acetyl-CoA and vanillin (Plaggenborg et al., 2001).
Rumen microbes exist as a complex symbiotic network of a diverse population, and are affected by various conditions such as temperature, probiotic and antibiotic treatment (Bhatt et al., 2013). Different strains showed different pathways to catabolize FA with different end products (Chesson et al., 1982). The individual's diet shows an effect on the type and the amount of phenolic substrates supplied to the intestinal bacteria, which may in turn cause fluctuations of these microbes in the gut (Hui et al., 2006). The anaerobic transformation is the main pathway to metabolize phenolic acids, but take longer than aerobic degradation (Besle et al., 2010). Theoretically, FA can be fully degraded in rumen, but the metabolism is limited by the low redox potential, the small populations of organisms able to metabolize them and their limited retention time. Cellulolytic bacteria could degrade phenolic acids but the total complete breakdown would require a ruminal retention time and large population of bacteria (Besle et al., 2010). Usually, the anaerobic bacteria catalyze specific substitution on aromatic ring through different strategies (Chamkha et al., 2001).
In the rumen, the reduction of side-chains, demethylation and dihydroxylation, are thought to be the main products of microbial action. But due to the mass of microbes in the rumen, the catabolization of FA is a complex process. According to Ohmiya's (1986) study, the ability of anaerobic microbe to catabolize FA act as synergistic reaction with cellulolytic anaerobes. Ferulic acids released from plant cell walls were provided as substrates for microbes. Cellulolytic anaerobes could be inhibited by FA. The catabolism of FA is a detoxication pathway (Adeboye et al., 2015). Thus, the FA-modifying microbes help the cellulolytic anaerobes from the toxic environment of FA. Additionally, H2 and formate produced during the fermentation of cellulose by R. albus can enhance the growth of W. succinogenes. These compounds would result in more electron donors for reduction of FA (Ohmiya et al., 1986). In another study, 3-phenylpropanoic acid, an intermediate of FA, can affect the rate of growth, cellulose digestion and in cell-bound cellulolytic enzymes of R. albus (Hungate and Stack, 1982; Stack and Hungate, 1984).
5. Recommendations for future work
In plant cell walls, FA forms ester and ether linkages between carbohydrates and lignin, which is a cross-linking agent. Due to the inhibition of microbes by FA and the limitation of ferulic acid-cross-linked complex cell walls, the existence of these linkages shows a negative effect on the digestibility of forage in ruminants. However, whether FA in diet level will inhibit the growth of rumen microbes and their enzymes still requires further research through in vitro trials. Bound FA in plant cell walls will influence the utilization of forage. The FAest in plant cell walls is generally positively correlated to the fiber degradation in rumen while FAeth mainly shows negative effects. However, relationships between FAest and FAeth in total-tract digestibility need further in vivo research. Breeding methods to reduce the cross linkages in plant cell walls are also ideal ways to increase the degradation of forages. FA undergoes several transformations when pass digestive tract. The authors wrote this review to draw attention to the release and catabolization of plant cell wall FA in rumen. After ingestion, FA is mainly released from plant cell walls by microbes in rumen. Both fungi and bacteria in rumen can release FA from plant cell walls through feruloyl esterases and p-coumaroyl esterases. The FAE hydrolyze the ester bonds between FA and sugar residues in plant cell wall polysaccharide. They cooperate with polysaccharide-degrading enzymes to improve microbe accessibility. Then, some of the released FA are absorbed in rumen, and others can be catabolized by microbes through reduction, demethylation, dihydroxylation or/and decarboxylation pathways. The product of metabolism may be further catabolized by microbes until the ring cleave. The catabolism of FA is affected by various factors. Thus, the isolation of rumen microbes may be essential for exploring the metabolism of FA in rumen.
