Abstract
Serum biomarkers are often insufficiently sensitive or specific to facilitate cancer screening or diagnostic testing. In ovarian cancer, the few established serum biomarkers are highly specific, yet insufficiently sensitive to detect early-stage disease and to impact the mortality rates of patients with this cancer. Here we show that a ‘disease fingerprint’ acquired via machine learning from the spectra of near-infrared fluorescence emissions of an array of carbon nanotubes functionalized with quantum defects detects high-grade serous ovarian carcinoma in serum samples from symptomatic individuals with 87% sensitivity at 98% specificity (compared with 84% sensitivity at 98% specificity for the current best clinical screening test, which uses measurements of cancer antigen 125 and transvaginal ultrasonography). We used 269 serum samples to train and validate several machine-learning classifiers for the discrimination of patients with ovarian cancer from those with other diseases and from healthy individuals. The predictive values of the best classifier could not be attained via known protein biomarkers, suggesting that the array of nanotube sensors responds to unidentified serum biomarkers.
Ovarian cancer, the second most common gynaecologic malignancy worldwide, is responsible for over 184,000 deaths each year1. If there is no sign that cancer has spread outside of the ovaries, 5-year survival rates are over 90%2. However, 59% of cases are diagnosed after they have metastasized to distant sites, for which the 5-year survival drops to only 29%2. The earlier detection of ovarian cancer and timely measurements of disease progression and recurrence would markedly improve outcomes.
Conventionally, serum biomarker measurements, such as those for cancer antigen 125 (CA125), are used as the first line test and/or to monitor high-risk women for ovarian cancer3,4. Other complementary serum biomarkers such as human epididymis protein 4 (HE4), chitinase-3-like protein 1 (YKL40) and mesothelin, or panels of biomarkers, have been reported to result in better discriminatory power over CA125-based screening4-6. However, stand-alone biomarker measurements have proven of little survival benefit due to limited specificity and low positive predictive value (PPV)7,8. Longitudinal CA125 measurements combined with transvaginal ultrasonography, result in improved PPV for ovarian cancer detection, but the benefit of screening is still elusive9,10. Currently, no screening strategy can identify disease at an early enough stage to reduce mortality10.
Major opportunities for improving patient outcomes from ovarian cancer include increasing the sensitivity of early-stage detection while maintaining high specificity and the detection of minimum-residual/low-volume disease in treated patients. However, accurate detection of known analytes does not always confer high sensitivity and specificity for disease. As established serum biomarkers do not sufficiently represent the ovarian cancer disease state and are not sensitive enough to achieve precise early diagnosis to benefit survival rates10, they only provide incremental value for improving treatment options and often do not reduce costs for patients11.
To seek an alternative approach to overcome diagnostic challenges, we investigated a perception-based strategy. Nature has evolved perception to identify and interpret multidimensional stimuli against target heterogeneity. Perception achieves target identification by using a number of sensory inputs wherein each encodes certain features of the target, and analysing these inputs against a pre-learned target pattern library. For instance, the perception of smell uses an array of non-specific olfactory receptors, whose pattern of responses is processed by the neural network in our brain to identify an odour12. Olfactory receptors are relatively small in number (100–200), yet through perception, they enable recognition of many different odours, far exceeding what is possible with one-to-one recognition. For these odours, although each signal produces relatively little predictive value, the full array of responses processed as a whole nevertheless leads to accurate identification.
Perception-based approaches have been used to classify various disease conditions on the basis of different patterns in methylation of DNA sequencing13, volatile organic compounds using electronic noses14, small metabolites using mass spectrometry15, and image analysis of pathology, computerized tomography scans and magnetic resonance imaging data16,17. Machine learning processes recognize disease-specific patterns that are too subtle or complex to be detected by human eyes or conventional analytical methods, and aid in the construction of robust diagnostic models17,18. Despite efforts to develop a generalizable method of perception-based diagnostic screening using pathology or radioimaging data, challenges remain in the identification of effective disease markers to achieve high sensitivity and selectivity, as well as practical feasibility in the clinic.
Semiconducting single-walled carbon nanotubes (SWCNTs) exhibit intrinsic near-infrared fluorescence19 with environmental responsivity down to the single-molecule level20. The emission of SWCNTs (E11) is sensitive to dielectric environments21,22, redox perturbations23 and electrostatic charge24,25. Non-covalent encapsulation with polymers, including short oligonucleotides, facilitates aqueous suspension and confers molecular selectivity to their optical responses via (1) contributing to a molecular masking effect that defines the shape and size of the exposed surface of SWCNTs26-28 and (2) modulating their optical bandgaps29.
Organic colour centres (OCCs) are molecularly tunable quantum defects on SWCNTs, which are produced by covalent functionalization of SWCNTs30,31. OCCs efficiently harvest mobile excitons through the SWCNT antenna, producing distinct fluorescence bands (E11−) at longer wavelengths from the E11 band. The E11− fluorescence introduces new biochemical sensitivities to SWCNTs determined by the chemical nature of the defect, making OCCs the molecular focal points for local environmental responses32.
Here we present a nanosensor array and a computational model that resulted in the perception-based detection of ovarian cancer from patient serum samples. To transduce broad types of physicochemical properties of a biofluid, we designed nanosensor arrays using OCC-functionalized, single-stranded DNA (ssDNA)-encapsulated SWCNTs (OCC-DNAs; Fig. 1). The emission of the OCC-DNA nanosensors exhibited diverse responses to serum samples collected from patients with high-grade serous ovarian carcinoma (HGSOC), other non-HGSOC diseases (including patients in remission, other gynaecologic processes such as endometriosis and low-grade ovarian carcinoma, non-gynaecologic cancers, and other conditions) and healthy individuals, but the optical responses did not provide substantial predictive value to differentiate these patients using conventional statistical analyses. We thus trained several machine learning models to classify HGSOC patients using the OCC-DNA sensor array responses. Support vector machine models resulted in excellent sensitivity and specificity of HGSOC detection, with an accuracy approaching 95%, outperforming conventional serum biomarker-based identification. Potential interferents, such as drug treatments, were accounted for. The sensors were then used to assess the degree of predictive value conferred by known ovarian cancer serum biomarkers, including CA125, HE4 and YKL40. Support vector regression models showed that the sensor elements responded quantitatively to these markers, but they did not account for all of the predictive value, suggesting that unknown biomarkers play an important role in the differentiation of HGSOC by the sensors.
Fig. 1 ∣. OCC-DNA nanosensor array.
a, Molecular model of an OCC-DNA nanosensor element. Shown is an ss(GT)15 DNA-wrapped (6,5)-SWCNT with 3,4,5-trifluoroaryl OCC. b, Construction of an OCC-DNA nanosensor array from OCC and ssDNA components.
Results
We synthesized an array of OCC-DNA nanosensors by introducing the OCCs to the (6,5)-SWCNT via diazonium chemistry33 and encapsulating them with a library of ssDNA to solubilize the nanosensors in biofluids. The ssDNA sequences were chosen on the basis of the recognition sequences of DNA that form specific wrapping patterns on the SWCNT surface34 to result in diverse, highly defined surface morphologies to confer disparate sensitivities to the local environment26,27. Ten different OCC-DNA nanosensors were synthesized from the combinations of three OCCs and four DNA sequences (Table 1). Each OCC-DNA nanosensor featured a pair of emission peaks that depend on the chemical nature of the OCC and DNA sequence. We used 575 nm excitation to selectively excite the (6,5)-SWCNT (Fig. 2a, and Supplementary Figs. 1 and 2), resulting in emission at ~1,000 nm from the (6,5) nanotube species E11 band and a peak falling between 1,110 to 1,170 nm, depending on the aryl functional group. The latter is denoted as the E11− band or ‘OCC peak’.
Table 1 ∣.
