Abstract
Recurrent waves of COVID19 remain a major global health concern. Repurposing either FDA-approved or clinically advanced drug candidates can save time and effort required for validating the safety profile and FDA approval. However, the selection of appropriate screening approaches is key to identifying novel candidate drugs with a higher probability of clinical success. Here, we report a rapid, stratified two-step screening approach using pseudovirus entry inhibition assay followed by an infectious prototypic SARS CoV2 cytotoxic effect inhibition assay in multiple cell lines. Using this approach, we screened a library of FDA-approved and clinical-stage drugs and identified four compounds, apilimod, berbamine, cepharanthine and (S)-crizotinib which potently inhibited SARS CoV2-induced cell death. Importantly, these drugs exerted similar inhibitory effect on the delta and omicron variants although they replicated less efficiently than the prototypic strain. Apilimod is currently under clinical trial (NCT04446377) for COVID19 supporting the validity and robustness of our screening approach.
Keywords: SARS CoV2, Delta, Omicron, Anti-viral drug repurposing
1. Introduction
COVID19, caused by SARS CoV2, continues to be a major global health concern. Despite the rapid development and relatively widespread administration of vaccines, we are into the third year of this pandemic. Vaccine hesitancy and unavailability in several parts of the world are enabling the emergence of newer variants despite the steady increase in vaccination rates. Furthermore, the emergence of highly transmissible variants which also possess the ability to evade the protection conferred by vaccines undermine our ability to effectively bring this pandemic to an end. For instance, the most widely administered vaccines such as Pfizer BioNtech, Moderna and AstraZeneca vaccines, which are directed against the spike protein of the prototypic Wuhan strain, have shown reduced neutralizing ability against the Delta variant which has 9 mutations in the spike protein region (Edara et al., 2021; Planas et al., 2021). The recently identified Omicron variant has 30 mutations in the spike region, further increasing the risk of immune escape. Consistent with this, most of the currently approved vaccines have proven to be less effective in neutralizing the Omicron variant. Thus, there is an acute need for broad-acting anti-viral drugs which can be produced and distributed widely. As of now, FDA has approved three drugs namely, Remdesivir, Molnupiravir and Paxlovid for treating COVID19 patients. Use of other drugs such as lopinavir, Baricitinib, chloroquine and hydroxychloroquine, which were initially thought to be effective (Consortium, 2021; Liu et al., 2020; Marconi et al., 2021; Wang et al., 2020) have now been proven to be ineffective in reducing COVID19 related illness, hospitalization or death, and FDA has now revoked the emergency use authorization for these drugs to treat COVID-19. Repurposing FDA-approved and late stage clinical drug candidates can save time and effort required for validating the safety profile and FDA-approval for novel candidate drugs. However, selection of appropriate screening approaches is key to identifying novel candidate drugs that will have higher probabilities of clinical success.
SARS CoV-2 is an enveloped single-stranded RNA beta coronavirus and its RNA genome encodes 4 structural proteins; i) Spike (S), ii) Membrane (M), iii) Envelope (E) and iv) Nucleocapsid (N) protein and 16 nonstructural proteins (NSPs).Two virus proteases namely, Papain–Like Protease (PL-Pro) and 3C-Like protease (3CL-Pro alias M-Pro) mediate the proteolytic processing of NSP1-4 and NSP5-16 respectively (Rajpoot et al., 2021). The life cycle of SARS CoV2 starts with the virus entry into host cells, fusion and uncoating, viral RNA replication, translation and assembly and exocytosis (V'Kovski et al., 2021). Human Angiotensin Converting Enzyme-2 (ACE2) is the most common receptor for SARS CoV2 which binds to the Receptor Binding Domain (RBD) of the SARS CoV2 spike protein to facilitate the attachment. Upon binding to ACE2 receptor, host proteases such as transmembrane serine protease 2 (TMPRSS2) (Hoffmann et al., 2020a) and lysosomal cysteine protease such as Cathepsin cleaves the spike protein into S1/S2 subunits followed by fusion of S2 subunit into the endosomes (Zhao et al., 2021; Zhu et al., 2021). While early studies with chloroquine and hydroxychloroquine inhibited SARS CoV2 entry and infection of Vero E6 cells (Liu et al., 2020) these drugs failed in clinical trials (Ho et al., 2021). Subsequently, it was shown that chloroquine did not inhibit SARS CoV2 infection in TMPRSS2 overexpressing Vero (Vero TMPRSS2) cells (Hoffmann et al., 2020b) which is consistent with a lack clinical efficacy of the drug. Interestingly, Camostat mesylate, a TMPRSS inhibitor, also failed in the clinical trials which reveals the insufficiency of targeting a single mode of action to prevent virus entry (Gunst et al., 2021). Collectively, these studies highlight the importance of using appropriate screening approaches to identify effective drug candidates with higher probability of clinical success.
We have developed a two-step approach for screening FDA approved and clinical candidate drug libraries using pseudovirus entry inhibition assay followed by an infectious prototypic SARS CoV2 (Isolate USA-WA1/2020, Lineage A) cytotoxic effect (cell death) inhibition assay. In the first stage, we screened the drug libraries for SARS CoV2 pseudovirus entry inhibitors using human ACE2 overexpressing HEK293T (293T-ACE2) cells which is a highly permissible host cell line for SARS CoV2 entry. In the second stage, we further screened selected hits for their ability to inhibit virus induced cell death using three different cell lines with varying susceptibility to SARS CoV2 infection such as 293T ACE2, Vero E6 and Vero TMPRSS2 cells. Using this approach, we screened a library of 2985 drug compounds including FDA-approved compounds and drugs that are under various phases of clinical trials. Our screening identified four compounds: apilimod, berbamine, cepharanthine and (S)-crizotinib that exerted potent inhibition of SARS CoV2-induced cell death in all three cell lines. Among these, apilimod is currently tested under clinical trial (NCT04446377) for COVID19, demonstrating the validity and robustness of our screening approach in identifying potentially clinically relevant COVID-19 treatments. More importantly, these compounds exerted similar efficacy against the other variants of concern (VOCs) such as delta (Lineage: B.1.617.2) and omicron variants (Lineage: B.1.1.529).
2. Materials and methods
-
1.
