Abstract
Elongating nuclear RNA polymerases (Pols) frequently pause, backtrack, and are then reactivated by endonucleolytic cleavage. Pol backtracking and RNA cleavage are also crucial for proofreading, which contributes to transcription fidelity. RNA polymerase I (Pol I) of the yeast Saccharomyces cerevisiae synthesizes exclusively 35S rRNA, the precursor transcript of mature ribosomal 5.8S, 18S, and 25S rRNA. Pol I contains the specific heterodimeric subunits Rpa34.5/49 and subunit Rpa12.2, which have been implicated in RNA cleavage and elongation activity, respectively. These subunits are associated with the Pol I lobe structure and encompass different structural domains, but the contribution of these domains to RNA elongation is unclear. Here, we used Pol I mutants or reconstituted Pol I enzymes to study the effects of these subunits and/or their distinct domains on RNA cleavage, backtracking, and transcription fidelity in defined in vitro systems. Our findings suggest that the presence of the intact C-terminal domain of Rpa12.2 is sufficient to support the cleavage reaction, but that the N-terminal domains of Rpa12.2 and the heterodimer facilitate backtracking and RNA cleavage. Since both N-terminal and C-terminal domains of Rpa12.2 were also required to faithfully incorporate NTPs in the growing RNA chain, efficient backtracking and RNA cleavage might be a prerequisite for transcription fidelity. We propose that RNA Pols containing efficient RNA cleavage activity are able to add and remove nucleotides until the matching nucleotide supports RNA chain elongation, whereas cleavage-deficient enzymes can escape this proofreading process by incorporating incorrect nucleotides.
Keywords: nucleolus, ribosome, backtracking, rRNA, RNA polymerase I, proofreading, transcription, Rpa34.5, Rpa49, TFIIS
Abbreviations: Pol, polymerase; Pol I, polymerase I; RT, room temperature; TEV, tobacco etch virus; YPD, yeast extract–peptone–dextrose
All three nuclear RNA polymerases (Pols) share a homologous core structure, which is responsible for their enzymatic activities but differ in Pol-specific subunits conferring distinct functional and structural features to the enzymes (1, 2, 3). The 14 subunits containing Pol I from the yeast Saccharomyces cerevisiae transcribe only one gene—the rRNA gene—with high rate and efficiency (1, 2). In Pol I, three specific subunits, the RNA cleavage–stimulating factor Rpa12.2, as well as subunit Rpa34.5 that forms via its N-terminal dimerization domain a heterodimer with subunit Rpa49, bind to a region within the second largest subunit Rpa135, which has been designated as lobe structure (Fig. 1A). In Pol II, only subunit Rpb9 binds to the corresponding lobe structure, which is formed by subunit Rpb2. Interestingly, Pol II transcription factors, TFIIE and the lobe-associated TFIIF and TFIIS, partly share functional and structural features with Pol I subunits Rpa49, Rpa34.5, and Rpa12.2, respectively. Thus, the subunits Rpa12.2 and Rpa34.5/49 were designated “built-in” transcription factors supporting the high transcriptional efficiency of Pol I (1, 4). Recent investigations suggested a dynamic interplay between the lobe-binding factors in many steps of the transcription cycle (reviewed in Ref. (5)) favoring the idea that peripheral Pol I subunits may change both their position and their association with the core enzyme during transcription elongation.
Figure 1.
Pol I mutants and recombinantly expressed Pol I subunits and subunit domains used for reconstitution assays.A, Pol I WT and Pol I mutants lacking distinct lobe-binding subunit domains used for the in vitro assays. The lobe-binding domains are highlighted in color. B, graphical outline of the Rpa34.5/49 heterodimer domains used for the reconstitution assays. The five different constructs used for the studies are indicated in black. The numbers represent the stretch of expressed amino acids, respectively. C, graphical outline of the Rpa12.2 domains used for the reconstitution assays. The 12 different constructs used for the studies are indicated in black. The numbers represent the stretch of expressed amino acids, respectively. Recombinant full-length Rpa12.2 is abbreviated Rpa12FL(1–125). All mutants containing exchanges of amino acids (fourth column) have 125 amino acids. Position and exchanged amino acid are indicated in brackets. Pol I, polymerase I.
Pol II counterparts of the Pol I lobe-binding subunits, TFIIS and TFIIF, cooperate to allow backtracking of arrested/stalled Pol II, TFIIS-assisted cleavage of the protruding RNA overhang, and relocation of the 3′end of the nascent RNA in the active center of the enzyme (6, 7, 8, 9, 10). In Pol I, the C-terminal part of subunit Rpa12.2 is involved to cleave 3′RNA overhangs after backtracking (4, 11, 12, 13, 14). This part of Rpa12.2 resembles the zinc ribbon domain III of TFIIS and ranges from amino acid 78 to 124 (Fig. 1C). Within the zinc ribbon at positions D105 and E106, Rpa12.2 contains two evolutionary conserved amino acids whose counterparts in domain III of TFIIS (D290 and E291) position a metal ion involved in hydrolytic cleavage of backtracked RNA (15). This suggests that D105 and E106 of Rpa12.2 have a similar function in Pol I transcription. In transcriptional inactive Pol I, backtracked Pol I, and Pol I with a partially expanded cleft conformation, the C-terminal domain of Rpa12.2 is inserted in the active site (16, 17, 18, 19). In contrast, the domain is displaced from the active site in initial transcribing complexes and elongation complexes correlating with the complete closure of the cleft (19, 20).
Deletion of either the full-length Rpa12.2 or lack of its N-terminal 69 amino acids results in growth inhibition at elevated temperature, sensitivity to drugs depleting the endogenous nucleotide pool, inefficient transcription termination, and hampers the assembly of Pol I (21, 22, 23). Furthermore, lack of Rpa12.2 or N-terminal truncation leads to the loss of Rpa34.5/49 and might influence the intrinsic stability of elongation and termination complexes (23, 24). In an RPA12.2 deletion strain, error rates in rRNA synthesis were increased (25). In contrast, deletion of the C-terminal 61 to 125 amino acids, which contain the RNA cleavage–supporting activity, has neither impact on growth at 30 °C nor on association of Rpa34.5/49. Kinetic studies on AT-rich templates indicated that the N-terminal domain of Rpa12.2 can stimulate transcription elongation suggesting that the N-terminal and C-terminal parts of Rpa12.2 contribute differently to (r)RNA synthesis (26).
In contrast to Rpa12.2, it is not clear if and how the Rpa34.5–Rpa49 heterodimeric complex or individual domains of the subunits participate in RNA cleavage (4, 11). Rpa34.5 contains an N-terminal dimerization domain (amino acids 1–144) and a disordered arm domain (amino acids 145–233) (Fig. 1B). In Rpa49, the N-terminal dimerization domain (amino acids 1–101) is connected to a TFIIE-like C-terminal tandem winged helix (amino acids 185–415) domain through a flexible linker domain (amino acids 102–184) (27, 28). The dimerization domain is attached to Pol I lobe at a similar position as TFIIF to Pol II lobe structure. Together with the unstructured Rpa49 linker and the Rpa34.5 arm domain, it contacts Pol I core enzyme. RNA cleavage by Pol I lacking Rpa34.5/49 was shown to be either less efficient (11) or completely absent (4). Accordingly, the exact role of Rpa34.5/49 to support RNA cleavage likely in cooperation with Rpa12.2 is still elusive (4).
RNA cleavage is also required for transcriptional proofreading (29, 30). Incorporation of a wrong nucleotide by Pol II may slow down RNA extension, which enables the enzyme to undergo backtracking. Pol II backtracking by one position enables nucleolytic cleavage of an RNA dinucleotide that contains the misincorporated nucleotide. RNA cleavage can occur in the absence of TFIIS (30). TFIIS recruitment to the backtracked complex stimulates fast cleavage of the transcript and the generation of a new 3′-RNA end, after which transcription resumes (29, 30). Pol II proofreading in vitro and in vivo is also supported by subunit Rpb9 (31, 32), which structurally resembles the N terminus of Rpa12.2. In Pol II, both nucleotide selectivity and incorporation kinetics as well as RNA cleavage in combination with RNA re-extension (RNA proofreading) contribute to transcription fidelity (29). In Pol I, subunit Rpa12.2 affects the kinetics and energetics of Pol I-catalyzed nucleotide incorporation (26, 33). Furthermore, lack of Rpa12.2 leads to NTP misincorporation in vivo (25). It remains to be determined how Rpa12.2 or its individual domains contribute to transcription fidelity by transcriptional proofreading and to investigate if subunits Rpa34.5/49 might participate in this process.
To analyze the function of Rpa12.2 and Rpa34.5/49 and/or individual domains of the proteins in distinct activities of Pol I, we purified enzymes lacking either all three subunits (Pol I ΔRpa12), Rpa34.5/49 (Pol I ΔRpa49), or the C-terminal part of Rpa12.2 (Pol I Rpa12.2ΔC) (Fig. 1A). We complemented the mutant Pols with different combinations of recombinantly expressed and purified proteins and/or domains of Rpa34.5, Rpa49, and Rpa12.2 and analyzed RNA cleavage, (re-)extension, and transcriptional proofreading activities (Fig. 1, B and C). Our results suggest a coordinated cooperativity between the different domains of the lobe-binding subunits to ensure efficient RNA cleavage and transcription fidelity.
Results
The N-terminal and C-terminal domains of Rpa12.2 are required to ensure efficient RNA cleavage
An overview about Pol I mutants and recombinant Pol I subunit domains used in this study is presented in Figure 1. Pol I WT and Pol I mutants can be efficiently purified from yeast cells expressing a protein A-tagged subunit Rpa135 (34). From a strain in which the gene coding for subunit Rpa12.2 is additionally deleted, a mutant enzyme lacking all three lobe-binding subunits (Pol I ΔRpa12) can be isolated (35, 36) (Fig. S1A). Affinity purification from a strain lacking Rpa49 results in an enzyme preparation without subunits Rpa34.5/49 (Pol I ΔRpa49) (18, 36) (Fig. S1A). Purification from a strain expressing a C-terminally truncated variant of Rpa12.2 (lacking amino acids 85–125) yielded a mutant Pol I (Pol I Rpa12ΔC) with stably associated truncated Rpa12.2 as well as Rpa34.5/49 (23, 36). The same was true for a mutant enzyme purified from a strain expressing an even further truncated Rpa12.2 lacking the C-terminal amino acids 70 to 125 (Pol I Rpa12ΔC) (Fig. S1A; see also Fig. S3 in Ref. (36)).
To find out which domains of Pol I lobe-binding subunits Rpa12.2, Rpa34.5/49 participate in RNA cleavage, recombinant purified subunits or their domains were first added to Pol I ΔRpa12 lacking all lobe-binding subunits. RNA cleavage was monitored using a RNA–DNA hybrid scaffold containing three nucleotide mismatches at the RNA 3′end (clv3) (Fig. 2A), which was similar to the previously used to analyze RNA cleavage activity of Pol I (11). The 19 nucleotide-long RNA contained a Cy5 fluorophore at its 5′end, which allowed to detect uncleaved and cleaved RNAs on a denaturing polyacrylamide gel.