OCC-DNA nanosensor elements
| Terminating group of aryl OCC |
ssDNA sequence |
OCC-DNA nanosensor |
|---|---|---|
| −4-N(C2H5)2 | CT2C3T2C | NEt2*CT2C3T2C |
| (TAT)4 | NEt2*(TAT)4 | |
| (GT)15 | NEt2*(GT)15 | |
| −3,4,5-F3 | CT2C3T2C | 3F* CT2C3T2C |
| (TAT)4 | 3F*(TAT)4 | |
| (AT)15 | 3F*(AT)15 | |
| (GT)15 | 3F*(GT)15 | |
| −3-F-4-CO2H | CT2C3T2C | F-CO2H*CT2C3T2C |
| (AT)15 | F-CO2H*(AT)15 | |
| (GT)15 | F-CO2H*(GT)15 |
Column 1: chemical diversity of OCCs with varying terminating moieties on the aryl functional group. Column 2: special oligonucleotide sequences that form molecular masks on SWCNTs. Column 3: synthesized OCC-DNA nanosensors. A sensor array comprising multiple OCC-DNA nanosensors (highlighted in bold) was used for the training of the machine learning models. NEt2, 3F and F-CO2H represent 4-N,N-diethylamino, 3,4,5-trifluoro and 3-fluoro-4-carboxy aryl organic colour centres, respectively. Asterisk (*) denotes the complexation of an OCC and ssDNA oligonucleotide, comprising each nanosensor.
Fig. 2 ∣. Spectroscopic responses of OCC-DNA sensors to patient serum samples.
a, Representative fluorescence spectra of the ss(GT)15-wrapped 3,4,5-trifluoroaryl OCC sensor, 3F*(GT)15, in PBS (grey), 20 v/v% serum from an HGSOC patient (orange) and serum from a healthy individual (blue). b, Spectral responses of the 3F*(GT)15 sensor to cancer and healthy individuals’ serum samples. Four spectral parameters—intensity and wavelength of the E11 and E11− peaks (int, int*, wl and wl*) were extracted from fluorescence spectra of 4 serum samples for each group. Data points represent the mean value of the spectroscopic variables. Each sample was measured in triplicate. Horizontal lines denote the median. Statistical significance was calculated via Welch’s t-test. c, E11 intensity change (dint) of each OCC-DNA sensor in response to 215 serum samples from individuals with HGSOC and other diseases, as well as healthy individuals at 2 h incubation. d, PCA of sensor responses to HGSOC (orange), other diseases (light blue) and healthy samples (blue). Source data.
To determine a minimal set of OCC-DNA combinations that provide the most diverse responses from the patient samples, we measured the fluorescence spectral responses of the OCC-DNAs to serum samples from HGSOC patients and healthy individuals. Four serum samples of the two conditions were incubated with ten different OCC-DNAs for 2 h, and the fluorescence spectra of the OCC-DNA complexes were acquired. For each OCC-DNA nanosensor, we analysed four different spectral features of the OCC-DNA nanosensors that were modulated in response to interactions with analytes in serum: E11 and E11− intensity (int and int*) and wavelength (wl and wl*). From these data, we identified the sensors that gave statistically significant differences in response to healthy versus cancer groups in parametric t-tests (quantified by P value; Fig. 2b and Extended Data Fig. 1). Our hypothesis was that OCC-DNAs that perform well independently would make good choices when used in combination. Six OCC-DNA nanosensors exhibiting E11 or E11− peak wavelengths with statistically significant differences between HGSOC and healthy groups (P < 0.10) were selected for the sensor array used in the subsequent parts of this study (highlighted in Table 1 and Extended Data Fig. 1). The selection/reduction of features improves the training speed and model performance by eliminating redundant features in the data set.
We initially exposed the OCC-DNA sensor array to 215 patient serum samples and constructed a data set comprising the spectral feature changes caused by the serum environment. Specifically, the size of the data matrix was Nsa × (Nf × NOCC-DNA), where Nsa is the number of serum samples, Nf is the number of features per OCC-DNA and NOCC-DNA is the number of different OCC-DNA complexes in the array. The set of serum samples was collected from 49 HGSOC, 51 other gynaecologic diseases (such as endometriosis and low-grade ovarian carcinoma), 29 non-gynaecologic cancer, 25 cancer patients in remission including 7 HGSOC, and 61 healthy donors (Supplementary Table 1). The fluorescence spectra were collected at three time points during incubation: 2 h, 24 h and 72 h.
The average of triplicate sensor responses was used for the data analysis. We note that the variation of each measurement from the averaged triplicates was small for all the OCC-DNA peaks (Supplementary Fig. 3). The variations in dwl and dwl* showed narrow Gaussian distributions, with standard deviations ranging from 3.72 to 5.37%. The maximum variation in the same sample was less than 15% (<0.3 nm). The analysis confirmed that our measurement can reliably identify the small spectral shifts. This is likely because OCC-DNAs exhibit relatively narrow bandwidths (35–80 meV), making small spectral shifts much easier to resolve compared with conventional fluorophores (>100 meV).
All four spectroscopic variables—int, int*, wl, wl*—measured from the OCC-DNA nanosensor array, exhibited statistically significant differentiation between HGSOC and healthy groups, but the data did not delineate a clear difference between HGSOC and other disease conditions (Fig. 2c and Extended Data Fig. 2). Principal component analysis (PCA) was performed on the spectroscopic data (Nf = 4) upon a 2 h incubation from all combinations of OCC-DNA sensors (NOCC-DNA = 6). The first two principal components accounted for 87.5% of the total variance (principal component loadings listed in Supplementary Table 2). Similar to Fig. 2c, healthy samples showed the differentiable signatures from the disease samples, denoted by their segregation into separate regions in the PCA plot (Fig. 2d), but HGSOC could not be separated from the other disease conditions.
To differentiate HGSOC from the other disease conditions, we next trained machine learning models using the sensor responses and clinical diagnostic results (Fig. 3 and Extended Data Fig. 3). The algorithms were used for binary classification of sensor responses: HGSOC vs other diseases + healthy (the differentiation of HGSOC from all other samples). The set of features chosen for the classification task were the spectroscopic variables dint, dint*, dwl and dwl* collected from the OCC-DNA sensor array. For robustness, we investigated five standard machine learning algorithms with nested levels of optimization processes: model hyperparameters, model choice and multilevel validation. We tested supervised machine learning algorithms—logistic regression, decision tree, artificial neural networks (ANN), random forest (RF) and support vector machine (SVM)—while tuning models’ hyperparameters with Bayesian optimization35 (Supplementary Table 2). The averaged F-score in 10-fold cross-validation was used to assess the model performance (see Methods).
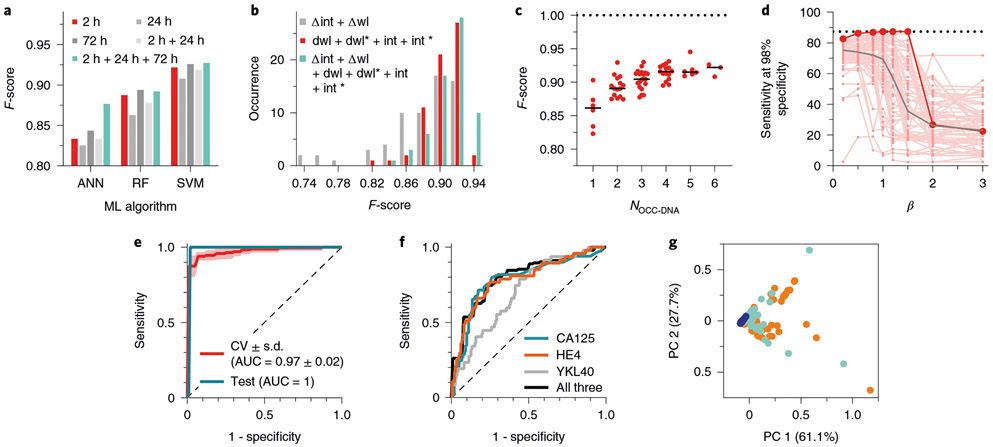
Fig. 3 ∣. Optimization of machine learning algorithms for HGSOC classification.
a, Comparison of F-scores of HGSOC identification with ANN, RF and SVM machine learning (ML) models, using sensor data collected with different serum incubation times. b, Distribution of F-scores obtained using data with different numbers of spectral variables: 2 variables (Δwl + Δint) vs 4 variables (dwl + dwl* + dint + dint*) vs 6 variables (dwl + dwl* + dint + dint* + Δwl + Δint). c, F-scores obtained with different numbers of OCC-DNA nanosensor types, via SVM. Dotted line indicates the upper limit of the F-score. The horizontal line is to guide the eye to the F-score of 1. d, Sensitivities of all possible sensor array combinations, composed of up to 6 OCC-DNAs, at 98% specificity, as a function of β in Fβ scoring via SVM. Small red dots denote sensitivities of each nanosensor array at varying β. Large red dots denote the overall best-performing nanosensor combination. The horizontal dashed line is the highest sensitivity at 98% specificity of the best-performing nanosensor combination at β=1. The grey line is the median of sensitivities for all optimized nanosensor arrays. e, Best ROC curves for binary classification of HGSOC, showing both cross-validated training set (CV) and test/validation set (Test). The shaded area is the standard deviation of 10-fold validation. AUC is the area under the curve. The dashed diagonal line is a ROC curve with no discrimination. f, ROC curves of HGSOC classification using individual serum biomarkers (CA125, blue; HE4, orange; YKL40, grey) and logistic regression of their combination (black). The dashed diagonal line is a ROC curve with no discrimination. g, PCA plot of three disease states: HGSOC (orange), other diseases (light blue) and healthy patients (blue), calculated using conventional serum measurements of CA125, HE4 and YKL40 levels from 215 patient sera. Source data.