Drug libraries
We combined two well-annotated drug libraries purchased from TargetMol (Wellesley Hills, MA). The L1000, approved drug library contained 2111 compounds which included drugs approved by FDA, EMA (Europe), PMDA (Japan), CFDA (China) and other countries. L3400 Clinical compound library contained 1336 compounds which were under phase I-IV clinical trials. The overlapping compounds between two libraries were eliminated from one of the libraries and our final drug library contained 2985 unique compounds.
-
2.
Pseudovirus entry inhibition assay
All experiments were approved and performed per the guidelines set forth by the Institutional Biosafety Committee (IBC) and the Environmental Health and Safety Office (EHSO) at the University of Illinois at Chicago. SARS CoV2 pseudotyped virus particles were produced by using the Wuhan-Hu-1 Spike-Pseudotyped Lentiviral Kit (NR-52948) obtained from BEI resources. The kit contained 1) pHDM expressing the SARS CoV2 Wuhan-Hu-1 Spike envelope Glycoprotein 2) lentiviral backbone plasmid-pHAGE-CMV-Luc2-IRES-ZsGreen that expresses luciferase and ZsGreen reporters.3) pHDM-Hgpm2-lentiviral helper plasmid expressing HIV Gag-Pol. 4) pHDM-tat1b: lentiviral helper plasmid expressing HIV Tat.5) pRC-CMV-Rev1b plasmids. Pseudovirus particles were produced by transfecting HEK293T cells with the above-mentioned plasmids using Lipofectamine 3.0 transfection reagent (Invitrogen, Thermo Fisher Scientific, MA). Mock pseudovirus particles were produced as described above without spike glycoprotein.
For pseudovirus entry inhibition screening, 25 × 103 human ACE2 expressing 293T cells in 100 μl of 5% DMEM medium containing 0.2% DMSO were seeded in a 96 well plate and pre-treated with 1 μM concentration of the library of drugs for 1 h followed by infection with mock pseudovirus and SARS CoV2 spike pseudotyped virus particles. 72 h post-infection, the cells were lysed, and the luciferase activity was determined. Briefly, 60 μl of the cell lysis buffer reagent (Promega Inc.) was added to each well after removing media and plates were incubated with shaking for 15 min 40 μl of luciferin substrate was added to each well and luminescence was read with 1 min integration and delay time. The Percentage inhibition was calculated as follows: 100 x [1 - (X - MIN)/(MAX - MIN)]. X = Test RLU; Min = RLU of Mock without envelope; Max = RLU of infected control. Assay was done in triplicates. Compounds that showed more than 50% inhibition were selected for the next stage of the screen. Cathepsin-L inhibitor (CAS 108005-94-3, Calbiochem, EMD Millipore Sigma, MA) was used as positive control in each plate.
-
3.
Cell death assay using infectious SARS CoV2
For the cell death screening, we used four cell lines 293T-ACE2 and Human ACE2 and TMPRSS2 overexpressing Vero E6 (Vero E6-TMPRSS2-T2A-ACE2) (BEI resources), Vero E6 (ATCC) and human TMPRSS2 expressing Vero E6 cells (Vero TMPRSS2) (Sekisui XenoTech, LLC, KS). Briefly, 25 × 103 cells/well were seeded into opaque 96 well plates in 100 μl of 5% DMEM medium containing 0.2% DMSO. SARS-CoV-2, Isolate USA-WA1/2020 (Lineage A; BEI Cat # NR-52281), Isolate hCoV-19/USA/PHC658/2021 (Lineage B.1.617.2; Delta Variant; NR-55611) and SARS-CoV-2, Isolate hCoV-19/USA/MD-HP20874/2021 (Lineage: B.1.1.529, Omicron Variant BEI Cat # NR-56461) obtained from BEI resources were used to infect the cells. 100 μl of the infection medium (2% DMEM) containing 10 x TCID50 virus was used to infect cells. Assay was performed in triplicates with uninfected/infected cells with or without inhibitors (200 nM, 1 μM and 5 μM). Cytopathic effect was quantified 72 h post-infection by an ATP-based cell viability assay. Briefly, 100 μl of spent medium was removed and 100 μl of the Cell Titer Glo assay reagent (Promega Inc.) was added to each well. Plates were incubated for 15 min with shaking. Luminescence was read with 1 min integration and delay time. The virus induced % cell death was calculated as follows: 100 x [1 - (X - MIN)/(MAX - MIN)].X = Test compound RLU; Min = RLU of infected control wells; Max = Mean RLU of uninfected control wells.
3. Nucleocapsid protein staining by flow cytometry
Vero-TMPRSS2 and Vero-ACE2-TMPRSS2 cells were infected with different variants of SARS CoV2, and 24 h post infection cells were fixed and permeabilized using nuclear antigen staining buffer set (Tonbo Bioscience, San Diego, CA). SARS CoV2 nucleocapsid positive cells were stained using anti-rabbit SARS CoV2–nucleocapsid monoclonal antibody at 1:400 dilution (ab271180) and Alexa Fluor® 488 conjugated secondary antibody at 1:2000 dilution (ab150077) from Abcam, MA, USA. Anti-rabbit IgG was used as isotype control. Samples were analyzed using CytoFLEX flow cytometer and Kaluza v2.1 software (Beckman and Coulter, CA, USA).