Author contributions
Yan-Lu Wang: Conceptualization, Writing- Reviewing and Editing. Wei-Kang Wang: Writing - Original Draft. Qi-Chao Wu: Investigation. Hong-Jian Yang: Coordinator, supervision, critical manuscript review.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
The authors appreciate the financial supports for Sheep Feed Evaluation & Feed Table Establishment from the Ministry of Agriculture and Rural Affairs of China (Project No. ZR20MAC10/9) and National Natural Science Foundation of China (Project No. 31072054).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
References
- Adeboye P.T., Bettiga M., Aldaeus F., Larsson P.T., Olsson L. Catabolism of coniferyl aldehyde, ferulic acid and p-coumaric acid by Saccharomyces cerevisiae yields less toxic products. Microb Cell Factories. 2015;14(1):1–14. doi: 10.1186/s12934-015-0338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adisakwattana S., Chantarasinlapin P., Thammarat H., Yibchok-Anun S. A series of cinnamic acid derivatives and their inhibitory activity on intestinal α-glucosidase. J Enzyme Inhib Med Chem. 2009;24:1194–1200. doi: 10.1080/14756360902779326. [DOI] [PubMed] [Google Scholar]
- Akin D.E., Borneman W.S., Rigsby L.L., Martin S.A. p-Coumaroyl and feruloyl arabinoxylans from plant cell walls as substrates for ruminal bacteria. Appl Environ Microbiol. 1993;59:644–647. doi: 10.1128/aem.59.2.644-647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen M.F., Kroon P.A., Williamson G., Garcia-Conesa M.-T. Intestinal release and uptake of phenolic antioxidant diferulic acids. Free Radic Biol Med. 2001;31:304–314. doi: 10.1016/s0891-5849(01)00585-8. [DOI] [PubMed] [Google Scholar]
- Andreasen M.F., Kroon P.A., Williamson G., Garcia-Conesa M.T. Esterase activity able to hydrolyze dietary antioxidant hydroxycinnamates is distributed along the intestine of mammals. J Agric Food Chem. 2001;49:5679–5684. doi: 10.1021/jf010668c. [DOI] [PubMed] [Google Scholar]
- Bache R., Pfennig N. Selective isolation of acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol. 1981:255–261. [Google Scholar]
- Badhan A., Jin L., Wang Y., Han S., Kowalczys K., Brown D.C., et al. Expression of a fungal ferulic acid esterase in alfalfa modifies cell wall digestibility. Biotechnol Biofuels. 2014;7:1–15. doi: 10.1186/1754-6834-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron C., Surget A., Rouau X. Relative amounts of tissues in mature wheat (Triticum aestivum L.) grain and their carbohydrate and phenolic acid composition. J Cereal Sci. 2007;45:88–96. [Google Scholar]
- Bartolome B., Faulds C.B., Kroon P.A., Waldron K., Gilbert H.J., Hazlewood G., et al. An Aspergillus niger esterase (ferulic acid esterase III) and a recombinant Pseudomonas fluorescens subsp. cellulosa Esterase (Xy1D) release a 5-5' ferulic dehydrodimer (diferulic acid) from barley and wheat cell walls. Appl Environ Microbiol. 1997;63:208–212. doi: 10.1128/aem.63.1.208-212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit I., Danchin E.G., Bleichrodt R.J., de Vries R.P. Biotechnological applications and potential of fungal feruloyl esterases based on prevalence, classification and biochemical diversity. Biotechnol Lett. 2008;30:387–396. doi: 10.1007/s10529-007-9564-6. [DOI] [PubMed] [Google Scholar]
- Bergman E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–570. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- Béra-Maillet C., Devillard E., Cezette M., Jouany J.-P., Forano E. Xylanases and carboxymethylcellulases of the rumen protozoa Polyplastron multivesiculatum, Eudiplodinium maggii and Entodinium sp. FEMS Microbiol Lett. 2005;244:149–156. doi: 10.1016/j.femsle.2005.01.035. [DOI] [PubMed] [Google Scholar]
- Besle J.M., Cornu As, Jouany J.I. Roles of structural phenylpropanoids in forage cell wall digestion. J Sci Food Agric. 1994;64:171–180. [Google Scholar]
- Besle J.M., Jouany J.P., Cornu As. Transformation of structural phenylpropanoids during cell wall digestion. FEMS Microbiol Rev. 2010;16:33–42. [Google Scholar]
- Betts W.B., Dart R.K. Screening of fungi and bacteria for their ability to degrade insoluble, lignin-related aromatic compounds. Microbios. 1988;55:85–93. [Google Scholar]
- Bhatt V.D., Dande S.S., Patil N.V., Joshi C.G. Molecular analysis of the bacterial microbiome in the forestomach fluid from the dromedary camel (Camelus dromedarius) Mol Biol Rep. 2013;40:3363–3371. doi: 10.1007/s11033-012-2411-4. [DOI] [PubMed] [Google Scholar]
- Borneman W.S., Akin D.E., VanEseltine W.P. Effect of phenolic monomers on ruminal bacteria. Appl Environ Microbiol. 1986;52:1331–1339. doi: 10.1128/aem.52.6.1331-1339.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman W.S., Hartley R.D., Morrison W.H., Akin D.E., Ljungdahl L.G. Feruloyl and p-coumaroyl esterase from anaerobic fungi in relation to plant cell wall degradation. Appl Microbiol Biotechnol. 1990;33:345–351. [Google Scholar]
