Abstract
Plant tannins belong to the antioxidant compound family, which includes chemicals responsible for protecting biological structures from the harmful effects of oxidative stress. A wide range of plants and crops are rich in antioxidant compounds, offering resistance to biotic, mainly against pathogens and herbivores, and abiotic stresses, such as light and wound stresses. These compounds are also related to human health benefits, offering protective effects against cardiovascular and neurodegenerative diseases in addition to providing anti-tumor, anti-inflammatory, and anti-bacterial characteristics. Most of these compounds are structurally and biosynthetically related, being synthesized through the shikimate-phenylpropanoid pathways, offering several classes of plant antioxidants: flavonoids, anthocyanins, and tannins. Tannins are divided into two major classes: condensed tannins or proanthocyanidins and hydrolysable tannins. Hydrolysable tannin synthesis branches directly from the shikimate pathway, while condensed tannins are derived from the flavonoid pathway, one of the branches of the phenylpropanoid pathway. Both types of tannins have been proposed as important molecules for taste perception of many fruits and beverages, especially wine, besides their well-known roles in plant defense and human health. Regulation at the gene level, biosynthesis and degradation have been extensively studied in condensed tannins in crops like grapevine (Vitis vinifera), persimmon (Diospyros kaki) and several berry species due to their high tannin content and their importance in the food and beverage industry. On the other hand, much less information is available regarding hydrolysable tannins, although some key aspects of their biosynthesis and regulation have been recently discovered. Here, we review recent findings about tannin metabolism, information that could be of high importance for crop breeding programs to obtain varieties with enhanced nutritional characteristics.
Keywords: antioxidants, flavonoids, proanthocyanidins, fruits, biosynthesis, ellagitannins (ETs)
Introduction
Antioxidant compounds are chemical species whose function is to inhibit or delay the process of oxidation (Mittler, 2017). Oxidation is the chemical process of losing electrons, but biologically speaking, it is the production of free radical (reactive) species, which can attack electron-rich molecules such as lipids, proteins, or even nucleic acids, damaging cells and tissues, and thereby causing the alteration of homeostasis (Engwa, 2018). To fight against these reactive species, plants have a myriad of chemical compounds known as bio-active molecules whose activities are based on their antioxidant properties. These compounds have the ability to protect biological structures by acting as scavengers of free radicals such as reactive oxygen species (ROS) and reactive nitrogen species (RNS) (Karadag et al., 2009; Wang et al., 2009).
One of the main chemical classes of plant antioxidants are polyphenol (phenolic) compounds. These metabolites are mainly composed of aromatic rings with hydroxyl groups capable of reacting with peroxide radicals to block degradation reactions (Hyun et al., 2010). They are major and ubiquitous secondary metabolites derived from phenylalanine and shikimate pathways (Quideau et al., 2011). Polyphenols range from simple molecules such as phenolic acids to more complex structures that are highly polymerized metabolites, including tannins, flavonoids, and lignans or stilbenes (Pott et al., 2019).
The study of plant polyphenols started thanks to the work of four scientists: Theodore White, E. C. Bate-Smith, Tony Swain, and Edwin Haslam. According to their definition of plant polyphenols, only monosubstituted phenols or compounds having di- and/or tri-hydroxyphenyl moieties can fit the definition of “true plant polyphenol” (Haslam and Cai, 1994). Given that, only proanthocyanidins and hydrolysable tannins, which exclude lignin polymers among others, are considered true plant polyphenols (Haslam and Cai, 1994). According to Quideau et al. (2011), a polyphenol is a plant secondary metabolite derived from the phenylpropanoid-shikimate and polyketide pathways that has more than one phenolic unit and is deprived of any nitrogen-based functional group. Nowadays, it is widely accepted that a polyphenol is a naturally occurring compound composed of several aromatic rings substituted with hydroxyl groups. This definition comprises four principal groups of polyphenols: flavonoids, phenolic acids, lignans, and stilbenes (Zhou et al., 2016).
An important class of polyphenols is tannins, which have been used for ages in the treatment of animal skins for leather manufacturing and to prevent putrefaction. The reason is the ability of tannins to interact with proteins, stabilizing them and turning the skin tanned in order to be transformed into leather (Baxter et al., 1997; Falcão and Araújo, 2018). The term “tannin” includes three families of compounds: condensed tannins (proanthocyanidins), hydrolysable tannins (gallotannins and ellagitannins) and phlorotannins (only found in marine red-brown algae). According to Zhou et al. (2016) classification, condensed tannins belong to flavonoid-related compounds, while ellagitannins belong to phenolic acid-related compounds. Tannins can be found naturally in plants and their main function is to act as a natural barrier against pathogens or herbivores (Haslam and Cai, 1994). The consumption of fruits enriched in polyphenols, such as berries, pomegranates (Punica granatum), persimmons (Diospyros kaki) and nuts, is related to health benefits (Basu et al., 2014; Cory et al., 2018). Of course, tannins, as polyphenols, play a role in crop health-promoting benefits.
The aim of this review is to summarize the genetic factors underlying the regulation of hydrolysable and condensed tannin biosynthesis, respectively. For this purpose, we focus on economically-relevant species, such as kaki, grape (Vitis vinifera) or strawberry (Fragaria x ananassa). These crops are known for their high tannin content and metabolites that significantly contribute to their antioxidant and organoleptic properties. In this sense, we also discuss how the recent findings about the genetic regulation of tannin synthesis may pave the way to breeding new varieties with enhanced nutritional value.
Biosynthetic Pathways Leading to the Formation of Polyphenol Compounds
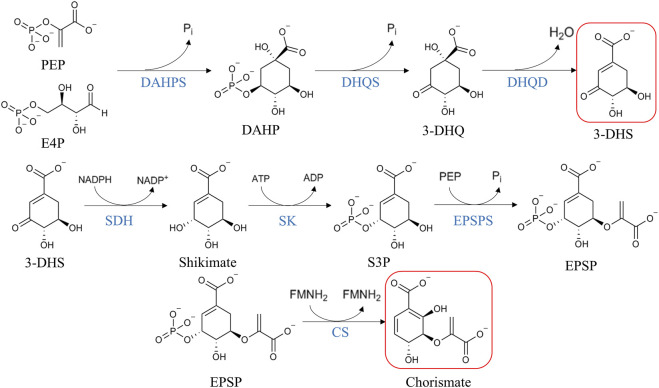
Many aromatic compounds, and by extension, many antioxidant compounds, are produced by de novo synthesis via the shikimate pathway in plants (Bentley and Haslam, 1990). They are produced in plastids, where this pathway is functional. Chorismate is synthetized through seven enzymatic reactions starting from phosphoenolpyruvate and erythrose-4-phosphate (Figure 1), and is finally transformed into the three aromatic amino acids, L-tyrosine, L-tryptophan, and L-phenylalanine (Mir et al., 2015). L-Phenylalanine is the precursor of many antioxidant compounds like flavonoids, isoflavonoids, phenylpropenes, aurones, coumarins, and phenylpropanoid esters (Stefanachi et al., 2018). Another interesting intermediate of the shikimate pathway is the 3-dehydroshikimate, which starts the biosynthesis of hydrolysable tannins (Vogt, 2010; Bontpart et al., 2016).
FIGURE 1.
Schematic of shikimate pathway reactions. 3-dehydroshikimate and chorismate are highlighted in red as the precursors of hydrolysable tannins and proanthocyanidins, respectively. Abbreviations: phosphoenolpyruvate (PEP), erythrose 4-phosphate (E4P), inorganic phosphate (Pi), 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHPS), 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP), dehydroquinate synthase (DHQS), 3-dehydroquinate (3-DHQ), 3-dehydroquinate dehydratase (DHQD), 3-dehydroshikimate (3-DHS), shikimate dehydrogenase (SDH), shikimate kinase (SK), shikimate-3-phosphate (S3P), 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS), 5-enolpyruvylshikimate-3-phosphate (EPSP), and chorismate synthase (CS). Adapted from Vogt, 2010; Akagi et al., 2011; Mir et al., 2015; Stefanachi et al., 2018.
The first step of the shikimate pathway is an aldol condensation catalyzed by 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, taking phosphoenolpyruvate (PEP) and erythrose-4-phosphate as substrates, resulting in the seven-carbon keto acid DAHP and inorganic phosphate (Mir et al., 2015). In plants, DAHP synthase is regulated transcriptionally by certain environmental stimuli like mechanical wounding or biotic stresses like insects’ attacks. Under these stress conditions, there is an accumulation of mRNA encoding DAHP synthase (Dyer et al., 1989), strengthening the idea of the role of aromatic compounds in plant defense. In addition, DAHP synthase is controlled by feedback inhibition loops, limiting the carbon flux from primary metabolism to polyphenol synthesis (Pott et al., 2019).
The next step is the transformation of DAHP into 3-dehydoquinate, catalyzed by dehydroquinate synthase (DHQS). This enzyme is a metalloenzyme nicotinamide adenine di-nucleotide (NAD+)-dependent that acts as an enzymatic complex in the five-step transformation of DAHP. In this multistep mechanism, several reactions occur spontaneously, but only a single site catalyzes five sequential reactions. NAD+ is required for the oxidation of C5 of DAHP and the NADH produced is then used in the reduction of the C5 carbonyl intermediate in DAHP (not shown in Figure 1) (Mir et al., 2015).
Then, 3-dehydroquinate is converted to 3-dehydroshikimate due to the presence of 3-dehydroquinate dehydratase (DHQase). This enzyme exists in two types: type I and type II, but with no sequential or structural homology and with no similar reaction mechanisms between them. Type I DHQase (susceptible to thermal denaturation) is found in plants, fungi and some bacteria (Chaudhuri et al., 1986) and this type only participates in biosynthetic reactions, whereas type II DHQase (resistant to thermal denaturation) can participate either in biosynthetic and catabolic reactions (White et al., 1990). Type I enzymes catalyze cis-dehydration of 3-dehydroquinate, involving the formation of a Schiff-base intermediate (Shneier et al., 1991), where the conserved amino acids lysine and histidine of the active site play a critical catalytic dyad in generating the carbanion intermediate essential for the course of the reaction (Mir et al., 2015).
The fourth enzyme of the shikimate pathway is the shikimate dehydrogenase (SDH) that acts in the reduction of 3-dehydroshikimate to shikimate by means of the transformation of NADPH to NADP+. This enzyme exists in a bifunctional complex in plants (Chaudhuri and Coggins, 1985; Coggins et al., 1987). The catalysis consists of an acid-base mechanism in which a lysine and an aspartate residue form a catalytic dyad. The lysine function is to deprotonate the 3-hydroxyl group of shikimate, its amino group acting as a base (Mir et al., 2015).
Shikimate kinase (SK) is the enzyme responsible for the fifth step of the shikimate pathway. This step is the phosphorylation of shikimate to shikimate-3-phosphate by converting ATP to ADP. The SK is a simple protein (26 kDa in Arabidopsis thaliana) that is composed of three domains: the core domain, the lid domain, and the shikimate binding domain. Within the shikimate binding domain, there are several highly conserved amino acids involved in the interaction with the substrate (Mir et al., 2015).
The penultimate step of the pathway is catalyzed by 5-enolpyruvylshikimate 3-phosphate (EPSPS). This is a monomer protein responsible for the reversible transfer of an enolpyruvoyl moiety from phosphoenolpyruvate (PEP) to shikimate-3-phosphate, giving EPSP and phosphate as reversible products. It is remarkable that in this reaction, which proceeds with the cleavage of the C-O bond of PEP, not with the cleavage of the high energy P-O bond (Mir et al., 2015).
Finally, the seventh and last reaction of the shikimate pathway is the one catalyzed by chorismate synthase. In this reaction, EPSPS is converted into chorismate. Although during the course of the reaction there is no redox change (Mir et al., 2015), this enzyme requires the presence of FMNH2. The presence of flavin mononucleotide may have two explanations: 1) the flavin has merely a structural role or 2) flavin restores the reduced form of sulfhydryl group structurally or catalytically important for the enzyme (Hasan and Nester, 1978).
Regulation of this pathway occurs mainly at gene expression. The cDNA of higher plants results in proteins with an amino-terminal signal sequence for plastid import. The expression of some plant genes encoding enzymes that participate in the shikimate pathway is regulated during development (Benfey and Chua, 1989; Maeda et al., 2010) and by certain stimuli like wounding (Dyer et al., 1989; Keith et al., 1991), elicitors or pathogens (Keith et al., 1991; Görlach et al., 1995). At a protein level, in higher plants the enzymes possess an amino-terminal sequence required for plastid import, suggesting that the synthesis of chorismate only takes place inside the plastids (Herrmann and Weaver, 1999).
Tannins and Their Properties: Antioxidants and Relationships with Human Health and Organoleptic Characteristics
The antioxidant properties of tannins have been thoroughly examined from natural sources or from food grade preparations. For example, Shin et al. (2014) studied the antioxidant activities of both hydrolysable and condensed tannins in three cultivars of persimmon. The radical scavenging activity (RSA) is often used to determine the antioxidant capacities and particularly, the 2,2′-azinobis-3-ethylbenzthiazolin-6-sulfonic acid (ABTS) RSA assay allows to measure the antioxidant activities of both hydrophilic and hydrophobic antioxidant chemicals (Floegel et al., 2011). Interestingly, 1,000 μg/ml of soluble tannins from three persimmon cultivars showed higher ABTS RSA than 100 μg/ml of L-ascorbic acid (Shin et al., 2014). The 2,2-diphenyl-1-picrylhydrazyl (DPPH) RSA assay (Blois, 1958) was also conducted in the same study to provide evidence that the DPPH RSA of unripe persimmons was significantly higher than those of ripe persimmons, mirroring the trend of tannin accumulation, which decreases during fruit ripening. Additionally, Ricci et al. (2016) studied the antioxidant activity of commercial food grade tannins (extracted from grape and Quercus tissues) in wine samples. Samples with higher tannin content (measured as (+)-catechin equivalents) showed, in most cases, the highest values of DPPH radical scavenging activity. The reducing power of the samples, estimated with the ferric-reducing antioxidant power (FRAP) assay (Benzie and Strain, 1999), ranged between 0.513 and 0.662 mM of FeSO4∙7H2O, providing effective reducing properties in some cases. Finally, and at least for wine, it seems that based on ABTS, FRAP, and DPPH assays, hydrolysable tannins show higher antioxidant activity than condensed tannins (Pascual et al., 2017; Vignault et al., 2018).
The use of tannins in food and beverages provides certain advantages, like protection from oxidation processes. In the industry, tannins are used as flavoring agents, and, for example, commercial tannins are added for the clarification of grape musts and wines (Ricci et al., 2016). This helps improve the color stability and avoid oxidation (Versari et al., 2013), offering a natural alternative to synthetic antioxidants. The oxidation of certain compounds plays a crucial role in the flavor of some beverages like beer, especially during aging processes (Vanderhaegen et al., 2006). To further confirm the effect of tannins on beverages, De Francesco et al. (2020) added various tannin-rich extracts to beer. This addition led to an improvement in turbidity, color formation, foam quality, citrus and spicy notes, and an increase in the body of the beer.
As previously mentioned, the antioxidant capacities of tannins are of high importance for human health, providing neuroprotective, cardioprotective, and even antitumoral activities. In a recent review, Maugeri et al. (2022) summarized the in vitro and in vivo effects of tannins in several experimental models and provided evidence of anti-bacterial and anti-inflammatory effects. It can be seen that the anti-inflammatory effects of tannins are related to palliating oxidative stress, reducing ROS and NOS, avoiding DNA damage, and taking part in some signaling routes (Maugeri et al., 2022). In addition, the health benefits of tannins have been thoroughly reviewed elsewhere (Basu et al., 2014; Cory et al., 2018; Lu et al., 2021).