4. Results
4.1. Pseudovirus entry inhibition screening of drug library in 293T ACE2 cells
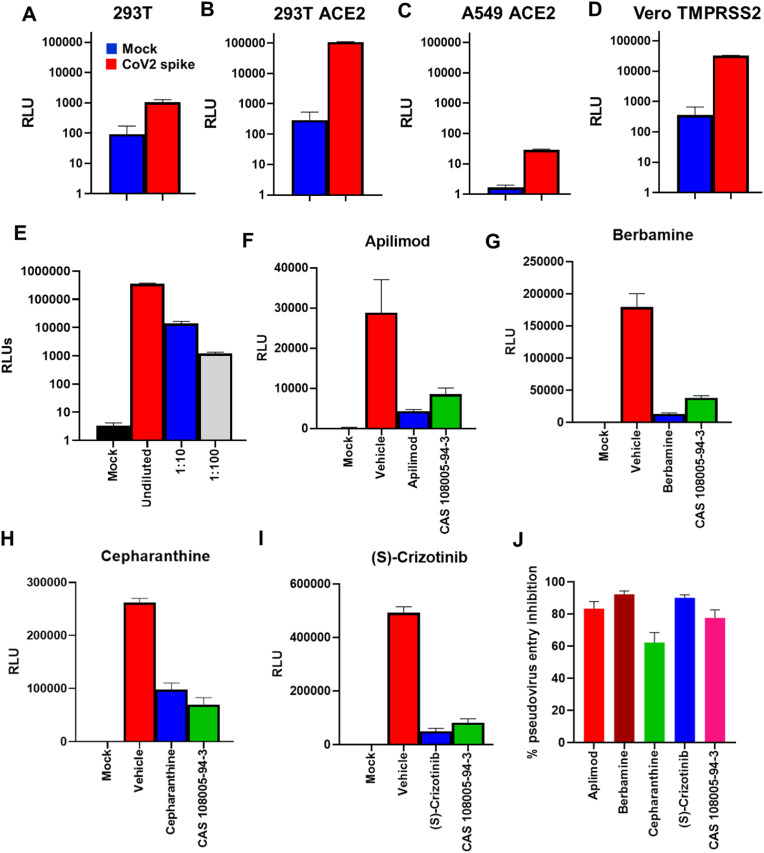
Initially, we tested HEK 293T (human embryonic kidney epithelial cells), human ACE2 expressing HEK 293T (293T-ACE2) cells, human ACE2 expressing A549 (human lung carcinoma epithelial cells), and human TMPRSS2 overexpressing Vero E6 (African monkey kidney epithelial Vero-TMPRSS2 cells) cell lines for their permissiveness to SARS CoV2 entry using SARS CoV2 spike protein pseudotyped virus particles (Fig. 1A-D). Among these, 293T-ACE2 and Vero-TMPRRS2 cells were found to be more permissive to SARS Cov2 pseudovirus entry. Between these two cell lines, the signal-to-noise ratio was better with 293T-ACE2 cells (Fig. 1 B & D) Moreover, expression of human ACE2 in 293T-ACE2 cells rendered them more suitable for studying the effect of the drugs to inhibit human ACE2-SARS CoV2 spike interaction. Furthermore, we validated the sensitivity of the assay by infecting 293T-ACE2 cells with 10-fold dilutions of the pseudovirus particles which resulted in a dose-dependent reduction of the RLU values (Fig. 1E).
Fig. 1.
Pseudovirus entry inhibition screening of drug library. A-D) 293T, 293T-ACE2, A549-ACE2 and Vero TMPRSS2 cells were transduced with SARS CoV2 pseudotyped virus particles containing luciferase reporter gene and luminescence was measured after 72 h. Bar graph shows respective Relative Light Units (RLUs) from Mock (Blue) without envelope and SARS CoV2 spike pseudotyped virus particle (Red) transduced cells. E) RLU values from 293T-ACE2 cells transduced with serially diluted SARS CoV2 spike pseudotyped virus particles. F–I) Bar graphs show RLU values from pseudovirus entry inhibition assay where 293T ACE2 cells were pre-treated with 1 μM of indicated drugs for 1 h and then transduced with SARS CoV2 spike pseudotyped virus particles. Cathepsin-L inhibitor CAS 108005943 was used as a positive control. J) Bar graph shows summary of percent pseudovirus entry inhibition values for each drug. Values represent Mean ± SEM, n = 3 and two independent experiments.
Using this system, we screened a library of 2985 compounds for the SARS CoV2 pseudovirus entry inhibitory activity. We found 231 compounds showing more than 60% mean inhibition of the pseudovirus entry. Respective RLU values of the compounds identified to have potential antiviral activity against SARS Cov2 in our subsequent studies such as apilimod, berbamine, cepharanthine and (S)-crizotinib are shown in Fig. 1F-I and the summary of the percent pseudovirus entry inhibition is shown in Fig. 1J. Cathepsin-L inhibitor was used as a positive control in our experiments, as it has been shown to be one of the key factors mediating SARS CoV entry by our previous studies and other groups (Elshabrawy et al., 2014; Simmons et al., 2005; Zhao et al., 2021).
4.2. SARS CoV2-induced cell death inhibition assay screening
SARS coronaviruses are known to replicate and induce cytopathy in several susceptible cell lines (Kaye, 2006) and Vero E6 is one of the first cell lines used to isolate SARS CoV and SARS CoV2(Ksiazek et al., 2003; Ogando et al., 2020). Many drug screening studies have used Vero E6 as the target cell line to determine the antiviral effects of the candidate drugs (Riva et al., 2020; Yan et al., 2021). As discussed above, the ability of chloroquine to inhibit SARS-CoV2 infection varied depending upon the cell line used for screening ((Liu et al., 2020), (Ho et al., 2021), and failed in clinical trials (Hoffmann et al., 2020b).