- Borneman W.S., Ljungdahl L.G., Hartley R.D., Akin D.E. Isolation and characterization of p-coumaroyl esterase from the anaerobic fungus Neocallimastix strain-Mc-2. Appl Environ Microbiol. 1991;57:2337–2344. doi: 10.1128/aem.57.8.2337-2344.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutigny A.-L., Atanasova-Pénichon V., Benet M., Barreau C., Richard-Forget F. Natural phenolic acids from wheat bran inhibit Fusarium culmorum trichothecene biosynthesis in vitro by repressing Tri gene expression. Eur J Plant Pathol. 2010;127:275–286. [Google Scholar]
- Buanafina M.M.D.O., Langdon T., Hauck B., Dalton S., Morris P. Expression of a fungal ferulic acid esterase increases cell wall digestibility of tall fescue (Festuca arundinacea) Plant Biotechnol J. 2008;6:264–270. doi: 10.1111/j.1467-7652.2007.00317.x. [DOI] [PubMed] [Google Scholar]
- Bunzel M., Ralph J., Brüning P., Steinhart H. Structural identification of dehydrotriferulic and dehydrotetraferulic acids isolated from insoluble maize bran fiber. J Agric Food Chem. 2006;54:6409–6418. doi: 10.1021/jf061196a. [DOI] [PubMed] [Google Scholar]
- Cao Y.C., Yang H.J., Zhang D.F. Enzymatic characteristics of crude feruloyl and acetyl esterases of rumen fungus Neocallimastix sp. YAK11 isolated from yak (Bos grunniens) J Anim Physiol Anim Nutr. 2013;97:363–373. doi: 10.1111/j.1439-0396.2012.01281.x. [DOI] [PubMed] [Google Scholar]
- Cai S., Li J., Hu F.Z., Zhang K., Luo Y., Janto B., et al. Cellulosilyticum ruminicola , a newly described rumen bacterium that possesses redundant fibrolytic-protein-encoding genes and degrades lignocellulose with multiple carbohydrate-borne fibrolytic enzymes. Appl Environ Microbiol. 2010;76:3818–3894. doi: 10.1128/AEM.03124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway T.R., Carr M.A., Edrington T.S., Anderson R.C., Nisbet D.J. Diet, Escherichia coli O157:H7, and cattle: a review after 10 years. Curr Issues Mol Biol. 2009;11:67–80. [PubMed] [Google Scholar]
- Cao B.B., Jin X., Yang H.J., Li S., Jiang L. Microbial release of ferulic and p-coumaric acids from forages and their digestibility in lactating cows fed total mixed rations with different forage combinations. J Sci Food Agric. 2016;96:650–655. doi: 10.1002/jsfa.7136. [DOI] [PubMed] [Google Scholar]
- Cao B.B., Wang R., Bo Y.K., Bai S., Yang H.J. In situ rumen digestibility of ester-linked ferulic and p-coumaric acids in crop stover or straws in comparison with alfalfa and Chinese wild ryegrass hays. Anim Feed Sci Technol. 2016;212:27–34. [Google Scholar]
- Cao B.B., Wang R., Yang H.J., Jiang L.S. In situ ruminal degradation of phenolic acid, cellulose and hemicellulose in crop brans and husks differing in ferulic and p-coumaric acid patterns. J Agric Sci. 2015;153:1312–1320. [Google Scholar]
- Casler M.D. Breeding forage crops for increased nutritional value. Adv Agron. 2001;71:51–57. [Google Scholar]
- Casler M.D., Jung H.-J.G. Selection and evaluation of smooth bromegrass clones with divergent lignin or etherified ferulic acid concentration. Crop Sci. 1999;39:1866–1873. [Google Scholar]
- Casler M.D., Jung H.-J.G. Relationships of fibre, lignin, and phenolics to in vitro fibre digestibility in three perennial grasses. Anim Feed Sci Technol. 2006;125:151–161. [Google Scholar]
- Castanares A., Wood T.M. Purification and characterization of a feruloyl/p-coumaroyl esterase from solid-state cultures of the aerobic fungus Penicillium pinophilum. Biochemical. 1992:275s. doi: 10.1042/bst020275s. [DOI] [PubMed] [Google Scholar]
- Chamkha M., Garcia J.L., Labat M. Metabolism of cinnamic acids by some Clostridiales and emendation of the descriptions of Clostridium aerotolerans, Clostridium celerecrescens and Clostridium xylanolyticum. Int J Syst Evol Microbiol. 2001;51:2105–2111. doi: 10.1099/00207713-51-6-2105. [DOI] [PubMed] [Google Scholar]
- Cheng F., Sheng J., Cai T., Jin J., Liu W., Lin Y., et al. A protease-insensitive feruloyl esterase from China Holstein cow rumen metagenomic library: expression, characterization, and utilization in ferulic acid release from wheat straw. J Agric Food Chem. 2012;60:2546–2553. doi: 10.1021/jf204556u. [DOI] [PubMed] [Google Scholar]
- Chesson A., Provan G.J., Russell W.R., Scobbie L., Richardson A.J., Stewart C. Hydroxycinnamic acids in the digestive tract of livestock and humans. J Sci Food Agric. 1999;79:373–378. [Google Scholar]