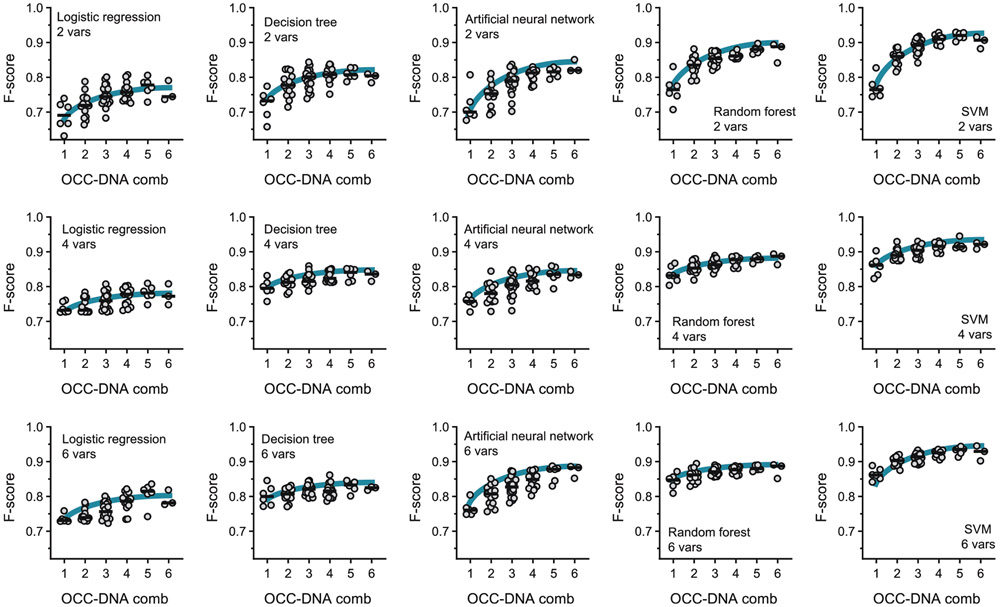
We first examined the machine learning algorithm that most accurately classifies HGSOC (Fig. 3a). We compared the averaged F-scores of the machine learning algorithms using OCC-DNA combinations within the sensor array. We assessed combinations of OCC-DNA nanosensor responses, up to 6 at a time, out of the 6 originally selected OCC-DNAs (1 ≤ NOCC-DNA ≤ 6), for 63 total possible combinations for each incubation duration (see Supplementary Table 3). We found that SVM resulted in the best F-scores among the 5 machine learning algorithms that we tested (Extended Data Fig. 3). Thus, we used SVM models for subsequent optimizations of the HGSOC classifier.
For our second optimization, we compared the differences in model performance using sensor responses measured under different durations of incubation with the serum samples (Nf = 3 × 63). In all the tested machine learning algorithms, there were no statistically significant differences between incubation times (Extended Data Fig. 3). We found that combining datasets obtained over multiple incubation durations could improve the model performance, but the performance was only marginally better than using 2 h of serum incubation (Fig. 3a and Supplementary Table 3). Thus, we used the 2 h data set for subsequent model development for simplicity.
Thirdly, we examined which spectroscopic variables in the set of feature vectors optimize F-scores. We compared three combinations of spectral variables, involving the E11− to E11 intensity ratio (Δint), the wavelength difference between E11− and E11 peaks (Δwl), dwl, dwl*, dint, dint*, and combinations thereof (Nf = (2, 4 or 6) × 63; Fig. 3b). The SVM models trained with the data set of 2 variables, Δint and Δwl, resulted in lower F-scores. We found no statistically significant difference between 4 and 6 variables in the F-scores of the optimized SVM models potentially because Δint and Δwl are derivatives of the others. Thus, we used the 4 variables for further investigations.
We then investigated the impact of the number of different OCC-DNA sensors in the array on the F-score (1 ≤ NOCC-DNA ≤ 6; Fig. 3c). When more OCC-DNA elements were added to the sensor array, the F-scores tended to increase systematically. The trend was the same regardless of which machine learning algorithm was used (Extended Data Fig. 3). The best SVM model was trained by the spectral response of 5 OCC-DNAs: NEt2*CTTC3TTC, NEt2*(TAT)4, 3F*(TAT)4, 3F*(AT)15 and 3F*(GT)15. The averaged cross-validation score of the SVM model was 93.9% sensitivity, 95.2% specificity, and an F-score of 0.945 differentiating HGSOC from all other disease + healthy samples. Small variances in F-score and sensitivity (<0.1) in cross-validations suggest that the optimized models are generalizable within the sample set.
Lastly, we examined whether tuning the hyperparameters to maximize the Fβ score can improve sensitivity at a high specificity (Fig. 3d). The Fβ score is the weighted harmonic mean of PPV and sensitivity, and β is chosen such that sensitivity is considered β-times as important as PPV. At decreasing β from 3 to 0.2, sensitivity at 98% specificity systematically increased, although the improvement was statistically non-significant between μ values of 0.2, 0.5, 0.8 and 1 in this study (Supplementary Table 4). The best-performing prediction model was the sensor array combination of 4-N(C2H5)2*C T2C3T2C, 4-N(C2H5)2*(TAT)4, 3,4,5-F3*(TAT)4, 3,4,5-F3*(AT)15 and 3,4,5-F3*(GT)15, and yielded 87% sensitivity at 98% specificity when PPV and sensitivity were equally weighted (β = 1).
To further assess the robustness of the sensor array and algorithm, we synthesized a new batch of OCC-DNAs under the same condition and collected the sensor array response data to an independent test set of 54 patient samples (Nsa = 54). To evaluate the model performance in various medical conditions, the test set was sampled from different patients, comprising 7 HGSOC, 5 other gynaecologic diseases, 32 non-gynaecologic diseases and 10 heathy patients. With this new sample set, the optimized SVM model resulted in 100% sensitivity at 98% specificity and an F-score of 0.978. These values are consistent with the cross-validation scores and gave a similar receiver operating characteristic (ROC) curve (Fig. 3e), indicating that the model did not overfit the data.
The risk of bias in the study was evaluated on the basis of Prediction model Risk Of Bias Assessment, PROBLAST36 (Appendix 1 in Supplementary Information). The risk of bias scored low in terms of predictors, outcomes and analyses. In participants, the tool resulted in the finding of no systematic differences between training and cross-validation sets. However, the limited medical record of healthy donors and the enriched fraction of breast cancers in the non-HGSOC group of the test set may introduce systematic bias in participant selection and the validation of machine learning models, respectively. For clinical translation of the technology, these risks of bias must be taken into account.
We also endeavoured to account for chemical interferents and background chronic conditions that could confer a bias in the sensor response. From a patient chart review, we identified chronic diseases and most common medications administered to the patients (Extended Data Fig. 4). We found that 75% of HGSOC and 68% of other disease patients suffered from at least one chronic condition, and the relative abundance between these disease groups was similar. Regarding the medications, we statistically assessed the contribution of each interferent to the sensor results using a multivariate regression model (Supplementary Table 5). The regression model determined a linear correlation between the sensor response and medication, using estimated parameters and errors. The adjusted R2 of the regression model ranged from −0.045 to 0.233, indicating weak linear correlations of each sensor response to medications. We confirmed that the sensor array technology accurately classified the disease status regardless of medication and chronic conditions, as evidenced by high F-scores of the HGSOC prediction models (Supplementary Table 3). The analysis suggested no indications that such interferents reduced the specificity of HGSOC detection.