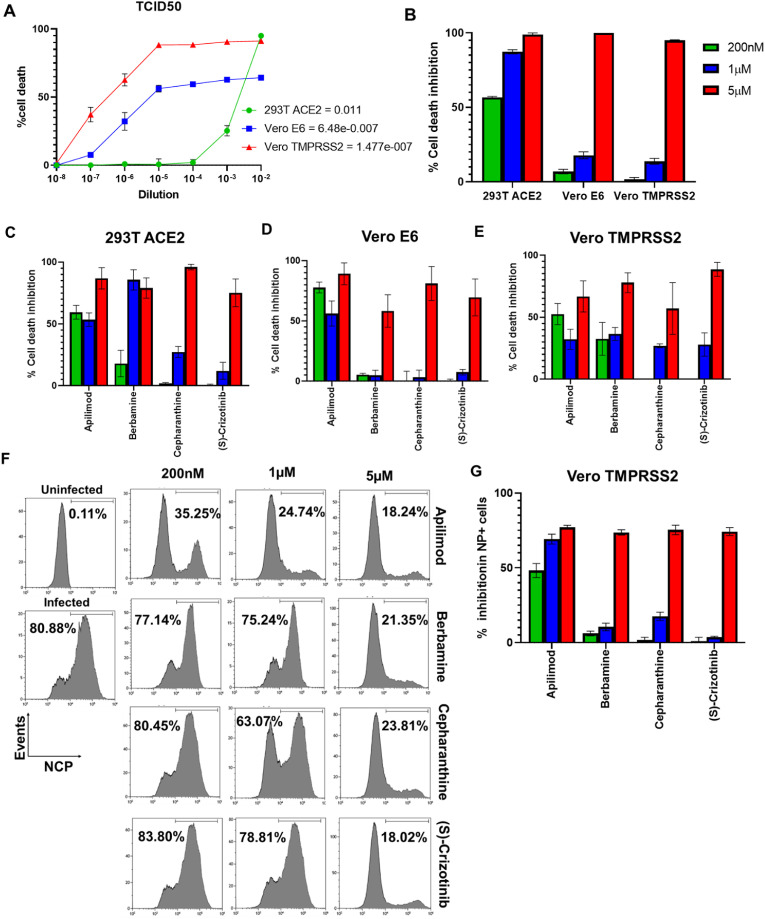
We determined the susceptibility of three different cell lines to SARS CoV2-induced cytotoxicity by infecting 293T-ACE2, Vero E6 and Vero-TMPRSS2 with different dilutions of the prototypic SARS CoV2, quantified cell death and determined TCID50 values. Interestingly, despite increased permissiveness to SARS CoV2 pseudovirus, TCID50 for 293T ACE2 was found to be least susceptible to cell death induction and required much higher amount of virus (5 logs10 more) relative to Vero E6 and Vero-TMPRSS2 cells (Fig. 2 A). Moreover, Vero-TMPRSS2 cells were more susceptible to cell death induction relative to (required ∼1 log TCID50 less virus) compared to Vero E6 cells (Fig. 2A). Interestingly, in spite of their high susceptibility to cell death induction, Vero E6 cells did not show more than 60–70% cell death even at the highest concentration of virus (10−2 dilution). We further validated this assay using the FDA-approved drug Remdesivir which is a known inhibitor of SARS CoV2 replication and exhibited dose-dependent effect on the inhibition of SARS CoV2-induced cell death in all three target cell lines. In line with the susceptibility of the cells line to SARS CoV2-induced cell death, the lowest concentration of Remdesivir (200 nM) showed strong inhibition in the least susceptible 293T ACE2 cells, followed by Vero E6 and Vero-TMPRSS2 cells (Fig. 2B).
Fig. 2.
Validation of cell death inhibition assay. A) Cells were infected with serial dilutions of SARS CoV2 (10^-2 to 10^-8) and cell death was quantified by Cell-Titer-Glo cell viability assay and TCID50 for each cell line was determined at 72 h. B) Cells were pre-treated with different concentrations of Remdesivir followed by infection with 10 TCID50 of SARS CoV2 and the effect on cell death inhibition was quantified. C-E) Dose response effect of candidate drugs apilimod, berbamine, cepharanthine and (S)-crizotinib on USA/WA1 SARS CoV2 induced cell death in 293T-ACE2, Vero E6 and Vero-TMPRSS2 cells was analyzed. F) Vero TMPRSS2 cells were infected with 10TCID50 SARS CoV2 and the nucleocapsid positive (NP+) cells were enumerated by flow cytometry 24 h post infection. Numbers in the histograms show the percentage of NP + cells. G) Bar graph shows the % inhibition in NP + cells in the presence of candidate drugs. Values represent Mean ± SEM, n = 3 and two independent experiments.
Next, we subjected the 231 hits from our pseudovirus entry inhibition assay that showed more than 60% pseudovirus entry inhibition to the cell death inhibition assay in three different cell lines. More than 50% SARS CoV2-induced cell death inhibition in at least one of the tested doses was seen with 32 compounds in 293T-ACE2 cells, 10 compounds in Vero E6 cells and 17 compounds in Vero-TMPRSS2 cells (Table 1 ). Moreover, four compounds, apilimod, berbamine, cepharanthine and (S)-crizotinib scored positive in all the three tested cell lines (Fig. 2C-E). Apilimod mesylate and berbamine dihydrochloride showed similar inhibition in our assays. Furthermore, we validated our results by staining the SARS CoV2 infected Vero-TMPRSS2 cells for SARS CoV2 nucleocapsid protein (NP). Consistent with our cell death inhibition assay results, we noted a marked reduction in SARS CoV2-NP + cells in the presence of apilimod, berbamine, cepharanthine and (S)-crizotinib compared to vehicle treated control cells (Fig. 2F&G).
Table 1.
Summary of CPE inhibition screening.