- Chesson A., Stewart C.S., Wallace R.J. Influence of plant phenolic acids on growth and cellulolytic activity of rumen bacteria. Appl Environ Microbiol. 1982;44:597–603. doi: 10.1128/aem.44.3.597-603.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepin V.F., Faulds C.B., Connerton I.F. Functional classification of the microbial feruloyl esterases. Appl Microbiol Biotechnol. 2004;6:647–652. doi: 10.1007/s00253-003-1476-3. [DOI] [PubMed] [Google Scholar]
- Curiel J.A., Rodriguez H., Landete J.M., Rivas B., Munoz R. Ability of Lactobacillus brevis strains to degrade food phenolic acids. Food Chem. 2010;120:225–229. [Google Scholar]
- Delmer D.P., Stone B.A. In: The biochemistry of plants. Preiss J., editor. Academic Press; San Diego: 1988. Biosynthesis of plant cell walls; pp. 373–380. [Google Scholar]
- Delneri D., Degrassi G., Rizzo R., Bruschi C.V. Degradation of trans ferulic and p-coumaric acid by Acinetobacter calcoaceticus DSM 586. Biochim Biophys Acta. 1995;1244:363–367. doi: 10.1016/0304-4165(95)00021-3. [DOI] [PubMed] [Google Scholar]
- Devillard E., Bera M.C., Flint H.J., Scott K.P., Newbold C.J., Wallace R.J. Characterization of xyn10b, a modular xylanase from the ruminal protozoan polyplastron multivesiculatum, with a family 22 carbohydrate-binding module that binds to cellulose. Biochem J. 2003;2:495–503. doi: 10.1042/BJ20021784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilokpimol A., Makela M.R., Aguilar-Pontes M.V., Benoit-Gelber I., Hilden K.S., de Vries R.P. Diversity of fungal feruloyl esterases: updated phylogenetic classification, properties, and industrial applications. Biotechnol Biofuels. 2016;9(1):1–18. doi: 10.1186/s13068-016-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Yu P. Relationship of physicochemical characteristics and hydrolyzed hydroxycinnamic acid profile of barley varieties and nutrient availability in ruminants. J Cereal Sci. 2011;53:178–187. [Google Scholar]
- Duncan S.H., Russell W.R., Quartieri A., Rossi M., Parkhill J., Walker A.W., et al. Wheat bran promotes enrichment within the human colonic microbiota of butyrate-producing bacteria that release ferulic acid. Environ Microbiol. 2016;18:2214–2225. doi: 10.1111/1462-2920.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconnier B., Lapierre C., Lesage-Meessen L., Yonnet G., Brunerie P., Colonna-Ceccaldi B., et al. Vanillin as a product of ferulic acid biotransformation by the white-rot fungus Pycnoporus cinnabarinus I-937: identification of metabolic pathways. J Biotechnol. 1994;37:123–132. [Google Scholar]
- Goldstone D.C., Villas-Bôas S.G., Till M., Kelly W.J., Arcus V.L. Structural and functional characterization of a promiscuous feruloyl esterase (Est1E) from the rumen bacterium Butyrivibrio proteoclasticus. Proteins: Struct Funct Bioinf. 2010;78:1457–1469. doi: 10.1002/prot.22662. [DOI] [PubMed] [Google Scholar]
- Gopalan N., Rodríguez-Duran L.V., Saucedo-Castaneda G., Nampoothiri K.M. Review on technological and scientific aspects of feruloyl esterases: a versatile enzyme for biorefining of biomass. Bioresour Technol. 2015;193:534–544. doi: 10.1016/j.biortech.2015.06.117. [DOI] [PubMed] [Google Scholar]
- Grabber J.H., Hatfield R.D., Ralph J., Zon J., Amrhein N. Ferulate cross-linking in cell walls isolated from maize cell suspensions. Phytochemistry. 1995;40:1077–1082. [Google Scholar]
- Grabber J.H., Mertens D.R., Kim H., Funk C., Lu F., Ralph J. Cell wall fermentation kinetics are impacted more by lignin content and ferulate cross-linking than by lignin composition. J Sci Food Agric. 2009;89:122–129. [Google Scholar]
- Grbić-Galić D. O-demethylation, dehydroxylation, ring-reduction and cleavage of aromatic substrates by enterobacteriaceae under anaerobic conditions. J Appl Microbiol. 1986;61:491–497. doi: 10.1111/j.1365-2672.1986.tb01721.x. [DOI] [PubMed] [Google Scholar]
- Grbić-Galić D., Pat-Polasko L.L. Enterobacter cloacae DG-6: a strain that transforms methoxylated aromatics under aerobic and anaerobic conditions. Curr Microbiol. 1985;12:321–324. [Google Scholar]
- Gu H., Zhang J., Bao J. High tolerance and physiological mechanism of Zymomonas mobilis to phenolic inhibitors in ethanol fermentation of corncob residue. Biotechnol Bioeng. 2015;112:1770–1782. doi: 10.1002/bit.25603. [DOI] [PubMed] [Google Scholar]
- Hartley R.D. p-Coumaric and ferulic acid components of cell walls of ryegrass and their relationships with lignin and digestibility. J Sci Food Agric. 1972;23:1347–1354. [Google Scholar]
- Hartley R.D., Akin D.E. Effect of forage cell wall phenolic acids and derivatives on rumen microflora. J Sci Food Agric. 1989;49:405–411. [Google Scholar]
- Hatfield R.D., Rancour D.M., Marita J.M. Grass cell walls: a story of cross-linking. Front Plant Sci. 2017;7:2056. doi: 10.3389/fpls.2016.02056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde S., Kavitha S., Varadaraj M.C., Muralikrishna G. Degradation of cereal bran polysaccharide-phenolic acid complexes by Aspergillus niger CFR 1105. Food Chem. 2006;96:14–19. [Google Scholar]