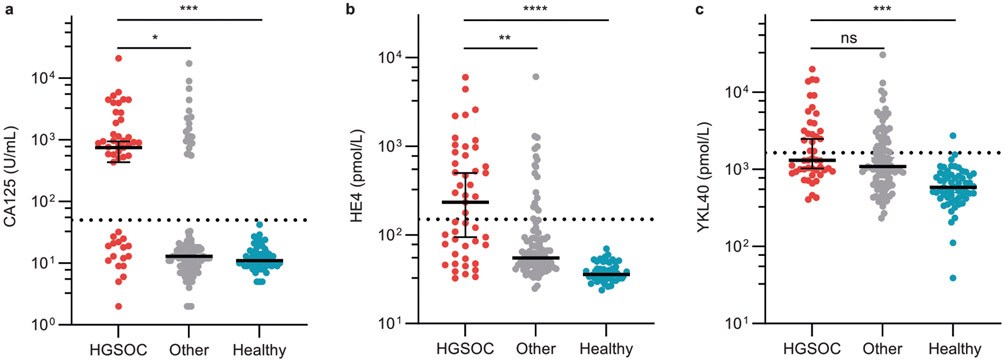
To test the utility of the SVM model relative to conventional diagnostic methods, we compared conventional biomarker-based HGSOC detection and histology results to the F-score predicted by the SVM model. We measured known biomarkers in the patient serum samples, including CA125, HE4 and YKL40, creatinine, and bilirubin by immunoassays (see Methods). We assessed the diagnostic accuracy of serum HGSOC biomarkers in these patients (Fig. 3f). Although the differences in serum CA125, HE4, and YKL40 levels, with respect to the clinical references, were statistically significant between HGSOC, healthy, and other (non-HGSOC) diseases (Extended Data Fig. 5), false-positive rates were high. For example, CA125-based screening with 50 U ml−1 cutoff resulted in 65.3% sensitivity, 88.3% specificity and an F-score of 0.621 in our patient sample set. The logistic regression of additional biomarkers marginally improved the HGSOC prediction (Fig. 3f). PCA plots of HGSOC biomarkers CA125, HE4 and YKL40 showed that these markers failed to differentiate HGSOC from other diseases, while the healthy individuals’ samples clustered together (Fig. 3g). Clinical trials using these biomarkers showed similar results4,37. These results confirmed that our perception/sensor-based technology substantially outperformed established serum biomarker-based classification, and the accuracy was much closer to diagnosis by a physician (using pathology, imaging and so on).
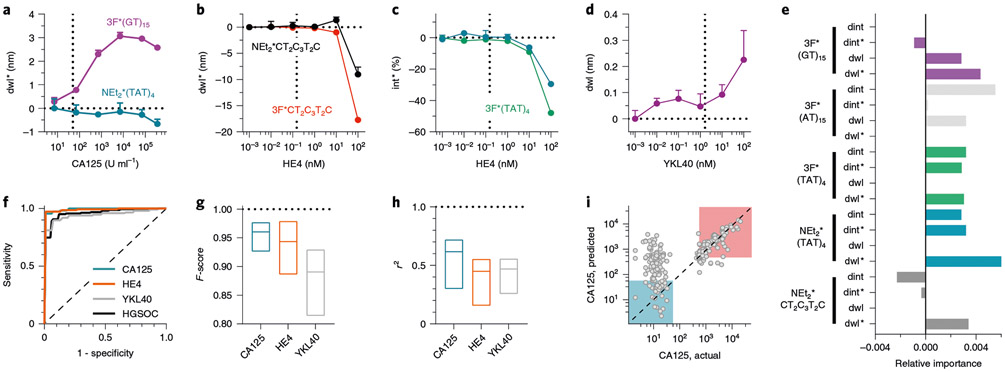
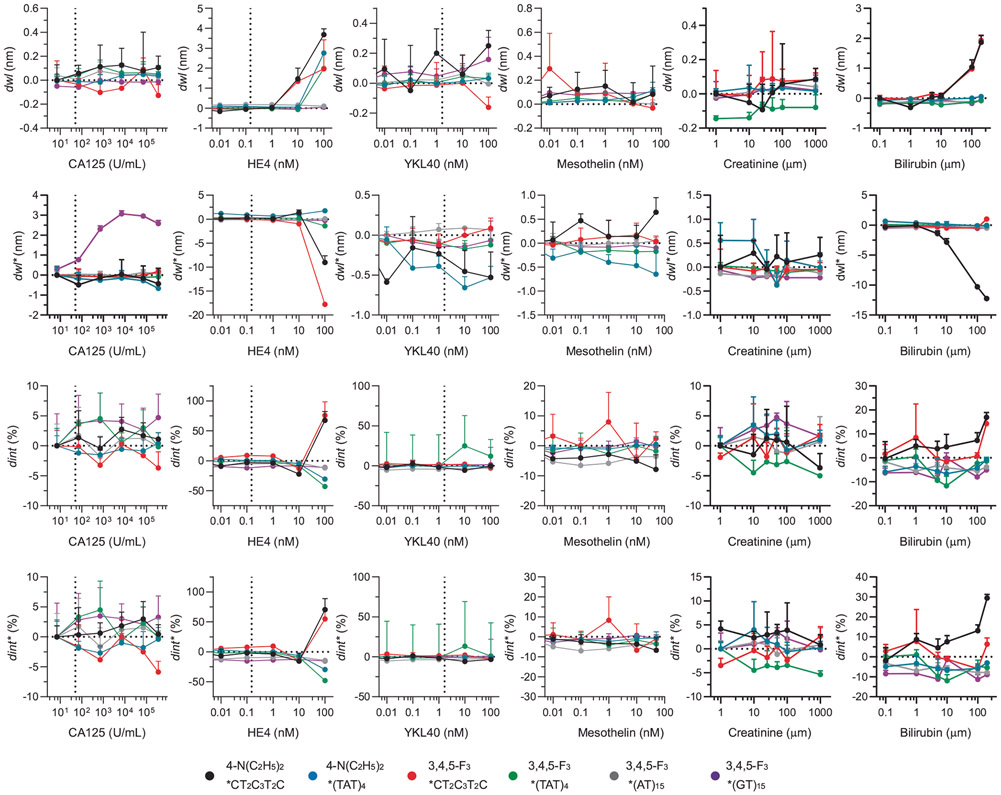
To better understand the molecular basis for the sensor-based HGSOC fingerprint, we investigated the sensor response to serum biomarkers (Fig. 4). We measured the spectral response of the OCC-DNA nanosensors upon single analyte titration with bilirubin, creatinine and HGSOC serum biomarkers, including CA125, HE4, YKL40 and mesothelin in 20% foetal bovine serum (FBS) (Fig. 4a-d and Extended Data Fig. 6). We found that several OCC-DNA spectral responses correlated with CA125, HE4, YKL40 and bilirubin concentrations, while mesothelin and creatinine showed no quantitative correlations with the sensor responses. We surmise that, because of this correlation, the inclusion of biomarker-dependent spectroscopic variables in the training data set improved F-scores for HGSOC identification. We then assessed the relative contribution of each spectral parameter to the model performance by an ablation study—individually dropping each spectroscopic variable from the analysis (Extended Data Fig. 7). On analysis of feature importance, we identified that 3F*(GT)15 and 3F*(TAT)4 were the most important OCC-DNA nanosensors. We also found that the same feature in different sensor arrays can improve or reduce the prediction scores (Extended Data Fig. 7). For instance, the E11− intensity (dint*) of NEt2*CTT has the highest positive feature importance (improved F-score by 0.067) in the sensor array of 4-N(C2H5)2*CT2C3T2C, while the same feature reduced the F-score by 0.018 in the sensor array combination of 4-N(C2H5)2*CT2C3T2C and 3,4,5-F3*(GT)15. Overall, the biomarker-dependent features scored highly, indicating that such features improved the SVM model performance (Fig. 4e). The observations confirmed that (1) OCC-DNA fluorescence transduces broad types of subtle differences in physicochemical properties of physisorbed molecules and (2) known serum biomarkers make up part of the disease fingerprint. However, the use of biomarker-dependent features exclusively did not result in optimal F-scores. The inclusion of certain features that showed no quantitative correlation with known biomarkers improved the model performance. These experiments suggest that the OCC-DNA nanosensor array results may be due, at least in part, to the transduction of heretofore unidentified biomarkers.
