Abstract
Phosphoinositide signaling lipids are crucial for eukaryotes and regulate many aspects of cell function. These signaling molecules are difficult to study because they are extremely low abundance. Here, we focus on two of the lowest abundance phosphoinositides, PI(3,5)P2 and PI(5)P, which play critical roles in cellular homeostasis, membrane trafficking and transcription. Their levels are tightly regulated by a protein complex that includes PIKfyve, Fig4 and Vac14. Importantly, mutations in this complex that decrease PI(3,5)P2 and PI(5)P are linked to human diseases, especially those of the nervous system. Paradoxically, PIKfyve inhibitors which decrease PI(3,5)P2 and PI(5)P, are currently being tested for some neurodegenerative diseases, as well as other diverse diseases including some cancers, and as a treatment for SARS-CoV2 infection. A more comprehensive picture of the pathways that are regulated by PIKfyve will be critical to understand the roles of PI(3,5)P2 and PI(5)P in normal human physiology and in disease.
Keywords: PIKfyve, Fig4, Vac14, phosphoinositide, endosomes, endomembrane trafficking
Abbreviations: Phosphatidylinositol 3-kinase C2α -, PI3KC2α; Myotubularin, MTM1; Phosphatidylinositol 4,5-bisphosphate, PI(4,5)P2
Introduction
Phosphoinositide lipids (PPIs) are signaling molecules that regulate critical cell processes. They are produced from phosphatidylinositol by phosphorylation of the hydroxyl groups in any combination at positions three, four or five of the inositol ring. A series of specific lipid kinases and phosphatases are responsible for the generation and turnover of the resultant seven PPI species. These enzymes are highly regulated both spatially and temporally and are generated and turned over at specific membrane subdomains. This establishes the distribution of PPIs, which act by recruiting a set of cognate effector proteins. Although a longstanding view in the field was that PPIs define organelle identity by restricting each species to one or few compartments, a growing body of evidence indicates that the localization of each PPI is more promiscuous than initially thought (reviewed in Choy et al. [1]).
Importantly, the synthesis of distinct PPIs are coordinated into cascades which control complicated multi-steps pathways. An example of such a cascade is how conversion of PPI lipids coordinates clathrin-mediated endocytosis. Plasma membrane-localized PI(4,5)P2 recruits proteins involved in the early steps of endocytosis including AP-2, dynamin and endophilin, as well as the lipid kinase PI3KC2α. Moreover, endophilin promotes the recruitment of the lipid phosphatase synaptojanin, which converts PI(4,5)P2 into PI(4)P. PI3KC2α subsequently transforms PI(4)P into PI(3,4)P2, which recruits proteins that act later in endocytosis (reviewed in the study by Posor et al. [2]). Another example is the ordered conversion of PI(3)P into PI(4)P at endosomes to promote protein recycling to the plasma membrane. This mechanism involves the formation of a complex composed of the lipid phosphatase MTM1 and the lipid kinase PI4K2A. MTM1 removes the phosphate from PI(3)P, while in parallel, a phosphate group is added to position 4 by PI4K2A, generating PI(4)P. PI(4)P, together with MTM1 and PI4K2A, promote the recruitment of the exocyst, a protein complex that is essential for the fusion of recycling endosomes with the plasma membrane [3].
The lipid kinase PIKfyve
Studies to date show that PIKfyve (Fab1 in yeast), which phosphorylates position 5 of PI(3)P, is the sole source of PI(3,5)P2. In addition, PIKfyve is responsible for virtually all of the PI(5)P pool. PI(5)P is generated from PI(3,5)P2 via 3-phosphatase activity, likely catalyzed by the myotubularin family of phosphatases [4]. There is also evidence that PIKfyve may generate PI(5)P directly from phosphatidylinositol [5]. In either scenario, knock-down or inhibition of PIKfyve results in the loss of both PI(3,5)P2 and PI(5)P. PI(3,5)P2 is particularly challenging to study as compared to other PPIs, due to its low abundance, and lack of a highly specific bioprobe. Indeed, PI(3,5)P2 accounts for approximately 0.1% and 0.04% of total PPIs in yeast and mouse embryonic fibroblasts (MEFs), respectively. This makes PI(3,5)P2 17-fold and 125-fold less abundant than PI(4,5)P2 in yeast and MEFs, respectively. PI(5)P is undetectable in yeast, and is present at levels similar to PI(3)P (approximately 0.5% of total PI) in MEFs, which means that PI(5)P is 16-fold lower than PI(4,5)P2 [6]. Note that PI(4,5)P2 is much more extensively characterized than either PI(3,5)P2 or PI(5)P.
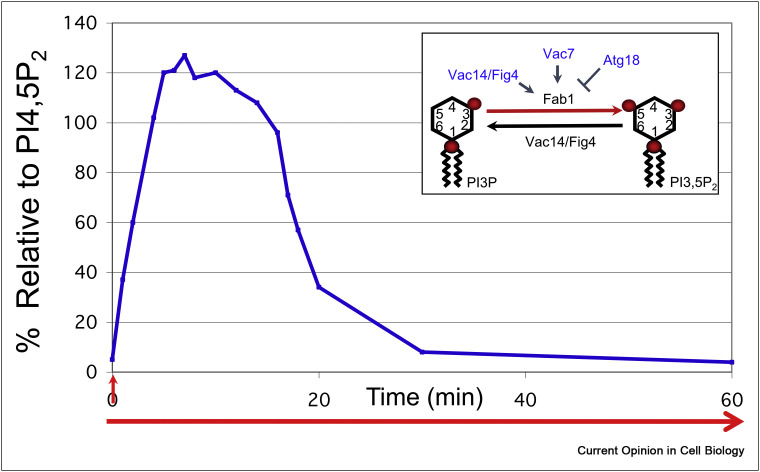
PI(3,5)P2 levels dynamically and transiently change in response to specific stimuli. The best characterized example is the transient elevation of PI(3,5)P2 that occurs during hyperosmotic shock in yeast. Within 5 min of exposure to hyperosmotic media, there is a 20-fold increase and then a rapid return to the normal levels within 30 min (Figure 1 ) [7]. In mammalian cells, physiological signals such as insulin and growth factors also cause an acute elevation of PI(3,5)P2 levels [4,8,9], which suggests that this lipid plays key roles in cellular homeostasis and in adaptation.
Figure 1.
PI(3,5)P2 levels acutely and transiently rise and fall in response to hyperosmotic stress. Yeast grown in normal media were transferred to hyperosmotic media (containing 0.9 M NaCl) at time zero (perpendicular arrow) and left in 0.9 M salt for the rest of the experiment (parallel arrow). Although there were no further perturbations to the external environment, the levels of PI(3,5)P2 (relative to PI(4,5)P2) rapidly increase, plateau and rapidly decrease. These findings indicate that the levels of PI(3,5)P2 are tightly controlled. Within 5 min, PI(3,5)P2 levels rise over 20-fold, plateau for 10 min, then rapidly return to basal levels. Multiple layers of regulations within the Fab1 complex, which includes both the lipid kinase Fab1/PIKfyve and lipid phosphatase Fig4 provide tight control of PI(3,5)P2 synthesis and turnover. Data modified from [7].
The dynamic and rapid changes in PI(3,5)P2 levels following these stimuli suggest that PIKfyve is tightly regulated. Genetic studies in yeast and mammalian cells revealed that PIKfyve activity requires the formation of a complex that includes the scaffold protein Vac14 and the lipid phosphatase Fig4, which converts PI(3,5)P2 to PI(3)P [10,11]. Paradoxically, Fig4 is also required to produce PI(3,5)P2 and mutations in the Fig4 catalytic site result in impaired Fab1 activation in yeast. At least three different regions of Fig4 are critical for the formation of the yeast Fab1-Vac14-Fig4 complex: the Fig4 catalytic site, a conserved N-terminal surface, and the C-terminus. The presence of three contact sites in Fig4 raises the possibility that Fig4 experiences conformational changes which regulate the assembly and/or activity of the complex [12].