- Heider J., Fuchs G. Microbial anaerobic aromatic metabolism. Anaerobe. 1997;3:1–22. doi: 10.1006/anae.1997.0073. [DOI] [PubMed] [Google Scholar]
- Hu H., Li L., Ding S. An organic solvent-tolerant phenolic acid decarboxylase from Bacillus licheniformis for the efficient bioconversion of hydroxycinnamic acids to vinyl phenol derivatives. Appl Microbiol Biotechnol. 2015;99:5071–5081. doi: 10.1007/s00253-014-6313-3. [DOI] [PubMed] [Google Scholar]
- Hui C.L., Jenner A.M., Low C.S., Lee Y.K. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res Microbiol. 2006;157:876–884. doi: 10.1016/j.resmic.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Hungate R.E., Stack R.J. Phenylpropanoic acid: growth factor for Ruminococcus albus. Appl Environ Microbiol. 1982;44:79–83. doi: 10.1128/aem.44.1.79-83.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iiyama K., Bach T., Lam T. Structural characteristics of cell walls of forage grasses--their nutritional evaluation for ruminants--a review. Asian-Australas J Anim Sci. 2001;14:862–879. [Google Scholar]
- Ishii T. Structure and functions of feruloylated polysaccharides. Plant Sci. 1997;127:111–127. [Google Scholar]
- Jugal K.G., Sven G.H., John A.B., Eriksson K.-E. Metabolism of trans-ferulic acid. Arch Microbiol. 1981:349–354. [Google Scholar]
- Jung H.-J.G. Maize stem tissues: ferulate deposition in developing internode cell walls. Phytochemistry. 2003;63:543–549. doi: 10.1016/s0031-9422(03)00221-8. [DOI] [PubMed] [Google Scholar]
- Jung H-oG., Vogel K.P. Lignification of switchgrass (Panicum virgatum) and big bluestem (Andropogon gerardii) plant parts during maturation and its effect on fibre degradability. J Sci Food Agric. 1992;59(2):169–176. [Google Scholar]
- Jung H.G., Allen M.S. Characteristics of plant cell walls affecting intake and digestibility of forages by ruminants. J Anim Sci. 1995;73:2774–2790. doi: 10.2527/1995.7392774x. [DOI] [PubMed] [Google Scholar]
- Jung H.G., Casler M.D. Maize stem tissues: impact of development on cell wall degradability. Crop Sci. 2006;46:1801–1809. [Google Scholar]
- Jung H.G., Mertens D.R., Phillips R.L. Effect of reduced ferulate-mediated lignin/arabinoxylan cross-linking in corn silage on feed intake, digestibility, and milk production. J Dairy Sci. 2011;94:5124–5137. doi: 10.3168/jds.2011-4495. [DOI] [PubMed] [Google Scholar]
- Jung H.G., Morrison T.A., Buxton D.R. Degradability of cell-wall polysaccharides in maize internodes during stalk development. Crop Sci. 1998;38:1047–1051. [Google Scholar]
- Jung H.J.G., Buxtono D.R. Forage quality variation among maize inbreds: relationships of cell-wall composition and in vitro degradability for stem internodes. J Sci Food Agric. 1994;66:313–322. [Google Scholar]
- Kern S.M., Bennett R.N., Needs P.W., Mellon F.A., Kroon P.A., Garcia-Conesa M.T. Characterization of metabolites of hydroxycinnamates in the in vitro model of human small intestinal epithelium caco-2 cells. J Agric Food Chem. 2003;51:7884–7891. doi: 10.1021/jf030470n. [DOI] [PubMed] [Google Scholar]
- Kothari R.K., Nathani N.M., Mootapally C., Rank J.K., Gosai H.B., Dave B.P., et al. Comprehensive exploration of the rumen microbial ecosystem with advancements in metagenomics. Metagenomics. 2018:215–229. [Google Scholar]
- Kühnel S., Pouvreau L., Appeldoorn M.M., Hinz S., Schols H.A., Gruppen H. The ferulic acid esterases of Chrysosporium lucknowense C1: purification, characterization and their potential application in biorefinery. Enzyme Microb Technol. 2012;50:77–85. doi: 10.1016/j.enzmictec.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Li J., Cai S., Luo Y., Dong X. Three feruloyl esterases in Cellulosilyticum ruminicola H1 act synergistically to hydrolyze esterified polysaccharides. Appl Environ Microbiol. 2011;77:6141–6147. doi: 10.1128/AEM.00657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Plant cell wall chemistry: implications for ruminant utilisation. J Appl Anim Res. 2021;9:31–36. [Google Scholar]
- Li X., Yang J., Li X., Gu W., Huang J., Zhang K.-Q. The metabolism of ferulic acid via 4-vinylguaiacol to vanillin by Enterobacter sp. Px6-4 isolated from vanilla root. Process Biochem. 2008;43:1132–1137. [Google Scholar]
- Mackenzie C.R., Bilous D., Schneider H., Johnson K.G. Induction of cellulolytic and xylanolytic enzyme systems in Streptomyces spp. Appl Environ Microbiol. 1987;53:2835–2839. doi: 10.1128/aem.53.12.2835-2839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandebvu P., West J.W., Hill G.M., Gates R.N., Hatfield R.D., Mullinix B.G., et al. Comparison of Tifton 85 and Coastal bermudagrasses for yield, nutrient traits, intake, and digestion by growing beef steers. J Anim Sci. 1999;77:1572–1586. doi: 10.2527/1999.7761572x. [DOI] [PubMed] [Google Scholar]