| Compound | 293T-ACE2 | Vero E6 | Vero-TMPRSS2 |
|---|---|---|---|
| Tilorone dihydrochloride | Yes | No | No |
| JNK–IN–7 | Yes | No | No |
| Berbamine | Yes | Yes | Yes |
| Succinobucol | Yes | No | No |
| Tetrandrine | Yes | No | No |
| Azithromycin | Yes | No | No |
| (S)-crizotinib | Yes | Yes | Yes |
| Melitracen hydrochloride | Yes | No | No |
| Apilimod mesylate | Yes | Yes | Yes |
| Apilimod | Yes | Yes | Yes |
| Olmutinib | Yes | No | Yes |
| Azithromycin dihydrate | Yes | No | No |
| Anidulafungin | Yes | No | No |
| Lazertinib | Yes | No | No |
| Fupentixol Dihydrochloride | Yes | No | No |
| Daurisoline | Yes | No | No |
| Chloroquine diphosphate | Yes | No | No |
| Bithionol | Yes | No | No |
| Halofuginone hydrobromide | Yes | No | No |
| SB1317 hydrochloride | Yes | No | Yes |
| Chlorpromazine hydrochloride | Yes | No | No |
| OTSSP167 | Yes | No | No |
| Sunitinib Malate | Yes | No | No |
| Berbamine dihydrochloride | Yes | Yes | No |
| Amiodarone hydrochloride | Yes | No | No |
| Balicatib | Yes | Yes | No |
| Omipalisib | Yes | Yes | No |
| Gilteritinib | Yes | No | No |
| XL019 | Yes | No | No |
| CBL0137 hydrochloride | Yes | No | No |
| Cepharanthine | Yes | Yes | Yes |
| Bardoxolone | No | Yes | Yes |
| Masitinib | No | Yes | Yes |
| Balicatib | No | Yes | No |
| LXS196 | No | No | Yes |
| HS-10296 hydrochloride | No | No | Yes |
| Dauricine | No | No | Yes |
| Quinacrine dihydrochloride | No | No | Yes |
| GSK369796 Dihydrochloride | No | No | Yes |
| Ozanimod | No | No | Yes |
| Dauricine | No | No | Yes |
| Crizotinib hydrochloride | No | No | Yes |
| Azeliragon | No | No | No |
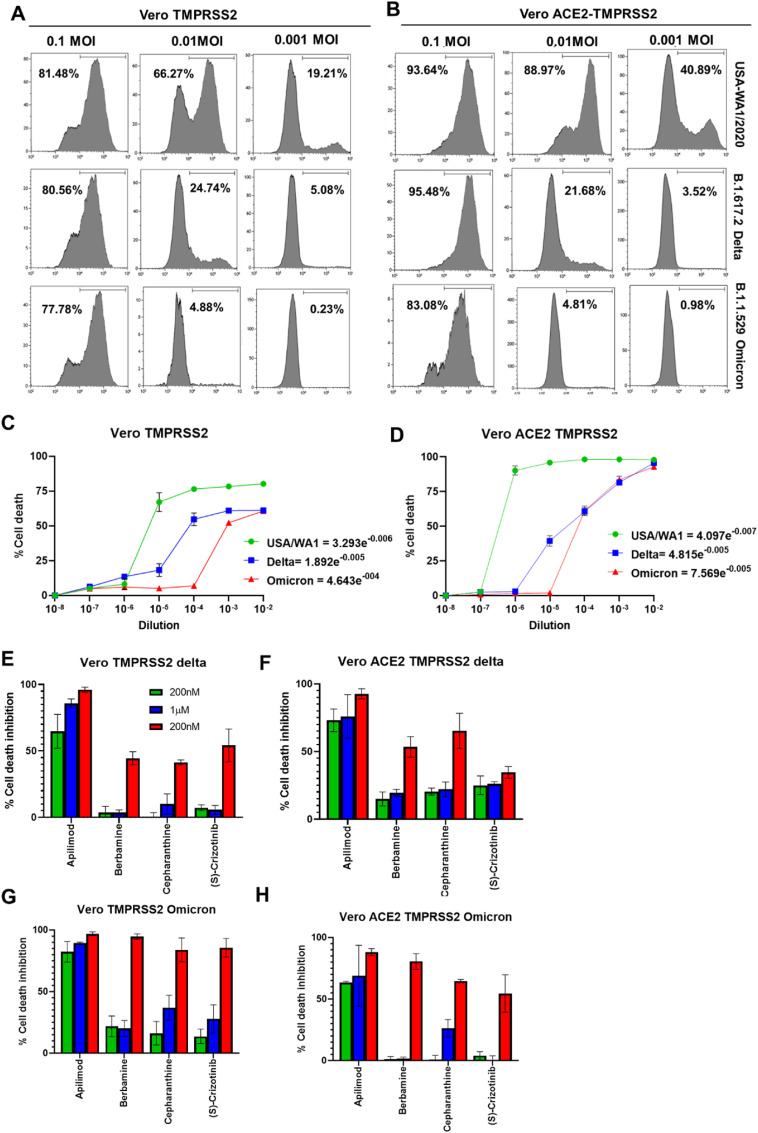
Next, we tested the efficacy of these compounds against other VOCs such as lineage B.1.617.2: Delta Variant and lineage: B.1.1.529, Omicron Variant. To compare the efficacy of virus entry and cell death inducing characteristics of these variants, we performed NP-staining and cell death assays in Vero-TMPRSS2 cells with different dilutions of the variants. Interestingly, both delta and omicron variants were less efficient in infecting these target cells compared to the prototypic USA/WA1 stain (Fig. 3 A) as evidenced by the differences in the percentage of NP + cells. To test if overexpression of human ACE2 in Vero-TMPRSS2 cells can increase the susceptibility to infection, we performed similar experiments using Vero-ACE2-TMPRSS2 cells (Fig. 3B). While the overexpression of human ACE2 increased susceptibility to infection and cell death in case of the prototypic strain compared to Vero-TMPRSS2 cells, no significant increase in infectivity or cytotoxicity was seen with delta and omicron variants (Fig. 3B). Consistent with these results, both delta and omicron variants had higher TCID50 values compared to the prototypic strain in Vero-TMPRSS2 cells (Fig. 3C). In contrast to no significant difference in the NP+ cells between Vero-TMPRSS2 vs Vero-ACE2-TMPRSS2 cells infected with delta and omicron variants, the cytotoxic effect of the variants was more pronounced in Vero-ACE2-TMPRSS2 cells compared to Vero-TMPRSS2 cells (Fig-3C-D). Furthermore, we tested the efficacy of the selected drug compounds to inhibit cytotoxic effects of the delta and omicron variants in Vero-TMPRSS2 and Vero-ACE2-TMPRSS2 cells. Among these compounds, apilimod showed superior efficacy in inhibiting both delta and omicron-induced cytotoxicity at all doses starting from 200 nM (Fig-3E-H). (S)-crizotinib was found to be the least effective drug against the delta variant especially in the highly susceptible Vero-ACE2-TMPRSS2 cells (Fig. 3E-H).
Fig. 3.
A-B) Representative histograms show the frequency of NP + cells in the Vero-TMPRSS2 (A) and Vero-ACE2-TMPRSS2 (B) cells infected with different doses of the USA-WA1, delta and omicron strains of SARS CoV2. C-D) Graphs show the TCID50 values for each strain of the virus in Vero TMPRSS2 (C) and Vero ACE2-TMPRSS2 cells (D). E-H) Dose response effect of the drugs on delta and omicron variant induced cell death in Vero TMPRSS2 and Vero-ACE2-TMPRSS2 cells. Values represent Mean ± SEM, n = 3 and two independent experiments.