Tannins are present in foods and beverages, providing them with the perception of astringency, which is felt as a sensation of dryness in the mouth and tongue. This event is thought to be due to the precipitation of oral proteins and mucopolysaccharides when they interact with tannins (Baxter et al., 1997). The mechanism of astringency perception has been widely discussed and some hypotheses have been proposed: 1) interaction between tannins and epithelial receptors (Ma et al., 2014), 2) physical movement of mouth musculature (Pires et al., 2020) or 3) increasing of the oral friction caused by protein-tannins aggregates (Rossetti et al., 2009). As tannins are widely distributed among plants, astringency is also present in many food-sources, such as rice (Shao et al., 2018), tea (Xu et al., 2018), grape (Sun et al., 2020), cocoa (Misnawi et al., 2005), and also in some cereals like sorghum and millet (Ahmad et al., 2018). Nevertheless, not all plant-sources have the same quantity of tannins. In the case of proanthocyanidins, the amount of (+)-catechin in some sorghum and millet varieties ranges from 172 to 179 mg/100 g for sorghum and from 172 to 174 mg/100 g (Ahmad et al., 2018), respectively. On the other hand, 11–54 mg/g of (+)-catechin were found in eight different cultivars of persimmon (Akagi et al., 2010b). Concerning ellagitannins, the content in strawberry fruits was around 2–18 mg/g (Karlińska et al., 2021).
Interestingly, astringency is often accompanied by bitterness, which is a source of confusion between both taste properties (Arnold et al., 1980; Green, 1993; Carpenter et al., 2019). However, the perception of astringency seems to be a secondary attribute when both astringency and bitterness are simultaneously perceived in the same product (Arnold et al., 1980; Green, 1993). Interestingly, the astringency/bitterness properties of tannins depend on their degree of polymerization. Molecules with low molecular weight tend to be bitterer, whereas tannins with a high degree of polymerization appear to be more astringent (Robichaud and Noble, 1990; Peleg et al., 1999; Hufnagel and Hofmann, 2008). There is a real challenge in enhancing the tannin content of foods and beverages because of their modulation in organoleptic characteristics, which can lead to non-pleasing foodstuffs. It is needed to better understand the food matrix effects on the perception of astringency and bitterness and to discover new ways to modulate tannins’ contributions to taste perception while maintaining or even increasing these health-promoting compounds.
Some research has already been carried out to solve this particular conflict. For example, the addition of some proteins leads to the removal of highly reactive tannins and the modification of astringency and bitterness (Siebert et al., 1996; Siebert, 1999). Proteins like β-casein, β-lactoglobullin, Ca-caseinate, Na-caseinate, and different types of gelatins have been assayed for their potential bitter-taste-masking (Bohin et al., 2012). For example, the addition of gelatin to red wines causes a decrease in astringency due to the specific removal of high molecular weight galloylated proanthocyanidins (Sarni-Manchado et al., 1999). The contribution of polysaccharides to astringency is also important. Soluble polysaccharides, derived, for example, from pectins during the ripening process, can compete with salivary proteins for polyphenolic substrates like tannins, which lead to a modification in astringency perception (Ozawa et al., 1987).
Proanthocyanidins or Condensed Tannins
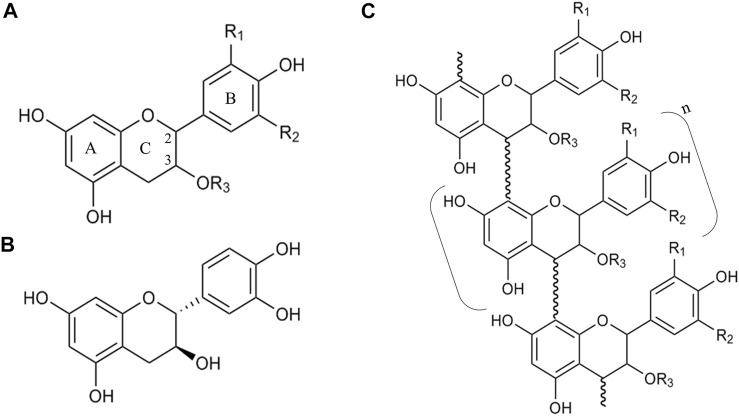
Condensed tannins, or proanthocyanidins, are a highly diverse class of polymers and/or oligomers of a flavonoid called flavan-3-ol. Structurally speaking, flavanols are composed of a carbon backbone of C6-C3-C6, where the C6 compounds correspond to aromatic rings (A- and B-ring) and the C3 is a dihydropyran ring (C-ring) (Aron and Kennedy, 2008). The A-ring is similar to a resorcinol moiety and the B-ring is similar to a catechol moiety (Figure 2A). The monomers of flavan-3-ols are called catechins, and they differ among them in the stereochemistry of the C2 and C3, the presence or absence of galloyl groups, and the hydroxylation of the B-ring (Watrelot and Norton, 2020). When the substituents are R1 = R3 = H and R2 = OH, then the molecule is (+)-catechin (Figure 2B). As flavan-3-ols have two chiral carbons, they possess four diastereisomers, so one isomer of (+)-catechin (2R, 3S configuration) is (+)-epicatechin (2S, 3S configuration). In nature, the configuration 2R is more common than the 2S configuration (Aron and Kennedy, 2008).
FIGURE 2.
Schematic of the structure of catechins and proanthocyanidins. (A) General structure of a catechin, where (A–C) indicate the distinct rings of the carbon backbone C6-C3-C6, numbers 2 and 3 indicate the chiral carbons (stereochemistry not shown), and R1, R2 and R3 are the possible substituent groups. (B) An example of a catechin (+)-catechin. (C) General structure of a proanthocyanidin, where n indicates the number of repetitions of the monomer in brackets. The bond between the monomers is called an interflavan linkage.
Aron and Kennedy (2008) described thirteen types of proanthocyanidins based upon the hydroxylation of A-ring and B-ring carbons and the C3 from the C-ring. Hydroxylation of C8 (A-ring) is the most uncommon modification of proanthocyanidins, present only in the cases of proteracacinidin and promelacacinidin. By contrast, in the case of C3 (C-ring), only in 5 types a proton is present instead of a hydroxyl group (Aron and Kennedy, 2008). In this classification, it is not contemplated that C3 substituents despite hydroxyl groups.
Proanthocyanidins Biosynthesis, Transport, and Accumulation
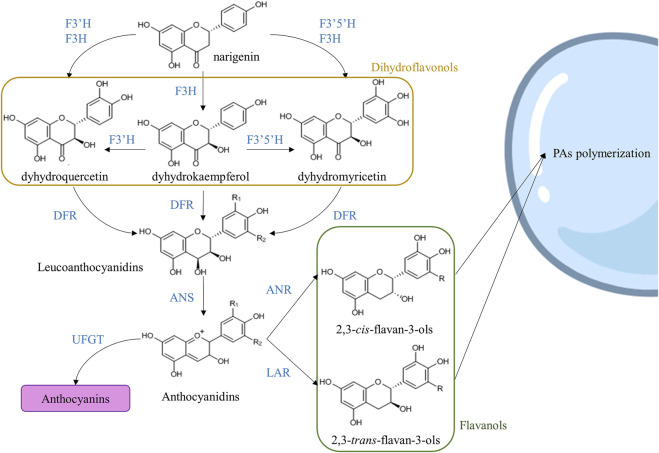
Proanthocyanidins are synthesized from L-phenylalanine through the phenylpropanoid and flavonoid pathways. First, L-phenylalanine is transformed into cinnamic acid or cinnamate by the phenylalanine ammonia lyase (PAL), and the cinnamate is hydroxylated to p-coumaric acid by means of cinnamate 4-hydroxylase (C4H). The next step is the addition of coenzyme A (CoA) to the p-coumaric acid by the catalysis of 4-coumaroyl-CoA ligase (4CL), yielding p-coumaroyl-CoA (Vogt, 2010; Deng and Lu, 2017). 4-coumaroyl-CoA and 3 molecules of malonyl-CoA are the substrates of chalcone synthase (CHS), which catalyzes the first step of the flavonoid pathway, yielding naringenin chalcone, and chalcone isomerase (CHI) then convertes this molecule to the flavanone naringenin. Naringenin serves as a substrate for flavonoid 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′5′-hydroxylase (F3′5′H). If naringenin is the substrate of F3′5′H and F3H, the product is a dihydroflavonol called dihydromyricetin, while the combined action of F3′H and F3H yields the production of dihydroquercetin. Dihydroflavonol 4-reductase (DFR) converts dihydromyricetin to leucodelphinidin (R1 = R2 = OH in Figure 3) and dihydroquercetin to leucocyanidin (R1 = OH and R2 = H in Figure 3). Then, the antocyanidin synthase (ANS), also called leucoanthocyanidin dioxygenase (LDOX), produced delphinidin and cyanidin from leucodelphinidin and leucocyanidin, respectively. Additionally, dihydrokaempferol is derived from naringenin and can be converted into leucopelargonidin (R1 = R2 = H in Figure 3) and pelargonidin by DFR and ANS catalysis, respectively. Cyanidin, delphinidin, and pelargonidin can be transformed into 2,3-cis-flavan-3ols (epi)catechin, (epi)gallocatechin, and (epi)afzelechin, respectively), by the action of anthocyanidin reductase (ANR). On the other hand, 2,3-trans-flavan-3-ols are synthetized from leucocyanidin, leucodelphinidin, and leucopelargonidin by the action of leucoanthocyanidin reductase (LAR) (Figure 3) (Akagi et al., 2011; Yu et al., 2019; Wei et al., 2021).
FIGURE 3.
Simplified flavonoid pathway. One branch of this pathway yields flavanols that can be transported to the vacuole by several proposed mechanisms. Abbreviations: flavonoid 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′5′-hydroxylase (F3′5′H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT), anthocyanidin reductase (ANR), and leucoanthocyanidin reductase (LAR). Modified from Akagi et al., 2011; Yu et al., 2020; Wei et al., 2021.
PAs are found to accumulate exclusively in the vacuoles, forming glycoside conjugates (Grotewold, 2004; Kitamura et al., 2010; Fontes et al., 2011). In particular, some vacuoles specialized in PA accumulation are known as “tannin vacuoles” (Terrier et al., 2009). The knowledge about biophysical and biochemical characteristics of vacuoles’ membranes is very elusive, converting tannin transport into these organelles in a poorly known mechanism. One mechanism proposed for the transport of PAs from the cytosol to the vacuole is membrane transporters. There are three kinds of putative membrane transporters for PAs: ATP binding cassette (ABC) transporters, mammalian bilitranslocase (BLT) transporters, and multidrug detoxification and extrusion (MATE) transporters (Rousserie et al., 2019). Only ABC transporters act as primary active transporters, using ATP for transport, while BLT and MATE transporters function as secondary active transporters, using an electrochemical gradient to move PAs.
The name “condensed tannins” for PAs is because the acid hydrolysis of these compounds releases anthocyanidins due to the breakage of interflavan linkages (Deng and Lu, 2017). As mentioned before, PAs are built of flavan-3-ol monomers linked by carbon-carbon bonds (Dixon et al., 2013). Actual knowledge indicates that (+)-catechins and (-)-epicatechins are thought to be starter units for PA polymerization (Xie et al., 2003; Kitamura et al., 2004; Dixon et al., 2005). Due to the interflavan linkages, two types of PAs can be distinguished. The A-type consists of subunits linked by C4-C8 and/or C4-C6 (A-ring carbons) and C2-O-C7 or C2-O-C5 bonds. On the other hand, B-type PAs presents only the two possible linkages C4-C8 and/or C4-C6 (Yu et al., 2020). The distribution of A-type and B-type PAs is different in nature. A-type PAs are mainly found in berries like bilberry (Vaccinium myrtillus) and cranberry (Vaccinium oxycoccus), cinnamon (Cinnamomum spp.) and peanuts (Arachis hypogaea), for example (Gabetta et al., 2000; Gu et al., 2002; Gu et al., 2003), while B-types are widely distributed (Gabetta et al., 2000; Gu et al., 2003; Suvanto et al., 2020). Condensed tannins are responsible for the bitterness and astringency of many fruits. For example, in apples, astringency is correlated to the mean degree of polymerization (mDP) and the galloylation degree of these compounds, but it is negatively correlated with the hydroxylation level of the B-ring of flavan-3-ols (Saxe et al., 2021).
In the next sections, we focus on the genetic factors known so far to be involved in PAs regulation, which are summarized in Table 1.
TABLE 1.
Summary of principal genes regulating tannins biosynthesis in crops species.