- Martin J.P., Haider K. Decomposition of specifically carbon-14-labeled ferulic acid: free and linked into model humic acid-type polymers1. Soil Sci Soc Am J. 1976;3:377–380. [Google Scholar]
- Mathew S., Abraham T.E. Ferulic acid: an antioxidant found naturally in plant cell walls and feruloyl esterases involved in its release and their applications. Crit Rev Biotechnol. 2004;24:59. doi: 10.1080/07388550490491467. [DOI] [PubMed] [Google Scholar]
- Mathew S., Abraham T.E. Bioconversions of ferulic acid, an hydroxycinnamic acid. Crit Rev Microbiol. 2006;32(3):115–125. doi: 10.1080/10408410600709628. [DOI] [PubMed] [Google Scholar]
- Mavinkurve S., Nazareth S. Degradation of ferulic acid via 4-vinylguaiacol by Fusarium solani (Mart.) Sacc. Can J Microbiol. 1986;32:494–497. [Google Scholar]
- McKinnon J.J., Christensen D.A. Hydroxycinnamic acids and ferulic acid esterase in relation to biodegradation of complex plant cell walls. Can J Anim Sci. 2005;85(3):255–267. [Google Scholar]
- McNeill D.M., Makkar H.P., Beever D. 2013. Forages for ruminants, cereals for human food and fuel. Optimization of feed use efficiency in ruminant production systems; pp. 15–32. [Google Scholar]
- Menke K.H., Raab L., Salewski A., Steingass H., Fritz D., Schneider W. The estimation of the digestibility and metabolizable energy content of ruminant feedingstuffs from the gas production when they are incubated with rumen liquor in vitro. J Agric Sci. 1979;93:217–222. [Google Scholar]
- Micard V., Landazuri T., Surget A., Moukha S., Labat M., Rouau X. Demethylation of ferulic acid and feruloyl-arabinoxylan by microbial cell extracts. LWT--Food Sci Technol. 2002;35:272–276. [Google Scholar]
- Mishra S., Sachan A., Vidyarthi A.S., Sachan S.G. Transformation of ferulic acid to 4-vinyl guaiacol as a major metabolite: a microbial approach. Rev Environ Sci Bio/Technol. 2014;13:377–385. [Google Scholar]
- Mogodiniyai Kasmaei K., Sundh J. Identification of novel putative bacterial feruloyl esterases from anaerobic ecosystems by use of whole-genome shotgun metagenomics and genome binning. Front Microbiol. 2019;10:2673. doi: 10.3389/fmicb.2019.02673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris P., Dalton S., Langdon T., Hauck B., Buanafina M.M.O.D. Expression of a fungal ferulic acid esterase in suspension cultures of tall fescue (Festuca arundinacea) decreases cell wall feruloylation and increases rates of cell wall digestion. Plant Cell Tissue Organ Cult. 2017;129:181–193. doi: 10.1007/s11240-017-1168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muheim A., Lerch K. Towards a high-yield bioconversion of ferulic acid to vanillin. Appl Microbiol Biotechnol. 1999;51:456–461. [Google Scholar]
- Nieter A., Kelle S., Linke D., Berger R.G. A p-coumaroyl esterase from Rhizoctonia solani with a pronounced chlorogenic acid esterase activity. N Biotechnol. 2017;37:153. doi: 10.1016/j.nbt.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Ohmiya K., Takeuchi M., Chen W., Shimizu S., Kawakami H. Anaerobic reduction of ferulic acid to dihydroferulic acid by Wolinella succinogenes from cow rumen. Appl Microbiol Biotechnol. 1986;23:274–279. [Google Scholar]
- Oliveira D.M., Mota T.R., Salatta FbV., de Almeida G.H.G., Olher V.G.A., Oliveira M.A.S., et al. Feruloyl esterase activity and its role in regulating the feruloylation of maize cell walls. Plant Physiol Biochem. 2020;156:49–54. doi: 10.1016/j.plaphy.2020.08.046. [DOI] [PubMed] [Google Scholar]
- Orpin C.G., Letcher A.J. Some factors controlling the attachment of the rumen holotrich protozoa Isotricha intestinalis and I. prostoma to plant particles in vitro. J Gen Microbiol. 1978;106:33–40. doi: 10.1099/00221287-106-1-33. [DOI] [PubMed] [Google Scholar]
- Overhage J., Priefert H., Steinbuchel A. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl Environ Microbiol. 1999;65:4837–4847. doi: 10.1128/aem.65.11.4837-4847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S.S., Kamra D.N., Sastry V.R., Sahu N.P., Kumar A. Effect of phenolic monomers on biomass and hydrolytic enzyme activities of an anaerobic fungus isolated from wild nil gai (Baselophus tragocamelus) Lett Appl Microbiol. 2003;36:377–381. doi: 10.1046/j.1472-765x.2003.01331.x. [DOI] [PubMed] [Google Scholar]
- Plaggenborg R., Steinbüchel A., Priefert H. The coenzyme A-dependent, non-β-oxidation pathway and not direct deacetylation is the major route for ferulic acid degradation in delftia acidovorans. FEMS Microbiol Lett. 2001;205:9–16. doi: 10.1111/j.1574-6968.2001.tb10918.x. [DOI] [PubMed] [Google Scholar]
