Figure 2.
HGT-enabled catabolic routes increase the likelihood of metabolic interactions
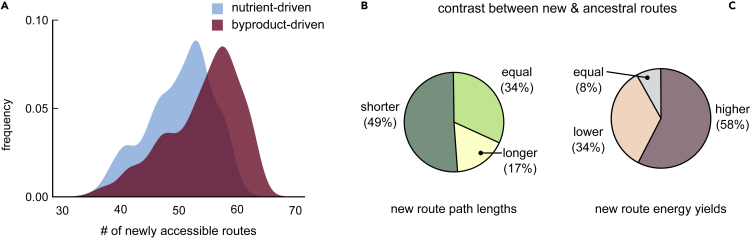
(A) Distribution of the number of newly accessible catabolic routes (routes gained along each phylogenetic branch) across all 1,669 branches. The number of routes starting from nutrients are shown in blue, and those starting from byproducts are in red. New byproduct-driven routes would increase the chance of metabolic interactions with other bacteria (via their byproducts). We find that new catabolic routes are more likely to be byproduct-driven (median 56, versus 51 for nutrient-driven routes; ; Kolmogorov-Smirnov test).
(B and C) Pie charts comparing new routes with their corresponding ancestral routes on each of the 1,669 branches. We compare new and ancestral routes based on their (B) path length (i.e., is the shortest new path shorter, longer, or the same length as the shortest ancestral path) and (C) energy yield (i.e., does the most energy-yielding new path have a higher, lower, or equal yield than the best ancestral path).