| Gene | Gene name | Species | References |
|---|---|---|---|
| Condensed tannins or proanthocyanidins | |||
| DkANR | Anthocyanidin reductase | Diospyros kaki | Ikegami et al. (2007) |
| DkLAC1 | Laccase | Diospyros kaki | Mo et al. (2015) |
| DkLAR | Leucoanthocyanidin reductase | Diospyros kaki | Ikegami et al. (2007) |
| DkMYB2/4 | MYB-family transcription factor | Diospyros kaki | Akagi et al. (2009b), Akagi et al. (2010a) |
| DkMYC1 | bHLH-family transcription factor | Diospyros kaki | Naval et al. (2016) |
| FabHLH3 | bHLH-family transcription factor | Fragaria x ananassa | Schaart et al. (2013) |
| FaMYB9/11 | MYB-family transcription factor | Fragaria x ananassa | Schaart et al. (2013) |
| MdMYB9/11 | MYB-family transcription factor | Malus domestica | Sun et al. (2019) |
| MdMYBPA1 | MYB-family transcription factor | Malus domestica | Wang et al. (2018) |
| PpMYB7 | MYB-family transcription factor | Prunus persica | Zhou M. et al. (2015) |
| PpMYB18 | MYB-family transcription factor | Prunus persica | Zhou et al. (2019) |
| VvLAR1/2 | Leucoanthocyanidin reductase | Vitis vinifera | Bogs et al. (2005) |
| VvMYB5a/5b | MYB-family transcription factor | Vitis vinifera | Deluc et al. (2006), Deluc et al. (2008) |
| VvMYBC2-L1/2/3 | MYB-family transcription factor | Vitis vinifera | Yu et al. (2020) |
| VvMYBPA1 | MYB-family transcription factor | Vitis vinifera | Huang et al. (2014) |
| VvMYBPA2 | MYB-family transcription factor | Vitis vinifera | Terrier et al. (2009) |
| Hydrolysable tannins | |||
| CsSDH | Shikimate dehydrogenase | Camellia sinensis | Huang et al. (2014) |
| DkSDH | Shikimate dehydrogenase | Diospyros kaki | Akagi et al. (2009a) |
| FaTA | Tannase | Fragaria x ananassa | Dai et al. (2020) |
| PgSDH3s/4 | Shikimate dehydrogenase | Punica granatum | Habashi et al. (2019) |
| PgUGT84A23/24 | UDP-glycosyltransferase | Punica granatum | Ono et al. (2016) |
| VvSDH3/4 | Shikimate dehydrogenase | Vitis vinifera | Bontpart et al. (2016) |
Proanthocyanidin Regulation
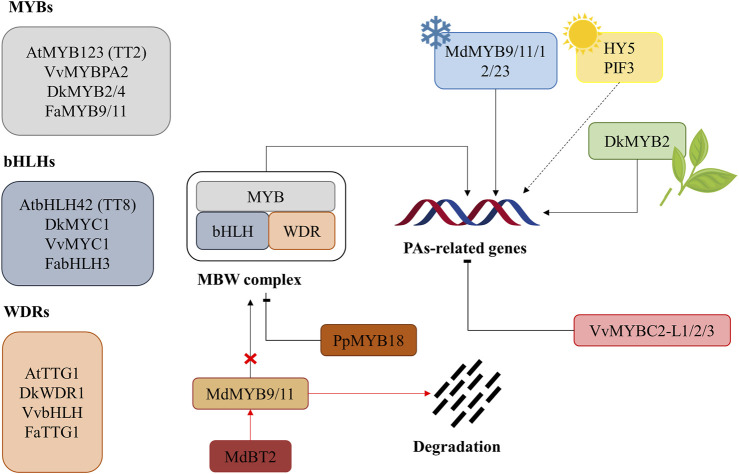
Regulation of proanthocyanidin biosynthesis occurs mainly at the transcriptional level, especially for the genes encoding biosynthetic enzymes (Koes et al., 2005; Xu et al., 2015). PAs synthesis is regulated during fruit growth, as their accumulation starts with the development of the fruit and stops when it starts ripening (Wei et al., 2021). The regulation of PAs is well-known in A. thaliana where there are 6 families of transcription factors thought to participate in PAs synthesis regulation: MYB, basic helix-loop-helix (bHLH, also known as MYC), tryptophan-aspartic acid repeat (WDR)-containing protein 40 (WD40), WRKY, and MADS-box proteins and zinc fingers (Terrier et al., 2009; Rousserie et al., 2019). The MYB-bHLH-WD40 (MBW) complexes are essential regulator elements for PAs accumulation and gene expression (Figure 4; Table 1) (Koes et al., 2005; Ramsay and Glover, 2005; Wei et al., 2021). Due to the ubiquity of bHLH and WD40 in transcriptional regulation, it is the C-terminal part of the MYB protein in the MBW-complex that is responsible for the specificity in activating target genes. Meanwhile, bHLH is responsible for binding to the nucleotide sequence (normally E-box or G-box cis-elements) (Xu et al., 2014), and WD40 is crucial for the stabilization of the complex (Huang et al., 2014; Yu et al., 2020). Both transcription factors, MYB and bHLH, can specifically interact with nucleotide sequences within the flavonoid promoter. MYBCORE and AC-rich elements are the two main binding sites for MYBs in the regulation of flavonoids and PAs, although the exact motif for MYB binding remains unknown in most PAs structural genes (Constabel, 2018). In A. thaliana, the MBW-complex is composed of AtMYB123 (or Transparent Testa, TT2), AtbHLH42 (or TT8), and Transparent Testa Glabra31 (or TTG3), which is the WD40 protein. This complex is responsible for the activation of ANR (Rousserie et al., 2019), LDOX and DFR genes, leading to the accumulation of PAs in the seed coat (Nesi et al., 2001; Baudry et al., 2004). In particular, TTG2-like proteins are thought to be crucial in PA transportation (Debeaujon et al., 2001; Gonzalez et al., 2016). Homologs of TT2, TT8, and TTG1 in other fruits and plants are often searched for their role in flavonoid and PA regulation (Yu et al., 2020).
FIGURE 4.
General regulation scheme of proanthocyanidin synthesis. The scheme shows regulation via MBW-complex formation and cold-, light- and wounding-induced regulation. The degradation of MdMYB9/11 through BT2 and the proteasome is also shown with the red arrows, resulting in the impossibility of these two transcription factors to form the MBW-complex. Arrows indicate positive regulation, while flat-ended lines mean negative regulation (repression). Dotted lines indicate a process not fully elucidated. At: Arabidopsis thaliana, Dk: Diospyros kaki, Fa: Fragaria x ananassa, Md: Malus domestica, Pp: Prunus persica, Vv: Vitis vinifera.
Besides regulation at the gene level, the biosynthesis of PAs is affected by biotic and abiotic stresses and plant hormones. Environmental or abiotic stresses, such as low temperature, drought, or wounding, among others, may lead to the production of ROS and the consequent oxidative stress responses (Suzuki et al., 2012). As described previously, flavonoids and other antioxidant compounds can palliate the action of ROS, and the genes responsible for their synthesis can be induced under these circumstances (Winkel-Shirley, 2002). Water quantity has been proven to influence the phenolic content of grapes (Castellarin et al., 2007). Light also plays a part in the regulation of PAs, and it has been suggested that light-signaling proteins, such as HY5 and PIF3, may interact with some promoters of PAs-related genes, like CHS, CHI, and DFR (Shin et al., 2007). The action of plant hormones courses through their interaction with MBW-complexes, thus regulating flavonoid genes at the transcriptional or post-transcriptional level (Jaakola, 2013). In addition to abiotic stresses, plants are also exposed to the influence of fungi, bacteria, viruses, and herbivores. In this context, accumulation of PAs is a plant defense strategy against these biotic stresses (Dixon et al., 2005; Ullah et al., 2017). The two main hormones related to biotic stresses are salicylic and jasmonic acids, suggesting a possible role for both molecules in PA regulation.
Proanthocyanidins in Persimmon Fruit (Diospyros kaki) and Its Regulation
Persimmon possesses PAs-rich fruits which make them inedible until physiological ripening or by artificial treatment, such as ethanol. In persimmons, PAs monomers are mainly prodelphinidins and they present a high degree of galloyl residues (Li et al., 2010). Pollination constant and non-astringent (PCNA) is a persimmon mutant spontaneously generated, whose principal characteristic is the total absence of astringency, making it a desirable fruit for consumption. The absence of astringency is explained by the reduced accumulation of epigallocatechin and epigallocatechin gallates at the early stage of fruit development.
In persimmon, several PAs-biosynthesis related genes have been identified: ANR (DkANR) and LAR (DkLAR) (Ikegami et al., 2007; Nakagawa et al., 2008; Akagi et al., 2009b). In vitro assays of DkANR in Escherichia coli showed a conversion of anthocyanidins to both isomers 2,3-cis-flavan-3-ols and 2,3-trans-flavan-3-ols (Akagi et al., 2009a), demonstrating the role of DkANR in PA accumulation in persimmon fruit. However, further steps in PA biosynthesis in persimmon are poorly known (Zhao et al., 2010), with the exception of DkLAC1, a laccase gene that participates in the polymerization of PAs (Mo et al., 2015). Several transcription factors crucial for PA accumulation in persimmon have been identified as DkMYB2/4, DkMYC1, and DkWDR1 as homologs of TT2, TT8, and TTG1, respectively (Akagi et al., 2009b; Akagi et al., 2010a; Naval et al., 2016). In yeast and Nicotiana benthamiana, it has been shown that DkMYB2, Shalini, and DkWDR1 interact together to act as a MBW-complex (Gil-Muñoz et al., 2020). Moreover, in PCNA fruits, DkMYB4 regulates the expression of ANR and F3′5′H (Akagi et al., 2009b). DkMYB4 is expressed mainly in fruit flesh and seed during development, and it was shown to bind to MYBCORE cis-motifs of the promoter region of three PAs-biosynthetic genes (Figure 3): DkANS, DkF3′5′H, and DkANR (Akagi et al., 2009b). However, in the same study, DkMYB4 was shown not to regulate the expression of DkLAR (Akagi et al., 2009b), supporting the possibility that in persimmon, like in other species, the expression level of LAR did not correlate with PA accumulation (Pang et al., 2007). Overexpression of DkMYB4 results in accumulation of PAs with toxic effects when expressed ectopically in persimmon and kiwifruit (Actinida deliciosa) calli (Akagi et al., 2009b), while RNAi knockdown results in a similar expression pattern to the one observed in PCNA fruits (Dixon et al., 2013). Of interest, DkMYB4 expression was in turn regulated by DkbZIP5, an abscisic acid (ABA)-responsive transcription factor (Akagi et al., 2012). The presence of ABA-responsive elements in the DkMYB4 promoter suggests that the phytohormone ABA may modulate PA biosynthesis and content (Akagi et al., 2012).
Another MYB gene has been described as playing a part in the regulation of PAs. DkMYB2 is a wound-inducible expression factor (see Figure 4) whose ectopic expression in persimmon and kiwifruit calli positively regulates the expression of ANR and LAR (Akagi et al., 2010a). Another interesting point is the regulation of PAs pathway genes by the action of ethanol treatment in order to remove astringency (Araki et al., 1975; Ikegami et al., 2007). Plants treated with ethanol (contained in plastic bags with 10 ml of 5% ethanol) (Ikegami et al., 2007) presented lower expression of two shikimate pathway genes, most PA pathway genes, and serin carboxypeptidase-like proteins, that are putatively involved in PA accumulation (Terrier et al., 2009), compared to the control non-treated plants (Ikegami et al., 2007). In this sense, the hypothesis of astringency removal is that ethanol both reduces PA biosynthesis and enhances PA solubility (Akagi et al., 2011).
Proanthocyanidins in Grapevine (Vitis vinifera) and Wine
Condensed tannins are highly studied in the case of grapes and wine due to their ability to offer astringency and change the organoleptic properties of the wine (Robinson et al., 2021). PAs are found mostly in red wines, a fact caused by maceration and skin contact, which does not occur in white wines. During alcoholic fermentation, both PAs extraction and mDP of tannins increase (Aron and Kennedy, 2008). In grapes, skin PAs are different from seeds PAs not only in their composition, but also in the mDP. Skin PAs are catechin-rich polymers, with epicatechin and epigallocatechin being the most common terminal units, and mDP being around 31–33, depending on the variety (Souquet et al., 1996; Hanlin and Downey, 2009). Seeds PAs are rich in epicatechin either as a monomer or as an extension unit (Dixon et al., 2013), and the mDP varies depending on the variety: 3.8–5.9 in Cabernet Sauvignon and 8–11 in Syrah before and after harvest maturity (Kyraleou et al., 2017; Blancquaert et al., 2019; Watrelot and Norton, 2020).
For V. vinifera, much more is known in the case of MYBs factors than in those of bHLH and WD proteins. In the case of bHLHs, VvMYC1 is thought to activate VvCHI and VvANR promoters by interacting with VvMYBPA1 (Huang et al., 2014). Indeed, it is required the interaction between VvMYC1 and VvMYBPA1 for the activation of the two promoters, leaving in evidence that VvMYC1 by its own cannot trigger PAs biosynthesis (Hichri et al., 2010). VvMYB5a and VvMYB5b were discovered to regulate positively PAs synthesis in skin, flesh and seeds of grapes during the early stages of fruit development (Deluc et al., 2006; Deluc et al., 2008) and they have been reported to regulate promoters of the flavonoid genes VvCHI and VvLAR1 together with AtEGL3 (bHLH partner). VvMYB5b overexpression in tobacco (Nicotiana tabacum) induced a rising in PAs concentration (Deluc et al., 2006; Deluc et al., 2008). On the other hand, overexpression of VvMYB5a caused a strong increase in several metabolites of the phenylpropanoid pathway, including anthocyanins, flavonols, and PAs, as well as an alteration in lignin metabolism, suggesting that this transcription factor may be involved in the regulation of the different branches of this pathway.
Interestingly, in V. vinifera, the transformation of 3-deoxy-leucocyanidin is controlled by two highly related genes: VvLAR1 and VvLAR2 (Bogs et al., 2005; Wei et al., 2021). Curiously, VvMYBPA1 and VvMYBPA2, another MYB factor identified in grapes, can activate ANR and LAR1, but not LAR2, affecting directly the flavonoid pathway (Bogs et al., 2007; Terrier et al., 2009; Wei et al., 2021). Silencing of ANR genes in V. vinifera resulted in an accumulation of anthocyanins and flavonols as a consequence of metabolic flux redirection towards other flavonoid compounds rather than proanthocyanidins (see Figure 3) (Robinson et al., 2021). Paolocci et al. (2007) demonstrated that ANR and LAR1 are regulated by the same bHLH factor that enhances the accumulation of PAs. Furthermore, the expression pattern of VvLAR1 and VvLAR2 is suggested to differ in skin and seed grapes, with VvLAR1 being specific of seed, while VvLAR2 can be found in both tissues (Wei et al., 2021). In addition, the LAR gene has been reported to be involved in determining tannin polymer length; indeed, when this gene is knocked-out, grape seed PAs have higher mDP in transgenic lines (Robinson et al., 2021). In addition, the expression of VvWRKY26 (a member of the WRKY family) in grape tissues participates in the regulation of PAs biosynthesis and vacuolar transportation (Wei et al., 2021).
On the other hand, several repressors of PA biosynthesis have been identified, such asVvMYBC2-L1, VvMYBC2-L2 or VvMYBC2-L3 (Yu et al., 2020). Overexpression of both VvMYBC2-L1 and VvMYBC2-L3 in grapevine hairy roots results in a significant decrease in PAs concentration. Interestingly, some microRNAs (miRNAs) may be important in the regulation of PAs and polyphenol biosynthesis (Rock, 2013). The miRNA TAS4/miR828 targets VvMYBA6 and VvMYBA7, two genes with homology withVvMYBPA1 and VvMYBPA2 (Rock, 2013). These repressors cause a reduction in the transcription of flavonoid and phenylpropanoid structural genes in addition to the inhibition of PA biosynthesis, as they also disrupt the transcriptional activation of the MBW complex (Yoshida et al., 2015; Ma et al., 2018).
Water conditions are determinant in grape characteristics, in fact water deficit can regulate the genes involved in the biosynthesis of PAs (Casassa et al., 2015). Full irrigated V. vinifera showed grape skin and seed extracts with high astringency with respect to non-irrigated plants (Kyraleou et al., 2016). One hypothesis is that water deficits may induce VvLAR2 and VvMYBPA1, increasing PAs levels and their polymerization in the Cabernet Sauvignon variety (Yu et al., 2020). Light can also affect gene expression; one example is that VvDFR expression, needed for the synthesis of PAs precursors (Figure 3), can be induced by light (Gollop, 2002; Wei et al., 2021). In addition, light can trigger the transcription of VvCHS2, VvDFR, and VvLDOX compared with shading treatment (Liu W. et al., 2017; Wei et al., 2021). On the other hand, the effect of temperature on gene expression is not conclusive because some studies do not find any correlation between high temperature and PAs concentration (Mori et al., 2004; Pastore et al., 2017), but some others show that high temperature causes PAs accumulation in grapes (Bonada et al., 2015; Poudel et al., 2020), due to a decrease in the levels of VvANR and VvLAR1 mRNA. Additionally, VvMYBA1 mRNA accumulated when treated with ABA, leading to increased levels of PAs but also higher levels of anthocyanins and PAs structural genes (Jeong et al., 2004).