5. Discussion
The development of antiviral drugs for SARS CoV2 is impeded by not only the novelty of this virus, but also by the requirement of using biosafety level-3 facilities to perform challenging viral infection screening experiments, and a lack of validated in vitro screening methods. There is a lack of clinically validated antiviral screening methods and therefore, drugs that show promising results in cell culture often fail in clinical trials. To an extent, this limitation can be overcome by deploying a more elaborate screening involving multiple screening tests, different cell lines with varying susceptibility to virus infection and different virus variants. Although these approaches cannot replace clinical trials, the stratified screening approach with highly susceptible host cell lines and viral variants that we used could identify potential candidate antiviral drugs with higher accuracy and probability of success in newer variants.
Here, we used a two-step screening strategy using FDA approved and clinical stage drug candidate libraries in a pseudovirus entry inhibition assay followed by further screening of selected hits for their SARS CoV2-induced cell death inhibitory effect on different cell lines that show varying degrees of susceptibility to SARS CoV2 induced cell death. Using this approach, we screened a library of 2985 compounds and identified 4 compounds, apilimod, cepharanthine, berbamine and S-crizotinib that inhibited pseudovirus entry and infectious virus induced cell death in all three cell lines i.e. 293T-ACE2, Vero E6, Vero-TMPRSS2 and Vero-ACE2-TMPRSS2 cells against VOCs such as delta and omicron.
Apilimod is an inhbitor of 5-phosphoinositide kinase (PIKfyve kinase) which mainly resides in early endosomes. Inhibition of PIKfyve kinase by apilimod can result in the blockade of trafficking between lysosomes and endosomes, and the trans-Golgi network (Rutherford et al., 2006). Lyso/endosomal pathway is important for SARS Cov2 entry and lysosomal proteases such as cathepsins play a key role in mediating SARS Cov2 entry. Previous studies have shown that apilimod can prevent viral infection by preventing release of the viral contents from endosomes by inhibiting cathepsin (Kang et al., 2020).
Berbamine, a bis-benzylisoquinoline alkaloid, has been shown to impair Ca2+ signaling required for viral entry. An earlier report has shown that berbamine inhibits SARS-CoV-2 entry into the target cells by impairing Transient Receptor Potential cation channel, MucoLipin subfamily (TRPMLs)-mediated endolysosomal trafficking of ACE2 (Huang et al., 2021) and could inhibit S-mediated cell-cell fusion.(Zhang et al., 2022). Cepharanthine, a naturally occurring alkaloid extracted from the plant Stephania cepharantha Hayata, has been previously reported to inhibit SARS-CoV-2 binding to target cells and thereby prevent virus entry (Ohashi et al., 2021). Cepharanthine in combination with Neflonvair, a HIV protease inhibitor, showed impressive antiviral activity against SARS CoV2 in Vero E6 and Vero TMPRSS2 cells (Ohashi et al., 2021).
(S)-Crizotinib, is the (S)-enantiomer of (R)-crizotinib which is an FDA-approved drug for treating non-small cell lung cancer. (S)-Crizonib is a selective inhibitor of human 7, 8-Dihydro-8-oxoguaninetriphosphatase MTH1 (NUDT1) (Dai et al., 2017; Huber et al., 2014). The molecular mechanism by which (S)-Crizotinib inhibited SARS CoV2 entry remains to be determined. We have used highly stringent approach by deploying different cell lines with varying sensitivity to SARS CoV2 induced cell death and a higher TCID50 dose of the virus. The variability in the sensitivity of different cell lines to different drugs could be related to both the virus entry and replication kinetics of the virus in those host cell lines.
Almost all the combinations of the previously FDA-approved mAbs such as imdevimab plus casirivimab, tixagevimab plus cilgavimab, and etesevimab plus bamlanivimab that were effective against the alpha and delta variants have proven to be less effective against the omicron variant (Takashita et al., 2022). This is due the novel mutations harbored in the spike and RBD regions of the omicron variant compared to the previous strains. Similarly, increased immune evasion and reduced neutralization of omicron variant by sera from vaccinated individuals and monoclonal antibodies have been reported (Dejnirattisai et al., 2021; Syed et al., 2022). The drugs identified in our screen inhibited delta and omicron variants-induced cell death to a similar degree to that of prototypic SARS CoV2, implying that the entry mechanisms utilized by these strains are conserved and these antiviral drugs could be effective in treating omicron variant infections.
Emerging studies showed that the omicron variant is more dependent on endosomal cathepsin pathway for the entry rather than TMPRSS2 (Hui et al., 2022; Pia and Rowland-Jones, 2022). Moreover, this variant showed weaker replication and cell-cell spread than delta and other strains in Calu3, Vero–TMPRSS2 cells (Meng et al., 2022; Zhao et al., 2022) and in the human lung parenchyma (Hui et al., 2022). These results are consistent with our observation of reduced NP + cells and cytotoxicity effect seen in Vero–TMPRSS2 and Vero-ACE2-TMPRSS2 cells caused by the omicron variant compared to the delta and the prototypic USA/WA1 strains. While the reduced severity of the COVID pathology caused by the omicron variant can be attributed to its weaker replication efficiency, the greater transmissibility could be attributed to the dependency of the endosomal pathway for entry (Hui et al., 2022; Meng et al., 2022; Pia and Rowland-Jones, 2022; Zhao et al., 2022). Hence, the observed superior inhibitory effect of the identified drug candidates against omicron could be because of their mechanism of action related to the endosomal pathway.
While repurposing of existing drugs for COVID saves considerable amount of time and efforts than de novo drug discovery efforts, a recent study has raised an admonitory tale on the efficacy of repurposed drugs and the screening approach. This study showed a strong correlation between the anti-SARS CoV2 activity and drug-induced phospholipidosis. (Tummino et al., 2021). More importantly, several cationic amphiphilic drugs (CADs) including cepharanthine, have been shown to accumulate in endosomes and inhibit endosomal lipid processing pathways which are critical for coronavirus replication (Muller et al., 2018). Therefore, the translational potential and in vivo efficacy of these drugs have been called in to question by this study (Tummino et al., 2021). Moreover, many of the protease inhibitors can play a critical role in the MHC-class II antigen-processing and presentation pathways and therefore, their interaction with host immune responses needs to be taken into consideration (Hsing and Rudensky, 2005). We believe future in vivo studies testing the safety and efficacy of these drugs are warranted given that they are effective against several VOCs even at higher doses of the viruses. Besides, further refinement/modification can be made to these candidate drugs to minimize the adverse effects while enhancing the antiviral properties.