Figure 2.
Efficient RNA cleavage requires the N terminus and the complete C terminus of Rpa12.2.A, DNA–RNA hybrid scaffold with three mismatched nucleotides at the RNA 3′end used for cleavage assays. Potential cleavage sites are indicated regarding the terminal hybrid base pair to the nucleotide insertion site (site 0). The RNA contains a fluorescent Cy5 label on the 5′RNA end. B, RNA cleavage requires the C terminus of Rpa12.2 and can be reconstituted with recombinant full-length Rpa12.2. About 0.15 pmol of either Pol I or Pol I mutants were added to 0.05 pmol cleavage scaffold and incubated for the indicated time intervals. Lanes 2 to 7, time course of the cleavage reaction using 0.15 pmol purified Pol I WT. Lane 8, 0.15 pmol Pol I lacking the C terminus of Rpa12.2 (Rpa12ΔC); lane 9, 0.15 pmol Pol I lacking subunits Rpa12.2 and Rpa34.5/49 (Pol I ΔRpa12) without, and lane 10, with 0.78 pmol recombinant full-length Rpa12.2 (Rpa12FL). Fluorescent transcripts were analyzed on a 20% denaturing polyacrylamide gel (20% [v/v] acrylamide/bisacrylamide [19:1], 6 M urea, 0.1% [v/v] TEMED, and 0.1% [w/v] ammonium persulfate). RNAs shortened by two (+2), four (0), or 8 (−4) nucleotides and uncleaved RNAs (+4) are indicated. Quantitation of two independent kinetics using Pol I WT is shown in the lower panel. Values are indicated as percentage of +4 RNA at time point 0. C, in the presence of Rpa12.2, Pol I removes predominantly four nucleotides from the RNA, which corresponds to cleavage site 0. Cleavage reactions were performed using 0.2 pmol WT Pol I or Pol I mutants. Increasing amounts of recombinant Rpa12.2 (0.1, 0.2, 1, and 2 pmol Rpa12FL) were added for 30 min to Pol I ΔRpa12 in reactions analyzed in lanes 5 to 8. Lanes 1 to 4, control cleavage reactions using 6 pmol recombinant Rpa12.2 (Rpa12FL) with Pol I ΔRpa12 (lane 1), 0.15 pmol Pol I WT (lane 2), 0.15 pmol Pol I Rpa12ΔC (lane 3), and 0.2 pmol Pol I ΔRpa12 (lane 4). Quantitation of two independent titration experiments using 0.2 pmol Pol I ΔRpa12 and increasing amounts of Rpa12FL (0.1–2 pmol) is shown in the lower panel. Values are indicated as percentage of uncleaved +4 RNA. D, regions of Rpa12.2 contribute differently to RNA cleavage. Cleavage reactions were performed using 0.2 pmol Pol I WT, 0.2 pmol Pol I ΔRpa49 (ΔRpa49), which lacks subunits Rpa34.5/49, but contains Rpa12.2, 0.2 pmol Pol I Rpa12ΔC, and 0.2 pmol Pol I ΔRpa12 (incubation time of 30 min). Reactions containing ΔRpa12 were substituted with 2 pmol Rpa12FL, 2 pmol of Rpa12.2 with C-terminal truncations (amino acids [1–69] [1–85] [1–112] [1–119]), or 1 pmol Rpa12.2 with C-terminal truncations (amino acids [1–112]), 20 pmol Rpa12.2 N-terminal truncations (amino acids [66–125] and [69–125]), and 2 pmol Rpa12.2 variants containing the point mutations as indicated. In the Rpa12.2 DE/AA subunit, the aspartate and glutamate at positions 105 and 106 are exchanged to alanine, which—according to the corresponding amino acids in TFIIS—should inhibit the cleavage reaction. E, efficient cleavage depends on the covalently linked N-terminal and C-terminal Rpa12.2 domains. Combinations of N-terminal and C-terminal Rpa12.2 domains were added to Pol I ΔRpa12, to see whether both domains can support backtracking/cleavage reactions without physical linkage. Preferentially dinucleotides are removed in the absence of covalent linkages. Pol I, polymerase I; TEMED, N, N, N', N'-tetramethylethylenediamine.
When Pol I WT was incubated with the clv3 scaffold, two additional RNA fragments appeared that were two or four nucleotides shorter than the original RNA indicating that Pol I backtracked and removed either two or four nucleotides from the 3′overhang (Fig. 2B, lanes 2–7 and quantitation below). Reduction of two nucleotides was predominant at earlier time points, whereas RNA shortened by four nucleotides became more prominent after 20 min of incubation indicating that nucleotides were removed in two base pair increments. In some experiments, Pol I WT also produced a minor RNA fraction, which was eight nucleotides shorter (position −4) (Fig. 2D, lane 2). Pol I Rpa12ΔC encompassing Rpa34.5/49 and Rpa12.2 lacking amino acids 70 to 125 (Fig. 2B, lane 8) or Pol I ΔRpa12 lacking all lobe-binding subunits (Fig. 2B, lane 9) were not able to cleave RNA. This confirms previous studies that showed that Pol I removes dinucleotides (11, 37) or four nucleotides and that the C terminus of Rpa12.2 (amino acids 79–125) is required for cleavage (11). Cleavage of four nucleotides from a 3-nucleotide overhang implies that the correct nucleotide preceding the mismatched 3′overhang and forming a correct hybrid with the DNA is also removed (cleavage position 0, Fig. 2A). In the presence of the recombinant full-length subunit Rpa12.2 (Rpa12FL), cleavage activity of Pol I ΔRpa12 was restored (Fig. 2B, lane 10). Shortening by four nucleotides (position 0) was predominant upon addition of recombinant Rpa12.2 to Pol I ΔRpa12 (see also time-course experiments in Fig. S4). There was also a minor fraction of RNAs shortened by eight nucleotides (position −4 Fig. 2C, lanes 6–8). Higher Rpa12.2 concentrations (Fig. 2C, compare lanes 5–8) did not significantly change the cleavage pattern. It should be noted that the occurrence of the RNA fragment cleaved at position −4 was not observed in all reactions and may depend on the respective enzyme preparation (compare Fig. 2B, lane 10 with Fig. 1C lanes 6–8).
Rpa12.2 encompasses two subdomains characterized by either an N-terminal or a C-terminal zinc ribbon (Fig. S1B). The N-terminal domain stretching from amino acids 1 to 60 is required for growth at higher temperature and for association of Rpa12.2 with the core enzyme (23). The C-terminal domain reaches from amino acids 80 to 125 and contains the evolutionary conserved amino acids D105 and E106, which—in analogy to residues D290 and E291 in TFIIS—might be required for hydrolytic cleavage of backtracked RNA. The linker between the N-terminal and C-terminal domain of TFIIS was suggested to transmit torsional stiffness, which facilitates the correct insertion of the C-terminal domain into the active center if RNA cleavage is required (15). Therefore, the Rpa12.2 linker area might also impact RNA cleavage by Pol I. Furthermore, the stretch from amino acids 69 to 85 including the Rpa12.2 N terminus has been reported to be required for the stable association of Rpa34.5/49 with Pol I (23). Nevertheless, Rpa34.5/49 were still associated with Pol I Rpa12ΔC encompassing only amino acids 1 to 69 (Fig. S1A). Several recombinant Rpa12.2 mutants were purified from bacteria (Figs. 1C and S1B) and analyzed in their ability to support cleavage by Pol I ΔRpa12 (Fig. 2D). Control reactions contained Pol I WT, Pol I ΔRpa49, Pol I Rpa12.2ΔC, Pol I ΔRpa12, and Pol I ΔRpa12 supplemented with full-length Rpa12.2 (Fig. 2D, lanes 2–6). The N-terminal Rpa12.2 domains 1 to 69 and 1 to 85 should in principal bind to the Pol I lobe but supported no cleavage (Fig. 2D, lanes 7 and 8). The same is true for the DE/AA mutant in which aspartate and glutamate at positions 105 and 106 were exchanged by alanine (Fig. 2D, lane 12).
Interestingly, truncation mutants that were lacking only a few C-terminal amino acids (110–125, 113–125, and 120–125) but still contained D105 and E106 were also not able to stimulate RNA cleavage (Fig. 2D, lanes 9–11) (Fig. S2, lanes 4–9). In contrast, single-point mutations in the final C-terminal β-strand (K120A and N124A) did not affect cleavage activity (lanes 16–17). Addition of the Rpa12.2 C-terminal domain including a part of the linker resulted in inefficient RNA cleavage (Fig. 2D, lanes 13 and 14). Transcripts were shortened not by four but by two nucleotides, and cleavage occurred only with an excess of the truncated subunit. This suggested that proper binding of the C-terminal Rpa12.2 domain to the core enzyme through the N terminus is important for efficient backtracking and cleavage. Increasing the concentration of the Rpa12.2 C terminus did not improve the cleavage activity (Fig. S2, lanes 10–17). Furthermore, the C and N terminus must be covalently linked since a combination of the individual domains did not further increase cleavage (Fig. 2E, lanes 1–4). Thus, the N-terminal region might help to position the C-terminal domain correctly in the active site of Pol I. Furthermore, the exchange of a serine at position 59 in the linker region—a target for phosphorylation in vivo (38)—with alanine (S59A) did not affect the cleavage reaction (Fig. 2D, lane 15).
In summary, our data underline the requirement of the evolutionary conserved Rpa12.2 C-terminal domain for the cleavage reaction and suggest that it requires the N-terminal lobe-binding domain to support efficient Pol I backtracking and cleavage.
The impact of the mismatched RNA 3′overhang and the influence of Rpa12.2 domains on backtracking, cleavage, and RNA re-extension
Using the RNA–DNA hybrid scaffold with three mismatched nucleotides at the RNA 3′end as an artificial substrate to analyze RNA cleavage by Pol I WT predominantly yielded a four-nucleotide shorter RNA (e.g., Fig. 2B, lane 7, Fig. 2C, lane 2, and Fig. 2D, lane 2). We next investigated if and how shorter 3′RNA mismatches might influence Pol I backtracking and cleavage. Cleavage reactions were performed using RNA–DNA scaffolds, which contained one, two, or three mismatched nucleotides at the RNA 3′end (Fig. 3A, clv1, clv2, and clv3). In addition, analogous scaffolds containing a pseudo transcription bubble were used to better reflect the in vivo situation (Fig. 3A, clv1 bu, clv2 bu, and clv3 bu). Cleavage efficiencies varied slightly between reactions using scaffolds with or without the pseudo transcription bubble (Figs. 3 and 4). Although no significant differences in the overall cleavage pattern were obtained, cleavage within the pseudo transcription bubble templates appeared to be generally less efficient. In general, Pol I WT cleaved dinucleotides until the mismatched nucleotide were removed. From a three-nucleotide mismatch scaffold, two and predominantly four nucleotides were removed (Fig. 3B, lanes 7 and 8), whereas from a two-and one-nucleotide mismatch scaffold, mainly two nucleotides were removed (Fig. 3B, lanes 9–12). Thus, cleavage occurs mainly in steps of dinucleotides until a correct 3′ RNA–DNA hybrid pair resides within the active center of the enzyme. Addition of recombinant Rpa12.2 to Pol I ΔRpa12 to the different RNA–DNA hybrid scaffolds resulted in a similar cleavage pattern as observed with Pol I WT (Fig. 3C lanes 7–12) confirming that addition of Rpa12.2 to Pol I ΔRpa12 lacking all lobe-binding subunits is sufficient for cleavage and backtracking.