Proanthocyanidins Regulations in Other Crops
Other crops have been the object of study in what pertains to PAs regulation. In reference to strawberries (Fragaria x ananassa), the major and common compounds related to PAs are catechin, proanthocyanidin B1, proanthocyanidin trimer, and proanthocyanidin B3 (Giampieri et al., 2012; Fierascu et al., 2020). In strawberry, epicatechins and catechins levels, as well as procyanidin levels, decrease with ripening (Aaby et al., 2012), specifically in the final maturation stage, this tendency has been observed in other species like bilberry (Suvanto et al., 2020). The functional orthologues of AtTT2, AtTT8, and AtTTG1 have been elucidated in strawberry, being FaMYB9/FaMYB11, FabHLH3, and FaTTG1, respectively (Schaart et al., 2013). In a yeast-two hybrid assay, FaMYC1 and FabHLH33 were also discovered to be related to PA synthesis, but their implication remains elusive. It is speculated that FaMYC1 and FabHLH33 could dimerize in the same way as other bHLHs (Feller et al., 2006). Derived from the same work, Schaart et al. (2013) hypothesized that FaMYB5 and FabHLH3Δ, a truncated version of FabHLH3 identified in strawberries, act as negative regulators of flavonoid biosynthesis. FaMYB5 may combine with FaMYB1, a confirmed repressor of PAs biosynthesis with high sequence homology to VvMYBC2-L1 (Aharoni et al., 2001; Paolocci et al., 2011), and they together interfere with the activity of MBW-complexes. On its part, FabHLH3Δ is capable of interacting with FaMYB9 or FaMYB11 and causing the blockage of their binding sites for bHLHs, avoiding the MBW-complex formation (Schaart et al., 2013). Interestingly, it was found that FaTT12-1 is a putative orthologue of AtTT12, which is light-sensitive. In fact red light can significantly activate its expression (Chen et al., 2018), suggesting a possible role in PA biosynthesis regulation. Delgado et al. (2018) demonstrated that treatment with methyl jasmonate caused upregulation of FaANS, FaLAR, and FaUFGT genes and a downregulation of the FaANR gene compared to the control, leading to increased anthocyanin levels and decreased PAs content. On the other hand, treatment with methyl jasmonate and jarin-1, an inhibitor of the bioactive form of jasmonate, jasmonoyl-isoleucine, caused the opposite effect. The increase of PAs in fruits treated with methyl jasmonate and jarin-1 could be explained by the upregulation of FaMYB9, FaMYB11, and FabHLH33 together with FaANR (Delgado et al., 2018). Together, these data suggest a connection between the phytohormone jasmonic acid and PA biosynthesis during strawberry fruit ripening. Inoculation experiments with Colletotrichum acutatum and Botrytis cinerea in strawberries at different maturity stages showed that fungal infection was much higher in the case of riper fruits, but almost absent in the case of unripe strawberries, where the expression levels of LAR and ANR were found to be higher (Guidarelli et al., 2011; Haile et al., 2019). This matches with the fact that flavan-3-ols and PA concentrations decrease as ripening progresses (Carbone et al., 2009; Nagpala et al., 2016), reinforcing the role of PAs in plant defense against biotic stresses.
The MYBPA1 ortholog in apple (Malus domestica) is MdMYBPA1 (Wang et al., 2018) found that PAs are not only regulated through the MBW-complex in this crop; indeed, a NAC transcription factor, NAC52, has been recently reported for its ability to bind to the promoters of MdLAR, MdMYB9, and MdMYB11, and this union leads to PAs accumulation when MdNAC52 is ectopically overexpressed (Sun et al., 2019). Interestingly, a negative regulation of PAs in apples occurs via the degradation of MdMYB9 protein by MdBT2, which substantially reduces PAs concentration (An et al., 2018). MdBT2 is a bric-à-brac, tramtrack, and broad complex protein, essential for the degradation of proteins through the ubiquitin proteasome system. Low temperatures trigger the expression of MdMYB9/11/12/23 and MdMYBPA1, a series of transcription factors capable of binding promoters of PAs structural genes, causing a rise in PAs levels, hypothetically to prevent damage due to cold stress (Gesell et al., 2014; An et al., 2018; Wang et al., 2018). MdMYB23 protein is induced by cold stress, which leads to PAs accumulation, but as MdMYB9, it is normally degraded by MdBT2 via ubiquitin-proteasome pathway (An et al., 2018). Jasmonic acid is reported to increase PA accumulation due to an upregulation of MdMYB9 and MdMYB11 (An et al., 2015). MdJAZ18 is a repressor of the jasmonate signaling pathway that is susceptible to phosphorylation by MdSnRK1.1. Once phosphorylated, MdHAZ18 is degraded by the 26 S proteasome, causing the release of MdbHLH3. MdbHLH3 can then act with MdMYB9 and MdMYB11 to increase anthocyanins and PA synthesis (Liu X.-J. et al., 2017). In peach (Prunus persica), PpMYB7 is reported to bind to the promoter of PpLAR, thus regulating PA synthesis (Zhou H. et al., 2015). A R2R3-MYB repressor, PpMYB18, downregulates PAs biosynthesis in transgenic tobacco (Nicotiana tabacum) leaves (Zhou et al., 2019). Interestingly, this transcription factor can be induced by PAs-related MYB activators (PpMYBA1 and PpMYB10.1), suggesting a role in the fine-tuning of the levels of these important metabolites by providing a feedback regulation of the MBW-complex during fruit ripening. PpMYB18 may act as both an active and passive repressor of PA biosynthesis by competing with PpMYB10.1 and PpMYBPA1 for the union with PpbHLHs (Zhou et al., 2019).
Hydrolysable Tannins: Ellagitannins and Gallotannins
The presence of hydrolysable tannins in food and beverages is more uncommon than the presence of condensed tannins. They can be found in black currant (Riges nigrum), blackberries (Rubus fructicosus), strawberries, raspberries (Rubus occidentalis), and pomegranate (Punica granatum), guava (Pisidium spp.), mango (Magnifera indica) and in nuts like almonds (Prunus dulcis), pecans (Carya illinoinensis), and walnuts (Juglans regia), with the total absence in species like legumes and cereal grains (Landete, 2011; Smeriglio et al., 2017).
On the opposite of proanthocyanidins, much less is known regarding hydrolysable tannins synthesis and regulation. Structurally, hydrolysable tannins are based on a central sugar core, mainly β-D-glucose, in which phenolic compounds are esterified through its hydroxyl groups (Grundhöfer et al., 2001; Quideau et al., 2011). There are two major classes of these compounds depending on the phenolic groups: gallotannins (gallic acid) and ellagitannins (hexahydroxydiphenic acid, HHDP) (Bar-Ya’akov et al., 2019). Acid hydrolysis of gallotannins yields sugar and gallic acid, whereas the hydrolysis of ellagitannins provides sugar, gallic acid, and ellagic acid (Figure 5) (Smeriglio et al., 2017). Gallotannins are the simplest hydrolysable tannins and are rarely found in nature. They are present in some woody species like mango and almonds; on the other hand, ellagitannins are more common in fruits, nuts and seeds (Landete, 2011; Smeriglio et al., 2017). In fact, a myriad of ellagitannin compounds have been identified to date, principally due to their ability to oligomerize (Okuda and Ito, 2011).
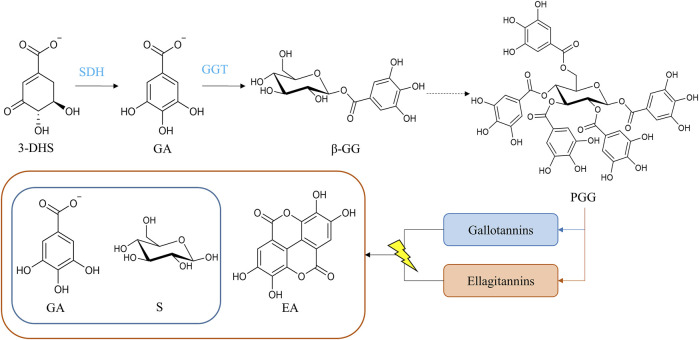
FIGURE 5.
Biosynthetic pathway of hydrolysable tannins. The hydrolysis of gallotannins and ellagitannins yields gallic acid and sugar (glucose represented) and gallic acid, ellagic acid, and sugar, respectively. Dotted lines represent multistep reactions. Abbreviations: 3-dehydroshikimate (3-DHS), gallic acid (GA), sugar unit (S), ellagic acid (EA), β-glucogallin (β-GG), pentagalloylglucose (PGG), shikimate dehydrogenase (SDH), and β-glucogallin-dependent galloyltransferases (GGT).
The key molecule for hydrolysable tannin biosynthesis is gallic acid, which is derived from 3-dehydroshikimate via a shikimate dehydrogenase (Niemetz and Gross, 2005; Muir et al., 2011; Bontpart et al., 2016). Furthermore, gallic acid is relevant for the formation of epigallocatechin and epicatechin gallate (Liu et al., 2012), which are proanthocyanidin monomers. Downregulation of the shikimate dehydrogenase, DkSDH, in persimmon resulted in a decrease in epigallocatechin content in PCNA-type mutants (Akagi et al., 2009a), which matches the implication of gallic acid in both types of tannins. Studies in Camellia sinensis and V. vinifera indicate the role of shikimate dehydrogenase in generating gallic acid (Bontpart et al., 2016; Huang et al., 2019). Gallic acid is then esterified with UDP-glucose (Gross, 1983), to form 1-O-galloyl-β-D-glucopyranose (also known as β-glucogallin), which is the simplest gallotannin. Ono et al. (2016) found two UDP-glycosyltransferases (UGTs) in P. granatum, PgUGT84A23 and PgUGHT84A24, related to β-glucogallin synthesis. According to studies in oak (Quercus rubur and Quercus rubra) and sumac (Rhus typhina), the next steps of galloylation occur with a remarkable specificity following the sequence: β-glucogallin, 1,6-digalloylglucose, 1,2,3-trigalloylglucose, 1,2,3,6-tetragalloylglucose and finally, 1,2,3,4,6-pentagalloyglucose (Schmidt et al., 1987; Gross and Denzel, 1991). These subsequent additions of galloyl moieties (Haslam and Cai, 1994) result in the formation of 1,2,3,4,6-pentagalloyl-D-glucose, the common precursor of both gallotannins and ellagitannins (Figure 5). Pentagalloylglucose can be oxidized to generate 3,4,5,3′,4′,5′-hexahydroxyphenoyl (HHDP) residues, a molecule that once liberated from the sugar core in the form of hexahydroxydiphenic acid, spontaneously lactonizes into ellagic acid (Niemetz and Gross, 2005; Smeriglio et al., 2017). The oxidation of pentagalloylglucose occurs via oxygen-dependent laccase-type enzyme catalyzation in Tellima grandiflora (Niemetz et al., 2001; Niemetz et al., 2003; Niemetz and Gross, 2003a; Niemetz and Gross, 2003b).
Besides β-D-glucose, which is by far the most common sugar core for gallotannins and ellagitannins, other exotic polyols may appear, such as xylose, sorbitol, fructose, or even not-sugar compounds like shikimic, and quinic acids. However, their abundance in nature is extraordinarily low and they are only found in maple (Acer negundo), chestnut (Castanea sativa) and witch-hazel (Hamamelis sp.) (Smeriglio et al., 2017). Galloylation reactions with β-glucogallin as substrate can yield a wide range of molecules from di-to octagalloylglucoses, although the most common is 1,2,3,4,6-pentagalloyl-D-glucose (Smeriglio et al., 2017). The enzyme β-glucogallin-dependent galloyltransferases (Niemetz and Gross, 2005) is thought to be involved in the modification of gallotannins, one example is the transformation of hexa-/heptagalloylglucoses into 3-O-trigalloyl-1,2,4,6-O-tetragalloyl-β-D-glucopyranose, and other gallotannins with high degree of galloylation (Niemetz and Gross, 2005). Two galloyl residues can form what is called a meta- or para-depside bond, which is the result of the esterification between the carbonyl group of gallic acid and the aromatic meta- or para-hydroxyl group and comprises one of the most common modifications of gallotannins (Smeriglio et al., 2017).
Ellagitannins can be present in monomeric forms (e.g. casuarticin, corilagin, castalagin, eugeniin, geraniin, corilagin, galloyl-HDDP-glucose, and potentillin), dimeric forms (e.g. sanguiin H-2, sanguiin H-6, agrimoniin, and lambertianin A), trimeric forms (e.g. nupharin C, nupharin E) or even oligomeric forms (e.g. hirtellin A). Ellagic acid is one of the most important hydrolysable tannins in F. x ananassa fruits (Ariza et al., 2018). It has been recently found that agrimoniin, which is a dimeric ellagitannin composed of two units of potentillin linked by a dehydrogalloyl group (Karlińska et al., 2021), is highly present in “Camino Real,” “San Andreas,” and “Festival” strawberry cultivars (Villamil-Galindo et al., 2021). In fact, agrimoniin has been proposed as a taxonomic marker for the Rosaceae family, to which strawberries belong (Grochowski et al., 2017). Other common ellagitannins in the Fragaria genus are sanguiin H-6, galloyl-bis-HHDP-glucose, and lambertianin C (Giampieri et al., 2012).
The information available in the literature about the genes involved in hydrolysable tannin synthesis is summarized in the next paragraphs and in Table 1.
Regarding ellagitannin biosynthesis, formation of HHDP through the C4-C6 bonding of two respective gallic acids leads to the synthesis of tellimagrandin II as the first product of the pathway in a reaction catalyzed by a laccase-type polyphenol oxidase, as described in strawberry (Karlińska et al., 2021). Esterification of two more gallic acids at C2 and C3 and a possible de-esterification in C1 of tellimagrandin II result in several monomers: pedunculagin, potentillin and casuartictin (Niemetz and Gross, 2005; Karlińska et al., 2021). Further steps implicate dimerization or condensation reactions, forming 3 types of C-O-C in the case of the Roseaceae family: GOD, DOG, and GOG, depending on the donor and the acceptor of the reaction. The GOG-type bond implies two galloyl groups, the DOG-type bond (most frequently found in oligomers) requires a hydroxyl group from an HHDP as donor and a galloyl group as acceptor, and finally, the GOD-type bond is created from a galloyl hydroxyl donor to form an ether linkage with a HDDP as acceptor (Quideau, 2009).
Pomegranate fruit is another example of a reservoir of hydrolysable tannins, with more than 30 chemical species identified in the fruit peel, juice, and seeds (Fischer et al., 2011; Mena et al., 2012; Ito et al., 2014; Ambigaipalan et al., 2017; Wu and Tian, 2017; Liu and Seeram, 2018). The most representative ellagitannins in pomegranate fruit peel are α- and β-punicalagin (Seeram et al., 2005), and gallagic acid, punicalagin, and ellagic acid glycosides count as hydrolysable tannins present in both juice and fruit peel. Only 3,3′-di-O-methylellagic acid and 3,3′,4′-tri-O-methylellagic acid are present exclusively in the seeds (Bar-Ya’akov et al., 2019). Once again, the accumulation of tannins can be modified by regulating the expression of genes encoding key enzymes in their biosynthesis. Under osmotic stress, two shikimate dehydrogenase transcripts, PgSDH3s and PgSDH4, were accumulated, which is thought to increase hydrolysable tannin concentration (Habashi et al., 2019). Moreover, Habashi et al. (2019) reported that sucrose and red light stress could affect the accumulation of gallic acid and hydrolysable tannins with the change of SDH expression in pomegranates.