Funding source
This work was supported by funding from the Vanda Pharmaceuticals, USA to Dr. BP.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Prabhakaran Kumar: Methodology, Investigation, Visualization, Writing – original draft. Manikannan Mathayan: Investigation. Sandra P. Smieszek: Conceptualization, Writing – review & editing. Bartlomiej P. Przychodzen: Conceptualization, Writing – review & editing. Vuk Koprivica: Conceptualization, Writing – review & editing. Gunther Birznieks: Writing – review & editing. Mihael H. Polymeropoulos: Writing – review & editing. Bellur S. Prabhakar: Conceptualization, Methodology, Funding acquisition, Writing – review & editing.
References
- Consortium W.S.T. Repurposed antiviral drugs for Covid-19-interim WHO solidarity trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Guo G., Zou P., Cui R., Chen W., Chen X., Yin C., He W., Vinothkumar R., Yang F., Zhang X., Liang G. (S)-crizotinib induces apoptosis in human non-small cell lung cancer cells by activating ROS independent of MTH1. J. Exp. Clin. Cancer Res. 2017;36:120. doi: 10.1186/s13046-017-0584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradnik J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., Nutalai R., Wang B., Dijokaite A., Khan S., Avinoam O., Bahar M., Skelly D., Adele S., Johnson S.A., Amini A., Ritter T., Mason C., Dold C., Pan D., Assadi S., Bellass A., Omo-Dare N., Koeckerling D., Flaxman A., Jenkin D., Aley P.K., Voysey M., Costa Clemens S.A., Naveca F.G., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Baillie V., Serafin N., Ditse Z., Silva K.D., Madhi S., Nunes M.C., Malik T., Openshaw P.J., Baillie J.K., Semple M.G., Townsend A.R., Huang K.A., Tan T.K., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Constantinides B., Webster H., Crook D., Pollard A.J., Lambe T., consortium O., consortium I.C., Paterson N.G., Williams M.A., Hall D.R., Fry E.E., Mongkolsapaya J., Ren J., Schreiber G., Stuart D.I., Screaton G.R. Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. bioRxiv. 2021 doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.V., Pinsky B.A., Suthar M.S., Lai L.L., Davis-Gardner M.E., Floyd K., Flowers M.W., Wrammert J., Hussaini L., Ciric C.R., Bechnak S., Stephens K., Graham B.S., Mokhtari E.B., Mudvari P., Boritz E., Creanga A., Pegu A., Derrien-Colemyn A., Henry A.R., Gagne M., Douek D.C., Sahoo M.K., Sibai M., Solis D., Webby R.J., Jeevan T., Fabrizio T.P. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N. Engl. J. Med. 2021;385:664–666. doi: 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshabrawy H.A., Fan J.L., Haddad C.S., Ratia K., Broder C.C., Caffrey M., Prabhakar B.S. Identification of a broad-spectrum antiviral small molecule against severe acute respiratory syndrome coronavirus and Ebola, Hendra, and Nipah viruses by using a novel high-throughput screening assay. J. Virol. 2014;88:4353–4365. doi: 10.1128/JVI.03050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunst J.D., Staerke N.B., Pahus M.H., Kristensen L.H., Bodilsen J., Lohse N., Dalgaard L.S., Bronnum D., Frobert O., Honge B., Johansen I.S., Monrad I., Erikstrup C., Rosendal R., Vilstrup E., Mariager T., Bove D.G., Offersen R., Shakar S., Cajander S., Jorgensen N.P., Sritharan S.S., Breining P., Jespersen S., Mortensen K.L., Jensen M.L., Kolte L., Frattari G.S., Larsen C.S., Storgaard M., Nielsen L.P., Tolstrup M., Saedder E.A., Ostergaard L.J., Ngo H.T.T., Jensen M.H., Hojen J.F., Kjolby M., Sogaard O.S. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with Covid-19-a double-blind randomized controlled trial. Eclinicalmedicine. 2021;35 doi: 10.1016/j.eclinm.2021.100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.C., Wang Y.H., Chen Y.L., Tsai W.C., Lee C.H., Chuang K.P., Chen Y.A., Yuan C.H., Ho S.Y., Yang M.H., Tyan Y.C. Chloroquine and hydroxychloroquine: efficacy in the treatment of the COVID-19. Pathogens. 2021;10 doi: 10.3390/pathogens10020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271. doi: 10.1016/j.cell.2020.02.052. + [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Mosbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Kruger N., Gassen N.C., Muller M.A., Drosten C., Pohlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- Hsing L.C., Rudensky A.Y. The lysosomal cysteine proteases in MHC class II antigen presentation. Immunol. Rev. 2005;207:229–241. doi: 10.1111/j.0105-2896.2005.00310.x. [DOI] [PubMed] [Google Scholar]
- Huang L., Yuen T.T., Ye Z., Liu S., Zhang G., Chu H., Yue J. Berbamine inhibits SARS-CoV-2 infection by compromising TRPMLs-mediated endolysosomal trafficking of ACE2. Signal Transduct. Targeted Ther. 2021;6:168. doi: 10.1038/s41392-021-00584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber K.V., Salah E., Radic B., Gridling M., Elkins J.M., Stukalov A., Jemth A.S., Gokturk C., Sanjiv K., Stromberg K., Pham T., Berglund U.W., Colinge J., Bennett K.L., Loizou J.I., Helleday T., Knapp S., Superti-Furga G. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508:222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K.P.Y., Ho J.C.W., Cheung M.C., Ng K.C., Ching R.H.H., Lai K.L., Kam T.T., Gu H., Sit K.Y., Hsin M.K.Y., Au T.W.K., Poon L.L.M., Peiris M., Nicholls J.M., Chan M.C.W. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603:715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- Kang Y.L., Chou Y.Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D., Case J.B., Chen R.E., Diamond M.S., Whelan S.P.J., Kirchhausen T. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye M. SARS-associated coronavirus replication in cell lines. Emerg. Infect. Dis. 2006;12:128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., Group S.W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Liu J., Cao R.Y., Xu M.Y., Wang X., Zhang H.Y., Hu H.R., Li Y.F., Hu Z.H., Zhong W., Wang M.L. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6 doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi V.C., Ramanan A.V., de Bono S., Kartman C.E., Krishnan V., Liao R., Piruzeli M.L.B., Goldman J.D., Alatorre-Alexander J., Pellegrini R.D., Estrada V., Som M., Cardoso A., Chakladar S., Crowe B., Reis P., Zhang X., Adams D.H., Ely E.W., Grp C.-B.S. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER) : a randomised, double-blind, parallel-group, placebo- controlled phase 3 trial. Lancet Respir. Med. 2021;9:1407–1418. doi: 10.1016/S2213-2600(21)00331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Abdullahi A., Ferreira I., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., Zepeda S.K., Papa G., Kemp S.A., Ikeda T., Toyoda M., Tan T.S., Kuramochi J., Mitsunaga S., Ueno T., Shirakawa K., Takaori-Kondo A., Brevini T., Mallery D.L., Charles O.J., Collaboration C.-N.B.C.-., Genotype to Phenotype Japan, C. Ecuador C.C., Bowen J.E., Joshi A., Walls A.C., Jackson L., Martin D., Smith K.G.C., Bradley J., Briggs J.A.G., Choi J., Madissoon E., Meyer K.B., Mlcochova P., Ceron-Gutierrez L., Doffinger R., Teichmann S.A., Fisher A.J., Pizzuto M.S., de Marco A., Corti D., Hosmillo M., Lee J.H., James L.C., Thukral L., Veesler D., Sigal A., Sampaziotis F., Goodfellow I.G., Matheson N.J., Sato K., Gupta R.K. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C., Hardt M., Schwudke D., Neuman B.W., Pleschka S., Ziebuhr J. Inhibition of Cytosolic phospholipase A2alpha impairs an early step of coronavirus replication in cell culture. J. Virol. 2018;92 doi: 10.1128/JVI.01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Barcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi H., Watashi K., Saso W., Shionoya K., Iwanami S., Hirokawa T., Shirai T., Kanaya S., Ito Y., Kim K.S., Nomura T., Suzuki T., Nishioka K., Ando S., Ejima K., Koizumi Y., Tanaka T., Aoki S., Kuramochi K., Suzuki T., Hashiguchi T., Maenaka K., Matano T., Muramatsu M., Saijo M., Aihara K., Iwami S., Takeda M., McKeating J.A., Wakita T. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience. 2021;24:102367. doi: 10.1016/j.isci.2021.102367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pia L., Rowland-Jones S. Omicron entry route. Nat. Rev. Immunol. 2022;22:145. doi: 10.1038/s41577-022-00681-9. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Pere H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Loriere E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Rajpoot S., Alagumuthu M., Baig M.S. Dual targeting of 3CL(pro) and PL(pro) of SARS-CoV-2: a novel structure-based design approach to treat COVID-19. Curr. Res. Struct. Biol. 2021;3:9–18. doi: 10.1016/j.crstbi.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., Chang M.W., Chan J.F., Cao J., Poon V.K., Herbert K.M., Cheng K., Nguyen T.H., Rubanov A., Pu Y., Nguyen C., Choi A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T.P., Sit K.Y., Martinez-Sobrido L., Liu W.C., White K.M., Chapman M.E., Lendy E.K., Glynne R.J., Albrecht R., Ruppin E., Mesecar A.D., Johnson J.R., Benner C., Sun R., Schultz P.G., Su A.I., Garcia-Sastre A., Chatterjee A.K., Yuen K.Y., Chanda S.K. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford A.C., Traer C., Wassmer T., Pattni K., Bujny M.V., Carlton J.G., Stenmark H., Cullen P.J. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 2006;119:3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. P Natl Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A.M., Ciling A., Khalid M.M., Sreekumar B., Chen P.Y., Kumar G.R., Silva I., Milbes B., Kojima N., Hess V., Shacreaw M., Lopez L., Brobeck M., Turner F., Spraggon L., Taha T.Y., Tabata T., Chen I.P., Ott M., Doudna J.A. medRxiv; 2022. Omicron Mutations Enhance Infectivity and Reduce Antibody Neutralization of SARS-CoV-2 Virus-like Particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., Iwatsuki-Horimoto K., Chiba S., Halfmann P., Nagai H., Saito M., Adachi E., Sullivan D., Pekosz A., Watanabe S., Maeda K., Imai M., Yotsuyanagi H., Mitsuya H., Ohmagari N., Takeda M., Hasegawa H., Kawaoka Y. Efficacy of antibodies and antiviral drugs against Covid-19 omicron variant. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2119407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummino T.A., Rezelj V.V., Fischer B., Fischer A., O'Meara M.J., Monel B., Vallet T., White K.M., Zhang Z., Alon A., Schadt H., O'Donnell H.R., Lyu J., Rosales R., McGovern B.L., Rathnasinghe R., Jangra S., Schotsaert M., Galarneau J.R., Krogan N.J., Urban L., Shokat K.M., Kruse A.C., Garcia-Sastre A., Schwartz O., Moretti F., Vignuzzi M., Pognan F., Shoichet B.K. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021;373:541–547. doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.L., Cao R.Y., Zhang L.K., Yang X.L., Liu J., Xu M.Y., Shi Z.L., Hu Z.H., Zhong W., Xiao G.F. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K., Rawle D.J., Le T.T., Suhrbier A. Simple rapid in vitro screening method for SARS-CoV-2 anti-virals that identifies potential cytomorbidity-associated false positives. Virol. J. 2021;18:123. doi: 10.1186/s12985-021-01587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.R., Zhang Y.N., Zhang H.Q., Zhang Q.Y., Li N., Li Q., Deng C.L., Zhang B., Li X.D., Ye H.Q. Berbamine hydrochloride potently inhibits SARS-CoV-2 infection by blocking S protein-mediated membrane fusion. PLoS Neglected Trop. Dis. 2022;16 doi: 10.1371/journal.pntd.0010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.J., Lu L., Peng Z., Chen L.L., Meng X.J., Zhang C.Y., Ip J.D., Chan W.M., Chu A.W.H., Chan W.H., Jin D.Y., Chen H.L., Yuen W.Y., To K.K.W. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microb. Infect. 2022;11:277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.M., Yang W.L., Yang F.Y., