Abstract
Genome editing technologies simplify our ability to rewrite genetic blueprints of life. However, CRISPR-Cas enzymes found in nature can only manipulate a fraction of the genome. Recently, new Cas variants have been developed that overcome this limitation, unlocking nearly the entire genome for editing.
Keywords: CRISPR, genome editing, PAM, targeting range
The adaptation of CRISPR-Cas enzymes for genome editing has profoundly altered the trajectory of science by enabling customizable DNA modifications. These new CRISPR-derived technologies have expanded research fields from cell biology to agriculture and have accelerated the application of genetic interventions in the clinic. Despite this progress, until recently, many regions of the genome remained inaccessible for precision editing.
These limitations can be ascribed to the evolution of Cas enzymes as part of a prokaryotic immune system, called CRISPR, that defends against foreign invading DNA molecules. Prokaryotes have developed a safeguard, called a protospacer adjacent motif (PAM), to distinguish their own ‘self’ DNA from foreign and potentially pathogenic nucleic acids (Fig. 1a). Most type II Cas enzymes proofread invading DNA fragments via the recognition of a short sequence known as a protospacer adjacent motif (PAM)[1] (Fig. 1b).The commonly used Cas9 enzyme from Streptococcus pyogenes (SpCas9) recognizes an NGG PAM (where N is any nucleotide) prior to pairing of the spacer segment of the guide RNA (gRNA) with the protospacer of the target. PAM recognition by Cas9 is the important first step for sanctioning binding of DNA substrates that encode this motif[2]. Prokaryotic genomes are usually protected from self-targeting by Cas enzymes because they lack PAMs adjacent to the protospacers within the CRISPR array. Thus, although the PAM plays an important biological role for CRISPR defense, genome editing applications can be hampered by this requirement (Fig. 1c). The targeting restriction imposed by the PAM limits the realization of accurate sequence manipulation with Cas enzymes to genomic sites harboring nearby PAMs.
Figure 1. The PAM requirement of CRISPR-Cas enzymes.
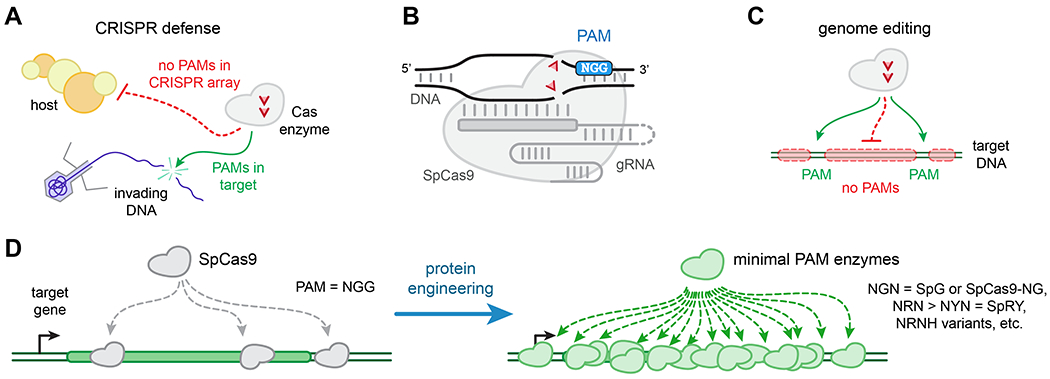
(A) The PAM permits CRISPR-Cas systems to distinguish self from non-self-DNA. (B) Schematic of the SpCas9-gRNA-target DNA complex, with the PAM highlighted in blue. (C) For genome editing applications, PAM availability dictates the genetic locations that Cas enzymes can target. (D) Protein engineering approaches have been utilized to relax or nearly remove the PAM requirement of Cas enzymes, improving PAM density and enabling more accurate single-base targeting of the genome.
Abbreviations: PAM, protospacer adjacent motif; gRNA, guide RNA.
Several approaches have been pursued to unlock the remainder of the genome for precision editing by overcoming the PAM constraint. Researchers have explored the natural repertoire of other less commonly used Cas enzymes that natively recognize different PAMs[3], and protein engineering has been utilized to generate variants with altered PAM profiles [4], [5]. Most recently, new Cas enzymes with relaxed PAMs have been developed[6]–[10], some that require only single nucleotide PAMs and others that are nearly PAMless. These new minimal PAM technologies enable access to a much greater fraction of the genome using a singular enzyme, permitting targeting of previously inaccessible regions (Fig. 1d).