Tannases are a family of enzymes also known as tannin acyl-hydrolases (EC 3.1.1.20) capable of breaking carboxylic ester bonds present in gallotannins and galloylated flavanols (Ramírez et al., 2008; Rodríguez et al., 2009). These enzymes have been extensively studied in bacteria and fungi (Banerjee et al., 2012), but there is still much to learn in plants. The work of Dai et al. (2020) revealed that transient overexpression and RNAi of a strawberry tannase (FaTA) provoke an alteration in ellagic acid content; in addition, several enzymes belonging to the class I carboxylesterase clade were identified in V. vinifera, J. regia, Citrus clementine, D. kaki, and C. sinensis, indicating the hypothetical existence of more tannase genes in plants. A recent work in Juglandaceae (Wang et al., 2021) reveals regulatory cis-elements found in tannase promoters: E-box and ARR1AT, involved in brassinolide and cytokine responsiveness, respectively, and W-box and WUN-motifs implicated in wound abiotic stress responses. Also, motifs related to flavonoid biosynthesis (MYB-related motifs) and MYC motifs, related to cell growth, were found in tannases from Chinese hickory (Carya cathayensis) and pecan (Carya illinoinensis). These last results suggest the possibility of studying the regulation of tannases belonging to the same clad described by Dai et al. (2020) in other species.
Future Perspectives in Metabolic Engineering of Crops Regarding Tannins
The role of tannins in providing health benefits has been extensively studied (Lu et al., 2021), a reason for which there is an increasing interest in selecting crops or varieties with enhanced levels of these therapeutic compounds. However, as tannins also participate in the organoleptic characteristics of foods and beverages by adding attributes of astringency and bitterness, precaution needs to be taken in order to maintain fruit palatability. As discussed in this review, several strategies can be followed to achieve fruit deastringency, such as ethanol treatment. Nevertheless, there is a need to increase our knowledge about how astringency and bitterness are perceived. In addition, an interesting way of decreasing crop astringency would be to reduce the ratio between high molecular weight and low molecular weight tannins. In this sense, a more precise insight of the molecular mechanisms underlying tannin synthesis may pave the way to develop new varieties more flavorsome. Further research will allow us to achieve foodstuffs that combine higher levels of these health-promoting compounds with a grade of astringency that makes them edible for the consumer. This could be the subject of genetic and biochemical regulation, taking into account the principal regulator genes of tannin biosynthetic pathways. The application of CRISPR/Cas9 technologies could be a solution in the improvement of new fruits by editing genes related to tannins (Zhang et al., 2020). These strategies can be coupled to genome-wide association studies (GWAS) or QTL mapping to find genes, alleles, or markers associated with a high content of tannins and other desirable traits for consumption. With all this knowledge, highly promising projects could be carried out, for example, breeding programs or marker-assisted selection with some elite cultivars.
Acknowledgments
JGV acknowledges the EMERGIA Programme (EMERGIA20_00309-Junta de Andalucía).
Author Contributions
All the authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by grants RTI 2018-099797-B-100 (Ministerio de Ciencia, Innovación y Universidades, Spain) and UMA18-FEDERJA-179 (FEDER-Junta Andalucía). In addition, we acknowledge partial funding by the European Union’s H2020 Programme (BreedingValue; grant number 101000747), and PY20_00408 (PAIDI 2020-Junta de Andalucia). JM has received a predoctoral grant from Ministerio de Ciencia e Innovación (grant PRE 2019-091188).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aaby K., Mazur S., Nes A., Skrede G. (2012). Phenolic Compounds in Strawberry (Fragaria x Ananassa Duch.) Fruits: Composition in 27 Cultivars and Changes during Ripening. Food Chem. 132, 86–97. 10.1016/j.foodchem.2011.10.037 [DOI] [PubMed] [Google Scholar]
- Aharoni A., De Vos C. H. R., Wein M., Sun Z., Greco R., Kroon A., et al. (2001). The Strawberry FaMYB1 Transcription Factor Suppresses Anthocyanin and Flavonol Accumulation in Transgenic Tobacco. Plant J. 28, 319–332. 10.1046/j.1365-313X.2001.01154.x [DOI] [PubMed] [Google Scholar]
- Ahmad F., Pasha I., Saeed M., Asgher M. (2018). Biochemical Profiling of Pakistani Sorghum and Millet Varieties with Special Reference to Anthocyanins and Condensed Tannins. Int. J. Food Properties 21, 1586–1597. 10.1080/10942912.2018.1502198 [DOI] [Google Scholar]
- Akagi T., Ikegami A., Suzuki Y., Yoshida J., Yamada M., Sato A., et al. (2009a). Expression Balances of Structural Genes in Shikimate and Flavonoid Biosynthesis Cause a Difference in Proanthocyanidin Accumulation in Persimmon (Diospyros Kaki Thunb.) Fruit. Planta 230, 899–915. 10.1007/s00425-009-0991-6 [DOI] [PubMed] [Google Scholar]
- Akagi T., Ikegami A., Tsujimoto T., Kobayashi S., Sato A., Kono A., et al. (2009b). DkMyb4 Is a MYB Transcription Factor Involved in Proanthocyanidin Biosynthesis in Persimmon Fruit. Plant Physiol. 151, 2028–2045. 10.1104/pp.109.146985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Ikegami A., Yonemori K. (2010a). DkMyb2 Wound-Induced Transcription Factor of Persimmon (Diospyros Kaki Thunb.), Contributes to Proanthocyanidin Regulation. Planta 232, 1045–1059. 10.1007/s00425-010-1241-7 [DOI] [PubMed] [Google Scholar]
- Akagi T., Katayama-Ikegami A., Kobayashi S., Sato A., Kono A., Yonemori K. (2012). Seasonal Abscisic Acid Signal and a Basic Leucine Zipper Transcription Factor, DkbZIP5, Regulate Proanthocyanidin Biosynthesis in Persimmon Fruit. Plant Physiol. 158, 1089–1102. 10.1104/pp.111.191205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Katayama-Ikegami A., Yonemori K. (2011). Proanthocyanidin Biosynthesis of Persimmon (Diospyros Kaki Thunb.) Fruit. Scientia Horticulturae 130, 373–380. 10.1016/j.scienta.2011.07.021 [DOI] [Google Scholar]
- Akagi T., Suzuki Y., Ikegami A., Kamitakahara H., Takano T., Nakatsubo F., et al. (2010b). Condensed Tannin Composition Analysis in Persimmon (Diospyros Kaki Thunb.) Fruit by Acid Catalysis in the Presence of Excess Phloroglucinol. J. Jpn. Soc. Hort. Sci. 79, 275–281. 10.2503/jjshs1.79.275 [DOI] [Google Scholar]
- Ambigaipalan P., de Camargo A. C., Shahidi F. (2017). Identification of Phenolic Antioxidants and Bioactives of Pomegranate Seeds Following Juice Extraction Using HPLC-DAD-ESI-MSn. Food Chem. 221, 1883–1894. 10.1016/j.foodchem.2016.10.058 [DOI] [PubMed] [Google Scholar]
- An J.-P., An X.-H., Yao J.-F., Wang X.-N., You C.-X., Wang X.-F., et al. (2018). BTB Protein MdBT2 Inhibits Anthocyanin and Proanthocyanidin Biosynthesis by Triggering MdMYB9 Degradation in Apple. Tree Physiol. 38, 1578–1587. 10.1093/treephys/tpy063 [DOI] [PubMed] [Google Scholar]
- An X.-H., Tian Y., Chen K.-Q., Liu X.-J., Liu D.-D., Xie X.-B., et al. (2015). MdMYB9 and MdMYB11 Are Involved in the Regulation of the JA-Induced Biosynthesis of Anthocyanin and Proanthocyanidin in Apples. Plant Cel Physiol 56, 650–662. 10.1093/pcp/pcu205 [DOI] [PubMed] [Google Scholar]
- Araki C., Furuta M., Kaneko K., Aketagawa T. (1975). Studies on the Removal of Astringency in Japanese Persimmon (Diospyros Kaki L.). Engei Gakkai zasshi 44, 183–191. 10.2503/jjshs.44.183 [DOI] [Google Scholar]
- Ariza M. T., Reboredo-Rodríguez P., Cervantes L., Soria C., Martínez-Ferri E., González-Barreiro C., et al. (2018). Bioaccessibility and Potential Bioavailability of Phenolic Compounds from Achenes as a New Target for Strawberry Breeding Programs. Food Chem. 248, 155–165. 10.1016/j.foodchem.2017.11.105 [DOI] [PubMed] [Google Scholar]
- Arnold R. A., Noble A. C., Singleton V. L. (1980). Bitterness and Astringency of Phenolic Fractions in Wine. J. Agric. Food Chem. 28, 675–678. 10.1021/jf60229a026 [DOI] [Google Scholar]
- Aron P. M., Kennedy J. A. (2008). Flavan-3-ols: Nature, Occurrence and Biological Activity. Mol. Nutr. Food Res. 52, 79–104. 10.1002/mnfr.200700137 [DOI] [PubMed] [Google Scholar]
- Banerjee A., Jana A., Pati B. R., Mondal K. C., Das Mohapatra P. K. (2012). Characterization of Tannase Protein Sequences of Bacteria and Fungi: An In Silico Study. Protein J. 31, 306–327. 10.1007/s10930-012-9405-x [DOI] [PubMed] [Google Scholar]
- Bar-Ya'akov I., Tian L., Amir R., Holland D. (2019). Primary Metabolites, Anthocyanins, and Hydrolyzable Tannins in the Pomegranate Fruit. Front. Plant Sci. 10, 620. 10.3389/fpls.2019.00620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Nguyen A., Betts N. M., Lyons T. J. (2014). Strawberry as a Functional Food: An Evidence-Based Review. Crit. Rev. Food Sci. Nutr. 54, 790–806. 10.1080/10408398.2011.608174 [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M. A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 Synergistically Specify the Expression of BANYULS and Proanthocyanidin Biosynthesis inArabidopsis Thaliana. Plant J. 39, 366–380. 10.1111/j.1365-313X.2004.02138.x [DOI] [PubMed] [Google Scholar]
- Baxter N. J., Lilley T. H., Haslam E., Williamson M. P. (1997). Multiple Interactions between Polyphenols and a Salivary Proline-Rich Protein Repeat Result in Complexation and Precipitation. Biochemistry 36, 5566–5577. 10.1021/bi9700328 [DOI] [PubMed] [Google Scholar]
- Benfey P. N., Chua N.-H. (1989). Regulated Genes in Transgenic Plants. Science 244, 174–181. 10.1126/science.244.4901.174 [DOI] [PubMed] [Google Scholar]
- Bentley R., Haslam E. (1990). The Shikimate Pathway - A Metabolic Tree with Many Branche. Crit. Rev. Biochem. Mol. Biol. 25, 307–384. 10.3109/10409239009090615 [DOI] [PubMed] [Google Scholar]
- Benzie I. F. F., Strain J. J. (1999). Ferric Reducing/Antioxidant Power Assay: Direct Measure of Total Antioxidant Activity of Biological Fluids and Modified Version for Simultaneous Measurement of Total Antioxidant Power and Ascorbic Acid Concentration. Methods Enzymol. 299, 15–27. 10.1016/S0076-6879(99)99005-5 [DOI] [PubMed] [Google Scholar]
- Blancquaert E. H., Oberholster A., Ricardo-da-Silva J. M., Deloire A. J. (2019). Grape Flavonoid Evolution and Composition under Altered Light and Temperature Conditions in Cabernet Sauvignon (Vitis vinifera L.). Front. Plant Sci. 10, 1062. 10.3389/fpls.2019.01062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blois M. S. (1958). Antioxidant Determinations by the Use of a Stable Free Radical. Nature 181, 1199–1200. 10.1038/1811199a0 [DOI] [Google Scholar]
- Bogs J., Downey M. O., Harvey J. S., Ashton A. R., Tanner G. J., Robinson S. P. (2005). Proanthocyanidin Synthesis and Expression of Genes Encoding Leucoanthocyanidin Reductase and Anthocyanidin Reductase in Developing Grape Berries and Grapevine Leaves. Plant Physiol. 139, 652–663. 10.1104/pp.105.064238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J., Jaffé F. W., Takos A. M., Walker A. R., Robinson S. P. (2007). The grapevine Transcription Factor VvMYBPA1 Regulates Proanthocyanidin Synthesis during Fruit Development. Plant Physiol. 143, 1347–1361. 10.1104/PP.106.093203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohin M. C., Vincken J.-P., van der Hijden H. T. W. M., Gruppen H. (2012). Efficacy of Food Proteins as Carriers for Flavonoids. J. Agric. Food Chem. 60, 4136–4143. 10.1021/jf205292r [DOI] [PubMed] [Google Scholar]
- Bonada M., Jeffery D. W., Petrie P. R., Moran M. A., Sadras V. O. (2015). Impact of Elevated Temperature and Water Deficit on the Chemical and Sensory Profiles of Barossa Shiraz Grapes and Wines. Aust. J. Grape Wine Res. 21, 240–253. 10.1111/ajgw.12142 [DOI] [Google Scholar]
- Bontpart T., Marlin T., Vialet S., Guiraud J.-L., Pinasseau L., Meudec E., et al. (2016). Two Shikimate Dehydrogenases,VvSDH3andVvSDH4, Are Involved in Gallic Acid Biosynthesis in Grapevine. J. Exp. Bot. 67, 3537–3550. 10.1093/jxb/erw184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone F., Preuss A., De Vos R. C. H., D'Amico E., Perrotta G., Bovy A. G., et al. (2009). Developmental, Genetic and Environmental Factors Affect the Expression of Flavonoid Genes, Enzymes and Metabolites in Strawberry Fruits. Plant Cel Environ. 32, 1117–1131. 10.1111/j.1365-3040.2009.01994.x [DOI] [PubMed] [Google Scholar]
- Carpenter G., Cleaver L., Blakeley M., Hasbullah N., Houghton J., Gardner A. (2019). Wine Astringency Reduces Flavor Intensity of Brussels Sprouts. J. Texture Stud. 50, 71–74. 10.1111/jtxs.12378 [DOI] [PubMed] [Google Scholar]
- Casassa L., Keller M., Harbertson J. (2015). Regulated Deficit Irrigation Alters Anthocyanins, Tannins and Sensory Properties of Cabernet Sauvignon Grapes and Wines. Molecules 20, 7820–7844. 10.3390/molecules20057820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin S. D., Matthews M. A., Di Gaspero G., Gambetta G. A. (2007). Water Deficits Accelerate Ripening and Induce Changes in Gene Expression Regulating Flavonoid Biosynthesis in Grape Berries. Planta 227, 101–112. 10.1007/s00425-007-0598-8 [DOI] [PubMed] [Google Scholar]