Fig. 4 ∣. Known serum biomarkers make up part of the disease fingerprint in the nanosensor array response.
a-d, Representative spectral responses of OCC-DNA in 20% FBS at increasing concentration of CA125 (a), HE4 (b,c) and YKL40 (d). Mean±s.d., n = 3 technical replicates. e, Feature importance analysis of the binary SVM model. f, ROC curves of binary biomarker classification (normal vs above clinical reference) using SVM of the OCC-DNA sensor responses. The dashed diagonal line indicates a ROC curve with no discrimination. g, F-score ranges of SVM classifications of HGSOC biomarkers or disease state. The line in each box indicates the median. h, r2 ranges of biomarker SVR. The line in each box indicates the median. The horizontal dotted line is to guide the eye to the F-score or r2 of 1. i, Serum CA125 levels predicted by SVR against immunoassay results. The prediction models were trained by the fluorescence response of NEt2*(TAT)4, 3F*(TAT)4 and 3F*(AT)15. The highlighted squares classify normal (<50 U ml−1, blue) and high CA125 (>250 U ml−1, red) groups. Source data. The dashed diagonal line represents where actual and predicted CA125 levels are the same.
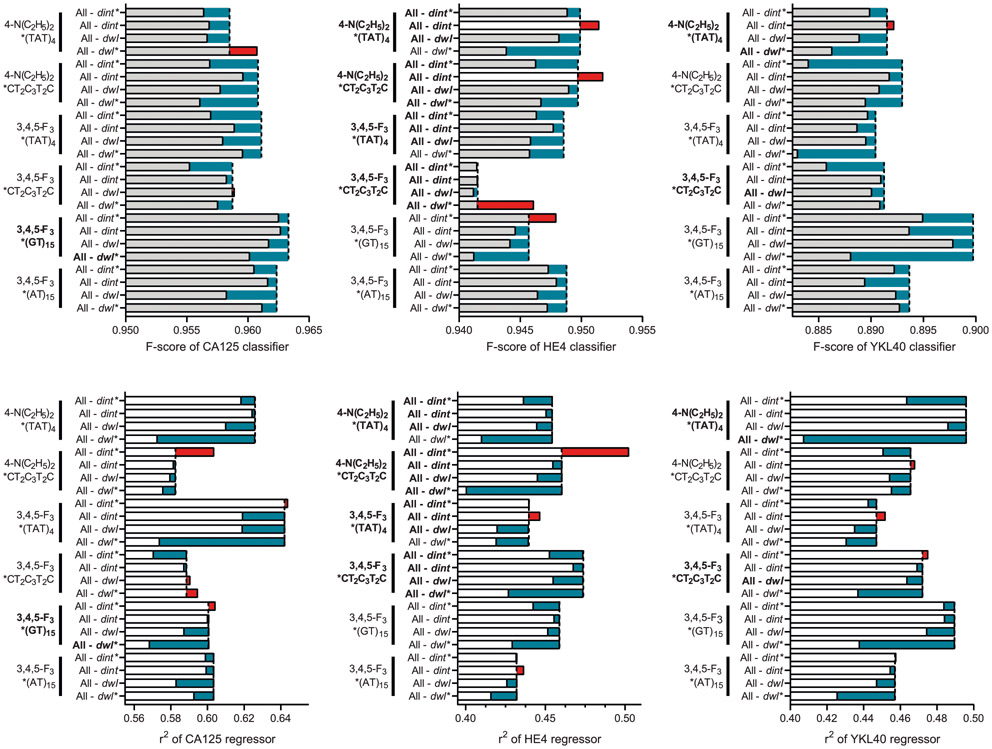
To further investigate the correlation between serum biomarker levels and the response of the nanosensor array, we assessed whether the sensor array responses could be used to train an SVM model to identify abnormal levels of known biomarkers in the patient samples. First, we trained an SVM classification model to detect elevated CA125 by dividing the patient sera into groups on the basis of threshold for suspicion of malignancy; normal (0–50 U ml−1) vs high (>50 U ml−1) CA125. The CA125 training resulted in high F-scores (>0.92) for all possible sensor array combinations (Fig. 4f,g and Supplementary Table 6). We similarly assessed HE4 and YKL40 with respect to their clinical references of 150 pM for HE4 and 1,650 pM for YKL40, and we developed binary classification models to differentiate abnormal levels using the SVM algorithm. Both HE4 and YKL40 classifications resulted in high F-scores (0.89–0.98 and 0.81–0.93, respectively) for the detection of abnormal biomarker levels.
We additionally investigated whether support vector regression (SVR) models can quantitatively predict serum biomarker levels using the sensor array (Fig. 4h). The best CA125 regression model, using the three OCC-DNAs NEt2*(TAT)4, 3F*(TAT)4 and 3F*(AT)15 resulted in an average R-squared (r2) value of 0.719 (Fig. 4i). We note that the prediction error in the normal concentration range (<50 U ml−1) was larger than in the high concentration range. This can be attributed to the fact that the detection limit in the single titration experiment was close to the clinical reference of CA125. The SVR models of HE4 and YKL40 were also constructed, resulting in r2 values of 0.55 and 0.56, respectively. The SVR models suggest that the known biomarkers influence the sensor responses, but the models were not sensitive enough to reliably predict the exact biomarker levels.
We assessed the contribution of each spectral parameter to the biomarker classification and regression models (Extended Data Fig. 8). Most of the spectral parameters had positive relative importance on average, indicating that including such features improved the positive predictive value and sensitivity of the biomarker identification. A positive correlation of the feature importance to F-score (for binary classification) and r2 (for regression) was stronger for the biomarker-dependent variables that were identified in the single-analyte experiments (Fig. 4a-d). Regarding bilirubin, however, although OCC-DNA fluorescence responses quantitatively correlated with its concentration over biologically relevant ranges (Extended Data Fig. 6), we failed to optimize a good SVR model for detection due to the small variance of the biomarker levels within the patient samples. The SVR model performance for serum creatinine was poor due to a lack of quantitative correlation between sensor response and creatinine concentrations in the single-titrant experiment (Extended Data Fig. 6).
Discussion
We constructed a nanosensor-array technology, composed of OCC-DNA elements and coupled with machine-learning algorithms, to investigate the potential to identify HGSOC in patient sera. The array was composed of multiple OCC moieties and DNA sequences, which together offer a rich design space for modulating the morphology and chemistry of the exposed nanotube surface. Our DNA sequence selection was based on the recognition sequences that form specific wrapping patterns on the nanotube surface. These sequences were originally selected to isolate individual (n,m) species/chiralities of nanotubes34. We reasoned that the recognition sequences of DNAs would confer the greatest diversity of interactions with the serum milieu, which is important to establish an OCC-DNA library for screening disease-specific sensor responses. We based this rationale on the findings that ssDNA encapsulates SWCNTs via π-π stacking interactions, and certain DNA sequences can behave like a ‘molecular mask’ that defines the shape and size of the exposed surface26. Their characteristic surface structures are responsible for diverse physicochemical properties of the OCC-DNAs27, leading to different protein corona compositions38,39. Different morphologies determined by OCCs and DNA thereby contribute to the selectivity of the nanotube surfaces to the serum milieu. The fluorescence modulation of SWCNTs is caused by several mechanisms, including Fermi level shifting through modulation of the immediate redox environment and exciton disruption in response to binding events, which change SWCNT intensity, and solvatochromic (wavelength) shifting due to perturbation of the local dielectric environment, including shifts due to modulation of the local electrostatic environment24,40. OCC fluorescence, on the other hand, is molecularly specific and extremely sensitive to the local chemical environment of the atomic defect sites32,41. Interactions between HGSOC serum biomarkers and OCC-DNA hybrids elicited diverse spectral responses of the sensor array that enabled sufficient differentiation of signals from other sera.