- Pedersen M.B., Bunzel M., SchFer J., Knudsen K., SRensen J.F., Yu S., et al. Ferulic acid dehydrodimer and dehydrotrimer profiles of distiller's dried grains with solubles from different cereal species. J Agric Food Chem. 2015;63(7):2006–2012. doi: 10.1021/jf505150g. [DOI] [PubMed] [Google Scholar]
- Poquet L., Clifford M.N., Williamson G. Investigation of the metabolic fate of dihydrocaffeic acid. Biochem Pharmacol. 2008;75:1218–1229. doi: 10.1016/j.bcp.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Qi M., Wang P., Selinger L.B., Yanke L.J., Forster R.J., McAllister T.A. Isolation and characterization of a ferulic acid esterase (Fae1A) from the rumen fungus Anaeromyces mucronatus. J Appl Microbiol. 2011;110:1341–1350. doi: 10.1111/j.1365-2672.2011.04990.x. [DOI] [PubMed] [Google Scholar]
- Quin J.I., Wath J., Myburgh S. Studies on the alimentary tract of Merino sheep in South Africa. IV. Description of experimental technique. Onderstepoort J Vet Sci. 1941;17:61–88. [Google Scholar]
- Raffrenato E., Fievisohn R., Cotanch K.W., Grant R.J., Chase L.E., Van Amburgh M.E. Effect of lignin linkages with other plant cell wall components on in vitro and in vivo neutral detergent fiber digestibility and rate of digestion of grass forages. J Dairy Sci. 2017;100:8119–8131. doi: 10.3168/jds.2016-12364. [DOI] [PubMed] [Google Scholar]
- Ralph J., Grabber J.H., Hatfield R.D. Lignin-ferulate cross-links in grasses: active incorporation of ferulate polysaccharide esters into ryegrass lignins. Carbohydr Res. 1995;275:167–178. [Google Scholar]
- Ralph J., Quideau S., Grabber J.H., Hatfield R.D. Identification and synthesis of new ferulic acid dehydrodimers present in grass cell walls. J Chem Soc. 1994;1(23):3485–3498. [Google Scholar]
- Rezaeian M., Beakes G.W., Parker D.S. Distribution and estimation of anaerobic zoosporic fungi along the digestive tracts of sheep. Mycol Res. 2004;108:1227–1233. doi: 10.1017/s0953756204000929. [DOI] [PubMed] [Google Scholar]
- Rodrigues M.A.M., Guedes C.M., Cone J.W., van Gelder A.H., Ferreira L.M.M., Sequeira C.A. Effects of phenolic acid structures on meadow hay digestibility. Anim Feed Sci Technol. 2007;136:297–311. [Google Scholar]
- Rodríguez H., Angulo I., Rivas B., Campillo N., Páez J., Munoz R., et al. p-Coumaric acid decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism. Proteins: Struct Funct Bioinf. 2010;78:1662–1666. doi: 10.1002/prot.22684. [DOI] [PubMed] [Google Scholar]
- Sauracalixto F. Dietary fiber as a carrier of dietary antioxidants: an essential physiological function. J Agric Food Chem. 2011;59:43–49. doi: 10.1021/jf1036596. [DOI] [PubMed] [Google Scholar]
- Schmidt C.G., Gonçalves L.M., Prietto L., Hackbart H.S., Furlong E.B. Antioxidant activity and enzyme inhibition of phenolic acids from fermented rice bran with fungus Rizhopus oryzae. Food Chem. 2014;146:371–377. doi: 10.1016/j.foodchem.2013.09.101. [DOI] [PubMed] [Google Scholar]
- Seigler D.S. 1998. Shikimic acid pathway; pp. 94–95. [Google Scholar]
- Sheng X., Lind M.E.S., Himo F., Stockholms U., Naturvetenskapliga F., Institutionen FrOK. Theoretical study of the reaction mechanism of phenolic acid decarboxylase. FEBS J. 2015;282:4703–4713. doi: 10.1111/febs.13525. [DOI] [PubMed] [Google Scholar]
- Shin H.D., McClendon S., Le T., Taylor F., Chen R.R. A complete enzymatic recovery of ferulic acid from corn residues with extracellular enzymes from Neosartorya spinosa NRRL185. Biotechnol Bioeng. 2006;95:1108–1115. doi: 10.1002/bit.21056. [DOI] [PubMed] [Google Scholar]
- Singh H.P., Batish D.R., Kohli R.K. Allelopathy in agroecosystems. J Crop Prod. 2001:1–11. [Google Scholar]
- Sirohi S.K., Choudhury P.K., Dagar S.S., Puniya A.K., Singh D. Isolation, characterization and fibre degradation potential of anaerobic rumen fungi from cattle. Ann Microbiol. 2013;63:1187–1194. [Google Scholar]
- Soberon M.A., Cherney D.J.R., Cherney J.H. Free ferulic acid uptake in ram lambs. J Anim Sci. 2012;90:1885–1891. doi: 10.2527/jas.2011-4356. [DOI] [PubMed] [Google Scholar]
- Stack R.J., Hungate R.E. Effect of 3-phenylpropanoic acid on capsule and cellulases of Ruminococcus albus 8. Appl Environ Microbiol. 1984;48:218–223. doi: 10.1128/aem.48.1.218-223.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C.S., Flint H.J., Bryant M.P. In: Hobson P.N., Stewart C.S., editors. Blackie Academic & Professional; 1988. The rumen bacteria; pp. 10–72. (Rumen Microbial Ecosystem). [Google Scholar]
- Tinikul R., Chenprakhon P., Maenpuen S., Chaiyen P. Biotransformation of plant-derived phenolic acids. Biotechnol J. 2018;13(6):1700632. doi: 10.1002/biot.201700632. [DOI] [PubMed] [Google Scholar]
- Toms A., Wood J.M. The degradation of trans-ferulic acid by Pseudomonas acidovorans. Biochemistry. 1970;9:337–343. doi: 10.1021/bi00804a021. [DOI] [PubMed] [Google Scholar]