- Chaudhuri S., Coggins J. R. (1985). The Purification of Shikimate Dehydrogenase from Escherichia Coli . Biochem. J. 226, 217–223. 10.1042/bj2260217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Lambert J. M., McColl L. A., Coggins J. R. (1986). Purification and Characterization of 3-Dehydroquinase from Escherichia Coli . Biochem. J. 239, 699–704. 10.1042/bj2390699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xiong J., Huang S., Li X., Zhang Y., Zhang L., et al. (2018). Analytical Profiling of Proanthocyanidins from Acacia Mearnsii Bark and In Vitro Assessment of Antioxidant and Antidiabetic Potential. Molecules 23, 2891. 10.3390/molecules23112891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins J. R., Boocock M. R., Chaudhuri S., Lambert J. M., Lumsden J., Nimmo G. A., et al. (1987). The Arom Multifunctional Enzyme from Neurospora Crassa. Methods Enzymol. 142, 325–341. 10.1016/S0076-6879(87)42044-2 [DOI] [PubMed] [Google Scholar]
- Constabel C. P. (2018). Molecular Controls of Proanthocyanidin Synthesis and Structure: Prospects for Genetic Engineering in Crop Plants. J. Agric. Food Chem. 66, 9882–9888. 10.1021/acs.jafc.8b02950 [DOI] [PubMed] [Google Scholar]
- Cory H., Passarelli S., Szeto J., Tamez M., Mattei J. (2018). The Role of Polyphenols in Human Health and Food Systems: A Mini-Review. Front. Nutr. 5, 87. 10.3389/fnut.2018.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Liu Y., Zhuang J., Yao S., Liu L., Jiang X., et al. (2020). Discovery and Characterization of Tannase Genes in Plants: Roles in Hydrolysis of Tannins. New Phytol. 226, 1104–1116. 10.1111/nph.16425 [DOI] [PubMed] [Google Scholar]
- De Francesco G., Bravi E., Sanarica E., Marconi O., Cappelletti F., Perretti G. (2020). Effect of Addition of Different Phenolic-Rich Extracts on Beer Flavour Stability. Foods 9, 1638. 10.3390/foods9111638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Peeters A. J. M., Leon-Kloosterziel K. M., Koornneef M. (2001). The TRANSPARENT TESTA12 Gene of Arabidopsis Encodes a Multidrug Secondary Transporter-Like Protein Required for Flavonoid Sequestration in Vacuoles of the Seed Coat Endothelium. The Plant Cell 13, 853. 10.2307/3871345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado L., Zúñiga P., Figueroa N., Pastene E., Escobar-Sepúlveda H., Figueroa P., et al. (2018). Application of a JA-Ile Biosynthesis Inhibitor to Methyl Jasmonate-Treated Strawberry Fruit Induces Upregulation of Specific MBW Complex-Related Genes and Accumulation of Proanthocyanidins. Molecules 23, 1433. 10.3390/molecules23061433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L., Barrieu F., Marchive C., Lauvergeat V., Decendit A., Richard T., et al. (2006). Characterization of a grapevine R2R3-MYB Transcription Factor that Regulates the Phenylpropanoid Pathway. Plant Physiol. 140, 499–511. 10.1104/pp.105.067231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L., Bogs J., Walker A. R., Ferrier T., Decendit A., Merillon J.-M., et al. (2008). The Transcription Factor VvMYB5b Contributes to the Regulation of Anthocyanin and Proanthocyanidin Biosynthesis in Developing Grape Berries. Plant Physiol. 147, 2041–2053. 10.1104/pp.108.118919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Lu S. (2017). Biosynthesis and Regulation of Phenylpropanoids in Plants. Crit. Rev. Plant Sci. 36, 257–290. 10.1080/07352689.2017.1402852 [DOI] [Google Scholar]
- Dixon R. A., Liu C., Jun J. H. (2013). Metabolic Engineering of Anthocyanins and Condensed Tannins in Plants. Curr. Opin. Biotechnol. 24, 329–335. 10.1016/j.copbio.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Xie D. Y., Sharma S. B. (2005). Proanthocyanidins - a Final Frontier in Flavonoid Research? New Phytol. 165, 9–28. 10.1111/j.1469-8137.2004.01217.x [DOI] [PubMed] [Google Scholar]
- Dyer W. E., Henstrand J. M., Handa A. K., Herrmann K. M. (1989). Wounding Induces the First Enzyme of the Shikimate Pathway in Solanaceae . Proc. Natl. Acad. Sci. U.S.A. 86, 7370–7373. 10.1073/pnas.86.19.7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engwa G. A. (2018). “Free Radicals and the Role of Plant Phytochemicals as Antioxidants against Oxidative Stress-Related Diseases,” in Phytochemicals - Source of Antioxidants and Role in Disease Prevention (Enugu, Nigeria: InTech; ). 10.5772/intechopen.76719 [DOI] [Google Scholar]
- Falcão L., Araújo M. (2018). Vegetable Tannins Used in the Manufacture of Historic Leathers. Molecules 23, 1081. 10.3390/molecules23051081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Hernandez J. M., Grotewold E. (2006). An ACT-Like Domain Participates in the Dimerization of Several Plant Basic-Helix-Loop-Helix Transcription Factors. J. Biol. Chem. 281, 28964–28974. 10.1074/jbc.M603262200 [DOI] [PubMed] [Google Scholar]
- Fierascu R. C., Temocico G., Fierascu I., Ortan A., Babeanu N. E. (2020). Fragaria Genus: Chemical Composition and Biological Activities. Molecules 25, 498. 10.3390/molecules25030498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U. A., Carle R., Kammerer D. R. (2011). Identification and Quantification of Phenolic Compounds from Pomegranate (Punica Granatum L.) Peel, Mesocarp, Aril and Differently Produced Juices by HPLC-DAD–ESI/MSn. Food Chem. 127, 807–821. 10.1016/j.foodchem.2010.12.156 [DOI] [PubMed] [Google Scholar]
- Floegel A., Kim D.-O., Chung S.-J., Koo S. I., Chun O. K. (2011). Comparison of ABTS/DPPH Assays to Measure Antioxidant Capacity in Popular Antioxidant-Rich US Foods. J. Food Compost. Anal. 24, 1043–1048. 10.1016/j.jfca.2011.01.008 [DOI] [Google Scholar]
- Fontes N., Gerós H., Delrot S. (2011). Grape Berry Vacuole: A Complex and Heterogeneous Membrane System Specialized in the Accumulation of Solutes. Am. J. Enol. Vitic. 62, 270–278. 10.5344/ajev.2011.10125 [DOI] [Google Scholar]
- Gabetta B., Fuzzati N., Griffini A., Lolla E., Pace R., Ruffilli T., et al. (2000). Characterization of Proanthocyanidins from Grape Seeds. Fitoterapia 71, 162–175. 10.1016/S0367-326X(99)00161-6 [DOI] [PubMed] [Google Scholar]
- Gesell A., Yoshida K., Tran L. T., Constabel C. P. (2014). Characterization of an Apple TT2-Type R2R3 MYB Transcription Factor Functionally Similar to the Poplar Proanthocyanidin Regulator PtMYB134 . Planta 240, 497–511. 10.1007/s00425-014-2098-y [DOI] [PubMed] [Google Scholar]
- Giampieri F., Tulipani S., Alvarez-Suarez J. M., Quiles J. L., Mezzetti B., Battino M. (2012). The Strawberry: Composition, Nutritional Quality, and Impact on Human Health. Nutrition 28, 9–19. 10.1016/j.nut.2011.08.009 [DOI] [PubMed] [Google Scholar]
- Gil-Muñoz F., Sánchez-Navarro J. A., Besada C., Salvador A., Badenes M. L., Naval M. D. M., et al. (2020). MBW Complexes Impinge on Anthocyanidin Reductase Gene Regulation for Proanthocyanidin Biosynthesis in Persimmon Fruit. Sci. Rep. 10, 3543. 10.1038/s41598-020-60635-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollop R. (2002). Expression of the Grape Dihydroflavonol Reductase Gene and Analysis of its Promoter Region. J. Exp. Bot. 53, 1397–1409. 10.1093/jexbot/53.373.1397 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Brown M., Hatlestad G., Akhavan N., Smith T., Hembd A., et al. (2016). TTG2 Controls the Developmental Regulation of Seed Coat Tannins in Arabidopsis by Regulating Vacuolar Transport Steps in the Proanthocyanidin Pathway. Develop. Biol. 419, 54–63. 10.1016/j.ydbio.2016.03.031 [DOI] [PubMed] [Google Scholar]
- Görlach J., Raesecke H. R., Rentsch D., Regenass M., Roy P., Zala M., et al. (1995). Temporally Distinct Accumulation of Transcripts Encoding Enzymes of the Prechorismate Pathway in Elicitor-Treated, Cultured Tomato Cells. Proc. Natl. Acad. Sci. U.S.A. 92, 3166–3170. 10.1073/PNAS.92.8.3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B. G. (1993). Oral Astringency: A Tactile Component of Flavor. Acta Psychologica 84, 119–125. 10.1016/0001-6918(93)90078-6 [DOI] [PubMed] [Google Scholar]
- Grochowski D. M., Skalicka-Woźniak K., Orhan I. E., Xiao J., Locatelli M., Piwowarski J. P., et al. (2017). A Comprehensive Review of Agrimoniin. Ann. N.Y. Acad. Sci. 1401, 166–180. 10.1111/nyas.13421 [DOI] [PubMed] [Google Scholar]
- Gross G. G., Denzel K. (1991). Biosynthesis of Gallotannins. ß-Glucogallin-Dependent Galloylation of 1,6-Digailoylglucose to 1,2,6-Trigalloylglucose. Z. für Naturforsch. C 46, 389–394. 10.1515/znc-1991-5-609 [DOI] [Google Scholar]
- Gross G. G. (1983). Partial Purification and Properties of UDP-Glucose: Vanillate 1-O-Glucosyl Transferase from Oak Leaves. Phytochemistry 22, 2179–2182. 10.1016/S0031-9422(00)80141-7 [DOI] [Google Scholar]
- Grotewold E. (2004). The Challenges of Moving Chemicals within and Out of Cells: Insights into the Transport of Plant Natural Products. Planta 219, 906–909. 10.1007/s00425-004-1336-0 [DOI] [PubMed] [Google Scholar]
- Grundhöfer P., Niemetz R., Schilling G., Gross G. G. (2001). Biosynthesis and Subcellular Distribution of Hydrolyzable Tannins. Phytochemistry 57, 915–927. 10.1016/S0031-9422(01)00099-1 [DOI] [PubMed] [Google Scholar]
- Gu L., Kelm M. A., Hammerstone J. F., Beecher G., Holden J., Haytowitz D., et al. (2003). Screening of Foods Containing Proanthocyanidins and Their Structural Characterization Using LC-MS/MS and Thiolytic Degradation. J. Agric. Food Chem. 51, 7513–7521. 10.1021/jf034815d [DOI] [PubMed] [Google Scholar]
- Gu L., Kelm M., Hammerstone J. F., Beecher G., Cunningham D., Vannozzi S., et al. (2002). Fractionation of Polymeric Procyanidins from Lowbush Blueberry and Quantification of Procyanidins in Selected Foods with an Optimized Normal-Phase HPLC−MS Fluorescent Detection Method. J. Agric. Food Chem. 50, 4852–4860. 10.1021/jf020214v [DOI] [PubMed] [Google Scholar]
- Guidarelli M., Carbone F., Mourgues F., Perrotta G., Rosati C., Bertolini P., et al. (2011). Colletotrichum Acutatum Interactions with Unripe and Ripe Strawberry Fruits and Differential Responses at Histological and Transcriptional Levels. Plant Pathol. 60, 685–697. 10.1111/j.1365-3059.2010.02423.x [DOI] [Google Scholar]
- Habashi R., Hacham Y., Dhakarey R., Matityahu I., Holland D., Tian L., et al. (2019). Elucidating the Role of Shikimate Dehydrogenase in Controlling the Production of Anthocyanins and Hydrolysable Tannins in the Outer Peels of Pomegranate. BMC Plant Biol. 19, 476. 10.1186/s12870-019-2042-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile Z. M., Nagpala-De Guzman E. G., Moretto M., Sonego P., Engelen K., Zoli L., et al. (2019). Transcriptome Profiles of Strawberry (Fragaria Vesca) Fruit Interacting with Botrytis Cinerea at Different Ripening Stages. Front. Plant Sci. 10, 1131. 10.3389/fpls.2019.01131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlin R., Downey M. (2009). Condensed Tannin Accumulation and Composition in Skin of Shiraz and Cabernet Sauvignon Grapes during Berry Development. Am. J. Enol. Vitic. 60, 13–23. [Google Scholar]
- Hasan N., Nester E. W. (1978). Purification and Properties of Chorismate Synthase from Bacillus Subtilis . J. Biol. Chem. 253, 4993–4998. 10.1016/S0021-9258(17)34646-X [DOI] [PubMed] [Google Scholar]
- Haslam E., Cai Y. (1994). Plant Polyphenols (Vegetable Tannins): Gallic Acid Metabolism. Nat. Prod. Rep. 11, 41. 10.1039/np9941100041 [DOI] [PubMed] [Google Scholar]
- Herrmann K. M., Weaver L. M. (1999). The Shikimate Pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 473–503. 10.1146/annurev.arplant.50.1.473 [DOI] [PubMed] [Google Scholar]
- Hichri I., Heppel S. C., Pillet J., Léon C., Czemmel S., Delrot S., et al. (2010). The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant 3, 509–523. 10.1093/mp/ssp118 [DOI] [PubMed] [Google Scholar]
- Huang K., Li M., Liu Y., Zhu M., Zhao G., Zhou Y., et al. (2019). Functional Analysis of 3-Dehydroquinate Dehydratase/Shikimate Dehydrogenases Involved in Shikimate Pathway in Camellia Sinensis . Front. Plant Sci. 10, 1268. 10.3389/fpls.2019.01268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. F., Vialet S., Guiraud J. L., Torregrosa L., Bertrand Y., Cheynier V., et al. (2014). A Negative MYB Regulator of Proanthocyanidin Accumulation, Identified through Expression Quantitative Locus Mapping in the Grape berry. New Phytol. 201, 795–809. 10.1111/nph.12557 [DOI] [PubMed] [Google Scholar]
- Hufnagel J. C., Hofmann T. (2008). Quantitative Reconstruction of the Nonvolatile Sensometabolome of a Red Wine. J. Agric. Food Chem. 56, 9190–9199. 10.1021/jf801742w [DOI] [PubMed] [Google Scholar]
- Hyun J., Woo Y., Hwang D.-s., Jo G., Eom S., Lee Y., et al. (2010). Relationships between Structures of Hydroxyflavones and Their Antioxidative Effects. Bioorg. Med. Chem. Lett. 20, 5510–5513. 10.1016/j.bmcl.2010.07.068 [DOI] [PubMed] [Google Scholar]
- Ikegami A., Eguchi S., Kitajima A., Inoue K., Yonemori K. (2007). Identification of Genes Involved in Proanthocyanidin Biosynthesis of Persimmon (Diospyros Kaki) Fruit. Plant Sci. 172, 1037–1047. 10.1016/j.plantsci.2007.02.010 [DOI] [Google Scholar]
- Ito H., Li P., Koreishi M., Nagatomo A., Nishida N., Yoshida T. (2014). Ellagitannin Oligomers and a Neolignan from Pomegranate Arils and Their Inhibitory Effects on the Formation of Advanced Glycation End Products. Food Chem. 152, 323–330. 10.1016/j.foodchem.2013.11.160 [DOI] [PubMed] [Google Scholar]
- Jaakola L. (2013). New Insights into the Regulation of Anthocyanin Biosynthesis in Fruits. Trends Plant Sci. 18, 477–483. 10.1016/j.tplants.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Jeong S. T., Goto-Yamamoto N., Kobayashi S., Esaka M. (2004). Effects of Plant Hormones and Shading on the Accumulation of Anthocyanins and the Expression of Anthocyanin Biosynthetic Genes in Grape berry Skins. Plant Sci. 167, 247–252. 10.1016/j.plantsci.2004.03.021 [DOI] [Google Scholar]