The expanded targeting ranges of these minimal PAM enzymes should improve many genome editing methods and applications, especially those that require precise positioning of edits. Some examples include the generation of disease model cell lines, base editing where the edit window should be carefully positioned, tiling screens that necessitate nucleotide-level perturbations, editing small genetic elements, prime editing, in vitro molecular biology, and many others. For example, with base editors (BEs), the PAM requirement is a particularly prominent limitation. The ‘edit window’ of C-to-T or A-to-G BEs is typically only a handful of nucleotides wide[11], prohibiting editing when PAMs are not available at the necessary distance upstream in the DNA sequence. Importantly, minimal PAM BEs have already been shown to be effective in various organisms. The ability to target sites with single nucleotide resolution by tiling gRNAs over the edit-of-interest allows meticulous phasing of the edit window, which can enhance on-target editing while also reducing or eliminating unwanted bystander edits. Beyond cell-based uses, this class of enzymes should also enable new in vitro applications, facilitating the digestion of DNA substrates freely without the requirement for defined restriction sites.
As genomes become more targetable with these unconstrained enzymes, there are important considerations for their use. Since PAM recognition is the primary licensing step for target site binding[2], minimal PAM Cas variants, in principle, will search a larger fraction of the genome. Not only does this have implications for the kinetics of genome editing (potentially altering the ability of Cas9 to unwind DNA and necessitating higher enzyme concentrations to achieve sufficient on-target editing), it can also lead to higher levels of off-target editing that result from exposure to a larger pool of closely matched DNA sequences[6]–[9]. Fortunately, high-fidelity variants of Cas9 that exhibit dramatically reduced levels of off-target editing were previously developed. These more specific variants have been shown to be compatible with minimal PAM enzymes, improving their genome-wide specificities and in many cases eliminating all off-target edits that result from the expanded PAM tolerance[6], [7], [9], [10]. Minimal PAM enzymes can also exhibit reduced efficiencies compared to canonical Cas enzymes. Thus, when possible, it is generally advisable to utilize the most efficient editor against any PAM to maximize on-target efficiency.
Another consideration arises when contemplating the biological role of the PAM. Cas enzymes utilize the PAM to distinguish the PAM-less spacer found in the host genome from the PAM-containing protospacer of foreign DNA (Fig. 1a). The virtual elimination of the PAM requirement with these Cas variants might impact applications where the gRNA must be constitutively and stably expressed in a cell. Potential self-targeting of the gRNA expression cassette could confound interpretation of CRISPR screens, lead to new classes of de novo secondary off-targets resulting from modified spacers of the gRNA or could attenuate on-target editing by disrupting expression of the intended gRNA. Although this may not be a concern for the majority of genome editing applications where the gRNA is delivered transiently (e.g. as a synthetic RNA or in excess from high titer plasmids), it still merits exploration for the remaining applications that necessitate stable gRNA expression. One approach that can mitigate gRNA self-targeting is to modify the crRNA scaffold[12], substituting the PAM within the gRNA to a poorly preferred sequence.
More detailed biochemical, biophysical, and structural studies will further illuminate the mechanisms by which these minimal PAM enzymes circumvent a critical biological feature. Such studies might also facilitate the continued engineering of these Cas variants to improve their balance of PAM recognition, potentially eliminating their major and minor PAM preferences. Whether the strategies used to reduce or eliminate the PAM requirement of Cas9 enzymes are extensible to other recently described Cas orthologs is an outstanding question, one that could fully realize the potential advantages of smaller Cas systems that are often beholden to longer PAMs.
With minimal PAM Cas enzymes it is now possible to edit many previously inaccessible regions of the genome. The improved flexibility of these enzymes will enable and improve a variety of editing applications. Further development of these technologies will improve the prospects of precision genome editing, bringing the field closer to high-resolution perturbations that help realize the promise of genomic medicines.
Acknowledgements
We thank R. A. Silverstein, L. T. Hille, and R. M. Doll for helpful suggestions. K.A.C. is supported by a Massachusetts General Hospital Fund for Medical Discovery (FMD) Fundamental Research Fellowship Award. B.P.K. acknowledges support from National Institutes of Health (NIH) R00 CA218870 and NIH P01 HL142494 (B.P.K).