Key insights into the complexity of the regulation of PIKfyve came from the determination of the structure of the complex using negative-stain and cryoelectron microscopy [13]. Intriguingly, Vac14 pentamerizes into a star-shaped structure, which binds a single copy each of PIKfyve and Fig4. PIKfyve binds one leg of the Vac14 pentamer, and Fig4 binds the opposite side of that leg, as well as an adjacent leg. In this solved structure, Fig4 is oriented such that its catalytic site would contact the membrane, thereby providing Fig4 with access to its substrate, PI(3,5)P2. However, paradoxically, the PIKfyve catalytic site is rotated away from the membrane. Moreover, mammalian Fig4 has protein phosphatase activity and acts on PIKfyve to stimulate its lipid kinase activity [13]. Yet, in the solved structure, the Fig4 catalytic site is far from PIKfyve. Together, these observations strongly suggests that during PIKfyve activation, the complex would undergo a large structural rearrangement that would place the PIKfyve catalytic site on the membrane and also facilitate Fig4 access to PIKfyve. This potential rearrangement is likely regulated via post-translational modifications of proteins within the complex.
In addition to Fig4 acting as a protein phosphatase, PIKfyve exhibits protein kinase activity, and autophosphorylation inhibits its lipid kinase activity [13,14]. This suggests extensive cross-talk between the PIKfyve lipid kinase and Fig4 lipid phosphatase.
Similar mechanisms within the PIKfyve-Vac14-Fig4 complex are likely to occur in yeast. Mutations of residues of Vac14, Fig4, and PIKfyve/Fab1 that are conserved between yeast and mammals display similar phenotypes [10,12,15]. The mouse Vac14-Ingls point mutation results in neurodegeneration and perinatal lethality in mice. The mutation is in a leucine that resides in a relatively conserved region of Vac14. Mutation of the same leucine in yeast causes a loss of the interaction of Vac14 with Fab1, and Vac14 interaction with PIKfyve, in mammalian cells [10]. Moreover, the Charcot-Marie-Tooth Type 4J (CMT4J) mutation, Fig4-I > T also shows similar mechanistic defects as the corresponding yeast mutant, with this mutation impairing the interaction between both mammalian and yeast Vac14 and Fig4 [16]. Furthermore, the six known autophosphorylation sites are conserved among vertebrate PIKfyve, and three of these sites are also conserved in yeast.
In addition to PIKfyve/Fab1, Vac14 and Fig4, yeast express two additional regulatory proteins, Atg18 and Vac7. It remains unclear whether there are analogous proteins which perform these same functions in mammals. Atg18 is best known for its roles in autophagy, where it is required for autophagosome formation. In addition, Atg18 negatively regulates yeast Fab1 activity [17,18]. Mammals have four proteins that show homology with Atg18: WIPI-1, WIPI-2, WIPI-3/WDR45L, and WIPI-4/WDR45. These function in autophagy [19,20], however it is not clear whether these proteins regulate PI(3,5)P2 levels in mammalian cells (reviewed in Jin et al. [21]). Vac7 is a key positive regulator of Fab1 activity. Deletion of Vac7 in yeast results in nearly undetectable levels of PI(3,5)P2 and an enlarged vacuole [22]. TMEM106B has been proposed as a mammalian homolog for Vac7 based on bioinformatics methods [23]. It is currently unknown whether this protein regulates the PIKfyve complex in mammals.
Animal models of PIKfyve deficiency and human diseases associated with defects in the PIKfyve complex
Fab1 is essential in yeast. Fab1 deletion yeast strains were initially thought to survive (Yamamoto et al., 1995). However, acute removal of Fab1 revealed that Fab1 is essential. Suppressor mutations in Fab1 deletion mutants are rapidly acquired, which allow the cells to survive [24,25]. Yeast deletion strains show defects in cell growth, with the Fab1 deletion presenting the most severe phenotype (probably due to a complete loss of PI(3,5)P2), followed by Vac7, Vac14 and Fig4 [22,25,26].
In mice, PIKfyve is critical during early embryonic development. Global deletion of the pikfyve gene results in the death of embryos before they reach the 32–64 cell stage [27] or by e8.5 due to a failure of the visceral endoderm to use maternal nutrients [28]. It is not clear whether these divergent early times of death were due to differences in mouse strain background. A hypomorphic PIKfyve mouse model expressing approximately 10% of the normal levels of PIKfyve protein is viable, but exhibits extensive neurodegeneration and dies perinatally due to defects in multiple organs, including neural tissues, heart, lung, kidney, thymus, and spleen [4]. In addition, several PIKfyve knockout mice were developed using the Cre-Lox system which demonstrate essential roles of PIKfyve in specific tissues or cell types, including the intestine, muscles, oligodendrocytes and myeloid cells [28, 29, 30, 31]. Mouse models where either Fig4 or Vac14 are deleted exhibit extensive neurodegeneration and die perinatally [32,33]. The fact that the absence of PIKfyve results in much earlier lethality than the lack of Fig4 or Vac14 is likely due to the fact that loss of PIKfyve causes total deficiency of PI(3,5)P2 and a loss of over 80% of PI(5)P, whereas the absence Vac14 or Fig4 results in a ∼50%–70% decrease in PI(3,5)P2 and PI(5)P [4,32,33].
Defects in the PIKfyve complex are linked to human diseases, especially those of the nervous system [32,34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60], although the underlying molecular mechanisms are not clear. For example, mutations in Fig4 predicted to have a modest effect in the regulation of PI(3,5)2 are linked to Charcot-Marie-Tooth syndrome (CMT4J), a peripheral neuropathy, as well as some cases of amyotrophic lateral sclerosis (ALS) and primary lateral sclerosis (PLS). More extensive mutations in either Vac14 or Fig4 cause Yunis–Varon syndrome, which results in infant mortality and severe pathological effects on multiple tissues, including the brain. One case of neurodegeneration with iron brain accumulation was linked to mutation of Vac14. A list of the mutations in the PIKfyve-Vac14-Fig4 complex that have been linked to diseases is provided in Table 1 . In addition, the extensive vacuolization observed in the brains of mouse models when PIKfyve activity is decreased is reminiscent of prion disorders. Notably, it was recently discovered that prion infection results in lower levels of PIKfyve [61], which suggests that the pathology associated with prion disease is due to lowered PI(3,5)P2, and PI(5)P.
Table 1.
Human diseases associated with mutations in PIKfyve, Vac14 and Fig4.
| Disease | Affected gene | References |
|---|---|---|
| Charcot-Marie–Tooth type 4J (CMT4J) | Homozygous or heterozygous mutations leading to reduced Fig4 expression | [32,34,45,54, 55, 56] |
| Amyotrophic lateral sclerosis (ALS) | Mutations in Fig4 | [34,57, 58, 59] |
| Primary lateral sclerosis (PLS) | Mutations in Fig4 | [34,60] |
| Yunis–Varon syndrome | Homozygous null mutation in Fig4 | [35,36] |
| Central nervous system white matter disorders | Mutations in Fig4 | [37] |
| Syndrome with severe neurological and psychiatric symptoms | Homozygous point mutation in Fig4 | [38] |
| Yunis–Varon syndrome | Biallelic Vac14 variants | [39] |
| Childhood onset striato-nigral degeneration | Mutations in Vac14 | [40, 41, 42, 43, 44,46, 47, 48, 49] |
| Francois–Mouchetee Fleck corneal dystrophy | Mutations in PIKfyve | [50, 51, 52] |
| Congenital cataract | Mutation in PIKfyve | [53] |
While human mutations in Fig4 or Vac14 have been linked to severe neurological syndromes, the main human disease associated with mutations in the PIKfyve gene is Fleck corneal dystrophy, characterized by the appearance of white flecks in the cornea which generally do not cause vision impairment [51,52]. These frameshift or nonsense mutations, which are predicted to result in the loss of PIKfyve function, are heterozygous, and the patients each have a wild-type copy of PIKfyve. This likely explains how these patients do not show phenotypes beyond defects in their cornea. It is plausible that the expected 50% decrease in PIKfyve levels is well tolerated. Note that while mice that are heterozygous for the hypomorphic PIKfyve allele have only 50% of the normal levels of PIKfyve, they have normal levels of PI(3,5)P2 and 70% of normal levels of PI(5)P. They do not have any observable defects in any of their tissues [4].