Figure 3.
RNAs containing one, two, or three terminal mismatched nucleotides in RNA–DNA hybrid scaffolds are shortened in the presence of Rpa12.2 by dinucleotides and can be extended after cleavage.A, RNA–DNA hybrid scaffolds used for the cleavage and re-extension assays. Cleavage sites are indicated. The scaffolds differ in the length of the mismatched 3′RNA overhang and/or the presence of a pseudo transcription bubble. B, RNAs in the different RNA–DNA hybrid scaffolds are preferentially shortened by two (+2), four (0), or eight (−4) nucleotides when incubated with Pol I WT. The positions of the cleavage sites are related to the terminal hybrid base pair in the nucleotide insertion site (position 0). RNA–DNA scaffolds were incubated with 0.2 pmol Pol I WT for 10 min on ice. The cleavage reaction was performed at 28 °C for 30 min, and fluorescent transcripts were analyzed on a 20% denaturing polyacrylamide gel (20% [v/v] acrylamide/bisacrylamide [19:1], 6 M urea, 0.1% [v/v] TEMED, and 0.1% [w/v] ammonium persulfate). C, adding of recombinant Rpa12.2 to Pol I lacking all lobe-binding subunits is sufficient for the dinucleotide-cleavage reaction and to restart elongation. RNA–DNA scaffolds were incubated with 0.2 pmol Pol I ΔRpa12 I in the presence of 2 pmol Rpa12FL for 10 min on ice. The cleavage reaction was performed at 28 °C for 30 min. Indicated samples were supplemented with NTPs (final concentration: 200 μM) after the cleavage reaction and incubated at 28 °C for 30 min to analyze extension of cleaved RNAs (lanes 13–18). Note that preferentially RNAs at position 0 (scaffolds clv3, clv3 bu and clv1 and clv1 bu) and position +1 (clv2 and clv2 bu) were extended. D, addition of 20 pmol recombinant mutant Rpa12(66–125) to Pol I ΔRpa12 restarted elongation from cleaved RNAs although they were shortened only by two nucleotides. Procedure according to Figure 2C. Pol I, polymerase I; TEMED, N, N, N', N'-tetramethylethylenediamine.
Figure 4.
In the absence of the cleavage reactions, a small fraction of RNAs with one or two mismatches at the 3′end can be elongated.A, Pol I lacking all lobe-binding subunits can elongate RNAs, which have one or two but not three nucleotides mismatch on the 3′end of the RNA–DNA hybrid. Cleavage reactions were performed as described for Figure 2, B and C. B, Pol I lacking the Rpa12.2 C terminus but containing the heterodimer can elongate RNAs, which have one or two but not three nucleotides mismatch on the 3′end of the RNA–DNA hybrid. Reactions were performed as described for Figure 2, B and C, but in the presence of 0.2 pmol Pol I Rpa12ΔC. Pol I, polymerase I.
Restarting transcription requires realigning the 3′ end of the RNA with the active site. To show that addition of Rpa12.2 to Pol I ΔRpa12 was also sufficient to re-extend shortened RNAs, NTPs were added after the cleavage reaction (Fig. 3C, lanes 13–18). In all reactions, fully extended transcripts (position +16) were barely detected. Instead, RNAs were re-extended at least up to position +6 from the “clv” templates, whereas RNAs from the “clv bu” templates were predominantly extended to position +7. Preferred targets of re-extension were RNAs cleaved at position 0 in reactions when the scaffolds clv3, clv3 bu and clv1 and clv1 bu were used (Fig. 3C, compare lanes 7, 8, 11, and 12 with lanes 13, 14, 17, and 18, respectively) and RNAs cleaved at position +1 when clv2 and clv2 bu were used (Fig. 3C, compare lanes 9 and 10 with lanes 15 and 16). Further shortened RNAs (position −4, compare lanes 13, 17, and 18 with lanes 7, 11, and 12) or shortened RNAs, which still contained a mismatched nucleotide (position +2, compare lanes 13 and 14 with lanes 7 and 8) were less appropriate substrates for extension. As observed before (Fig. 2, D and E), when N-terminal truncated Rpa12.2 (Rpa12 [66–125]) was added to Pol I ΔRpa12, the template “clv3” and also “clv3 bu” were only shortened by two nucleotides (position +2, Fig. 3D, lanes 7 and 8). Although these RNAs obviously contained an incorrect hybrid base pair, they could be extended (Fig. 3D, compare lanes 7 and 8 with lanes 13 and 14). Similar observations were made with the “clv1,2” and “clv1,2 bu” templates, where cleavage was relatively inefficient while a substantial fraction of RNAs could be extended (Fig. 3D, compare lanes 9–12 with lanes 15–18). This indicates that Pol I ΔRpa12 containing truncated (or lacking) Rpa12.2 tolerates RNA–DNA mismatches as substrates for elongation.
Pol I without cleavage activity extends RNA–DNA hybrids containing one and two but not three mismatches at the 3′ RNA end
Based on the aforementioned observations, we investigated if mutant Pol I enzymes lacking cleavage and backtracking activity could extend mismatched RNA–DNA hybrids. The scaffolds clv1–3 with and without pseudo transcription bubble were incubated and re-extended in the presence of NTPs with Pol I ΔRpa12, Pol I Rpa12ΔC, or Pol I ΔRpa12 supplemented with cleavage-inactive Rpa12.2 DE/AA mutant (Figs. 4 and S3). As expected, none of the Pol I mutants was able to significantly remove nucleotides from RNA–DNA hybrids containing three terminal base mismatches (Figs. 3 and S3, lanes 7–12, respectively). However, after addition of NTPs, extended RNAs could be observed with scaffolds containing one or two mismatched hybrid bases at the RNA 3′end. This suggested that Pol I lacking cleavage activity can tolerate templates with one or two incorrect hybrid base pairs as substrates for elongation under the used experimental conditions.
The role of the heterodimer Rpa34.5/49 for Pol I cleavage
It was previously suggested that the dimerization module of the heterodimer Rpa34.5/49 supports RNA cleavage (4). On the other hand, cleavage activity was well detectable in Pol I preparations lacking the heterodimer (11). Using the reconstituted Pol I system, we reanalyzed the role of Rpa34.5/49 as well as individual domains of the proteins to support cleavage by Pol I ΔRpa12 complemented with recombinant full-length Rpa12.2 or distinct domains. To this end, Rpa34.5/49 as well as individual domains of the proteins were recombinantly expressed in Escherichia coli and purified (Figs. 1B and S1C). Specifically, reconstitution experiments were performed using the complete heterodimer (Rpa34.5/49), the dimerization module including Rpa34.5 and the N-terminal domain of Rpa49 with the linker (Rpa34.5/49NTL), the dimerization module including Rpa34.5 and the N-terminal domain of Rpa49 without the linker (Rpa34.5/49NT), and the C-terminal domain of RPA49 with the linker (Rpa49LCT) and without the linker (Rpa49CT). In combination with Pol I ΔRpa12 lacking all lobe-binding subunits, neither the heterodimer nor the individual domains of the heterodimer supported Pol I cleavage (Fig. 5A, lanes 8–11). However, both the complete heterodimer and the dimerization module including the linker increased cleavage by Pol I ΔRpa12 complemented with Rpa12.2, whereas addition of the C-terminal domain of Rpa49 did not have a significant effect (Fig. 5A, compare lanes 4 and 6 with lanes 3, 5, and 7 and quantification aside) (see also Fig. S5, A and B). Interestingly, both heterodimer and dimerization module including the linker, but not the C-terminal part of Rpa49 or the dimerization module without the linker, stimulated the removal of further four nucleotides to position −4 (Fig. 5A, lanes 4–7 and quantification aside) (Fig. S5). Both increased cleavage activity and removal of further four nucleotides were dependent on the duration of the cleavage reaction and on the concentration of the heterodimer or the dimerization module (Fig. S5, A and B). Addition of the heterodimer stimulated neither cleavage nor backtracking if the N-terminal or C-terminal domain of Rpa12.2 was absent or inactive (Fig. 5B, compare lanes 5–9 with lanes 11–15). Even in the presence of the Rpa12.2 N terminus (residues 1–69 and 1–85) including the linker that should bind the heterodimer or the Rpa12.2 C terminus (residues 66–125 and 69–125) including the part of the linker interacting with Rpa34.5/49, the heterodimer was not able to support cleavage (Fig. 5B, compare lanes 11 and 12 with lanes 5 and 6; lanes 13 and 14 with lanes 7 and 8). In summary, RNA cleavage was strictly dependent on the presence of the structured C-ribbon of Rpa12.2. The Rpa12.2 N terminus including the linker stimulated backtracking and cleavage. The presence of the dimerization module further increased cleavage and backtracking, whereas the C-terminal part of Rpa49 was dispensable for this effect. This suggests that the interaction of domains of Rpa12.2 and Rpa34.5/49, which physically associate with the Pol I lobe structure, contributes to backtracking and cleavage mediated by the Rpa12.2 C-ribbon.
Figure 5.
The heterodimer Rpa34.5/49 and the dimerization domain Rpa34.5/49NTL reinforces RNA cleavage only if full-length Rpa12.2 is present.A, 0.2 pmol Pol I ΔRpa12 was complemented with 2 pmol full-length Rpa12.2 (Rpa12FL) (lanes 3–7) or without Rpa12.2 (lanes 8–11). About 2 pmol recombinant heterodimer (Rpa34.5/49) (lanes 4 and 8), 2 pmol dimerization domain without linker (Rpa34.5/49NT) (lanes 5 and 9), 2 pmol dimerization domain with linker (Rpa34.5/49NTL) (lanes 6 and 10), or 2 pmol C-terminal Rpa49 domain including linker (Rpa49LCT) (lanes 7 and 11) were added and analyzed in cleavage reactions using scaffold template clv3. In the absence of Rpa12FL, no heterodimer variant could support cleavage. Note that addition of both the complete heterodimer and the dimerization domain including the linker supported longer backtracks, which are visualized by an increasing amount of −4 RNAs. Quantitation of cleavage efficiency (+4 RNA) and the accumulation of −4 RNA from two independent assays is depicted on the right. Values are indicated as percentage of +4 RNA after incubation of scaffold template clv3 with Pol I ΔRpa12. B, deep backtracking and cleavage depend on the covalently linked two Rpa12.2 domains and on the heterodimer. Cleavage reactions were performed as described for Figure 1D with 0.4 pmol Pol I ΔRpa12, 4 pmol Rpa12FL, or 4 pmol of Rpa12.2 with C-terminal truncations (1–69, 1–85, 1–112, 1–119), 40 pmol Rpa12.2 N-terminal truncations (66–125, 69–125), 4 pmol Rpa12.2 variant (DE/AA), and 0.1 pmol of the template clv3. In lanes 10 to 15, heterodimer was added to Pol I ΔRpa12 in the presence of full-length Rpa12.2 (Rpa12FL) or of its N-terminal or C-terminal truncated variants. Note: only full-length Rpa12.2 together with the heterodimer was able to support strong cleavage and long backtracks. Pol I, polymerase I.