The sensor technology was used to identify HGSOC with high positive and negative predictive values. Model performance of the sensor technology exceeds the results of the current best clinical screening test using longitudinal CA125 and second-line transvaginal ultrasonography9 (87% vs 84% clinical sensitivities at 98% specificity). However, considering the fact that specimens obtained from symptomatic individuals at diagnosis were used for the development and assessment of the technology, prediction outcomes will be lower in the clinical screening setting in which specimens are obtained in asymptomatic individuals before clinical diagnosis. Further studies, in the context of screening the general population, are warranted to evaluate the ability of the technology to identify pre-invasive and early-invasive disease.
This sensor technology exhibits several unique potential advantages for clinical applications. First, this method could be rapidly adapted to the detection of many diseases/conditions. The array could be used to train an algorithm to recognize nearly any disease when given enough data from the sensor responses to the appropriate patient serum samples. Second, this technology could supplement or replace the use of known biomarkers when there are issues with selectivity in conventional multi-analyte tests. Due to the potential to iteratively modify the sensor array and machine learning algorithms and to additively augment training set size, the selectivity may be increasingly optimized. Third, this sensor technology can be used in a high-throughput fashion to facilitate the screening of large populations. Fourth, because the technology does not rely on antibody-based molecular recognition elements, the sensors could be more robust than existing methods (Supplementary Fig. 4), enabling use in resource-limited settings and in technologies such as point-of-care and wearable/implantable devices42. Lastly, the sensor technology has the potential to be developed into an inexpensive and rapid screening tool that produces a single, easy-to-interpret test result in primary-care settings. The materials needed for the sensor cost approximately $5 per sample because of the small amount of OCC-DNAs needed for screening (<5 ng). The cost of the sensor measurement would also diminish if measured via high-throughput instruments, and the potential for the use of very low sample volumes is substantial.
This work employed machine perception to detect disease fingerprints using an array of optical nanosensors. The study carefully investigated the attributes and molecular mechanism that resulted in the excellent accuracy of the machine-learning-aided nanosensor array. It is important to note that the best-performing HGSOC prediction model (Fig. 3d) included the spectroscopic variables that were not sensitive to the known biomarkers, and their relative importance was much more significant than the biomarker-related variables (Extended Data Fig. 7). This suggests that there exist potential biomarkers or combinations thereof that are either unknown or not part of conventional screening approaches but were captured by the OCC-DNA sensor array. Information detailing which biomarkers and molecular interactions primarily result in the disease fingerprint is unknown and largely cannot be determined by current machine-learning methods43. We believe that it may be possible, with extensive investigations, to use quantitative proteomics aided by the nanosensor array as a discovery tool44-46. Such investigation could be used to facilitate biomarker-discovery efforts47 and to uncover new information related to disease pathophysiology.
Methods
Large-scale synthesis of OCC-DNAs.
Raw SWCNT material, CoMoCAT SG65 and SG65i (Sigma-Aldrich) was used for the large-scale preparation of OCC-SWCNTs. The SWCNTs were dissolved in chlorosulfonic acid (Sigma-Aldrich, 99.9%) at a concentration of ~4 mg ml−1 with magnetic stirring, followed by the addition of an aniline derivative at different molar ratios relative to the SWCNT carbon, and equimolar amounts of sodium nitrite (≥97.0%, Sigma-Aldrich). The aniline derivatives tested for these experiments include 4-amino-2-fluorobenzoic acid (97%, Sigma-Aldrich), 3,4,5-trifluoroaniline (98%, Sigma-Aldrich) and N,N-diethyl-p-phenylenediamine (97%, Sigma-Aldrich). The SWCNT–superacid mixture was then added drop-by-drop into Nanopure water with vigorous stirring (Safety note: the neutralization process is aggressive; a significant amount of heat and acidic smog can be generated. Personal protective equipment, including goggles/facial mask, lab coats, and acid-resistant gloves, are necessary. The neutralization must be performed in a fume hood). The resulting OCC-SWCNTs instantly precipitate out from the solution. The precipitates were then filtered on an anodic aluminum oxide filtration membrane with a pore size of 0.02 μm (Whatman Anodisc inorganic filter membrane), thoroughly rinsed with Nanopure water and then dried in a vacuum oven.
The OCC-SWCNTs were stabilized by 3.5 mg ml−1 ssDNA in phosphate buffered saline (PBS). The OCC-SWCNT were individually dispersed by ultrasonication at 6 W for 60 min using a probe-tip sonicator (Sonics & Materials) at 4 °C for 1 h. The DNA to SWCNT mass ratio is 5 to 1. Then the OCC-DNA solutions were centrifuged at 100,000 g and 4 °C for 30 min. The 80% supernatant was dialysed against PBS for 36 h to remove free DNA (Spectra-Por, Float-A-Lyzer, MWCO = 1MDa). The absorption spectra of the dialysed solutions were collected with a UV-Vis-NIR spectrophotometer (Jasco). After subtracting absorption background, the optical density at (6,5) E11 (~1,000 nm) was used to estimate the relative OCC-DNA concentration48. The OCC-DNAs were kept at 4 °C until used (up to 6 months) as the OCC-DNAs remained colloidally stable.
OCC-DNA and serum/recombinant protein handling.
For the training set data collection, we used the OCC-DNAs that were synthesized within 6 months prior to testing with patient serum samples (1 week to 6 months old). For the test set, we used freshly prepared OCC-DNAs (less than 2 weeks old). The OCC-DNA concentration was adjusted to 0.325 mg l−1 in PBS. We introduced 20 μl of a patient serum sample to 80 μl of OCC-DNAs in a 96-well plate (Corning) to make the OCC-DNA concentration of 0.26 mg l−1 in each well. OCC-DNAs in 100 μl PBS (0.26 mg l−1) was also prepared to compare the relative changes in sensor response in serum for feature vector construction (see Data preprocessing in Methods). The OCC-DNA was incubated at room temperature for 2 h and in a cold room (4 °C) after the spectral acquisition at 2 h time point. Data were taken at three time points during incubation: 2 h, 24 h and 72 h.
To test sensor sensitivity to serum biomarkers, OCC-DNA complexes were added to a 96-well plate at a concentration of 0.26 mg l−1 in a 100 μl total volume of 20% FBS (Gibco). In triplicate, the following were added into wells at biologically relevant concentrations: 0–352,000 U ml−1 recombinant human CA125/MUC16 (R&D Systems), 0–100 nM recombinant human HE4 (RayBiotech), 0–100 nM recombinant human YKL40 (R&D Systems), 0–50 nM recombinant human mesothelin (BioLegend), 0–1,000 μM creatinine (≥98%, anhydrous, Fisher Scientific) or 0–200 μM bilirubin (≥97%, Fisher Scientific). Experiments were performed with the same time points as above. All experiments were performed in triplicate.
High-throughput near-infrared spectroscopy.
Fluorescence emission spectra of OCC-DNAs were acquired using a home-built near-infrared fluorescence spectroscopy apparatus consisting of a tunable white-light laser source, inverted microscope and InGaAs NIR detector. A SuperK EXTREME supercontinuum white-light laser source (NKT Photonics) was used with a VARIA variable bandpass filter accessory, capable of tuning the output to 500–825 nm, set to a bandwidth of 20 nm centred at 575 nm. The light path was shaped and fed into the back of an inverted IX-71 microscope (Olympus), where it passed through a ×20 NIR objective (Olympus) and illuminated the samples in a 96-well plate. Emission from the OCC-DNAs was collected through the ×20 objective and passed through a dichroic mirror (875 nm cutoff, Semrock). The light was f/# matched to the spectrometer using several lenses and injected into a Shamrock 303i spectrograph (Andor, Oxford Instruments) with a slit width of 100 μm, which dispersed the emission using an 86 g mm−1 grating with 1.35 μm blaze wavelength. The spectral range was 723–1,694 nm with a resolution of 1.89 nm. The light was collected by an iDus 1.7 μm InGaAs (Andor, Oxford Instruments) with an exposure time of 10 s. An HL-3-CAL-EXT halogen calibration light source (Ocean Optics) was used to correct for wavelength-dependent features in the emission intensity arising from the spectrometer, detector and other optics. An Hg/Ne pencil-style calibration lamp (Newport) was used to calibrate the spectrometer wavelength. Background subtraction was conducted using a well in a 96-well plate filled with PBS or 20% FBS, depending on the experiment. Following acquisition, the data were processed with custom code written in Matlab that applied the aforementioned spectral corrections and background subtraction and was used to fit the data with Lorentzian functions.