Interest in PIKfyve has recently increased because apilimod, a selective PIKfyve inhibitor, has been tested as a treatment for several diseases both in cell and animal disease models. These include COVID-19, some neurodegenerative diseases and some types of cancer (see references in Huang et al. [62]). In addition to apilimod, other PIKfyve inhibitors have been developed. YM201636 [63], another commonly used PIKfyve inhibitor, may also inhibit Akt phosphorylation, suggesting that it may also be a PI3K inhibitor [64]; APY0201 [65] is more potent than apilimod, although there are not yet published reports on its impact on the seven PIP species, including PI(3,5)P2. WX8 [66], a more recently published inhibitor, may have less specificity than the other inhibitors. Importantly, apilimod is the most extensively characterized inhibitor and the only one that has been approved for use in clinical trials.
Apilimod, initially discovered as a small molecule inhibitor of IL-12 and IL-23 with an unknown target, was used in clinical trials of Crohn's disease and rheumatoid arthritis [67]. Although it was ineffective against these autoimmune disorders, it proved to be safe and well tolerated. PIKfyve plays a role in viral entry and infection of some types of viruses. Apilimod prevents the infection of cultured cells with Ebola, Marburg and SARS-CoV-2 [68,69]. Based on a large scale screen in mammalian cell lines [70], as well as several targeted studies [71, 72, 73], apilimod was proposed as a treatment for COVID-19 and is currently undergoing a phase II clinical trial.
Apilimod was also found to have antiproliferative effects against B-cell non-Hodgkin lymphoma [74], and is currently in phase II clinical trials. Moreover, a role for PIKfyve in tumor cell invasiveness has been described in mouse embryonic fibroblasts expressing the oncogenic fusion protein nucleophosmin–anaplastic lymphoma kinase [75] and in cancer cell lines of three different origins (lung, rhabdomyosarcoma, and osteosarcoma) [76]. In addition, PIKfyve inhibition mitigated several types of cancers in animal models (reviewed by Ikonomov et al. [77]) including metastatic liver cancer in a mouse model [78]. These findings raise the possibility of using PIKfyve inhibitors to prevent hematological cancers, as well as cancers that cause solid tumors and metastasis.
Paradoxically, PIKfyve inhibition has shown beneficial effects in several cellular and in vivo models of neurodegeneration. For example, a genome-wide CRISPR interference screen showed that PIKfyve inhibition reduced α-synuclein aggregation induced with both recombinant α-synuclein fibrils and fibrils isolated from patients [79]. PIKfyve inhibition also improved the survival of motor neurons derived from iPSC from patients with ALS [80]. In addition, apilimod treatment improved the deterioration of motor neurons with mutations in the ALS-related protein C9Orf72 in mouse models in vivo [81]. Furthermore, PIKfyve inhibition also reduced the formation of tau aggregates in neurons [82]. The potential of PIKfyve inhibition as a treatment for neurodegeneration is surprising given that decreased PIKfyve activity in mouse models causes a spongiform phenotype in the brain and that mutations in the PIKfyve complex are associated with neurodegenerative diseases. However, the fact that individuals with just one copy of the PIKfyve gene do not develop neurodegeneration, indicate that a partial decrease of PIKfyve activity is well tolerated. However, time and dose may need to be carefully adjusted to prevent the deleterious consequences of inhibiting PIKfyve.
Functions of PIKfyve
To understand the processes affected in diseases associated with PIKfyve, it is critical to identify the functions of PIKfyve. Although PIKfyve was initially thought to act solely on lysosomes, a growing body of evidence indicates that it plays several additional roles that are critical for cell function. Some of these pathways are directly regulated by PI(3,5)P2, as PI(3,5)P2 protein effectors have been identified. However, in other cases, the role of PIKfyve may be due to PI(3,5)P2, PI(5)P or both lipids. Due to the lack of suitable and/or sufficiently sensitive bioprobes, the localization of endogenous levels of PI(3,5)P2 and PI(5)P is inferred from the localization of PIKfyve (Figure 2 ). Although a bioprobe that specifically recognizes PI(3,5)P2, ML1N-2∗-GFP, has been described, it is important to note that in the presence of YM201636, ML1N-2∗-GFP signal was still observed on some vacuoles, raising the possibility that in the absence of PI(3,5)P2, the probe may recognize another epitope [83].
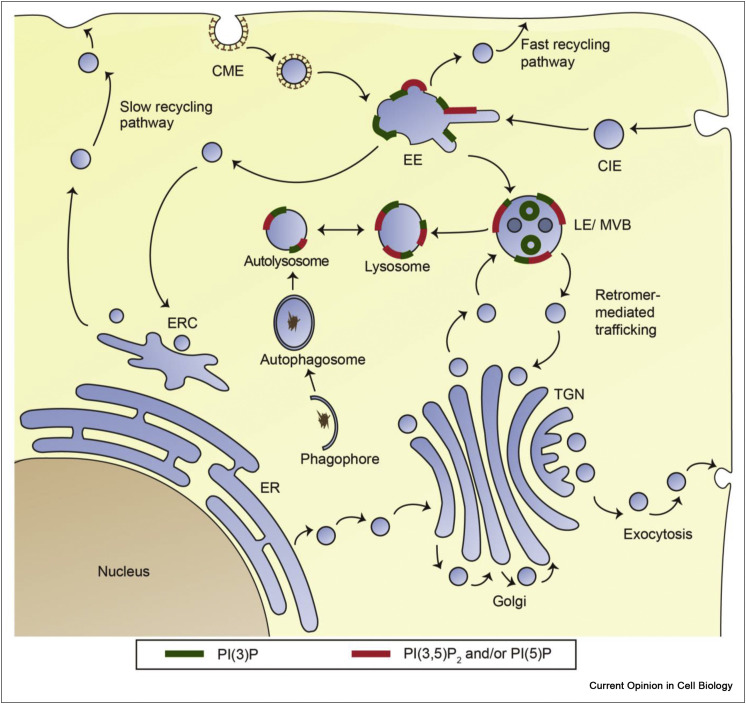
Figure 2.
Schematic of the cellular localization of the two lipids products downstream of PIKfyve activity, PI(3,5)P2 and PI(5)P, as well as the precursor PI(3)P. Due to the lack of suitable and/or sufficiently sensitive bioprobes, the localization of endogenous levels of PI(3,5)P2 and PI(5)P is inferred from the localization of PIKfyve. PI(3,5)P2 localizes on early endosomes, late endosomes and lysosomes. PI(5)P may also be present at all or some of these locations. CME: Clathrin-mediated endocytosis. CIE: Clathrin-independent endocytosis. EE: Early endosome. LE: Late endosome. MVB: Multivesicular body. EA: Endoplasmic reticulum. ERC: Early recycling compartment. TGN: Trans-Golgi network.
Roles at the lysosome
Inhibition or knockdown of PIKfyve results in the appearance of enlarged vacuoles in mammalian cells and an enlarged vacuole/lysosome in yeast [15,84, 85, 86]. These enlarged vacuoles are likely due to the combination of two different defects: alterations in ion homeostasis and pH within the endomembrane system, and alterations in membrane dynamics, particularly membrane fission and formation of tubules.
PIKfyve is critical to the maintenance of ion homeostasis in lysosomes. It is clear that these events are driven by PI(3,5)P2, which has been shown in atomic resolution structures to bind directly to key lysosomal ion channels, including the mucolipin TRP channel (TRPML1) and the Na+ two-pore channels, TPC1 and TPC2 [87]. Changes in ion homeostasis due to channel inactivation during PIKfyve inhibition, may also indirectly lead to swelling of the lysosome due to an accompanying influx of water. Similarly in yeast, PI(3,5)P2 was shown to be required for a vacuole/lysosome monovalent cation/proton antiporter [24].
Luminal pH is also closely tied to vacuolar function, as the acidic environment of lysosomes and vacuoles is critical for the activity of hydrolytic enzymes and to maintain a pH gradient that drives the transport of ions and metabolites. Notably, PIKfvye/Fab1 has a role in the regulation of the pH of the yeast vacuole and mammalian lysosome. Yeast mutants deficient in PI(3,5)P2 synthesis have less acidified vacuoles. This effect is mediated in part by interaction of PI(3,5)P2 with the vacuolar ATPase (v-ATPase), which is key for organelle acidification and cellular pH control (reviewed in [88]). Indeed, hyperosmotic shock generated an almost two-fold increase in V-ATPase activity, accompanied by increased levels of V1 subunits in wild-type but not Vac14 deficient yeasts [89]. Similarly, using a stably expressed genetically encoded ratiometric probe, it was found that treatment with a low dose of the PIKfyve inhibitor, apilimod for 3 h, resulted in an elevation of lysosomal pH in a mammalian cell line [90]. It is tempting to speculate that this may be due in part to PI(3,5)P2 regulation of mammalian v-ATPase.
Membrane trafficking defects in the endolysosomal system due to impaired PIKfyve activity likely also contribute to vacuole and lysosome enlargement. Indeed, PIKfyve activity is required to drive lysosome reformation from endolysosomes [91], which suggests that PIKfyve has a role in membrane fission. Moreover, the large vacuoles in PIKfyve inhibited cells failed to undergo fission, an effect that is partly mediated by TRPML1 and is critical to redistribute degraded cargo back to the endosomal network [92]. The block in lysosome fission during PIKfyve inhibition may contribute to lysosomal enlargement via an overall steady-state increase in lysosomal fusion [93]. The full mechanism for how PI(3,5)P2 plays a role in membrane fission is not known. However, WIPI-1, a known PI(3,5)P2 binding protein, with an independent role in autophagy, participates in fission of endosomal tubules [94]. In yeast, PI(3,5)P2 also plays a role in vacuole fusion [95].
Roles at endosomes
PIKfyve is necessary for the generation of Stage I melanosomes, which are derived from early endosomes [96]. Moreover, PIKfyve is required for retrograde traffic of proteins from endosomes to the trans-Golgi network (TGN). Knockdown or inhibition of PIKfyve impairs the trafficking of several proteins to the TGN, including the cation-independent mannose-6-phosphate receptor, sortilin, furin, and Shiga Toxin B, although the mechanistic basis of these defects remains unknown [97,98].
In addition, PIKfyve plays a role in endocytic recycling to the plasma membrane. Early evidence comes from studies at synapses, where decreased PIKfyve activity results in an increase in the surface levels of AMPA-type glutamate receptors (AMPARs). This is in part due to increased recycling from endosomes to the plasma membrane, as apilimod treatment for 10 min promoted the recycling of AMPARs, although an additional effect of endocytosis cannot be excluded [15,99].
Our recent studies gained mechanistic insight into the role of PIKfyve in endocytic recycling. We found that PIKfyve is a key regulator of the recently discovered Retriever pathway. PIKfyve inhibition causes a loss of Retriever and the CCC complex from endosomes, and mutation of the lipid binding site on COMMD1, a CCC subunit, impairs its endosomal localization and delays the recycling of several SNX17 cargoes including α5-integrin, β1-integrin and LRP1. We also found that PIKfyve coordinates with VPS34, the enzyme that generates PI(3)P from PIP, in a phosphoinositide cascade to promote the ordered assembly of SNX17, WASH, Retriever and the CCC complex [100].
Intriguingly, PIKfyve has a positive role in β1-integrin recycling but has a negative regulatory role in AMPAR localization to the plasma membrane. This could be due to the fact that AMPAR recycling is dependent on the SNX27-Retromer pathway, while β1-integrin depends on the SNX17-Retriever pathway. This would open the possibility that PIKfyve differentially regulates different recycling pathways that emerge from endosomes.
The role of PIKfyve in recycling from endosomes to the plasma membrane may also explain the reduction in steady-state levels of exogenously expressed FLAG-TGFβ-R2 at the cell surface, as well as the accumulation of the tight junction proteins Claudin-1 and Claudin-2 in intracellular compartments upon inhibition of PIKfyve activity [101,102]. Note that the recycling pathway utilized by these proteins are currently unknown.
While PIKfyve may separately regulate ion homeostasis and membrane dynamics, a recent study suggests that observed changes in membrane dynamics upon PIKfyve inhibition, might be solely accounted for by defects in ion flux [103]. This study proposes that two-pore channels, which are regulated by PI(3,5)P2, release sodium ions accompanied by water, and as the organelle loses volume, it crenelates, which recruits curvature sensing protein required for membrane tubulation and fission.
Regulation of transcription
Although the majority of studies on PIKfyve have focused on its roles in the endomembrane system, PIKfyve also plays roles in regulating the transcription of some genes. PI(3,5)P2 modulates transcription in yeast by promoting the assembly of the Tup1/Cyc8/Cti6 transcription complex on the yeast vacuole via direct interaction of Tup1 and Cti6 [104]. In mammalian cells, PIKfyve also plays roles in the regulation of transcription. Apilimod inhibits IL-12 expression by transcriptional upregulation of the repressor ATF3, although the molecular mechanism is unknown [105]. Moreover, PIKfyve is a key regulator of Transcription factor EB (TFEB), a master regulator of autophagy and lysosomal biogenesis, and inhibition of PIKfyve induces the nuclear translocation of TFEB [93]. PIKfyve acts by positively regulating mTORC1-dependent phosphorylation of Ser-211 on TFEB [106]. Upon PIKfyve inhibition, protein phosphatase 2A dephosphorylates TFEB Ser-211, which results in TFEB translocation into the nucleus. While positive regulation of mTORC1 by PIKfyve is required for TFEB phosphorylation, and retention on the lysosome, mTORC1-dependent phosphorylation of other downstream targets does not require PIKfyve activity. Importantly, PIKfyve inhibition results in TFEB translocation to the nucleus, and the upregulation of the TFEB targets tested. These findings support a role for PIKfyve in the regulation of transcription. Future studies will be necessary to determine the PIKfyve-dependent transcriptome, which may provide important insights into the roles of PIKfyve in cellular physiology [106].
Concluding remarks
Studies on PIKfyve have increased our knowledge of PPIs. That PIKfyve is present in multiple places within the cell supports the view that PPIs do not define organelle identity, and instead, the transient appearance of PPIs in specific compartments allows for the regulation and coordination of signaling and trafficking pathways. PPI formation is tightly controlled in space and time and is triggered by specific stimuli. A clear example is the acute elevation of PI(3,5)P2 upon hyperosmotic shock in yeast. Since PIKfyve is the only source of this lipid, it is easier to observe a more dramatic effect upon genetic manipulation of PIKfyve levels. Similar acute responses may be true for other PPIs but will be more difficult to detect due to the existence of multiple sources of some PPI, which may provide a compensatory effect. An important property of PPIs is that they often participate in phosphoinositide cascades, in which the ordered appearance of PPIs at one compartment allows for the progression of a cellular pathway, as shown by the coordinated action of VPS34 and PIKfyve to regulate endocytic recycling.
PIKfyve has gained attention from different sources in recent years, when it became a promising drug target for neurodegeneration, cancer, and COVID-19. However, the difficulty of measuring the low levels of PI(3,5)P2 and the lack of specific biosensors to monitor its spatial and temporal dynamics have been major challenges to elucidate the functions of PIKfyve. Although considerable progress has been made, a more comprehensive picture of the pathways that are regulated by PIKfyve will be critical to understand the roles of PI(3,5)P2 and PI(5)P in disease. In addition, since ablation of PIKfyve activity results in very early embryonic lethality, the successful development of PIKfyve inhibitors as therapies for some diseases necessitates a deeper understanding of the diverse pathways that require PIKfyve.
Author contributions
PRR and LSW co-wrote the manuscript.
Conflict of interest statement
Nothing declared.
Acknowledgements
This work was supported by R01-NS099340 and R01 NS064015 to LSW, and by the University of Michigan Protein Folding Diseases Fast Forward Initiative. PRR was funded in part by a Michigan Life Sciences Postdoctoral Fellowship.
This review comes from a themed issue on Membrane Trafficking 2022
Edited by Chiara Zurzolo and Benjamin Glick
References
- 1.Choy C.H., Han B.-K., Botelho R.J. Phosphoinositide diversity, distribution, and effector function: stepping out of the box. Bioessays. 2017;39:1700121. doi: 10.1002/bies.201700121. [DOI] [PubMed] [Google Scholar]
- 2.Posor Y., Eichhorn-Grünig M., Haucke V. Phosphoinositides in endocytosis. Biochim Biophys Acta Mol Cell Biol Lipids. 2015;1851:794–804. doi: 10.1016/j.bbalip.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Ketel K., Krauss M., Nicot A.-S., Puchkov D., Wieffer M., Müller R., Subramanian D., Schultz C., Laporte J., Haucke V. A phosphoinositide conversion mechanism for exit from endosomes. Nature. 2016;529:408–412. doi: 10.1038/nature16516. [DOI] [PubMed] [Google Scholar]
- 4.Zolov S.N., Bridges D., Zhang Y., Lee W.W., Riehle E., Verma R., Lenk G.M., Converso-Baran K., Weide T., Albin R.L., et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci U S A. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sbrissa D., Ikonomov O.C., Filios C., Delvecchio K., Shisheva A. Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am J Physiol Cell Physiol. 2012;303:C436–C446. doi: 10.1152/ajpcell.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinfeld N., Giridharan S.S.P., Kauffman E.J., Weisman L.S. Simultaneous detection of phosphoinositide lipids by radioactive metabolic labeling. Methods Mol Biol. 2021;2251:1–17. doi: 10.1007/978-1-0716-1142-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duex J.E., Nau J.J., Kauffman E.J., Weisman L.S. Phosphoinositide 5-phosphatase Fig4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell. 2006;5:723–731. doi: 10.1128/EC.5.4.723-731.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bridges D., Ma J.-T., Park S., Inoki K., Weisman L.S., Saltiel A.R. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–2962. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sbrissa D., Ikonomov O.C., Shisheva A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides: effect OF insulin. J Biol Chem. 1999;274:21589–21597. doi: 10.1074/jbc.274.31.21589. [DOI] [PubMed] [Google Scholar]
- 10.Jin N., Chow C.Y., Liu L., Zolov S.N., Bronson R., Davisson M., Petersen J.L., Zhang Y., Park S., Duex J.E., et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P2 in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botelho R.J., Efe J.A., Teis D., Emr S.D. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell. 2008;19:4273–4286. doi: 10.1091/mbc.E08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strunk B.S., Steinfeld N., Lee S., Jin N., Muñoz-Rivera C., Meeks G., Thomas A., Akemann C., Mapp A.K., MacGurn J.A., et al. Roles for a lipid phosphatase in the activation of its opposing lipid kinase. Mol Biol Cell. 2020;31:1835–1845. doi: 10.1091/mbc.E18-09-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J.A., Li P., Kumar N., Weisman L.S. Reinisch KM: insights into lysosomal PI(3,5)P(2) homeostasis from a structural-biochemical analysis of the PIKfyve lipid kinase complex. Mol Cell. 2020;80:736–743. doi: 10.1016/j.molcel.2020.10.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper combines negative stain- and cryo-EM studies to elucidate the architecture of the human PIKfyve complex, and provides insights into how the antagonistic activities or PIKfyve and Fig4 are regulated within the complex.
- 14.Sbrissa D., Ikonomov O.C., Shisheva A. PIKfyve lipid kinase is a protein Kinase: downregulation of 5‘-phosphoinositide product formation by autophosphorylation. Biochemistry. 2000;39:15980–15989. doi: 10.1021/bi001897f. [DOI] [PubMed] [Google Scholar]
- 15.McCartney A.J., Zolov S.N., Kauffman E.J., Zhang Y., Strunk B.S., Weisman L.S., Sutton M.A. Activity-dependent PI(3,5)P2 synthesis controls AMPA receptor trafficking during synaptic depression. Proc Natl Acad Sci U S A. 2014;111:E4896–E4905. doi: 10.1073/pnas.1411117111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenk G.M., Ferguson C.J., Chow C.Y., Jin N., Jones J.M., Grant A.E., Zolov S.N., Winters J.J., Giger R.J., Dowling J.J., et al. Pathogenic mechanism of the Fig4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. 2011;7:e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dove S.K., Piper R.C., McEwen R.K., Yu J.W., King M.C., Hughes D.C., Thuring J., Holmes A.B., Cooke F.T., Michell R.H., et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Efe J.A., Botelho R.J., Emr S.D. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell. 2007;18:4232–4244. doi: 10.1091/mbc.E07-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Proikas-Cezanne T., Takacs Z., Dönnes P., Kohlbacher O. WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci. 2015;128:207–217. doi: 10.1242/jcs.146258. [DOI] [PubMed] [Google Scholar]
- 20.Ji C., Zhao H., Chen D., Zhang H., Zhao Y.G. β-propeller proteins WDR45 and WDR45B regulate autophagosome maturation into autolysosomes in neural cells. Curr Biol. 2021;31:1666–1677. doi: 10.1016/j.cub.2021.01.081. e6. [DOI] [PubMed] [Google Scholar]
- 21.Jin N., Lang M.J., Weisman L.S. Phosphatidylinositol 3,5-bisphosphate: regulation of cellular events in space and time. Biochem Soc Trans. 2016;44:177–184. doi: 10.1042/BST20150174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gary J.D., Sato T.K., Stefan C.J., Bonangelino C.J., Weisman L.S., Emr S.D. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell. 2002;13:1238–1251. doi: 10.1091/mbc.01-10-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine T.P. TMEM106B in humans and Vac7 and Tag1 in yeast are predicted to be lipid transfer proteins. Proteins Struct Funct Bioinforma. 2022;90:164–175. doi: 10.1002/prot.26201. [DOI] [PubMed] [Google Scholar]
- 24.Wilson Z.N., Scott A.L., Dowell R.D., Odorizzi G. PI(3,5)P(2) controls vacuole potassium transport to support cellular osmoregulation. Mol Biol Cell. 2018;29:1718–1731. doi: 10.1091/mbc.E18-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin N., Jin Y., Weisman L.S. Early protection to stress mediated by CDK-dependent PI3,5P2 signaling from the vacuole/lysosome. J Cell Biol. 2017;216:2075–2090. doi: 10.1083/jcb.201611144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonangelino C.J., Nau J.J., Duex J.E., Brinkman M., Wurmser A.E., Gary J.D., Emr S.D., Weisman L.S. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol. 2002;156:1015–1028. doi: 10.1083/jcb.200201002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikonomov O.C., Sbrissa D., Delvecchio K., Xie Y., Jin J.-P., Rappolee D., Shisheva A. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve-/- embryos but normality of PIKfyve+/- mice. J Biol Chem. 2011;286:13404–13413. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takasuga S., Horie Y., Sasaki J., Sun-Wada G.-H., Kawamura N., Iizuka R., Mizuno K., Eguchi S., Kofuji S., Kimura H., et al. Critical roles of type III phosphatidylinositol phosphate kinase in murine embryonic visceral endoderm and adult intestine. Proc Natl Acad Sci Unit States Am. 2013;110:1726–1731. doi: 10.1073/pnas.1213212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikonomov O.C., Sbrissa D., Delvecchio K., Feng H.-Z., Cartee G.D., Jin J.-P., Shisheva A. Muscle-specific Pikfyve gene disruption causes glucose intolerance, insulin resistance, adiposity, and hyperinsulinemia but not muscle fiber-type switching. Am J Physiol Endocrinol Metab. 2013;305:E119–E131. doi: 10.1152/ajpendo.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mironova Y.A., Lenk G.M., Lin J.-P., Lee S.J., Twiss J.L., Vaccari I., Bolino A., Havton L.A., Min S.H., Abrams C.S., et al. PI(3,5)P2 biosynthesis regulates oligodendrocyte differentiation by intrinsic and extrinsic mechanisms. Elife. 2016;5:e13023. doi: 10.7554/eLife.13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hee M.S., Aae S., Lehn W., Jessica G., Yutein C., Huiyan J., Francina G., Claire T., Liang Z., A. Sl, et al. PIKfyve deficiency in myeloid cells impairs lysosomal homeostasis in macrophages and promotes systemic inflammation in mice. Mol Cell Biol. 2022;39 doi: 10.1128/MCB.00158-19. e00158–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chow C.Y., Zhang Y., Dowling J.J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M.E., Li J., Zhang X., et al. Mutation of Fig4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Zolov S.N., Chow C.Y., Slutsky S.G., Richardson S.C., Piper R.C., Yang B., Nau J.J., Westrick R.J., Morrison S.J., et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci U S A. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow C.Y., Landers J.E., Bergren S.K., Sapp P.C., Grant A.E., Jones J.M., Everett L., Lenk G.M., McKenna-Yasek D.M., Weisman L.S., et al. Deleterious variants of Fig4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campeau P.M., Lenk G.M., Lu J.T., Bae Y., Burrage L., Turnpenny P., Román Corona-Rivera J., Morandi L., Mora M., Reutter H., et al. Yunis-Varón syndrome is caused by mutations in Fig4, encoding a phosphoinositide phosphatase. Am J Hum Genet. 2013;92:781–791. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umair M., Alkharfy T.M., Sajjad S., Alfadhel M. FIG4-Associated yunis-varon syndrome: identification of a novel missense variant. Mol Syndromol. 2021;12:386–392. doi: 10.1159/000516971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenk G.M., Berry I.R., Stutterd C.A., Blyth M., Green L., Vadlamani G., Warren D., Craven I., Fanjul-Fernandez M., Rodriguez-Casero V., et al. Cerebral hypomyelination associated with biallelic variants of Fig4. Hum Mutat. 2019;40:619–630. doi: 10.1002/humu.23720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baulac S., Lenk G.M., Dufresnois B., Ouled Amar Bencheikh B., Couarch P., Renard J., Larson P.A., Ferguson C.J., Noé E., Poirier K., et al. Role of the phosphoinositide phosphatase Fig4 gene in familial epilepsy with polymicrogyria. Neurology. 2014;82:1068–1075. doi: 10.1212/WNL.0000000000000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lines M.A., Ito Y., Kernohan K.D., Mears W., Hurteau-Miller J., Venkateswaran S., Ward L., Khatchadourian K., McClintock J., Bhola P., et al. Yunis-Varón syndrome caused by biallelic VAC14 mutations. Eur J Hum Genet. 2017;25:1049–1054. doi: 10.1038/ejhg.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lenk G.M., Szymanska K., Debska-Vielhaber G., Rydzanicz M., Walczak A., Bekiesinska-Figatowska M., Vielhaber S., Hallmann K., Stawinski P., Buehring S., et al. Biallelic mutations of VAC14 in pediatric-onset neurological disease. Am J Hum Genet. 2016;99:188–194. doi: 10.1016/j.ajhg.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stutterd C., Diakumis P., Bahlo M., Fanjul Fernandez M., Leventer R.J., Delatycki M., Amor D., Chow C.W., Stephenson S., Meisler M.H., et al. Neuropathology of childhood-onset basal ganglia degeneration caused by mutation of VAC14. Ann Clin Transl Neurol. 2017;4:859–864. doi: 10.1002/acn3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taghavi S., Chaouni R., Tafakhori A., Azcona L.J., Firouzabadi S.G., Omrani M.D., Jamshidi J., Emamalizadeh B., Shahidi G.A., Ahmadi M., et al. A clinical and molecular genetic study of 50 families with autosomal recessive parkinsonism revealed known and novel gene mutations. Mol Neurobiol. 2018;55:3477–3489. doi: 10.1007/s12035-017-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Gusmao C.M., Stone S., Waugh J.L., Yang E., Lenk G.M., Rodan L.H. VAC14 gene-related parkinsonism-dystonia with response to deep brain stimulation. Mov Disord Clin Pract. 2019;6:494–497. doi: 10.1002/mdc3.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyon G.J., Marchi E., Ekstein J., Meiner V., Hirsch Y., Scher S., Yang E., De Vivo D.C., Madrid R., Li Q., et al. VAC14 syndrome in two siblings with retinitis pigmentosa and neurodegeneration with brain iron accumulation. Cold Spring Harb Mol case Stud. 2019:5. doi: 10.1101/mcs.a003715. a003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X., Chow C.Y., Sahenk Z., Shy M.E., Meisler M.H., Li J. Mutation of Fig4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain. 2008;131:1990–2001. doi: 10.1093/brain/awn114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaur P., Bhavani G.S., Raj A., Girisha K.M., Shukla A. Homozygous variant, p. (Arg643Trp) in VAC14 causes striatonigral degeneration: report of a novel variant and review of VAC14-related disorders. J Hum Genet. 2019;64:1237–1242. doi: 10.1038/s10038-019-0678-1. [DOI] [PubMed] [Google Scholar]
- 47.Liao S., Chen T., Dai Y., Wang Y., Wu F., Zhong M. Novel VAC14 variants identified in two Chinese siblings with childhood-onset striatonigral degeneration. Mol Genet genomic Med. 2020;8:e1101. doi: 10.1002/mgg3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baumann H., Tunc S., Günther A., Münchau A., Lohmann K., Brüggemann N. Altered homodimer formation and increased iron accumulation in VAC14-related disease: case report and review of the literature. Park Relat Disord. 2020;80:41–46. doi: 10.1016/j.parkreldis.2020.09.012. [DOI] [PubMed] [Google Scholar]
- 49.Karaoğlu P., Köse M. Expanding the spectrum of VAC14 related pediatric-onset neurological disease; striatonigral degeneration with brainstem involvement. Eur J Med Genet. 2021;64:104117. doi: 10.1016/j.ejmg.2020.104117. [DOI] [PubMed] [Google Scholar]
- 50.Li S., Tiab L., Jiao X., Munier F.L., Zografos L., Frueh B.E., Sergeev Y., Smith J., Rubin B., Meallet M.A., et al. Mutations in PIP5K3 are associated with françois-neetens mouchetée fleck corneal dystrophy. Am J Hum Genet. 2005;77:54–63. doi: 10.1086/431346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotoulas A., Kokotas H., Kopsidas K., Droutsas K., Grigoriadou M., Bajrami H., Schorderet D.F., Petersen M.B. A novel PIKFYVE mutation in fleck corneal dystrophy. Mol Vis. 2011;17:2776–2781. [PMC free article] [PubMed] [Google Scholar]
- 52.Gee J.A., Frausto R.F., Chung D.-W.D., Tangmonkongvoragul C., Le D.J., Wang C., Han J., Aldave A.J. Identification of novel PIKFYVE gene mutations associated with Fleck corneal dystrophy. Mol Vis. 2015;21:1093–1100. [PMC free article] [PubMed] [Google Scholar]
- 53.Mei S., Wu Y., Wang Y., Cui Y., Zhang M., Zhang T., Huang X., Yu S., Yu T., Zhao J. Disruption of PIKFYVE causes congenital cataract in human and zebrafish. Elife. 2022;11:e71256. doi: 10.7554/eLife.71256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lafontaine M., Lia A.-S., Bourthoumieu S., Beauvais-Dzugan H., Derouault P., Arné-Bes M.-C., Sarret C., Laffargue F., Magot A., Sturtz F., et al. Clinical features of homozygous FIG4-p.Ile41Thr Charcot-Marie-Tooth 4J patients. Ann Clin Transl Neurol. 2021;8:471–476. doi: 10.1002/acn3.51175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gentil B.J., O'Ferrall E., Chalk C., Santana L.F., Durham H.D., Massie R. A new mutation in Fig4 causes a severe form of CMT4J involving TRPV4 in the pathogenic cascade. J Neuropathol Exp Neurol. 2017;76:789–799. doi: 10.1093/jnen/nlx062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orengo J.P., Khemani P., Day J.W., Li J., Siskind C.E. Charcot Marie Tooth disease type 4J with complex central nervous system features. Ann Clin Transl Neurol. 2018;5:222–225. doi: 10.1002/acn3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osmanovic A., Rangnau I., Kosfeld A., Abdulla S., Janssen C., Auber B., Raab P., Preller M., Petri S., Weber R.G. Fig4 variants in central European patients with amyotrophic lateral sclerosis: a whole-exome and targeted sequencing study. Eur J Hum Genet. 2017;25:324–331. doi: 10.1038/ejhg.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertolin C., Querin G., Bozzoni V., Martinelli I., De Bortoli M., Rampazzo A., Gellera C., Pegoraro E., Sorarù G. New Fig4 gene mutations causing aggressive ALS. Eur J Neurol. 2018;25:e41–e42. doi: 10.1111/ene.13559. [DOI] [PubMed] [Google Scholar]
- 59.Liu C.-Y., Lin J.-L., Feng S.-Y., Che C.-H., Huang H.-P., Zou Z.-Y. Novel variants in the Fig4 gene associated with Chinese sporadic amyotrophic lateral sclerosis with slow progression. J Clin Neurol. 2022;18:41–47. doi: 10.3988/jcn.2022.18.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bergner C.G., Neuhofer C.M., Funke C., Biskup S., von Gottberg P., Bartels C., Koch J.C., Radenbach K. Case report: association of a variant of unknown significance in the fig4 gene with frontotemporal dementia and slowly progressing motoneuron disease: a case report depicting common challenges in clinical and genetic diagnostics of rare neuropsychiatric a. Front Neurosci. 2020;14:559670. doi: 10.3389/fnins.2020.559670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju A.K.K., Frontzek K., Lemes E., Herrmann U., Losa M., Marpakwar R. Aguzzi A: loss of PIKfyve drives the spongiform degeneration in prion diseases. EMBO Mol Med. 2021;13:e14714. doi: 10.15252/emmm.202114714. [DOI] [PMC free article] [PubMed] [Google Scholar]; Loss of PIKfyve activity in mammals results in a spongioform-like phenotype in the brain. This article demonstrates that prion infection results in the degradation of PIKfyve although the levels of Vac14 and Fig4 are not perturbed. In addition, the authors show that prion infection of cells results in additional phenotypes associated with loss of PIKfyve activity including enlarged lysosomes and translocation of TFEB to the nucleus.