The role of the Rpa12.2 subunit in RNA proofreading
For Pol II, it was reported that RNA cleavage in response to nucleotide misincorporation is important to maintain transcription fidelity (for review, see Refs. (29, 39)). It can be anticipated that the same may hold true for Pol I. Thus, we tested the individual roles of different Rpa12.2 domains as well as of the heterodimer to support the correct addition of nucleotides. Single nucleotides were added to RNA–DNA hybrid scaffolds with (extP) or without pseudo transcription bubble (ext1) (Fig. 6A) in the presence of Pol I WT or Pol I mutants. In these templates, the first two nucleotides to be incorporated were GTP and UTP. Stepwise elongation of transcripts was monitored (Fig. 6, B–D). No significant differences were observed using scaffolds with or without pseudo transcription bubble. Addition of GTP and GTP/UTP to Pol I WT and scaffold extP resulted in extension of one or two nucleotides, respectively, with no evidence for nucleotide misincorporation (Fig. 6B, lanes 2–4). The same was observed for Pol I ΔRpa49 (Fig. 6B, lanes 5–7). In contrast, Pol I ΔRpa12 and Pol I Rpa12ΔC did not precisely stop after incorporation of GTP (Fig. 6B, lanes 9 and 12) or GTP/UTP (lanes 10 and 13). This suggests that in these Pol I mutants, either the lack of cleavage activity to remove erroneously added NTPs or an altered NTP specificity results in the misincorporation of nucleotides. We also investigated if the misincorporation of NTPs might depend on the sequence of the DNA template. Pol I ΔRpa12 and Pol I Rpa12ΔC did also not stop precisely using template ext2 or ext3 in which the first three nucleotides to be incorporated were CTP/ATP/UTP and UTP/GTP/CTP, respectively (Fig. S6, A and B). Addition of recombinant Rpa12.2 to Pol I ΔRpa12 was sufficient to prevent significant nucleotide misincorporation (Fig. 6C, compare lanes 5–7 with lanes 2–4). In contrast addition of either N-terminal (Fig. 6D, lanes 5–16) or C-terminal truncated (Fig. 6C, lanes 11–16) Rpa12.2 variants or the mutant Rpa12 DE/AA (Fig. 6C, lanes 8–10) revealed NTP misincorporation. This suggests that the addition of the complete Rpa12.2 subunit, but neither of the C-terminal domain nor of the N-terminal domain alone, is sufficient for correct NTP addition. In analogy, purified Pol II (36), which lacks the RNA cleavage supporting activity of TFIIS, was also not precise in the extension assay (Fig. 7A compare lanes 8–11 with lanes 2–5) (Fig. S7, A and B) confirming previous reports that Pol II without TFIIS leads to NTP misincorporation (40, 41). As expected, addition of recombinant TFIIS to Pol II improved significantly transcription fidelity (Fig. 7A, compare lanes 14 to 16 with lanes 1–9).
Figure 6.
Pol I lacking Rpa12.2 domains exhibits NTP promiscuity.A, RNA–DNA scaffolds used for misincorporation assays. B, Pol I WT and Pol I lacking the heterodimer incorporate predominantly matching nucleotides in contrast to Pol I without cleaving activity. About 0.2 pmol Pol I WT, 0.2 pmol Pol I ΔRpa49, 0.2 pmol Pol I ΔRpa12, and 0.2 pmol Pol I Rpa12ΔC were incubated with 0.066 pmol RNA–DNA scaffold extP for 20 min on ice. Mixtures of nucleotides (200 μmol) were added, and samples were incubated at 28 °C for 30 min as indicated and resulting transcripts were analyzed on a 20% denaturing polyacrylamide gel (20% [v/v] acrylamide/bisacrylamide [19:1], 6 M urea, 0.1% [v/v] TEMED, and 0.1% [w/v] ammonium persulfate). Expected extensions are indicated at the right. Pol I ΔRpa12 and Pol I Rpa12ΔC incorporated wrong nucleotides. C, Pol I lacking the N-terminal domain of Rpa12.2 and Pol I complemented with Rpa12 DE/AA incorporate wrong nucleotides. Reactions were performed as for Figure 5B with 0.066 pmol extension template ext1, 0.2 pmol Pol I WT (lanes 17–19), 0.2 pmol Pol I ΔRpa12 (lanes 2–4), and 0.2 pmol Pol I ΔRpa12 supplemented with 2 pmol Rpa12(DE/AA) (lanes 8–10), 40 pmol Rpa12(66–125) (lanes 11–13), and 40 pmol Rpa12(69–125) (lanes 14–16). Note that the third nucleotide to be incorporated should be CTP and not ATP. D, the lacking parts of Pol I of the C-terminal domain of Rpa12.2 incorporate wrong nucleotides. Reactions were performed as for Figure 5B with 0.066 pmol extension template ext1, 0.2 pmol Pol I Rpa12ΔC (lanes 17–19), 0.2 pmol Pol I ΔRpa12 (lanes 2–4), 0.2 pmol Pol I ΔRpa12 supplemented with 2 pmol Rpa12 (1–69) (lanes 5–7), 2 pmol Rpa12 (1–85) (lanes 8–10), 2 pmol Rpa12 (1–112) (lanes 11–13), or 2 pmol Rpa12 (1–119) (lanes 14–16). Note that the third nucleotide to be incorporated should be CTP and not ATP. Pol I, polymerase I; TEMED, N, N, N', N'-tetramethylethylenediamine.
Figure 7.
The Pol II elongation factor TFIIS resembles Rpa12.2 function in the NTP extension and RNA cleavage assays.A, in contrast to Pol I WT, purified Pol II incorporates wrong nucleotides, which can be revised upon addition of the Pol II elongation factor TFIIS. Reactions were performed as for Figure 6B with 0.066 pmol extension template ext1, 0.2 pmol Pol I WT (lanes 2–6), 0.2 pmol Pol II (lanes 8–12), and 0.2 pmol Pol II including 0.2 pmol recombinant TFIIS. B, the C-terminal part of Rpa12.2 can enhance Pol II-dependent cleavage reactions, but TFIIS shows no effect in Pol I ΔRpa12-dependent reactions. About 2 pmol full-length Rpa12.2 (lane 4), 540 pmol Rpa12 (66–125), or 4 pmol TFIIS was added to either 0.2 pmol Pol I ΔRpa12 (lanes 4–6) or 0.2 pmol Pol II (lanes 8–11) and analyzed for RNA cleavage. DNA–RNA scaffold clv3 was used as template. Pol I, polymerase I.
The C-terminal domain of Rpa12.2 stimulates Pol II-dependent RNA cleavage
To find out whether the cleaving supporting subunit Rpa12.2 and transcription factor TFIIS can substitute each other in RNA cleavage, swap experiments with purified Pol II and Pol I ΔRpa12 were performed. Both full-length Rpa12.2 and its C-terminal domain clearly stimulated Pol II RNA cleavage (Fig. 7B, compare lanes 9 and 10 with lane 8, and Fig. S7C, compare lanes 9 and 10 with lane 8). Whereas TFIIS removed up to eight nucleotides from the DNA–RNA hybrid, both Rpa12.2 and its C-terminal domain enhanced only removal of two nucleotides from the 4 bp overhang. On the other hand, TFIIS was not able to support RNA cleavage of Pol I ΔRpa12 (Fig. 7B, compare lane 6 with lanes 4 and 5, Fig. S7B). Simple explanations for these effects are that the N-terminal domains of RPA12.2 and TFIIS are structurally too different that Rpa12.2 can fully complement for TFIIS and that the active center of Pol I is structurally not compatible with TFIIS-mediated cleavage.
Misincorporation of nucleotides by Pol I Rpa12.2 mutants is rather because of impaired cleavage activity than to altered NTP specificity
Time-course experiments with extension scaffold ext1 were performed to distinguish whether NTP misincorporation by Rpa12.2 mutants was due to impaired cleavage activity or altered NTP specificity (Fig. 8). Incubation of both Pol I WT and all analyzed Pol I mutants with the matching nucleotide GTP resulted in addition of one nucleotide (+1) within 1 min (Fig. 8B). In reactions with Pol I WT, Pol I ΔRpa49, and Pol I ΔRpa12 supplemented with recombinant Rpa12.2, a truncated RNA appeared after 5 min, which was one nucleotide shorter (−1) than the original RNA suggesting that these enzymes started to remove nucleotides because of their cleavage activity. In contrast, all enzymes with mutated Rpa12.2 removed no nucleotides but added a second nonmatching nucleotide starting after 5 min of incubation. A straightforward explanation is that Pols without cleavage activity can escape their arrested (probably backtracked) state and incorporate an additional mismatching nucleotide. Incubation of all Pol I enzymes with the mismatching ATP resulted in incorporation of one additional nucleotide, however, apparently with different efficiency (Fig. 8C). Whereas enzymes without cleavage activity incorporate a second mismatching nucleotide after 5 min of incubation, Rpa12.2 containing enzymes remove nucleotides within the same time frame. To better distinguish how Pol I WT and Pol I Rpa12 mutants deal with matching and mismatching nucleotides in the herein analyzed time frame, time-course experiments of a matching (first, GTP) and a mismatching (second, ATP) nucleotide were performed (Fig. 8D). All enzymes add the matching GTP at the +1 position and certain amounts of the mismatching ATP at the +2 position. However, Pol I enzymes without cleavage activity significantly misincorporate ATP at the second position, whereas enzymes containing cleavage activity misincorporate only a minor amount of ATP but generate cleaved RNA substrates within the same time frame (compare Fig. 8D, lanes 7–9 and 15–17 with lanes 3–5 and 11–13; see quantitations aside). Apparently, Pol I with cleavage activity keeps a balance between NMP removal and NTP incorporation until the correct nucleotide is offered. If an incorrect nucleotide was added, the cleavage reaction is faster than the addition of a further nucleotide. In contrast, cleavage-deficient enzymes can escape this proofreading process and stably incorporate nonmatching nucleotides.
Figure 8.