- Karadag A., Ozcelik B., Saner S. (2009). Review of Methods to Determine Antioxidant Capacities. Food Anal. Methods 2, 41–60. 10.1007/s12161-008-9067-7 [DOI] [Google Scholar]
- Karlińska E., Masny A., Cieślak M., Macierzyński J., Pecio Ł., Stochmal A., et al. (2021). Ellagitannins in Roots, Leaves, and Fruits of Strawberry (Fragaria × Ananassa Duch.) Vary with Developmental Stage and Cultivar. Scientia Horticulturae 275, 109665. 10.1016/j.scienta.2020.109665 [DOI] [Google Scholar]
- Keith B., Dong X. N., Ausubel F. M., Fink G. R. (1991). Differential Induction of 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase Genes in Arabidopsis Thaliana by Wounding and Pathogenic Attack. Proc. Natl. Acad. Sci. U.S.A. 88, 8821–8825. 10.1073/pnas.88.19.8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S., Matsuda F., Tohge T., Yonekura-Sakakibara K., Yamazaki M., Saito K., et al. (2010). Metabolic Profiling and Cytological Analysis of Proanthocyanidins in Immature Seeds of Arabidopsis Thaliana Flavonoid Accumulation Mutants. Plant J. 62, 549–559. 10.1111/j.1365-313X.2010.04174.x [DOI] [PubMed] [Google Scholar]
- Kitamura S., Shikazono N., Tanaka A. (2004). TRANSPARENT TESTA 19 Is Involved in the Accumulation of Both Anthocyanins and Proanthocyanidins in Arabidopsis . Plant J. 37, 104–114. 10.1046/j.1365-313X.2003.01943.x [DOI] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F. (2005). Flavonoids: A Colorful Model for the Regulation and Evolution of Biochemical Pathways. Trends Plant Sci. 10, 236–242. 10.1016/j.tplants.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Kyraleou M., Kallithraka S., Theodorou N., Teissedre P.-L., Kotseridis Y., Koundouras S. (2017). Changes in Tannin Composition of Syrah Grape Skins and Seeds during Fruit Ripening under Contrasting Water Conditions. Molecules 22, 1453. 10.3390/molecules22091453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyraleou M., Kotseridis Y., Koundouras S., Chira K., Teissedre P.-L., Kallithraka S. (2016). Effect of Irrigation Regime on Perceived Astringency and Proanthocyanidin Composition of Skins and Seeds of Vitis Vinifera L. Cv. Syrah Grapes under Semiarid Conditions. Food Chem. 203, 292–300. 10.1016/j.foodchem.2016.02.052 [DOI] [PubMed] [Google Scholar]
- Landete J. M. (2011). Ellagitannins, Ellagic Acid and Their Derived Metabolites: A Review about Source, Metabolism, Functions and Health. Food Res. Int. 44, 1150–1160. 10.1016/j.foodres.2011.04.027 [DOI] [Google Scholar]
- Li C., Leverence R., Trombley J. D., Xu S., Yang J., Tian Y., et al. (2010). High Molecular Weight Persimmon (Diospyros Kaki L.) Proanthocyanidin: A Highly Galloylated, A-Linked Tannin with an Unusual Flavonol Terminal Unit, Myricetin. J. Agric. Food Chem. 58, 9033–9042. 10.1021/jf102552b [DOI] [PubMed] [Google Scholar]
- Liu W., Zhao S., Wang J., Shi J., Sun Y., Wang W., et al. (2017a). Grape Seed Proanthocyanidin Extract Ameliorates Inflammation and Adiposity by Modulating Gut Microbiota in High-Fat Diet Mice. Mol. Nutr. Food Res. 61, 1601082. 10.1002/mnfr.201601082 [DOI] [PubMed] [Google Scholar]
- Liu X.-J., An X.-H., Liu X., Hu D.-G., Wang X.-F., You C.-X., et al. (2017b). MdSnRK1.1 Interacts with MdJAZ18 to Regulate Sucrose-Induced Anthocyanin and Proanthocyanidin Accumulation in Apple. J. Exp. Bot. 68, 2977–2990. 10.1093/jxb/erx150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gao L., Liu L., Yang Q., Lu Z., Nie Z., et al. (2012). Purification and Characterization of a Novel Galloyltransferase Involved in Catechin Galloylation in the tea Plant (Camellia Sinensis). J. Biol. Chem. 287, 44406–44417. 10.1074/jbc.M112.403071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Seeram N. P. (2018). Liquid Chromatography Coupled with Time-Of-Flight Tandem Mass Spectrometry for Comprehensive Phenolic Characterization of Pomegranate Fruit and Flower Extracts Used as Ingredients in Botanical Dietary Supplements. J. Sep. Sci. 41, 3022–3033. 10.1002/jssc.201800480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Shi Y., Wang R., Su D., Tang M., Liu Y., et al. (2021). Antioxidant Activity and Healthy Benefits of Natural Pigments in Fruits: A Review. Int. J. Mol. Sci. 22, 4945. 10.3390/ijms22094945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Reichelt M., Yoshida K., Gershenzon J., Constabel C. P. (2018). Two R2R3-MYB Proteins Are Broad Repressors of Flavonoid and Phenylpropanoid Metabolism in Poplar. Plant J. 96, 949–965. 10.1111/tpj.14081 [DOI] [PubMed] [Google Scholar]
- Ma W., Guo A., Zhang Y., Wang H., Liu Y., Li H. (2014). A Review on Astringency and Bitterness Perception of Tannins in Wine. Trends Food Sci. Technol. 40, 6–19. 10.1016/j.tifs.2014.08.001 [DOI] [Google Scholar]
- Maeda H., Shasany A. K., Schnepp J., Orlova I., Taguchi G., Cooper B. R., et al. (2010). RNAi Suppression of Arogenate Dehydratase1 Reveals that Phenylalanine Is Synthesized Predominantly via the Arogenate Pathway in Petunia Petals. Plant Cell 22, 832–849. 10.1105/tpc.109.073247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugeri A., Lombardo G. E., Cirmi S., Süntar I., Barreca D., Laganà G., et al. (2022). Pharmacology and Toxicology of Tannins. Arch. Toxicol. 1, 1–21. 10.1007/s00204-022-03250-0 [DOI] [PubMed] [Google Scholar]
- Mena P., Calani L., Dall'Asta C., Galaverna G., García-Viguera C., Bruni R., et al. (2012). Rapid and Comprehensive Evaluation of (Poly)phenolic Compounds in Pomegranate (Punica Granatum L.) Juice by UHPLC-MSn. Molecules 17, 14821–14840. 10.3390/molecules171214821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir R., Jallu S., Singh T. P. (2015). The Shikimate Pathway: Review of Amino Acid Sequence, Function and Three-Dimensional Structures of the Enzymes. Crit. Rev. Microbiol. 41, 172–189. 10.3109/1040841X.2013.813901 [DOI] [PubMed] [Google Scholar]
- Misnawi J., Jinap S., Jamilah B., Nazamid S. (2005). Changes in Polyphenol Ability to Produce Astringency during Roasting of cocoa Liquor. J. Sci. Food Agric. 85, 917–924. 10.1002/jsfa.1954 [DOI] [Google Scholar]
- Mittler R. (2017). ROS Are Good. Trends Plant Sci. 22, 11–19. 10.1016/j.tplants.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Mo R., Huang Y., Yang S., Zhang Q., Luo Z. (2015). Development of Agrobacterium-Mediated Transient Transformation in Persimmon (Diospyros Kaki Thunb.). Scientia Horticulturae 192, 29–37. 10.1016/j.scienta.2015.05.013 [DOI] [Google Scholar]
- Mori K., Sugaya S., Gemma H. (2004). Regulatory Mechanism of Anthocyanin Biosynthesis in 'Kyoho' Grape Berries Grown under Different Temperature Conditions. ecb 42, 21–30. 10.2525/ecb1963.42.21 [DOI] [Google Scholar]
- Muir R. M., Ibáñez A. M., Uratsu S. L., Ingham E. S., Leslie C. A., McGranahan G. H., et al. (2011). Mechanism of Gallic Acid Biosynthesis in Bacteria (Escherichia Coli) and walnut (Juglans Regia). Plant Mol. Biol. 75, 555–565. 10.1007/s11103-011-9739-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpala E. G., Guidarelli M., Gasperotti M., Masuero D., Bertolini P., Vrhovsek U., et al. (2016). Polyphenols Variation in Fruits of the Susceptible Strawberry Cultivar Alba during Ripening and upon Fungal Pathogen Interaction and Possible Involvement in Unripe Fruit Tolerance. J. Agric. Food Chem. 64, 1869–1878. 10.1021/acs.jafc.5b06005 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nakatsuka A., Yano K., Yasugahira S., Nakamura R., Sun N., et al. (2008). Expressed Sequence Tags from Persimmon at Different Developmental Stages. Plant Cel Rep 27, 931–938. 10.1007/s00299-008-0518-9 [DOI] [PubMed] [Google Scholar]
- Naval M. D. M., Gil-Muñoz F., Lloret A., Besada C., Salvador A., Badenes M. L., et al. (2016). A WD40-Repeat Protein from Persimmon Interacts with the Regulators of Proanthocyanidin Biosynthesis DkMYB2 and DkMYB4 . Tree Genet. Genomes 12, 13. 10.1007/s11295-016-0969-z [DOI] [Google Scholar]
- Nesi N., Jond C., Debeaujon I., Caboche M., Lepiniec L. (2001). The Arabidopsis TT2 Gene Encodes an R2R3 MYB Domain Protein that Acts as a Key Determinant for Proanthocyanidin Accumulation in Developing Seed. Plant Cell 13, 2099–2114. 10.1105/TPC.010098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemetz R., Gross G. G. (2003a). Ellagitannin Biosynthesis: Laccase-Catalyzed Dimerization of Tellimagrandin II to Cornusiin E in Tellima Grandiflora . Phytochemistry 64, 1197–1201. 10.1016/j.phytochem.2003.08.013 [DOI] [PubMed] [Google Scholar]
- Niemetz R., Gross G. G. (2005). Enzymology of Gallotannin and Ellagitannin Biosynthesis. Phytochemistry 66, 2001–2011. 10.1016/j.phytochem.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Niemetz R., Gross G. G. (2003b). Oxidation of Pentagalloylglucose to the Ellagitannin, Tellimagrandin II, by a Phenol Oxidase from Tellima Grandiflora Leaves. Phytochemistry 62, 301–306. 10.1016/S0031-9422(02)00557-5 [DOI] [PubMed] [Google Scholar]
- Niemetz R., Gross G. G., Schilling G. (2001). Ellagitannin Biosynthesis: Oxidation of Pentagalloylglucose to Tellimagrandin II by an Enzyme from Tellima Grandiflora Leaves. Chem. Commun. 62 (3), 35–36. 10.1039/b006270g [DOI] [Google Scholar]
- Niemetz R., Schilling G., Gross G. G. (2003). Biosynthesis of the Dimeric Ellagitannin, Cornusiin E, in Tellima Grandiflora . Phytochemistry 64, 109–114. 10.1016/S0031-9422(03)00280-2 [DOI] [PubMed] [Google Scholar]
- Okuda T., Ito H. (2011). Tannins of Constant Structure in Medicinal and Food Plants-Hydrolyzable Tannins and Polyphenols Related to Tannins. Molecules 16, 2191–2217. 10.3390/molecules16032191 [DOI] [Google Scholar]
- Ono N. N., Qin X., Wilson A. E., Li G., Tian L. (2016). Two UGT84 Family Glycosyltransferases Catalyze a Critical Reaction of Hydrolyzable Tannin Biosynthesis in Pomegranate (Punica Granatum). PLoS One 11, e0156319. 10.1371/journal.pone.0156319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T., Lilley T. H., Haslam E. (1987). Polyphenol Interactions: Astringency and the Loss of Astringency in Ripening Fruit. Phytochemistry 26, 2937–2942. 10.1016/S0031-9422(00)84566-5 [DOI] [Google Scholar]
- Pang Y., Peel G. J., Wright E., Wang Z., Dixon R. A. (2007). Early Steps in Proanthocyanidin Biosynthesis in the Model Legume. Medicago truncatula. Plant Physiol. 145, 601–615. 10.1104/pp.107.107326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolocci F., Robbins M. P., Madeo L., Arcioni S., Martens S., Damiani F. (2007). Ectopic Expression of a Basic Helix-Loop-Helix Gene Transactivates Parallel Pathways of Proanthocyanidin Biosynthesis. Structure, Expression Analysis, and Genetic Control of Leucoanthocyanidin 4-Reductase and Anthocyanidin Reductase Genes in Lotus Corniculatus . Plant Physiol. 143, 504–516. 10.1104/pp.106.090886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolocci F., Robbins M. P., Passeri V., Hauck B., Morris P., Rubini A., et al. (2011). The Strawberry Transcription Factor FaMYB1 Inhibits the Biosynthesis of Proanthocyanidins in Lotus Corniculatus Leaves. J. Exp. Bot. 62, 1189–1200. 10.1093/jxb/erq344 [DOI] [PubMed] [Google Scholar]
- Pascual O., Vignault A., Gombau J., Navarro M., Gómez-Alonso S., García-Romero E., et al. (2017). Oxygen Consumption Rates by Different Oenological Tannins in a Model Wine Solution. Food Chem. 234, 26–32. 10.1016/j.foodchem.2017.04.148 [DOI] [PubMed] [Google Scholar]
- Pastore C., Dal Santo S., Zenoni S., Movahed N., Allegro G., Valentini G., et al. (2017). Whole Plant Temperature Manipulation Affects Flavonoid Metabolism and the Transcriptome of Grapevine Berries. Front. Plant Sci. 8, 929. 10.3389/fpls.2017.00929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg H., Gacon K., Schlich P., Noble A. C. (1999). Bitterness and Astringency of Flavan-3-Ol Monomers, Dimers and Trimers. J. Sci. Food Agric. 79, 1123–1128. [DOI] [Google Scholar]
- Pires M. A., Pastrana L. M., Fuciños P., Abreu C. S., Oliveira S. M. (2020). Sensorial Perception of Astringency: Oral Mechanisms and Current Analysis Methods. Foods 9, 1124. 10.3390/foods9081124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott D. M., Osorio S., Vallarino J. G. (2019). From central to Specialized Metabolism: An Overview of Some Secondary Compounds Derived from the Primary Metabolism for Their Role in Conferring Nutritional and Organoleptic Characteristics to Fruit. Front. Plant Sci. 10, 835. 10.3389/fpls.2019.00835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel P. R., Koyama K., Goto-Yamamoto N. (2020). Evaluating the Influence of Temperature on Proanthocyanidin Biosynthesis in Developing Grape Berries (Vitis Vinifera L.). Mol. Biol. Rep. 47, 3501–3510. 10.1007/s11033-020-05440-4 [DOI] [PubMed] [Google Scholar]
- Quideau S. (2009). Chemistry and Biology of Ellagitannins. Bordeaux, France: World Scientific. 10.1142/6795 [DOI] [Google Scholar]
- Quideau S., Deffieux D., Douat-Casassus C., Pouységu L. (2011). Plant Polyphenols: Chemical Properties, Biological Activities, and Synthesis. Angew. Chem. Int. Ed. 50, 586–621. 10.1002/anie.201000044 [DOI] [PubMed] [Google Scholar]
- Ramírez L., Arrizon J., Sandoval G., Cardador A., Bello-Mendoza R., Lappe P., et al. (2008). A New Microplate Screening Method for the Simultaneous Activity Quantification of Feruloyl Esterases, Tannases, and Chlorogenate Esterases. Appl. Biochem. Biotechnol. 151, 711–723. 10.1007/s12010-008-8319-8 [DOI] [PubMed] [Google Scholar]
- Ramsay N. A., Glover B. J. (2005). MYB-bHLH-WD40 Protein Complex and the Evolution of Cellular Diversity. Trends Plant Sci. 10, 63–70. 10.1016/j.tplants.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Ricci A., Olejar K. J., Parpinello G. P., Mattioli A. U., Teslić N., Kilmartin P. A., et al. (2016). Antioxidant Activity of Commercial Food Grade Tannins Exemplified in a Wine Model. Food Addit. Contam. Part. A. 33, 1761–1774. 10.1080/19440049.2016.1241901 [DOI] [PubMed] [Google Scholar]
- Robichaud J. L., Noble A. C. (1990). Astringency and Bitterness of Selected Phenolics in Wine. J. Sci. Food Agric. 53, 343–353. 10.1002/jsfa.2740530307 [DOI] [Google Scholar]
- Robinson S. P., Bogs J., McDavid D. A. J., Hooper L. C., Speirs J., Walker A. R. (2021). Transgenic Grapevines with Decreased Expression of Tannin Synthesis Genes Have Altered Grape and Wine Flavonoid Composition. Aust. J. Grape Wine Res. 27, 106–117. 10.1111/ajgw.12468 [DOI] [Google Scholar]
- Rock C. D. (2013). Trans-acting Small Interfering RNA4: Key to Nutraceutical Synthesis in Grape Development? Trends Plant Sci. 18, 601–610. 10.1016/j.tplants.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez H., Curiel J. A., Landete J. M., de las Rivas B., de Felipe F. L., Gómez-Cordovés C., et al. (2009). Food Phenolics and Lactic Acid Bacteria. Int. J. Food Microbiol. 132, 79–90. 10.1016/j.ijfoodmicro.2009.03.025 [DOI] [PubMed] [Google Scholar]
- Rossetti D., Bongaerts J. H. H., Wantling E., Stokes J. R., Williamson A.-M. (2009). Astringency of tea Catechins: More Than an Oral Lubrication Tactile Percept. Food Hydrocoll 23, 1984–1992. 10.1016/j.foodhyd.2009.03.001 [DOI] [Google Scholar]
- Rousserie P., Rabot A., Geny-Denis L. (2019). From Flavanols Biosynthesis to Wine Tannins: What Place for Grape Seeds? J. Agric. Food Chem. 67, 1325–1343. 10.1021/acs.jafc.8b05768 [DOI] [PubMed] [Google Scholar]
- Sarni-Manchado P., Deleris A., Avallone S., Cheynier V., Moutounet M. (1999). Analysis and Characterization of Wine Condensed Tannins Precipitated by Proteins Used as Fining Agent in Enology. Am. J. Enol. Vitic. 50, 81–86. [Google Scholar]
- Saxe H. J., Horibe T., Balan B., Butterfield T. S., Feinberg N. G., Zabaneh C. M., et al. (2021). Two UGT84A Family Glycosyltransferases Regulate Phenol, Flavonoid, and Tannin Metabolism in Juglans Regia (English Walnut). Front. Plant Sci. 12, 178. 10.3389/fpls.2021.626483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaart J. G., Dubos C., Romero De La Fuente I., Houwelingen A. M. M. L., Vos R. C. H., Jonker H. H., et al. (2013). Identification and Characterization of MYB‐bHLH ‐ WD 40 Regulatory Complexes Controlling Proanthocyanidin Biosynthesis in Strawberry (F Ragaria × Ananassa) Fruits. New Phytol. 197, 454–467. 10.1111/nph.12017 [DOI] [PubMed] [Google Scholar]
- Schmidt S. W., Denzel K., Schilling G., Gross G. G. (1987). Enzymatic Synthesis of 1,6-Digalloylglucose from β-Glucogallin by β-Glucogallin: β-Glucogallin 6-O-Galloyltransferase from Oak Leaves. Z. für Naturforsch. C 42, 87–92. 10.1515/znc-1987-1-215 [DOI] [Google Scholar]
- Seeram N., Lee R., Hardy M., Heber D. (2005). Rapid Large Scale Purification of Ellagitannins from Pomegranate Husk, a By-Product of the Commercial Juice Industry. Separat. Purif. Technol. 41, 49–55. 10.1016/j.seppur.2004.04.003 [DOI] [Google Scholar]
- Shao Y., Hu Z., Yu Y., Mou R., Zhu Z., Beta T. (2018). Phenolic Acids, Anthocyanins, Proanthocyanidins, Antioxidant Activity, Minerals and Their Correlations in Non-Pigmented, Red, and Black rice. Food Chem. 239, 733–741. 10.1016/j.foodchem.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 Regulates Anthocyanin Biosynthesis in an HY5-Dependent Manner with Both Factors Directly Binding Anthocyanin Biosynthetic Gene Promoters in Arabidopsis . Plant J. 49, 981–994. 10.1111/j.1365-313X.2006.03021.x [DOI] [PubMed] [Google Scholar]
- Shin Y.-J., Shon M.-S., Kim G.-N., Lee S.-C. (2014). Antioxidant and Anti-Adipogenic Activities of Persimmon Tannins. Food Sci. Biotechnol. 23, 1689–1694. 10.1007/s10068-014-0230-1 [DOI] [Google Scholar]
- Shneier A., Kleanthous C., Deka R., Coggins J. R., Abell C. (1991). Observation of an Imine Intermediate on Dehydroquinase by Electrospray Mass Spectrometry. J. Am. Chem. Soc. 113, 9416–9418. 10.1021/ja00024a085 [DOI] [Google Scholar]
- Siebert K. J. (1999). Effects of Protein−Polyphenol Interactions on Beverage Haze, Stabilization, and Analysis. J. Agric. Food Chem. 47, 353–362. 10.1021/jf980703o [DOI] [PubMed] [Google Scholar]
- Siebert K. J., Troukhanova N. V., Lynn P. Y. (1996). Nature of Polyphenol−Protein Interactions. J. Agric. Food Chem. 44, 80–85. 10.1021/jf9502459 [DOI] [Google Scholar]
- Smeriglio A., Barreca D., Bellocco E., Trombetta D. (2017). Proanthocyanidins and Hydrolysable Tannins: Occurrence, Dietary Intake and Pharmacological Effects. Br. J. Pharmacol. 174, 1244–1262. 10.1111/bph.13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquet J.-M., Cheynier V., Brossaud F., Moutounet M. (1996). Polymeric Proanthocyanidins from Grape Skins. Phytochemistry 43, 509–512. 10.1016/0031-9422(96)00301-9 [DOI] [Google Scholar]
- Stefanachi A., Leonetti F., Pisani L., Catto M., Carotti A. (2018). Coumarin: A Natural, Privileged and Versatile Scaffold for Bioactive Compounds. Molecules 23, 250. 10.3390/molecules23020250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Jiang S., Zhang T., Xu H., Fang H., Zhang J., et al. (2019). Apple NAC Transcription Factor MdNAC52 Regulates Biosynthesis of Anthocyanin and Proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 289, 110286. 10.1016/j.plantsci.2019.110286 [DOI] [PubMed] [Google Scholar]
- Sun X., Cheng X., Zhang J., Ju Y., Que Z., Liao X., et al. (2020). Letting Wine Polyphenols Functional: Estimation of Wine Polyphenols Bioaccessibility under Different Drinking Amount and Drinking Patterns. Food Res. Int. 127, 108704. 10.1016/j.foodres.2019.108704 [DOI] [PubMed] [Google Scholar]
- Suvanto J., Karppinen K., Riihinen K., Jaakola L., Salminen J.-P. (2020). Changes in the Proanthocyanidin Composition and Related Gene Expression in Bilberry (Vaccinium Myrtillus L.) Tissues. J. Agric. Food Chem. 68, 7378–7386. 10.1021/acs.jafc.0c02158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Koussevitzky S., Mittler R., Miller G. (2012). ROS and Redox Signalling in the Response of Plants to Abiotic Stress. Plant Cel Environ. 35, 259–270. 10.1111/j.1365-3040.2011.02336.x [DOI] [PubMed] [Google Scholar]
- Terrier N., Torregrosa L., Ageorges A., Vialet S., Verriès C., Cheynier V., et al. (2009). Ectopic Expression of VvMybPA2 Promotes Proanthocyanidin Biosynthesis in Grapevine and Suggests Additional Targets in the Pathway. Plant Physiol. 149, 1028–1041. 10.1104/pp.108.131862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah C., Unsicker S. B., Fellenberg C., Constabel C. P., Schmidt A., Gershenzon J., et al. (2017). Flavan-3-ols Are an Effective Chemical Defense against Rust Infection. Plant Physiol. 175, 1560–1578. 10.1104/pp.17.00842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaegen B., Neven H., Verachtert H., Derdelinckx G. (2006). The Chemistry of Beer Aging - a Critical Review. Food Chem. 95, 357–381. 10.1016/j.foodchem.2005.01.006 [DOI] [Google Scholar]
- Versari A., du Toit W., Parpinello G. P. (2013). Oenological Tannins: A Review. Aust. J. Grape Wine Res. 19, 1–10. 10.1111/ajgw.12002 [DOI] [Google Scholar]
- Vignault A., González-Centeno M. R., Pascual O., Gombau J., Jourdes M., Moine V., et al. (2018). Chemical Characterization, Antioxidant Properties and Oxygen Consumption Rate of 36 Commercial Oenological Tannins in a Model Wine Solution. Food Chem. 268, 210–219. 10.1016/j.foodchem.2018.06.031 [DOI] [PubMed] [Google Scholar]
- Villamil-Galindo E., Van de Velde F., Piagentini A. M. (2021). Strawberry Agro-Industrial By-Products as a Source of Bioactive Compounds: Effect of Cultivar on the Phenolic Profile and the Antioxidant Capacity. Bioresour. Bioproc. 8, 61. 10.1186/s40643-021-00416-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid Biosynthesis. Mol. Plant 3, 2–20. 10.1093/mp/ssp106 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang K., Lyu S., Huang J., Huang C., Xing Y., et al. (2021). Genome-wide Identification of Tannase Genes and Their Function of Wound Response and Astringent Substances Accumulation in Juglandaceae. Front. Plant Sci. 12, 739. 10.3389/fpls.2021.664470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Qu C., Jiang S., Chen Z., Xu H., Fang H., et al. (2018). The Proanthocyanidin-Specific Transcription Factor MdMYBPA1 Initiates Anthocyanin Synthesis under Low-Temperature Conditions in Red-Fleshed Apples. Plant J. 96, 39–55. 10.1111/tpj.14013 [DOI] [PubMed] [Google Scholar]
- Wang T., Jónsdóttir R., Ólafsdóttir G. (2009). Total Phenolic Compounds, Radical Scavenging and Metal Chelation of Extracts From Icelandic Seaweeds. Food Chem. 116, 240–248. 10.1016/j.foodchem.2009.02.041 [DOI] [Google Scholar]
- Watrelot A. A., Norton E. L. (2020). Chemistry and Reactivity of Tannins in Vitis spp: A Review. Molecules 25, 2110. 10.3390/molecules25092110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Ju Y., Ma T., Zhang J., Fang Y., Sun X. (2021). New Perspectives on the Biosynthesis, Transportation, Astringency Perception and Detection Methods of Grape Proanthocyanidins. Crit. Rev. Food Sci. Nutr. 61, 2372–2398. 10.1080/10408398.2020.1777527 [DOI] [PubMed] [Google Scholar]
- White P. J., Young J., Hunter I. S., Nimmo H. G., Coggins J. R. (1990). The Purification and Characterization of 3-Dehydroquinase from Streptomyces Coelicolor. Biochem. J. 265, 735–738. 10.1042/bj2650735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002). Biosynthesis of Flavonoids and Effects of Stress. Curr. Opin. Plant Biol. 5, 218–223. 10.1016/S1369-5266(02)00256-X [DOI] [PubMed] [Google Scholar]
- Wu S., Tian L. (2017). Diverse Phytochemicals and Bioactivities in the Ancient Fruit and Modern Functional Food Pomegranate (Punica Granatum). Molecules 22, 1606. 10.3390/molecules22101606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.-Y., Sharma S. B., Paiva N. L., Ferreira D., Dixon R. A. (2003). Role of Anthocyanidin Reductase, Encoded by BANYULS in Plant Flavonoid Biosynthesis. Science 299, 396–399. 10.1126/science.1078540 [DOI] [PubMed] [Google Scholar]
- Xu W., Dubos C., Lepiniec L. (2015). Transcriptional Control of Flavonoid Biosynthesis by MYB-bHLH-WDR Complexes. Trends Plant Sci. 20, 176–185. 10.1016/j.tplants.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Xu W., Grain D., Bobet S., Le Gourrierec J., Thévenin J., Kelemen Z., et al. (2014). Complexity and Robustness of the Flavonoid Transcriptional Regulatory Network Revealed by Comprehensive Analyses of MYB-bHLH-WDR Complexes and Their Targets in Arabidopsis Seed. New Phytol. 202, 132–144. 10.1111/nph.12620 [DOI] [PubMed] [Google Scholar]
- Xu Y.-Q., Zhang Y.-N., Chen J.-X., Wang F., Du Q.-Z., Yin J.-F. (2018). Quantitative Analyses of the Bitterness and Astringency of Catechins from green tea. Food Chem. 258, 16–24. 10.1016/j.foodchem.2018.03.042 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Ma D., Constabel C. P. (2015). The MYB182 Protein Down-Regulates Proanthocyanidin and Anthocyanin Biosynthesis in Poplar by Repressing Both Structural and Regulatory Flavonoid Genes. Plant Physiol. 167, 693–710. 10.1104/pp.114.253674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Huang T., Tian B., Zhan J. (2020). Advances in Biosynthesis and Biological Functions of Proanthocyanidins in Horticultural Plants. Foods 9, 1774. 10.3390/foods9121774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Jun J. H., Duan C., Dixon R. A. (2019). VvLAR1 and VvLAR2 Are Bifunctional Enzymes for Proanthocyanidin Biosynthesis in grapevine. Plant Physiol. 180, 1362–1374. 10.1104/pp.19.00447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pribil M., Palmgren M., Gao C. (2020). A CRISPR Way for Accelerating Improvement of Food Crops. Nat. Food 1, 200–205. 10.1038/s43016-020-0051-8 [DOI] [Google Scholar]
- Zhao J., Pang Y., Dixon R. A. (2010). The Mysteries of Proanthocyanidin Transport and Polymerization. Plant Physiol. 153, 437–443. 10.1104/pp.110.155432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Lin‐Wang K., Wang F., Espley R. V., Ren F., Zhao J., et al. (2019). Activator‐type R2R3‐MYB Genes Induce a Repressor‐type R2R3‐MYB Gene to Balance Anthocyanin and Proanthocyanidin Accumulation. New Phytol. 221, 1919–1934. 10.1111/nph.15486 [DOI] [PubMed] [Google Scholar]
- Zhou H., Lin-Wang K., Liao L., Gu C., Lu Z., Allan A. C., et al. (2015a). Peach MYB7 Activates Transcription of the Proanthocyanidin Pathway Gene Encoding Leucoanthocyanidin Reductase, but Not Anthocyanidin Reductase. Front. Plant Sci. 6, 908. 10.3389/fpls.2015.00908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Wei L., Sun Z., Gao L., Meng Y., Tang Y., et al. (2015b). Production and Transcriptional Regulation of Proanthocyanidin Biosynthesis in Forage Legumes. Appl. Microbiol. Biotechnol. 99, 3797–3806. 10.1007/s00253-015-6533-1 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zheng J., Li Y., Xu D.-P., Li S., Chen Y.-M., et al. (2016). Natural Polyphenols for Prevention and Treatment of Cancer. Nutrients 8, 515. 10.3390/nu8080515 [DOI] [PMC free article] [PubMed] [Google Scholar]