- 62.Huang P., Einav S., Asquith C. PIKfyve: a lipid kinase target for COVID-19, cancer and neurodegenerative disorders. Nat Rev Drug Discov. 2021;10:730. doi: 10.1038/d41573-021-00158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayakawa M., Kaizawa H., Moritomo H., Koizumi T., Ohishi T., Okada M., Ohta M., Tsukamoto S., Parker P., Workman P., et al. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3 kinase p110α inhibitors. Bioorg Med Chem. 2006;14:6847–6858. doi: 10.1016/j.bmc.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 64.Ikonomov O.C., Sbrissa D., Shisheva A. YM201636, an inhibitor of retroviral budding and PIKfyve-catalyzed PtdIns(3,5)P2 synthesis, halts glucose entry by insulin in adipocytes. Biochem Biophys Res Commun. 2009;382:566–570. doi: 10.1016/j.bbrc.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayakawa N., Noguchi M., Takeshita S., Eviryanti A., Seki Y., Nishio H., Yokoyama R., Noguchi M., Shuto M., Shima Y., et al. Structure–activity relationship study, target identification, and pharmacological characterization of a small molecular IL-12/23 inhibitor, APY0201. Bioorg Med Chem. 2014;22:3021–3029. doi: 10.1016/j.bmc.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 66.Sharma G., Guardia C.M., Roy A., Vassilev A., Saric A., Griner L.N., Marugan J., Ferrer M., Bonifacino J.S., DePamphilis M.L. A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy. 2019;15:1694–1718. doi: 10.1080/15548627.2019.1586257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai X., Xu Y., Cheung A.K., Tomlinson R.C., Alcázar-Román A., Murphy L., Billich A., Zhang B., Feng Y., Klumpp M., et al. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem Biol. 2013;20:912–921. doi: 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nelson E.A., Dyall J., Hoenen T., Barnes A.B., Zhou H., Liang J.Y., Michelotti J., Dewey W.H., DeWald L.E., Bennett R.S., et al. The phosphatidylinositol-3-phosphate 5-kinase inhibitor apilimod blocks filoviral entry and infection. PLoS Neglected Trop Dis. 2017:11. doi: 10.1371/journal.pntd.0005540. e0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang Y.-L., Chou Y.-Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D., Case J.B., Chen R.E., Diamond M.S., Whelan S.P.J., et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identified around 100 molecules that inhibit viral replication of SARS-CoV-2 among a library of clinical-stage or FDA-approved drugs. The PIKfyve inhibitor apilimod was one of the most promising candidates, and inhibited viral replication in different cell lines at concentrations that could be achievable therapeutic doses.
- 71.Kreutzberger A.J.B., Sanyal A., Ojha R., Pyle J.D., Vapalahti O., Balistreri G., Kirchhausen T. Synergistic block of SARS-CoV-2 infection by combined drug inhibition of the host entry factors PIKfyve kinase and TMPRSS2 protease. J Virol. 2021:95. doi: 10.1128/JVI.00975-21. e0097521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Puray-Chavez M., LaPak K.M., Schrank T.P., Elliott J.L., Bhatt D.P., Agajanian M.J., Jasuja R., Lawson D.Q., Davis K., Rothlauf P.W., et al. Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep. 2021;36:109364. doi: 10.1016/j.celrep.2021.109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gayle S., Landrette S., Beeharry N., Conrad C., Hernandez M., Beckett P., Ferguson S.M., Mandelkern T., Zheng M., Xu T., et al. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood. 2017;129:1768–1778. doi: 10.1182/blood-2016-09-736892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dupuis-Coronas S., Lagarrigue F., Ramel D., Chicanne G., Saland E., Gaits-Iacovoni F., Payrastre B., Tronchère H. The nucleophosmin-anaplastic lymphoma kinase oncogene interacts, activates, and uses the kinase PIKfyve to increase invasiveness. J Biol Chem. 2011;286:32105–32114. doi: 10.1074/jbc.M111.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oppelt A., Haugsten E.M., Zech T., Danielsen H.E., Sveen A., Lobert V.H., Skotheim R.I., Wesche J. PIKfyve, MTMR3 and their product PtdIns5P regulate cancer cell migration and invasion through activation of Rac1. Biochem J. 2014;461:383–390. doi: 10.1042/BJ20140132. [DOI] [PubMed] [Google Scholar]
- 77.Ikonomov O.C., Sbrissa D., Shisheva A. Small molecule PIKfyve inhibitors as cancer therapeutics: translational promises and limitations. Toxicol Appl Pharmacol. 2019;383:114771. doi: 10.1016/j.taap.2019.114771. [DOI] [PubMed] [Google Scholar]
- 78.Hou J., Xi Z., Niu J., Li W., Wang X., Liang C., Sun H., Fang D., Xie S. Inhibition of PIKfyve using YM201636 suppresses the growth of liver cancer via the induction of autophagy. Oncol Rep. 2019;41:1971–1979. doi: 10.3892/or.2018.6928. [DOI] [PubMed] [Google Scholar]
- See S.K., Chen M., Bax S., Tian R., Woerman A., Tse E., Johnson I.E., Nowotny C., Muñoz E.N., Sengstack J., et al. PIKfyve inhibition blocks endolysosomal escape of α-synuclein fibrils and spread of α-synuclein aggregation. bioRxiv. 2021 doi: 10.1101/2021.01.21.427704. [DOI] [Google Scholar]; This manuscript performed a genome-wide CRISPR interference screen with a FRET readout to identify compounds that could prevent the propagation of α-synuclein aggregation, This manuscript performed a genome-wide CRISPR interference screen with a FRET readout to identify compounds that could prevent the propagation of α-synuclein aggregation. PIKfyve inhibition decreased α-synuclein aggregation by interfering with the trafficking of α-synuclein from the early endosome to the lysosome.
- 80.Shi Y., Lin S., Staats K.A., Li Y., Chang W.-H., Hung S.-T., Hendricks E., Linares G.R., Wang Y., Son E.Y., et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. 2018;24:313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Staats K.A., Seah C., Sahimi A., Wang Y., Koutsodendris N., Lin S., Kim D., Chang W.-H., Gray K.A., Shi Y., et al. Small molecule inhibition of PIKFYVE kinase rescues gain- and loss-of-function C9ORF72 ALS/FTD disease processes in vivo. bioRxiv. 2019 doi: 10.1101/685800. [DOI] [Google Scholar]
- 82.Soares A.C., Ferreira A., Mariën J., Delay C., Lee E., Trojanowski J.Q., Moechars D., Annaert W., De Muynck L. PIKfyve activity is required for lysosomal trafficking of tau aggregates and tau seeding. J Biol Chem. 2021;296:100636. doi: 10.1016/j.jbc.2021.100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X., Wang X., Zhang X., Zhao M., Tsang W.L., Zhang Y., Yau R.G., Weisman L.S., Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc Natl Acad Sci Unit States Am. 2013;110:21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ikonomov O.C., Sbrissa D., Shisheva A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J Biol Chem. 2001;276:26141–26147. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- 85.Gary J.D., Wurmser A.E., Bonangelino C.J., Weisman L.S., Emr S.D. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho C.Y., Alghamdi T.A., Botelho R.J. Phosphatidylinositol-3,5-Bisphosphate: No longer the poor PIP2. Traffic. 2012;13:1–8. doi: 10.1111/j.1600-0854.2011.01246.x. [DOI] [PubMed] [Google Scholar]
- 87.Li P., Hu M., Wang C., Feng X., Zhao Z., Yang Y., Sahoo N., Gu M., Yang Y., Xiao S., et al. LRRC8 family proteins within lysosomes regulate cellular osmoregulation and enhance cell survival to multiple physiological stresses. Proc Natl Acad Sci U S A. 2020;117:29155–29165. doi: 10.1073/pnas.2016539117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Banerjee S., Kane P.M. Regulation of V-ATPase activity and organelle pH by phosphatidylinositol phosphate lipids. Front Cell Dev Biol. 2020;8:510. doi: 10.3389/fcell.2020.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li S.C., Diakov T.T., Xu T., Tarsio M., Zhu W., Couoh-Cardel S., Weisman L.S., Kane P.M. The signaling lipid PI(3,5)P₂ stabilizes V₁-V(o) sector interactions and activates the V-ATPase. Mol Biol Cell. 2014;25:1251–1262. doi: 10.1091/mbc.E13-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsford A.H., Ryan T.A., Raimondi A., Cocucci E., Wycislo S.A., Fröhlich F., Swan L.E. Stagi M: live imaging of intra-lysosome pH in cell lines and primary neuronal culture using a novel genetically encoded biosensor. Autophagy. 2021;17:1500–1518. doi: 10.1080/15548627.2020.1771858. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors built a genetically-encoded biosensor, based on the fusion of a ratiometric fluorophore to the lysosomal protein LAMP1, to study different aspects of lysosomal signaling and pH. They found that in unperturbed cells, lysosomal pH is remarkably stable. They tested several types of lysosomal stressors and found that distinct stressors changed lysosomal pH, lysosomal size and/or resulted in TFEB translocation to the nucleus. However, these three types of changes were not necessarily correlated with each other. With regards to PIKfyve, they found that treatment of cells with low levels of apilimod for a short period of time resulted in elevated lysosomal pH, an increase in lysosomal size, and translocation of TFEB to the nucleus.
- 91.Bissig C., Hurbain I., Raposo G., van Niel G. PIKfyve activity regulates reformation of terminal storage lysosomes from endolysosomes. Traffic. 2017;18:747–757. doi: 10.1111/tra.12525. [DOI] [PubMed] [Google Scholar]
- 92.Krishna S., Palm W., Lee Y., Yang W., Bandyopadhyay U., Xu H., Florey O., Thompson C.B., Overholtzer M. PIKfyve regulates vacuole maturation and nutrient recovery following engulfment. Dev Cell. 2016;38:536–547. doi: 10.1016/j.devcel.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choy C.H., Saffi G., Gray M.A., Wallace C., Dayam R.M., Ou Z.-Y.A., Lenk G., Puertollano R., Watkins S.C., Botelho R.J. Lysosome enlargement during inhibition of the lipid kinase PIKfyve proceeds through lysosome coalescence. J Cell Sci. 2018;131 doi: 10.1242/jcs.213587. jcs213587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.De Leo M.G., Berger P., Mayer A. WIPI1 promotes fission of endosomal transport carriers and formation of autophagosomes through distinct mechanisms. Autophagy. 2021;17:3644–3670. doi: 10.1080/15548627.2021.1886830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miner G.E., Sullivan K.D., Guo A., Jones B.C., Hurst L.R., Ellis E.C., Starr M.L., Fratti R.A. Phosphatidylinositol 3,5-bisphosphate regulates the transition between trans-SNARE complex formation and vacuole membrane fusion. Mol Biol Cell. 2019;30:201–208. doi: 10.1091/mbc.E18-08-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bissig C., Croisé P., Heiligenstein X., Hurbain I., Lenk G.M., Kaufman E., Sannerud R., Annaert W., Meisler M.H., Weisman L.S., et al. The PIKfyve complex regulates the early melanosome homeostasis required for physiological amyloid formation. J Cell Sci. 2019;132 doi: 10.1242/jcs.229500. jcs229500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rutherford A.C., Traer C., Wassmer T., Pattni K., Bujny M.V., Carlton J.G., Stenmark H., Cullen P.J. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci. 2006;119:3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Lartigue J., Polson H., Feldman M., Shokat K., Tooze S.A., Urbé S., Clague M.J. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y., McCartney A.J., Zolov S.N., Ferguson C.J., Meisler M.H., Sutton M.A., Weisman L.S. Modulation of synaptic function by VAC14, a protein that regulates the phosphoinositides PI(3,5)P 2 and PI(5)P. EMBO J. 2012;31:3442–3456. doi: 10.1038/emboj.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giridharan S.S.P., Luo G., Rivero-Rios P., Steinfeld N., Tronchere H., Singla A., Burstein E., Billadeau D.D., Sutton M.A. Weisman LS: lipid kinases VPS34 and PIKfyve coordinate a phosphoinositide cascade to regulate Retriever-mediated recycling on endosomes. Elife. 2022;11:e69709. doi: 10.7554/eLife.69709. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides mechanistic insight into the role of PIKfyve in endocytic recycling and cell migration. It describes a phosphoinositide cascade coordinated by the lipid kinases VPS34 and PIKfyve, which is critical to regulate Retriever-dependent recycling from endosomes. This cascade is necessary for the recycling of β1-integrin, a protein that acts in cell migration.
- 101.Dukes J.D., Whitley P., Chalmers A.D. The PIKfyve inhibitor YM201636 blocks the continuous recycling of the tight junction proteins claudin-1 and claudin-2 in MDCK cells. PLoS One. 2012;7:e28659. doi: 10.1371/journal.pone.0028659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cinato M., Guitou L., Saidi A., Timotin A., Sperazza E., Duparc T., Zolov S.N., Giridharan S.S.P., Weisman L.S., Martinez L.O., et al. Apilimod alters TGFβ signaling pathway and prevents cardiac fibrotic remodeling. Theranostics. 2021;11:6491–6506. doi: 10.7150/thno.55821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman S.A., Uderhardt S., Saric A., Collins R.F., Buckley C.M., Mylvaganam S., Boroumand P., Plumb J., Germain R.N., Ren D., et al. Lipid-gated monovalent ion fluxes regulate endocytic traffic and support immune surveillance. Science. 2020;367:301–305. doi: 10.1126/science.aaw9544. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article supports the hypothesis that ion fluxes directly and specifically regulate endomembrane trafficking. The authors suggest that the exit of ions and osmotically-coupled water causes the shrinkage and subsequent crenation of endosomal compartments, which promotes the recruitment of curvature-sensing proteins that drive different trafficking pathways. This flux of ions occurs through TPC channels and is regulated by PIKfyve activity.
- 104.Han B.-K., Emr S.D. Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 2011;25:984–995. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cai X., Xu Y., Kim Y.-M., Loureiro J., Huang Q. PIKfyve, a class III lipid kinase, is required for TLR-induced type I IFN production via modulation of ATF3. J Immunol. 2014;192:3383–3389. doi: 10.4049/jimmunol.1302411. [DOI] [PubMed] [Google Scholar]
- Hasegawa J., Tokuda E., Yao Y., Sasaki T., Inoki K. Weisman LS: PP2A-dependent TFEB activation is blocked by PIKfyve-induced mTORC1 activity. Mol Biol Cell. 2022 doi: 10.1091/mbc.E21-06-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper found that PIKfyve facilitates mTORC1-dependent phosphorylation of TFEB which prevents TFEB activity. When PIKfyve is inhibited, the association between mTORC1 and TFEB is lost, PP2A dephosphorylates the mTORC1 dependent site on TFEB, which then translocates to the nucleus where it promotes TFEB-dependent transcription. This paper combined with previous studies indicate that PIKfyve plays regulatory roles in transcription.