Time course of NTP addition using matching and nonmatching nucleotides and Pol I enzymes with and without cleavage activity.A, upper panel, RNA–DNA scaffold used for misincorporation assays (ext1). B, Pol I enzymes containing cleavage activity incorporate and remove matching nucleotides during longer incubation times, whereas Pol I Rpa12 mutants add also mismatching nucleotides. About 0.2 pmol Pol I WT, 0.2 pmol Pol I ΔRpa49, 0.2 pmol Pol I ΔRpa12, 0.2 pmol Pol I Rpa12ΔC, and 0.2 pmol Pol I ΔRpa12 supplemented with 2 pmol Rpa12FL, 2 pmol Rpa12 (1–69), or 40 pmol Rpa12 (69–125) were incubated with RNA–DNA scaffold ext1 for 20 min on ice. GTP (final concentration: 200 μmol) was added for the indicated time points. The resulting transcripts were analyzed on a 20% denaturing polyacrylamide gel (20% [v/v] acrylamide/bisacrylamide [19:1], 6 M urea, 0.1% [v/v] TEMED, and 0.1% [w/v] ammonium persulfate). C, incorporation kinetics of a mismatching nucleotide by Pol I enzymes with and without cleavage activity. Same experiment as for (B), but with 200 μmol ATP instead of GTP. Note that Pol I enzymes incorporate in general ATP less efficiently than GTP. Pol I enzymes without RNA cleavage activity misincorporate a second ATP, whereas enzymes with cleavage activity end up with an equilibrium of one (wrongly) added nucleotide and its cleaved substrate. D, time-course experiment of a matching (first, GTP) and a mismatching (second, ATP) nucleotide. Same experiment as for (B), but with 200 μmol GTP and 200 μmol ATP. Pol I enzymes without cleavage activity misincorporate ATP at the second position, whereas enzymes containing cleavage activity misincorporate a minor amount of ATP but generate distinct cleaved substrates. Quantitation of two independent experiments is shown in the graphic representations. Pol I, polymerase I; TEMED, N, N, N', N'-tetramethylethylenediamine.
Discussion
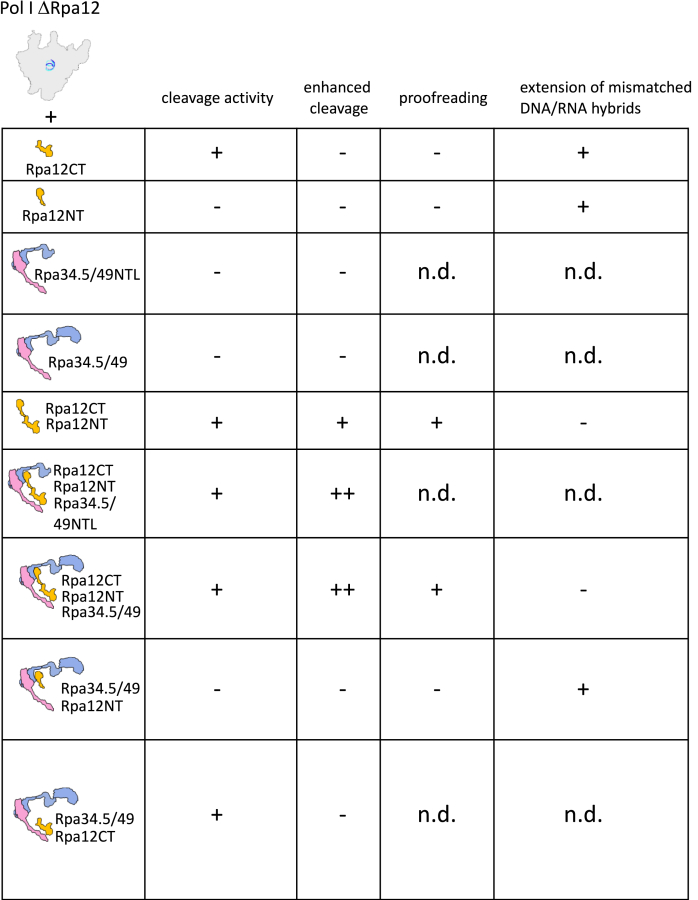
Nuclear RNA Pols need RNA-synthesizing and RNA-hydrolyzing activities to transcribe efficiently and faithfully their target genes. Transcript cleavage in response to nucleotide misincorporation is considered to play an important role in proofreading. Goal of this study was to find out how the domains of the lobe-binding subunits of Pol I participate in RNA cleavage and contribute to transcription fidelity. An overview of the obtained data is summarized in Figure 9.
Figure 9.
Summary of how lobe-binding domains contribute to RNA cleavage, cleavage efficiency, proofreading, and extension of mismatched DNA–RNA hybrids. n.d., not determined.
The role of lobe-binding subunit domains in RNA cleavage and Pol I backtracking
The C terminus of Rpa12.2 resembles the C-ribbon domain of the Pol II transcription factor TFIIS and contains the two conserved amino acids Asp105 and Glu106, the TFIIS counterparts of which (Asp290 and Glu291) probably support RNA hydrolysis by positioning a metal ion and water in the active center of Pol II (42). As anticipated, both exchange of the two amino acids to alanine and deletion of the complete C-ribbon domain abolished RNA hydrolysis confirming the crucial role of both acidic amino acids for RNA cleavage. Exchange of the amino acids K120 and N124 to alanine at the very C-terminal tip of Rpa12.2 did not influence cleavage activity. In contrast, deletion of the last six amino acids, which represent the terminal β-strand of the C-ribbon, resulted in impairment of RNA hydrolysis, which indicates the importance of the 3D structure of the C-ribbon for cleavage. During initiation and elongation, the C-ribbon of Rpa12.2 is not positioned in the active Pol I site (17, 18) but detached from the surface of Pol I probably because of steric clashes provoked by the complete closure of the cleft (19, 20). According to these structural data, conformational changes including reopening of the cleft are required to insert the C-ribbon back into the active site for cleavage. It is possible that a complete and space-filling C-ribbon is required to either structurally reorganize or stabilize the active center of Pol I and to allow the correct positioning of the metal ion for RNA hydrolysis. This resembles Pol II-dependent RNA cleavage, where a conformational change in Pol II upon TFIIS binding was proposed by functional and structural studies (15, 43).
The C-terminal part of Rpa12.2 was not sufficient for efficient RNA cleavage and needed a covalent connection to the lobe-binding N-terminal ribbon domain of Rpa12.2. The absence of the N-terminal ribbon or its physical separation from the C-terminal part led to weaker cleavage efficiency and apparently a shallow backtracking, which allowed only the removal of two nucleotides. In TFIIS, the linker region is important for opening the crevice in the Pol II funnel and thereby responsible for major structural changes in Pol II (15). Shortening the TFIIS linker length and isolated TFIIS domains II and III abolish TFIIS function (31), but influence of the linker region on the backtracking depth was not yet reported. If backtracking is not deep enough, the 3′end of the transcript cannot be realigned to the active Pol I site. This was the case if three mismatched RNA nucleotides were at the 3′end of the transcript and the Rpa12.2 N-terminal domain was separated from the C-terminal part. Consequently, an incorrect RNA–DNA hybrid pair remained, which causes a transcription error if elongation proceeds. Indeed error-prone transcription was significantly increased in the absence of the N-terminal Rpa12.2 domain.
The N-terminal ribbon of Rpa12.2 is homologous to the N-terminal ribbon of the Pol II lobe-binding subunit Rpb9 (12). This Pol II subunit was suggested to stimulate the cleavage reaction by enhancing the response of Pol II to TFIIS (31, 44). Rpb9 is important for transcriptional fidelity in vivo (32) and in vitro (31). Rpb9 is involved in accurate Pol II start site selection (45) and regulates elongation (46). Apparently, the N-terminal Rpa12.2 ribbon domain adopts at least some of the Rpb9 functions for Pol I transcription indicating that proper assembly of lobe-binding subunits contributes in general to efficient transcription elongation.
The other Pol I lobe-binding subunits, Rpa34.5/49, were also suggested to function in RNA cleavage (4, 11). According to our reconstituted transcription system, the heterodimer can only contribute to RNA hydrolysis if Rpa12.2 is complete and functional. In particular, the Rpa49 dimerization domain including the linker, but neither the dimerization domain lacking the linker nor the isolated tandem winged helix domain, increased cleavage efficiency and backtrack depth in the presence of full-length Rpa12.2. Cleavage and backtracking stimulation was not obtained if the N-terminal ribbon of Rpa12.2 was absent. An ordered Rpa49 linker structure correlates with cleft contraction (47). It is possible that the linker fused to the dimerization domain is bridging the Pol I cleft, which leads to its partly constriction. Together with the N-terminal domain of Rpa12.2, this assembly could be the prerequisite to stimulate deep backtracking and efficient RNA cleavage mediated by the Rpa12.2 C-ribbon.
The role of lobe-binding subunit domains in resumption of elongation and proofreading
RNA Pols are frequently paused during elongation, which results either in forward transcription or stable arrest (48). Incorporation of wrong nucleotides slows down elongation, which apparently promotes pausing (40, 41, 49, 50). According to Pol II-dependent transcription, mismatched 3′-RNA termini of the herein used RNA–DNA scaffolds should cause a distortion in the RNA–DNA duplex and either adopt a frayed state or promote backtracking of the enzyme (50, 51, 52). The enzymes can overcome the arrest and restart transcription either by reversion of the backtracked state or RNA cleavage, which relocates the 3′end of the RNA at the active site. In principal, the choice of backtrack recovery mechanism is determined by a kinetic competition between 1D diffusion and RNA cleavage (14).
The RNA–DNA scaffolds for the herein used cleavage assays had mismatched RNA–DNA hybrids of different length and require backtracking and removal of either one (clv1 and clv2) or two dinucleotides (clv3) to relocate the active site with a matching base pair. Pol I WT and Pol I ΔRpa12 substituted with full-length Rpa12.2 were able to resume elongation after the cleavage reaction. Since RNA cleavage was described as a stochastic resetting process of backtracked Pols (53), backtracking in combination with cleavage and reactivation probably dominated the 1D diffusion kinetics resulting in an error-free transcript. In cleavage-deficient Pol I mutants, the backtracked state apparently could not be resolved by removal of nucleotides. Therefore, 1D diffusion kinetics might become more pronounced and a small fraction of transcripts with one or two RNA–DNA base mismatches could escape the backtracked state and was extended over time. However, more than two mismatched RNA–DNA nucleotides were not tolerated by the 1D diffusion mechanism. Interestingly, N-terminal truncated Rpa12.2 mutants shortened RNAs with three terminal mismatched RNA–DNA nucleotides only by two nucleotides resulting in one terminal mismatched RNA–DNA hybrid. Apparently, the remaining single unpaired nucleotide was tolerated, and elongation could be reactivated by 1D diffusion.
Proper backtracking and RNA cleavage serve also as editing function if a wrong nucleotide was incorporated in the nascent Pol II-transcribed RNA chain (reviewed in Refs. (29, 30)). However, in principle, NTP promiscuity in mutant enzymes can be evoked either by deficient cleavage properties or by different NTP selectivity and/or incorporation kinetics. In fact, it was suggested that in addition to RNA cleavage, subunit Rpa12.2 affects the kinetics and energetics of nucleotide incorporation (33), destabilizes elongation complexes (24), and that its N terminus contributes to core transcription elongation properties rather than stimulating cleavage activity (26). Accordingly, Rpa12.2 domains might contribute differently to transcription fidelity and chain elongation. In our experimental setup, misincorporated nucleotides by all analyzed Rpa12.2 mutants were observed after several minutes of incorporation times, which were slower than RNA cleavage reactions mediated by Pol I WT. In contrast, nucleotide addition rate constants of Pol I WT and Pol I ΔRpa12 were in the range of milliseconds (33, 37) although the polymerization mechanism was affected in Rpa12.2-depleted Pol I (33). RNA cleavage reactions were in the same time span though slightly delayed (37). In consideration of the much longer time scale in the herein presented extension assays, our data support the explanation that NTP promiscuity of Rpa12.2 mutants is due to their missing proofreading, which is caused by inefficient RNA cleavage.