Glossary
- Base editors
Modified versions of Cas enzymes that harbor fusions to deaminase domains, permitting the directed conversion of DNA or RNA bases.
- CRISPR-associated (Cas)
Cas enzymes, encoded by CRISPR systems, are an essential component of CRISPR immune systems that protect bacteria and archaea from invading nucleic acids. They can be reprogrammed via a gRNA to bind and cleave DNA targets.
- Guide RNA (gRNA)
a short RNA molecule that directs Cas enzymes to target sites. The gRNA can be comprised of a single crRNA or a dual crRNA with tracrRNA, that together can be joined to form a single composite gRNA. Complementarity between the spacer of the gRNA and the protospacer of the target site is required for DNA or RNA binding.
- Protospacer-adjacent motif (PAM)
a short DNA sequence next to protospacers of DNA targets, which permits most Cas enzymes to proofread invading DNA segments (distinguishing ‘self’ DNA from foreign and potentially pathogenic nucleic acids)
- Prime editors
A genome editing technology built on the Cas enzyme chassis that contains a fusions to a reverse transcriptase domain, which enables the introduction of insertions, deletions, and base conversions
Footnotes
Competing Interests
K.A.C. and B.P.K are inventors on patents and/or patent applications filed by Mass General Brigham that describe genome engineering technologies. B.P.K. is a consultant for Avectas Inc., EcoR1 capital, and ElevateBio, and is an advisor to Acrigen Biosciences and Life Edit Therapeutics.
References
- [1].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E, “A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity,” Science, vol. 337, no. 6096, pp. 816–821, Aug. 2012, doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sternberg SH, Redding S, Jinek M, Greene EC, and Doudna JA, “DNA interrogation by the CRISPR RNA-guided endonuclease Cas9,” Nature, vol. 507, no. 7490, Art. no. 7490, Mar. 2014, doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Collias D and Beisel CL, “CRISPR technologies and the search for the PAM-free nuclease,” Nature Communications, vol. 12, no. 1, Art. no. 1, Jan. 2021, doi: 10.1038/s41467-020-20633-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kleinstiver BP et al. , “Engineered CRISPR-Cas9 nucleases with altered PAM specificities,” Nature, vol. 523, no. 7561, Art. no. 7561, Jul. 2015, doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kleinstiver BP et al. , “Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition,” Nature Biotechnology, vol. 33, no. 12, Art. no. 12, Dec. 2015, doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nishimasu H et al. , “Engineered CRISPR-Cas9 nuclease with expanded targeting space,” Science, vol. 361, no. 6408, pp. 1259–1262, Sep. 2018, doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Walton RT, Christie KA, Whittaker MN, and Kleinstiver BP, “Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants,” Science, vol. 368, no. 6488, pp. 290–296, Apr. 2020, doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Miller SM et al. , “Continuous evolution of SpCas9 variants compatible with non-G PAMs,” Nat Biotechnol, vol. 38, no. 4, pp. 471–481, Apr. 2020, doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chatterjee P et al. , “An engineered ScCas9 with broad PAM range and high specificity and activity,” Nat Biotechnol, vol. 38, no. 10, pp. 1154–1158, Oct. 2020, doi: 10.1038/s41587-020-0517-0. [DOI] [PubMed] [Google Scholar]
- [10].Kleinstiver BP et al. , “Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing,” Nature Biotechnology, vol. 37, no. 3, Art. no. 3, Mar. 2019, doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huang TP, Newby GA, and Liu DR, “Precision genome editing using cytosine and adenine base editors in mammalian cells,” Nat Protoc, vol. 16, no. 2, pp. 1089–1128, Feb. 2021, doi: 10.1038/s41596-020-00450-9. [DOI] [PubMed] [Google Scholar]
- [12].Qin R, Li J, Liu X, Xu R, Yang J, and Wei P, “SpCas9-NG self-targets the sgRNA sequence in plant genome editing,” Nat. Plants, vol. 6, no. 3, pp. 197–201, Mar. 2020, doi: 10.1038/s41477-020-0603-9. [DOI] [PubMed] [Google Scholar]