- Topakas E., Paul C. Springer Netherlands; 2007. Microbial xylanolytic carbohydrate esterases; pp. 83–87. [Google Scholar]
- Tripathi U., Rao S.R., Ravishankar G.A. Biotransformation of phenylpropanoid compounds to vanilla flavor metabolites in cultures of Haematococcus pluvialis. Process Biochem. 2002;38:419–426. [Google Scholar]
- Uraji M., Tamura H., Mizohata E., Arima J., Wan K., Ogawa Ki, et al. Loop of Streptomyces feruloyl esterase plays an important role in the enzyme's catalyzing the release of ferulic acid from biomass. Appl Environ Microbiol. 2018;84(3):e02300–e02317. doi: 10.1128/AEM.02300-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardakou M., Nueno Palop C., Gasson M., Narbad A., Christakopoulos P. In vitro three-stage continuous fermentation of wheat arabinoxylan fractions and induction of hydrolase activity by the gut microflora. Int J Biol Macromol. 2007;41:584–589. doi: 10.1016/j.ijbiomac.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Varel V.H., Jung H.J. Influence of forage phenolics on ruminal fibrolytic bacteria and in vitro fiber degradation. Appl Environ Microbiol. 1986;52:275–280. doi: 10.1128/aem.52.2.275-280.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai A., Costa T.H., Fa Ulds C.B., Milagres A.M., Siika-Aho M., Ferraz A. Effects of enzymatic removal of plant cell wall acylation (acetylation, p-coumaroylation, and feruloylation) on accessibility of cellulose and xylan in natural (non-pretreated) sugar cane fractions. Biotechnol Biofuels. 2014;7 doi: 10.1186/s13068-014-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Geng X., Egashira Y., Sanada H. Release of ferulic acid from wheat bran by an inducible feruloyl esterase from an intestinal bacterium Lactobacillus acidophilus. Food Sci Technol Res. 2005;11:241–247. [Google Scholar]
- Wang R., Yang H., Yang X., Cao B. Four phenolic acids determined by an improved hplc method with a programmed ultraviolet wavelength detection and their relationships with lignin content in 13 agricultural residue feeds. J Sci Food Agric. 2012;93(1):53–60. doi: 10.1002/jsfa.5727. [DOI] [PubMed] [Google Scholar]
- Wen G., Jinkui Y., Zhiyong L., Lianming L., Yuna S., Jingwen H., et al. Structural basis of enzymatic activity for the ferulic acid decarboxylase (FADase) from Enterobacter sp. Px6-4. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.G. The selectivity of carbohydrate assimilation by the anaerobic rumen Ciliate dasytricha ruminantium. J Appl Microbiol. 1979;47(3):511–520. doi: 10.1111/j.1365-2672.1979.tb01212.x. [DOI] [PubMed] [Google Scholar]
- Williams A.G., Coleman G.S. 1997. The rumen protozoa; pp. 73–79. (Rumen Microbial Ecosystem). [Google Scholar]
- Wong D.W.S. Feruloyl esterase a key enzyme in biomass degradation. Appl Biochem Biotechnol Part A. 2006;2:87–92. doi: 10.1385/abab:133:2:87. [DOI] [PubMed] [Google Scholar]
- Wong D.W.S., Chan V.J., Liao H. Metagenomic discovery of feruloyl esterases from rumen microflora. Appl Microbiol Biotechnol. 2019 doi: 10.1007/s00253-019-10102-y. [DOI] [PubMed] [Google Scholar]
- Wong D.W.S., Chan V.J., Liao H., Zidwick M.J. Cloning of a novel feruloyl esterase gene from rumen microbial metagenome and enzyme characterization in synergism with endoxylanases. J Ind Microbiol Biotechnol. 2013;40:287–295. doi: 10.1007/s10295-013-1234-1. [DOI] [PubMed] [Google Scholar]
- Wubah D.A., Kim D.S.H. Chemoattraction of anaerobic ruminai fungi zoospores to selected phenolic acids. Microbiol Res. 1996;151(3):257–262. doi: 10.1016/s0944-5013(96)80022-x. [DOI] [PubMed] [Google Scholar]
- Yang H.J., Yue Q., Cao Y.C., Zhang D.F., Wang J.Q. Effects of crude feruloyl and acetyl esterase solutions of Neocallimastix sp. YQ1 and Anaeromyces sp. YQ3 isolated from Holstein steers on hydrolysis of Chinese wildrye grass hay, wheat bran, maize bran, wheat straw and corn stalks. Anim Feed Sci Technol. 2009;154:218–227. [Google Scholar]
- Yu P., Mckinnon J.J., Maenz D.D., Racz V.J., Christensen D.A. The interactive effects of enriched sources of Aspergillus ferulic acid esterase and Trichoderma xylanase on the quantitative release of hydroxycinnamic acids from oat hulls. Can J Anim Sci. 2002;82:251–257. [Google Scholar]
- Zuhainis Saad W., Abdullah N., Alimon A.R., Ho Y.W. Effects of phenolic monomers on the enzymes activities and volatile fatty acids production of Neocallimastix frontalis B9. Anaerobe. 2008;14:118–122. doi: 10.1016/j.anaerobe.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Zuhainis S.W., Abdullah N., Alimon A.R., Ho Y.W. Effects of phenolic monomers on the degradation of 14C-cellulose by rumen fungi. Res J Microbiol. 2007;2:918–925. [Google Scholar]