Certainly, Rpa12.2 domains might contribute differently to elongation properties. The C terminus stimulates RNA cleavage, whereas the N terminus might modulate elongation including acceleration of nucleotide incorporation and support of backtracking which, in turn, influences RNA cleavage. Future structural analyses are necessary to clarify the specific Rpa12.2 functions.
Although the Rpa34.5/49 heterodimer was involved in efficiency of backtracking and cleavage, it seemed not to influence resumption of elongation and proofreading under the herein used conditions. It was previously shown that the heterodimer supports transcription elongation in vivo and in vitro (4, 11, 18, 35). It is possible that the heterodimer supports elongation in a phase of the transcription process, which is not covered by the experimental setup in the present study. This is presently under investigation. Pausing, backtracking, RNA cleavage, and resumption of elongation are crucial elements for Pol II to pass through nucleosomes (see for reviews, Refs. (54, 55)). Since RNA cleavage activity and the presence of the heterodimer were recently reported to support Pol I passage through nucleosomes (36), it will be interesting to see how the domains of the lobe-binding subunits cooperate when Pol I transcribes in the chromatin context.
Experimental procedures
Yeast strains, plasmids, and oligonucleotides
Yeast strains, plasmids, and oligonucleotides used in this study are listed in Tables S1–S3. Molecular biological methods and transformation of yeast cells were performed according to standard protocols (56, 57, 58). Transcription templates were generated by PCR using the indicated oligonucleotides and plasmids. Plasmid sequences are available upon request. To purify Pol I from yeast, cells were grown at 30 °C in either yeast extract–peptone–dextrose (YPD) (2% [w/v] peptone, 1% [w/v] yeast extract, and 2% [w/v] glucose), in yeast extract–peptone–galactose (2% [w/v] peptone, 1% [w/v] yeast extract, 2% [w/v] galactose), in 2× SCG-LEU (1.34% [w/v] YNB + nitrogen, 0.134% [w/v] CSM His-Leu-Trp, 50 μg/ml l-tryptophan, 20 μg/ml l-histidine, 2% [w/v] galactose), or in 2× SCD-LEU (1.34% [w/v] YNB + nitrogen, 0.134% [w/v] CSM His-Leu-Trp, 50 μg/ml l-tryptophan, 20 μg/ml l-histidine, and 2% [w/v] galactose).
Template generation
The minimal transcription templates used in this study are listed in Table S4. The oligonucleotides for template generation are listed in Table S1. About 20 μl of the template strand oligo (100 μM) and 20 μl of the nontemplate strand oligo (100 μM) were incubated at 95 °C for 5 min. Afterward, the sample was stepwise cooled to 25 °C in 60 min (ramp rate: 1 °C/s, −1 °C per cycle, 70 cycles per 50 s) resulting in a preannealed scaffold (50 μM) containing the template and nontemplate strand oligos. About 10 μl of the preannealed template and nontemplate strand oligos (50 μM) were incubated with 5 μl 5′fluorescently labeled RNA (100 μM) and 35 μl melting point water at 45 °C for 5 min. Then, the sample was stepwise cooled to 4 °C in 60 min (ramp rate: 1 °C/s, −1 °C per cycle, 41 cycles per 80 s) resulting in a minimal transcription cleavage or extension scaffold (10 μM) containing the template and nontemplate strand oligos and a Cy5-labeled RNA. The scaffold was diluted to 0.1 μM for subsequent RNA cleavage or extension assays.
RNA cleavage assay
Unless otherwise stated, 0.2 pmol of Pol was incubated with 0.066 pmol of the respective preannealed minimal transcription cleavage scaffold in 1× transcription buffer (20 mM Hepes/KOH [pH 7.8], 10 mM MgCl2, 5 mM EGTA/KOH [pH 8.0], 5 μM ZnCl2, and 100 mM KOAc) at 4 °C for 10 to 20 min. For RNA cleavage, the samples were incubated at 28 °C for 30 min. The reactions were stopped by addition of an equal amount of 2× TBE loading dye (8 M urea, 0.08% [w/v] bromophenol blue, 4 mM EDTA/NaOH [pH 8.0], 178 mM Tris–HCl, and 178 mM boric acid). For re-extension analysis, NTPs (final concentration of each nucleotide: 200 μM) were added, and the samples were incubated for further 30 min at 28 °C. Then, the reactions were stopped by addition of an equal amount of 2× TBE loading dye. The samples were incubated at 95 °C for 3 min and then chilled on ice. The 5′Cy5-labeled RNA was size-separated on a 20% denaturing polyacrylamide gel (20% [w/v] acrylamide/bisacrylamide [19:1], 6 M urea, 0.1% [v/v] N, N, N', N'-tetramethylethylenediamine, 0.1% [w/v] ammonium persulfate, 2 mM EDTA/NaOH [pH 8.0], 89 mM Tris–HCl, and 89 mM boric acid) using 1× TBE running buffer (89 mM Tris–HCl, 89 mM boric acid, and 2 mM EDTA/NaOH [pH 8.0]) and visualized with a Typhoon imaging system (GE Healthcare).
RNA extension assay
Unless otherwise stated, 0.2 pmol of Pol was incubated with 0.066 pmol of the respective preannealed minimal transcription extension scaffold in 1× transcription buffer (20 mM Hepes/KOH [pH 7.8], 10 mM MgCl2, 5 mM EGTA/KOH [pH 8.0], 5 μM ZnCl2, and 100 mM KOAc) at 4 °C for 10 to 20 min. For RNA extension, NTPs (final concentration of each nucleotide: 200 μM) were added, and the samples were incubated at 28 °C for 30 min. The reactions were stopped by addition of an equal amount of 2× TBE loading dye (8 M urea, 0.08% [w/v] bromophenol blue, 4 mM EDTA/NaOH [pH 8.0], 178 mM Tris–HCl, and 178 mM boric acid). The samples were incubated at 95 °C for 3 min and then chilled on ice. The 5′Cy5-labeled RNA was size-separated on a 20% denaturing polyacrylamide gel (20% [w/v] acrylamide/bisacrylamide [19:1], 6 M urea, 0.1% [v/v] N, N, N', N'-tetramethylethylenediamine, 0.1% [w/v] ammonium persulfate, 2 mM EDTA/NaOH [pH 8.0], 89 mM Tris–HCl, and 89 mM boric acid) using 1× TBE running buffer (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA/NaOH [pH 8.0]) and visualized with a Typhoon imaging system (GE Healthcare).
Purification of RPA12 variants
The coding sequences of A12.2 and A12.2 variants were cloned sequentially into vector pOPIN-B, resulting in a human rhinovirus–cleavable N-terminal hexahistidine tag on A12.2 and A12.2 variants. pOPIN-B was a gift from Ray Owens (Addgene plasmid #41142; http://n2t.net/addgene:41142; Research Resource Identifier: Addgene_41142). The resulting plasmids contain a T7 promotor, a lac operon for induction of protein expression, a ribosome-binding site to enable bicistronic expression, the coding sequence of a 6×His-tag for protein purification, the coding sequence of RPA12 variants, and a T7 terminator sequence, respectively. The plasmids also possess a kanamycin resistance gene for selection of positive clones. For the overexpression of the respective target proteins, the generated plasmids (K2720–2725, K2931–2936; listed in Table S4) were transformed into chemically competent E. coli BL21 (DE3) pRARE cells and plated on LB plates supplemented with kanamycin (final concentration: 50 μg/ml) and chloramphenicol (final concentration: 30 μg/ml). About 50 ml LB medium supplemented with the respective antibiotics was inoculated with a single clone, and cells were incubated overnight at 37 °C, shaking at 120 rpm. Next day, 1 l LB medium supplemented with kanamycin (final concentration: 50 μg/ml) and chloramphenicol (final concentration: 30 μg/ml) was inoculated with the overnight culture to a final absorbance of 0.1 at 600 nm. The cells were incubated at 37 °C, shaking at 120 rpm. At absorbance of ∼1 at 600 nm, the cells were cold shocked for 15 min on ice. The protein expression was induced by the addition of IPTG (final concentration: 0.1 mM), and cells were further incubated at 18 °C for 3 h, shaking at 120 rpm. Cells were harvested at 9000g and room temperature (RT) for 8 min. Cell pellets were resuspended in 20 ml ice-cold sterile 1× PBS (137 mM NaCl, 2.7 mM KCl, 18 mM KH2PO4, and 1 mM Na2PO4) and centrifuged at 9000g and 4 °C for 8 min. The pellets were frozen in liquid nitrogen and stored at −20 °C until usage. The cells were thawed on ice and resuspended in equal amounts of ice-cold lysis buffer L1 (for plasmid K2725; 50 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 10 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF, 2 mM benzamidine, 10 μM ZnCl2, and 500 mM KCl), L6 (for plasmids K2721, K2722; 50 mM MES/NaOH [pH 6.0], 10% [v/v] glycerol, 10 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF, 2 mM benzamidine, 10 μM ZnCl2, 500 mM KCl, and 1% [v/v] NP-40) or L0 (for plasmids K2722, K2723, K2724, K2931–K2936; 50 mM Hepes/KOH [pH 7.0], 10% [v/v] glycerol, 500 mM KCl, 10 mM imidazole, 10 μM ZnCl2, 5 mM β-mercaptoethanol, 0.5% [v/v] NP-40, 1 mM PMSF, and 2 mM benzamidine). Then, the cell suspension was centrifuged at 9000g and 4 °C for 8 min. After discarding the supernatant, the cells were resuspended in 25 ml lysis buffer. The cells were lysed by ultrasonication. Therefore, the cells were lysed for 5 min in a water bath followed by a 5 min pause. The procedure was repeated twice. Afterward, the lysate was cleared by centrifugation at 15,000g and 4 °C for 20 min. The supernatant was centrifuged at 146,500g and 4 °C for 40 min. Meanwhile, 500 μl TALON Metal Affinity Resin beads (TaKaRa) were equilibrated with the respective lysis buffer. Then, the supernatant was incubated with pre-equilibrated TALON beads for 1 h at 4 °C on a turning wheel. To remove unbound proteins, the beads were centrifuged at 800 rpm and 4 °C for 2 min, and the supernatant was discarded. The beads were resuspended with 10 ml lysis buffer and transferred in a polypropylene column resin. Then, the beads were washed three times with 10 ml wash buffers W1 (for plasmid K2725; 20 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 20 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF, 2 mM benzamidine, 10 μM ZnCl2, and 500 mM KCl), W6 (for plasmids K2721, K2722; 20 mM MES/NaOH [pH 6.0], 10% [v/v] glycerol, 20 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF, 2 mM benzamidine, 10 μM ZnCl2, 500 mM KCl, and 1% [v/v] NP-40) or W0 (for plasmids K2722, K2723, K2724, K2931–K2936; 20 mM Hepes/KOH [pH 7.0], 10% [v/v] glycerol, 500 mM KCl, 10 mM imidazole, 10 μM ZnCl2, 5 mM β-mercaptoethanol, 0.5% [v/v] NP-40, 1 mM PMSF, and 2 mM benzamidine). Then, the recombinant proteins were eluted with 5× 250 μl elution buffer E1 (for plasmid K2725; 20 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 150 mM imidazole, 5 mM β-mercaptoethanol, 10 μM ZnCl2, and 500 mM KCl), E6 (20 mM MES/NaOH [pH 6.0], 10% [v/v] glycerol, 150 mM imidazole, 5 mM β-mercaptoethanol, 10 μM ZnCl2, 500 mM KCl, and 1% [v/v] NP-40) or E0 (for plasmids K2722, K2723, K2724, K2931–K2936; 20 mM Hepes/KOH [pH 7.0], 10% [v/v] glycerol, 500 mM KCl, 150 mM imidazole, 10 μM ZnCl2, 5 mM β-mercaptoethanol, and 0.5% [v/v] NP-40). The purified protein fractions were analyzed by SDS-PAGE and Coomassie staining.
Pol I preparation
Pol I and Pol II were purified according to Ref. (36) with changes. All RNA Pols were purified from yeast strains (Table S3) via the protein A affinity tag, which was fused to the second largest subunit of Pol I (RPA135) or Pol II (RPB2). The tag and the respective subunit were separated by a tobacco etch virus (TEV) cleavage site.
The yeast strains Y4094 (WT Pol I), Y2423 (WT Pol I), and Y2424 (Pol II) were grown in YPD (2% [w/v] peptone, 2% [w/v] glucose, 1% [w/v] yeast extract) at 30 °C. Precultures of yeast strains Y4005 (Δ12 Pol I), Y4006 (WT Pol I), and Y4007 (12ΔC Pol I) were grown in 2× SCG-LEU (1.34% [w/v] YNB + nitrogen, 0.134% [w/v] CSM His-Leu-Trp, 2% [w/v] galactose, 50 μg/ml l-tryptophan, and 20 μg/ml l-histidine) at 30 °C. For depletion of A12.2, the cells were grown 2× SCD-LEU (1.34% [w/v] YNB + nitrogen, 0.134% [w/v] CSM His-Leu-Trp, 2% [w/v] glucose, 50 μg/ml l-tryptophan, and 20 μg/ml l-histidine) at 30 °C for 18 h. Precultures of yeast strains Y2670 (Δ49 Pol I) and Y2679 (Δ12 Pol I) were grown in yeast extract–peptone–galactose (2% [w/v] peptone, 2% [w/v] galactose, and 1% [w/v] yeast extract) at 30 °C. For depletion of A12.2 or A49, the cells were grown in YPD (2% [w/v] peptone, 2% [w/v] glucose, and 1% [w/v] yeast extract) at 30 °C for 18 h. The cells were harvested at 9000g and RT for 8 min. Cells were washed with ice-cold melting point grade water and centrifuged at 9000g and RT for 8 min. The cell pellets were frozen in liquid nitrogen and stored at −20 °C until usage. For RNA Pol purification, the cells were thawed on ice and resuspended in equal amounts of ice-cold lysis buffer L2 (50 mM Hepes/KOH [pH 7.8], 20% [v/v] glycerol, 40 mM MgCl2, 400 mM (NH4)2SO4, 3 mM DTT, 1 mM PMSF, and 2 mM benzamidine). The cells were centrifuged at 4500 rpm (Heraeus Megafuge 16R) and 4 °C for 10 min. The cells were resuspended in lysis buffer L2 (1.5 ml per 1 g cells). About 6 ml of the cell suspension was transferred in a precooled 15 ml precellys tube (Precellys Lysing kit) filled with 12 g glass beads (Ø 0.75–1 mm). The cells were lysed with Precellys Evolution (Bertin Technologies) using 6× pulse at 6000 rpm for 30 s followed by 30 s pause in between. The process was repeated 2 to 3 times. The cell lysate was transferred in a 50 ml falcon and centrifuged at 15,000g and 4 °C for 15 min. The supernatant was ultracentrifuged at 146,500g and 4 °C for 40 min. Meanwhile, magnetic immunoglobulin G affinity beads (1 μl bead suspension per 2 mg protein) were equilibrated with lysis buffer L2. Then, the supernatant was incubated with pre-equilibrated magnetic immunoglobulin G beads for 2 h at 4 °C on a turning wheel. To remove unbound proteins, the beads were separated via a magnetic separator. The beads were washed four times with 1 ml wash buffer W2 (20 mM Hepes/KOH [pH 7.8], 20% [v/v] glycerol, 5 mM MgCl2, 1500 mM KOAc, and 0.15% [v/v] NP-40) for 10 min at 4 °C on a turning wheel. Afterward, the beads were equilibrated with elution buffer E2.1 (for Pol I; 20 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 5 mM MgCl2, 200 mM KOAc, and 5 μM ZnCl2) or E2.2 (for Pol II; 20 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 5 mM MgCl2, and 5 μM ZnCl2). Therefore, the beads were incubated three times in 1 ml of the respective elution buffer at 4 °C for 3 min on a turning wheel. The beads were resuspended in the respective elution buffer (0.5 μl elution buffer per 1 μl of bead suspension), and TEV protease (final concentration: 115 ng/μl) was added. The suspension was incubated at 16 °C overnight, shaking at 1200 rpm (Eppendorf incubator). The eluate was transferred in a new tube, and the beads were resuspended in elution buffer (0.25 μl elution buffer per 1 μl of bead suspension). Afterward, TEV protease (final concentration: 115 ng/μl) was added, and the suspension was incubated at 16 °C for 2 h, shaking at 1200 rpm (Eppendorf incubator). After magnetic separation, the eluate (eluate 2) was transferred in a new tube. The purified Pol fractions were analyzed by SDS-PAGE and Coomassie staining. The presence of the subunits A12.2, A49, and A190 was verified by Western blotting using the respective antibodies.
Purification of heterodimer variants
For the overexpression of the respective target proteins, the plasmids (K2250, K2404, K2406, K2403, and K2407; listed in Table S4) were transformed into chemically competent E. coli BL21 (DE3) pRARE cells and plated on LB plates supplemented with kanamycin (final concentration: 50 μg/ml) and chloramphenicol (final concentration: 30 μg/ml). About 50 ml LB medium supplemented with the respective antibiotics was inoculated with a single clone, and cells were incubated overnight at 37 °C, shaking at 120 rpm. Next day, 2 l prewarmed Terrific Broth medium supplemented with kanamycin (final concentration: 50 μg/ml) and chloramphenicol (final concentration: 30 μg/ml) was inoculated with the overnight culture to a final absorbance of 0.1 at 600 nm. The cells were incubated at 37 °C, shaking at 120 rpm. At absorbance of ∼0.5 at 600 nm, the cells were cold shocked for 15 min on ice. The cells were incubated at 18 °C overnight, shaking at 120 rpm. Cells were harvested at 9000g and RT for 8 min. Cell pellets were resuspended in 20 ml ice-cold sterile 1× PBS and centrifuged at 9000g and 4 °C for 8 min. The pellets were frozen in liquid nitrogen and stored at −20 °C until usage. The cells were thawed on ice and resuspended in equal amounts of ice-cold lysis buffer L5 (50 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 5 mM MgAc2, 200 mM KCl, 10 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF, and 2 mM benzamidine). Then, the cell suspension was centrifuged at 9000g and 4 °C for 8 min. After discarding the supernatant, the cells were resuspended in lysis buffer L5 (1–1.5 ml buffer per 1 g cells). The cells were lysed by ultrasonication. Therefore, the cells were lysed for 5 min in a water bath followed by a 5 min pause. The procedure was repeated twice. Afterward, the lysate was cleared by centrifugation at 15,000g and 4 °C for 20 min. The supernatant was ultracentrifuged at 146,500g and 4 °C for 40 min. Meanwhile, 1000 μl TALON Metal Affinity Resin beads were equilibrated with lysis buffer L5. Then, the supernatant was incubated with pre-equilibrated TALON beads for 1 h at 4 °C on a turning wheel. To remove unbound proteins, the beads were centrifuged at 800 rpm and 4 °C for 2 min, and the supernatant was discarded. The beads were resuspended with 10 ml lysis buffer and transferred in a polypropylene column resin. Then, the beads were washed three times with 10 ml wash buffers W5 (50 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 5 mM MgAc2, 10 mM imidazole, 200 mM KCl, 5 mM β-mercaptoethanol, 1 mM PMSF, and 2 mM benzamidine). Then, the recombinant proteins were with 5× 500 μl elution buffer E5 (20 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 5 mM MgAc2, 150 mM imidazole, 200 mM KCl, and 5 mM β-mercaptoethanol). The purified protein fractions were analyzed by SDS-PAGE and Coomassie staining. Fractions with the respective proteins were pooled and diluted with equal volume of buffer A (20 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 1 mM MgAc2, and 3 mM DTT), loaded onto a MonoS anion exchange column (MonoS GL 5/50; GE Healthcare). The recombinant proteins were eluted with a linear gradient of 10 to 100% buffer B (20 mM Hepes/KOH [pH 7.8], 10% [v/v] glycerol, 1 mM MgAc2, 3 mM DTT, and 1 M KCl). The purified protein fractions were analyzed by SDS-PAGE and Coomassie staining.
Data availability
Underlying data for quantitations are available upon request. All other data are contained within the article.
Supporting information
This article contains supporting information (18, 36, 59).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to members of the chair Biochemistry III and especially to Christoph Engel and the members of his group for critical discussion. Michael Pilsl's advice for development and improvement of methods is greatly appreciated. This work was supported through grants of the Deutsche Forschungsgemeinschaft (grant no.: SFB 960).
Author contributions
K. S., P. M., J. G., and H. T. conceptualization; K. S. and T. F. methodology; K. S. and C. S. investigation; H. T. writing–original draft; K. S., P. M., and J. G. writing–review & editing; H. T. visualization; H. T. supervision; P. M., J. G., and H. T. project administration; P. M., J. G., and H. T. funding acquisition.
Edited by Karin Musier-Forsyth
Contributor Information
Philipp Milkereit, Email: philipp.milkereit@ur.de.
Joachim Griesenbeck, Email: joachim.griesenbeck@ur.de.
Herbert Tschochner, Email: herbert.tschochner@ur.de.