Serum sample set.
Waste samples (269) were collected from female patients diagnosed with ovarian and other cancers under a Memorial Sloan Kettering Cancer Center Institutional Review Board approved protocol. From this sample set, 56 specimens were collected from patients diagnosed with high-grade serous ovarian cancer, 71 from healthy donors, 56 from patients with other gynaecologic diseases, 61 from patients with non-gynaecologic diseases and 25 from patients in remission. There was no statistically significant difference in age distribution for each group. Diagnoses were identified from a chart review of each patient; all diagnoses included histology and were confirmed by a gynaecologic oncology attending physician. Patient demographics, diagnosis and biomarker levels are available in Supplementary Information.
Serum assays.
Serum concentrations of CA125 and HE4 were determined on the Abbott Architect i2000 analyser (Abbott Diagnostics) using a chemiluminescent microparticle immunoassay. YKL40 was analysed using a singleplex immunoassay on the Protein Simple Ella system. The Abbott C8000 analyser was used to determine the concentrations of creatinine by quantitating the formation of creatinine picrate in alkaline conditions, and bilirubin was analysed by the formation of azobilirubin using the diazo reagent under specified conditions.
Data preprocessing.
Quantities representing the sensor response to patient serum were acquired by the Lorentzian fitting of OCC-DNA fluorescence spectra: E11 intensity, E11− intensity, E11 wavelength and E11− wavelength. The average value of triplicates was used as feature data for machine learning processes. Feature values were defined as a difference in sensor response acquired from patient serum and PBS. Specifically, the E11 peak position feature, dwl, was defined as the wavelength difference between the E11 peak in the patient sample (wl) and PBS (wl0): dwl = wl – wl0. The E11 peak intensity feature, dint, was normalized as dint = int/int0, where int and int0 are the E11 peak intensity in serum and PBS, respectively. Similarly, we defined E11− peak related features, dwl* and dint*, indicating the relative E11− peak position and intensity. We additionally considered the relative change in E11− to E11 intensity: Δint = (int*/int)(int0*/int0)−1 – 1, and the wavelength difference between two peaks: Δwl = dwl* – dwl, to check whether the addition of these features would create a larger variance in HGSOC prediction.
We normalized each feature vector to be in the range of −1 and 1 to balance the feature contribution to the model. The imbalance in the size of each group was corrected by upscaling minority species (SMOTE: Synthetic Minority Oversampling Technique)49 so that the prediction models were not biased by groups with larger sample sizes. For the biomarker prediction models, we divided the data into normal versus high biomarker level groups on the basis of clinical references (CA125, 50 U ml−1; HE4, 150 pM; YKL40, 1,650 pM) and corrected the group size using SMOTE.
Model training and performance assessment.
Using algorithms implemented in ‘Scikit-Learn’50, we created models on the basis of decision tree, logistic regression, artificial neural networks, random forest and SVM for binary classification. Hyperparameters for each model were optimized using Bayesian optimization, implemented in the HyperOpt library35. The loss function to minimize in the hyperparameter optimization was set to (1 – F-score). F-score (or F1-score) is a measure of accuracy in binary classification and is calculated from the harmonic mean of the positive predictive value (PPV) and sensitivity: 2/(sensitivity−1 + PPV−1). To rule out possible overfitting in the machine learning process, model performance was evaluated using 10-fold cross-validation. In the cross-validation process, stratified shuffle split validation was used to randomly partition the data set into 10 subsamples. In each partition, 9 of the 10 subsamples were used to train the model, while a single subsample was used to test the trained model. The average F-score of the 10-fold cross-validation was used to assess model performance. The trained models were then tested with an independent set of patient sera (N = 54), sampled from different patients (test set), as external validation. SVR was used to construct the regression models of HGSOC serum biomarkers with 10-fold cross-validation. The loss function in the hyperparameter optimization was (1 – r2). For SVM and SVR, a radial basis function kernel was used and the hyperparameter optimization was performed for the regularization parameter (cost) and the kernel coefficient (gamma), with the maximum iteration of 1,000. The hyperparameter space of each machine learning algorithm for model optimization is noted in Supplementary Information.
Extended Data
Extended Data Fig. 1 ∣. Spectral responses of OCC-DNAs to a small set of HGSOC and benign serum samples.

Four spectral parameters –intensity and wavelength changes of the E11 and E11– peaks– were extracted from fluorescence spectra of four serum samples in each group. Each sample was measured in triplicate. Horizontal lines denote the median. Six OCC-DNA nanosensors, with p-values of the spectroscopic features lower than 0.10, were selected for the sensor array.
Extended Data Fig. 2 ∣. Spectral responses of the nanosensor array to training and validation sets of patient serum samples (Nsa = 215).
Four spectral parameters, a, dint, b, dint*, c, dwl, and d, dwl*, were extracted from fluorescence spectra of the sensor array after 2-hour serum incubation. Each sample was measured in triplicate.
Extended Data Fig. 3 ∣. Averaged F-scores of optimized machine learning models with 10-fold validation.
The classification was divided as HGSOC versus other gynecologic diseases and benign groups. The blue line is the logarithmic regression of the median F-score.
Extended Data Fig. 4 ∣. Assessment of medications as potential interferents to nanosensor prediction.
a, Fraction of medication dose for HGSOC and other disease patients. b, Chronic conditions, and prevalence thereof, in patients measured in this study. Comorbidity was identified based on the patients’ medication information. c, Anti-cancer drugs or prescription drugs whose occurrence differed by 0.1 or higher between HGSOC and other disease groups.
Extended Data Fig. 5 ∣. Serum levels of known ovarian cancer biomarkers in the model study population.
a, CA125, b, HE4, and c, YKL40. The serum protein levels were quantified by automated immunoassay. Dotted lines indicate the clinical reference of each biomarker for HGSOC diagnosis. The error bars denote median ± 95% CI.
Extended Data Fig. 6 ∣. Response of OCC-DNA nanosensors to protein HGSOC biomarkers, creatinine, and bilirubin in 20% fetal bovine serum.
The fluorescence spectra were obtained 2 hours after the incubation. Vertical dashed lines indicate the clinical reference of each serum biomarker for HGSOC screening.
Extended Data Fig. 7 ∣. Relative feature importance of each spectroscopic variable in the HGSOC binary classification models.
a, Feature importance of each spectral parameter, used to train the SVM models, of all OCC-DNA sensors in the arrays tested in this work. Solid lines indicate the median feature importance. b, Correlation of averaged F-score with the averaged feature importance of each spectroscopic variable. Vertical dashed lines indicate F-score when all four spectroscopic variables (dint, dint*, dwl, and dwl*) of the OCC-DNA were included as feature vectors in the model development.
Extended Data Fig. 8 ∣. Correlation of F-score and r2 of the biomarker prediction models with the relative feature importance of each spectroscopic variable.
For the binary classification models (top rows), samples were divided into two groups–abnormal vs. normal levels of serum biomarkers–based on the clinical references (CA125: 50 U/mL, HE4: 150 pM, YKL40: 1650 pM) and assessed the prediction accuracy of abnormal levels of each biomarker. Feature importance of the prediction models shows which spectral parameters most impacted the model performance using an ablation study. Biomarker dependent variables that were identified in Extended Data Fig. 4 are highlighted in bold. Vertical dashed lines indicate F-score when all four spectroscopic variables (dint, dint*, dwl, and dwl*) of the OCC-DNA were included as feature vectors in the model development.
