Figure 3.
HGT can affect dependency evolution via coupled gains and losses of genes
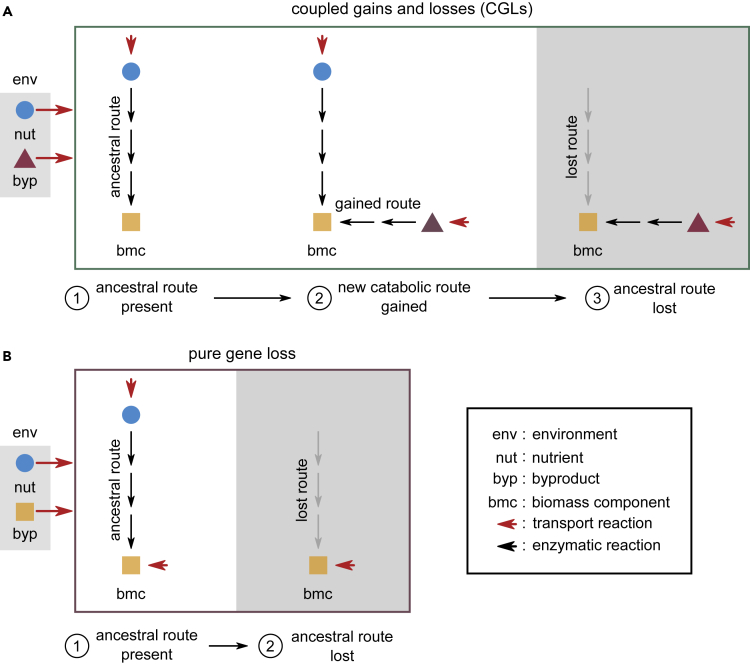
(A) Schematic illustration of coupled gains and losses (CGLs), a new, alternate mechanism for dependency evolution driven by HGT. The gray box on the left indicates the environment, with a nutrient (nut, blue circle) and a byproduct (byp, purple triangle). Red arrows indicate the secretion and import of metabolites by bacteria. The three steps in the frame show how a bacterial species can evolve a dependency on another species that donates byp in the environment; by step 3, the acceptor species eventually depends on byp. Each step follows the modification of a part of the acceptor’s metabolic network. At step 1, the acceptor uses an ancestral route (black arrows) to convert nut in the environment to a key biomass component (bmc, yellow square). At step 2, it can alternatively use a newly gained route to convert byp to the same bmc. The ancestral and gained routes are coupled to each other, because they both produce bmc, which is crucial for survival and growth. At step 3, the acceptor loses the ancestral route (gray arrows) but can still produce bmc through the gained route. It thus becomes dependent on donors of byp for survival.
(B) Schematic illustration of dependency evolution via pure gene loss, not driven by HGT. The environment now has bmc available as a byproduct, instead of byp (gray box on the left). In this mechanism, step 1 is the same as (A), but unlike (A), at step 2, the acceptor can lose the ancestral route straight away, without requiring an alternate coupled route. However, this requires a particular environment where bmc is available.