Supporting information
References
- 1.Engel C., Neyer S., Cramer P. Distinct mechanisms of transcription initiation by RNA polymerases I and II. Annu. Rev. Biophys. 2018;47:425–446. doi: 10.1146/annurev-biophys-070317-033058. [DOI] [PubMed] [Google Scholar]
- 2.Khatter H., Vorlander M.K., Muller C.W. RNA polymerase I and III: Similar yet unique. Curr. Opin. Struct. Biol. 2017;47:88–94. doi: 10.1016/j.sbi.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Vannini A., Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 4.Geiger S.R., Lorenzen K., Schreieck A., Hanecker P., Kostrewa D., Heck A.J., Cramer P. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell. 2010;39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Knutson B.A., McNamar R., Rothblum L.I. Dynamics of the RNA polymerase I TFIIF/TFIIE-like subcomplex: A mini-review. Biochem. Soc. Trans. 2020;48:1917–1927. doi: 10.1042/BST20190848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luse D.S., Studitsky V.M. The mechanism of nucleosome traversal by RNA polymerase II: Roles for template uncoiling and transcript elongation factors. RNA Biol. 2011;8:581–585. doi: 10.4161/rna.8.4.15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schweikhard V., Meng C., Murakami K., Kaplan C.D., Kornberg R.D., Block S.M. Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6642–6647. doi: 10.1073/pnas.1405181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C., Burton Z.F. Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J. Mol. Biol. 2004;342:1085–1099. doi: 10.1016/j.jmb.2004.07.070. [DOI] [PubMed] [Google Scholar]
- 9.Zhang C., Yan H., Burton Z.F. Combinatorial control of human RNA polymerase II (RNAP II) pausing and transcript cleavage by transcription factor IIF, hepatitis delta antigen, and stimulatory factor II. J. Biol. Chem. 2003;278:50101–50111. doi: 10.1074/jbc.M307590200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C., Zobeck K.L., Burton Z.F. Human RNA polymerase II elongation in slow motion: Role of the TFIIF RAP74 alpha1 helix in nucleoside triphosphate-driven translocation. Mol. Cell Biol. 2005;25:3583–3595. doi: 10.1128/MCB.25.9.3583-3595.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn C.D., Geiger S.R., Baumli S., Gartmann M., Gerber J., Jennebach S., Mielke T., Tschochner H., Beckmann R., Cramer P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 12.Ruan W., Lehmann E., Thomm M., Kostrewa D., Cramer P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J. Biol. Chem. 2011;286:18701–18707. doi: 10.1074/jbc.M111.222273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jennebach S., Herzog F., Aebersold R., Cramer P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. 2012;40:5591–5601. doi: 10.1093/nar/gks220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lisica A., Engel C., Jahnel M., Roldan E., Galburt E.A., Cramer P., Grill S.W. Mechanisms of backtrack recovery by RNA polymerases I and II. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2946–2951. doi: 10.1073/pnas.1517011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kettenberger H., Armache K.J., Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 16.Zhao D., Liu W., Chen K., Wu Z., Yang H., Xu Y. Structure of the human RNA polymerase I elongation complex. Cell Discov. 2021;7:97. doi: 10.1038/s41421-021-00335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel C., Plitzko J., Cramer P. RNA polymerase I-Rrn3 complex at 4.8 A resolution. Nat. Commun. 2016;7:12129. doi: 10.1038/ncomms12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilsl M., Crucifix C., Papai G., Krupp F., Steinbauer R., Griesenbeck J., Milkereit P., Tschochner H., Schultz P. Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat. Commun. 2016;7:12126. doi: 10.1038/ncomms12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tafur L., Sadian Y., Hoffmann N.A., Jakobi A.J., Wetzel R., Hagen W.J.H., Sachse C., Muller C.W. Molecular structures of transcribing RNA polymerase I. Mol. Cell. 2016;64:1135–1143. doi: 10.1016/j.molcel.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neyer S., Kunz M., Geiss C., Hantsche M., Hodirnau V.V., Seybert A., Engel C., Scheffer M.P., Cramer P., Frangakis A.S. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016;540:607–610. doi: 10.1038/nature20561. [DOI] [PubMed] [Google Scholar]
- 21.Nogi Y., Yano R., Dodd J., Carles C., Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol. Cell Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prescott E.M., Osheim Y.N., Jones H.S., Alen C.M., Roan J.G., Reeder R.H., Beyer A.L., Proudfoot N.J. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Mullem V., Landrieux E., Vandenhaute J., Thuriaux P. Rpa12p, a conserved RNA polymerase I subunit with two functional domains. Mol. Microbiol. 2002;43:1105–1113. doi: 10.1046/j.1365-2958.2002.02824.x. [DOI] [PubMed] [Google Scholar]
- 24.Appling F.D., Scull C.E., Lucius A.L., Schneider D.A. The A12.2 subunit is an intrinsic destabilizer of the RNA polymerase I elongation complex. Biophys. J. 2018;114:2507–2515. doi: 10.1016/j.bpj.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gout J.F., Li W., Fritsch C., Li A., Haroon S., Singh L., Hua D., Fazelinia H., Smith Z., Seeholzer S., Thomas K., Lynch M., Vermulst M. The landscape of transcription errors in eukaryotic cells. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1701484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scull C.E., Lucius A.L., Schneider D.A. The N-terminal domain of the A12.2 subunit stimulates RNA polymerase I transcription elongation. Biophys. J. 2021;120:1883–1893. doi: 10.1016/j.bpj.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel C., Sainsbury S., Cheung A.C., Kostrewa D., Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Tornero C., Moreno-Morcillo M., Rashid U.J., Taylor N.M., Ruiz F.M., Gruene T., Legrand P., Steuerwald U., Muller C.W. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 29.Sydow J.F., Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr. Opin. Struct. Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Xu L., Da L., Plouffe S.W., Chong J., Kool E., Wang D. Molecular basis of transcriptional fidelity and DNA lesion-induced transcriptional mutagenesis. DNA Repair. 2014;19:71–83. doi: 10.1016/j.dnarep.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knippa K., Peterson D.O. Fidelity of RNA polymerase II transcription: Role of Rpb9 [corrected] in error detection and proofreading. Biochemistry. 2013;52:7807–7817. doi: 10.1021/bi4009566. [DOI] [PubMed] [Google Scholar]
- 32.Nesser N.K., Peterson D.O., Hawley D.K. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3268–3273. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Appling F.D., Schneider D.A., Lucius A.L. Multisubunit RNA polymerase cleavage factors modulate the kinetics and energetics of nucleotide incorporation: An RNA polymerase I case study. Biochemistry. 2017;56:5654–5662. doi: 10.1021/acs.biochem.7b00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merkl P., Perez-Fernandez J., Pilsl M., Reiter A., Williams L., Gerber J., Bohm M., Deutzmann R., Griesenbeck J., Milkereit P., Tschochner H. Binding of the termination factor nsi1 to its cognate DNA site is sufficient to terminate RNA polymerase I transcription in vitro and to induce termination in vivo. Mol. Cell Biol. 2014;34:3817–3827. doi: 10.1128/MCB.00395-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckouet F., Labarre-Mariotte S., Albert B., Imazawa Y., Werner M., Gadal O., Nogi Y., Thuriaux P. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol. Cell Biol. 2008;28:1596–1605. doi: 10.1128/MCB.01464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merkl P.E., Pilsl M., Fremter T., Schwank K., Engel C., Längst G., Milkereit P., Griesenbeck J., Tschochner H. RNA polymerase I (Pol I) passage through nucleosomes depends on Pol I subunits binding its lobe structure. J. Biol. Chem. 2020;295:4782–4795. doi: 10.1074/jbc.RA119.011827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Appling F.D., Lucius A.L., Schneider D.A. Transient-state kinetic analysis of the RNA polymerase I nucleotide incorporation mechanism. Biophysical J. 2015;109:2382–2393. doi: 10.1016/j.bpj.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L., Wang W., Chong J., Shin J.H., Xu J., Wang D. RNA polymerase II transcriptional fidelity control and its functional interplay with DNA modifications. Crit. Rev. Biochem. Mol. Biol. 2015;50:503–519. doi: 10.3109/10409238.2015.1087960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeon C., Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas M.J., Platas A.A., Hawley D.K. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 42.Jeon C., Yoon H., Agarwal K. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9106–9110. doi: 10.1073/pnas.91.19.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cipres Palacin G., Kane C.M. Cleavage of the nascent transcript induced by TFIIS is insufficient to promote read-through of intrinsic blocks to elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8087–8091. doi: 10.1073/pnas.91.17.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Awrey D.E., Weilbaecher R.G., Hemming S.A., Orlicky S.M., Kane C.M., Edwards A.M. Transcription elongation through DNA arrest sites: A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS ∗. J. Biol. Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 45.Hull M.W., McKune K., Woychik N.A. RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev. 1995;9:481–490. doi: 10.1101/gad.9.4.481. [DOI] [PubMed] [Google Scholar]
- 46.Hemming S.A., Jansma D.B., Macgregor P.F., Goryachev A., Friesen J.D., Edwards A.M. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J. Biol. Chem. 2000;275:35506–35511. doi: 10.1074/jbc.M004721200. [DOI] [PubMed] [Google Scholar]
- 47.Han Y., Yan C., Nguyen T.H.D., Jackobel A.J., Ivanov I., Knutson B.A., He Y. Structural mechanism of ATP-independent transcription initiation by RNA polymerase I. Elife. 2017;6 doi: 10.7554/eLife.27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu W., Reines D. Identification of a decay in transcription potential that results in elongation factor dependence of RNA polymerase II. J. Biol. Chem. 1995;270:11238–11244. doi: 10.1074/jbc.270.19.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erie D.A., Hajiseyedjavadi O., Young M.C., von Hippel P.H. Multiple RNA polymerase conformations and GreA: Control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 50.Sydow J.F., Brueckner F., Cheung A.C., Damsma G.E., Dengl S., Lehmann E., Vassylyev D., Cramer P. Structural basis of transcription: Mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol. Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Wang D., Bushnell D.A., Huang X., Westover K.D., Levitt M., Kornberg R.D. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheung A.C., Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 53.Roldán É., Lisica A., Sánchez-Taltavull D., Grill S.W. Stochastic resetting in backtrack recovery by RNA polymerases. Phys. Rev. E. 2016;93 doi: 10.1103/PhysRevE.93.062411. [DOI] [PubMed] [Google Scholar]
- 54.Noe Gonzalez M., Blears D., Svejstrup J.Q. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat. Rev. Mol. Cell Biol. 2021;22:3–21. doi: 10.1038/s41580-020-00308-8. [DOI] [PubMed] [Google Scholar]
- 55.Kujirai T., Kurumizaka H. Transcription through the nucleosome. Curr. Opin. Struct. Biol. 2020;61:42–49. doi: 10.1016/j.sbi.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook J., Fritsch E.F., Maniatis T. 2. ed. Cold Spring Harbour Laboratory Press; NY: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 57.Burke D., Dawson D., Stearns T. Cold Spring Harbor; Plainview, NY: 2000. Methods In Yeast Genetics, A Cold Spring Harbor Lanoratory Course Manual. [Google Scholar]
- 58.Schiestl R.H., Gietz R.D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 59.Hierlmeier T., Merl J., Sauert M., Perez-Fernandez J., Schultz P., Bruckmann A., Hamperl S., Ohmayer U., Rachel R., Jacob A., Hergert K., Deutzmann R., Griesenbeck J., Hurt E., Milkereit P., et al. Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae. Nucleic Acids Res. 2013;41:1191–1210. doi: 10.1093/nar/gks1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Underlying data for quantitations are available upon request. All other data are contained within the article.