Supplementary Material
Acknowledgements
We thank B. Kwon, S. Chatterjee, A. Chatterjee, M. Fleisher, B. D. Davison, S. David and N. Osiroff for helpful discussions. This work was supported in part by NIH grants R01-CA215719, U54-CA137788, U54-CA132378 and P30-CA008748; the National Science Foundation CAREER Award (1752506); the Honorable Tina Brozman Foundation for Ovarian Cancer Research; the Tina Brozman Ovarian Cancer Research Consortium 2.0; the Kelly Auletta Fund for Ovarian Cancer Research; the American Cancer Society Research Scholar Grant (GC230452); the Pershing Square Sohn Cancer Research Alliance; the Expect Miracles Foundation – Financial Services Against Cancer; the Experimental Therapeutics Center; W. H. Goodwin and A. Goodwin and the Commonwealth Foundation for Cancer Research. M.K. was supported by the Marie-Josée Kravis Women in Science Endeavor Postdoctoral Fellowship. Y.H.W. gratefully acknowledges support from the National Science Foundation (CHE-1904488) and NIH grant (R01-GM114167). H.-B.L. acknowledges the support provided by the China Scholarships Council (CSC No. 201708320366) during his visit to the University of Maryland. P.W. gratefully acknowledges the Millard and Lee Alexander Fellowship from the University of Maryland. M.Z.’s work was NIST internally funded. Y.Y. was supported by a Dean’s Fellowship at Lehigh University. A.J. acknowledges the NHI initiative at Lehigh University.
Footnotes
Competing interests
D.A.H. is a co-founder and officer, with an equity interest, of Goldilocks Therapeutics Inc., Lime Therapeutics Inc. and Resident Diagnostics Inc., and is a member of the scientific advisory board of Concarlo Holdings LLC, Nanorobotics Inc. and Mediphage Bioceuticals Inc. The other authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41551-022-00860-y.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41551-022-00860-y.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
The custom Python and MATLAB codes for the machine learning and the data analyses reported in this study are not yet publicly available owing to intellectual-property-filing issues, yet they are available for research purposes from the corresponding author on reasonable request.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. Source data for the figures are provided with this paper. The raw datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding author on reasonable request. Source data are provided with this paper.
References
- 1.Bray F et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD & Jemal A Cancer statistics, 2020. CA Cancer J. Clin 70, 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Blyuss O et al. Comparison of longitudinal CA125 algorithms as a first-line screen for ovarian cancer in the general population. Clin. Cancer Res 24, 4726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cramer DW et al. Ovarian cancer biomarker performance in prostate, lung, colorectal, and ovarian cancer screening trial specimens. Cancer Prev. Res 4, 365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupont J et al. Early detection and prognosis of ovarian cancer using serum YKL-40. J. Clin. Oncol 22, 3330–3339 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Han C et al. A novel multiple biomarker panel for the early detection of high-grade serous ovarian carcinoma. Gynecol. Oncol 149, 585–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertlein L et al. Human epididymis protein 4 (HE4) in benign and malignant diseases. Clin. Chem. Lab. Med 50, 2181–2188 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Pinsky PF et al. Extended mortality results for ovarian cancer screening in the PLCO trial with median 15 years follow-up. Gynecol. Oncol 143, 270–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs IJ et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet 387, 945–956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menon U et al. Ovarian cancer population screening and mortality after long-term follow-up in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet 397, 2182–2193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamandis EP The failure of protein cancer biomarkers to reach the clinic: why, and what can be done to address the problem? BMC Med. 10, 87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su C-Y, Menuz K & Carlson JR Olfactory perception: receptors, cells, and circuits. Cell 139, 45–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu MC et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol 31, 745–759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao Y et al. Detection of volatile organic compounds in breath as markers of lung cancer using a novel electronic nose. Proc. IEEE Sens 2, 1333–1337 (2003). [Google Scholar]
- 15.Zhang J et al. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci. Transl. Med 9, eaan3968 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu K-H et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun 7, 12474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bera K, Schalper KA, Rimm DL, Velcheti V & Madabhushi A Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol 16, 703–715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajkomar A et al. Scalable and accurate deep learning with electronic health records. NPJ Digit. Med 1, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachilo SM et al. Structure-assigned optical spectra of single-walled carbon nanotubes. Science 298, 2361 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Cognet L et al. Stepwise quenching of exciton fluorescence in carbon nanotubes by single-molecule reactions. Science 316, 1465 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Heller DA et al. Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science 311, 508 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Jena PV et al. A carbon nanotube optical reporter maps endolysosomal lipid flux. ACS Nano 11, 10689–10703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heller DA et al. Multimodal optical sensing and analyte specificity using single-walled carbon nanotubes. Nat. Nanotech 4, 114–120 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Roxbury D, Jena PV, Shamay Y, Horoszko CP & Heller DA Cell membrane proteins modulate the carbon nanotube optical bandgap via surface charge accumulation. ACS Nano 10, 499–506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams RM et al. Noninvasive ovarian cancer biomarker detection via an optical nanosensor implant. Sci. Adv 4, eaaq1090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roxbury D, Mittal J & Jagota A Molecular-basis of single-walled carbon nanotube recognition by single-stranded DNA. Nano Lett. 12, 1464–1469 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Roxbury D, Jagota A & Mittal J Structural characteristics of oligomeric DNA strands adsorbed onto single-walled carbon nanotubes. J. Phys. Chem. B 117, 132–140 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Roxbury D, Tu X, Zheng M & Jagota A Recognition ability of DNA for carbon nanotubes correlates with their binding affinity. Langmuir 27, 8282–8293 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Horoszko CP, Jena PV, Roxbury D, Rotkin SV & Heller DA Optical voltammetry of polymer-encapsulated single-walled carbon nanotubes. J. Phys. Chem. C 123, 24200–24208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brozena AH, Kim M, Powell LR & Wang Y Controlling the optical properties of carbon nanotubes with organic colour-centre quantum defects. Nat. Rev. Chem 3, 375–392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piao YM et al. Brightening of carbon nanotube photoluminescence through the incorporation of sp3 defects. Nat. Chem 5, 840–845 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Kwon H et al. Optical probing of local pH and temperature in complex fluids with covalently functionalized, semiconducting carbon nanotubes. J. Phys. Chem. C 119, 3733–3739 (2015). [Google Scholar]
- 33.Luo H-B et al. One-pot, large-scale synthesis of organic color center-tailored semiconducting carbon nanotubes. ACS Nano 13, 8417–8424 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Ao G, Streit JK, Fagan JA & Zheng M Differentiating left- and right-handed carbon nanotubes by DNA. J. Am. Chem. Soc 138, 16677–16685 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Shahriari B, Swersky K, Wang Z, Adams RP & de Freitas N Taking the human out of the loop: a review of Bayesian optimization. Proc. IEEE 104, 148–175 (2016). [Google Scholar]
- 36.Wolff RF et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann. Intern. Med 170, 51–58 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Moore LE et al. Proteomic biomarkers in combination with CA 125 for detection of epithelial ovarian cancer using prediagnostic serum samples from the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. Cancer 118, 91–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinals RL, Yang D, Lui A, Cao W & Landry MP Corona exchange dynamics on carbon nanotubes by multiplexed fluorescence monitoring. J. Am. Chem. Soc 142, 1254–1264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenzer S et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotech 8, 772–781 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Heller DA et al. Peptide secondary structure modulates single-walled carbon nanotube fluorescence as a chaperone sensor for nitroaromatics. Proc. Natl Acad. Sci. USA 108, 8544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Kim M, Qu H & Wang Y Single-defect spectroscopy in the shortwave infrared. Nat. Commun 10, 2672 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee MA et al. Can fish and cell phones teach us about our health? ACS Sens. 4, 2566–2570 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Zednik C Solving the Black Box Problem: a normative framework for explainable artificial intelligence. Philos. Technol 34, 265–288 (2021). [Google Scholar]
- 44.Docter D et al. Quantitative profiling of the protein coronas that form around nanoparticles. Nat. Protoc 9, 2030–2044 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Pinals RL et al. Quantitative protein corona composition and dynamics on carbon nanotubes in biological environments. Angew. Chem. Int. Ed 59, 23668–23677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai ZW, Yan Y, Caruso F & Nice EC Emerging techniques in proteomics for probing nano–bio interactions. ACS Nano 6, 10438–10448 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Hadjidemetriou M et al. Nano-scavengers for blood biomarker discovery in ovarian carcinoma. Nano Today 34, 100901 (2020). [Google Scholar]
- 48.Zheng M & Diner BA Solution redox chemistry of carbon nanotubes. J. Am. Chem. Soc 126, 15490–15494 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Chawla NV, Bowyer KW, Hall LO & Kegelmeyer WP SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res 16, 321–357 (2002). [Google Scholar]
- 50.Pedregosa F et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res 12, 2825–2830 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. Source data for the figures are provided with this paper. The raw datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding author on reasonable request. Source data are provided with this paper.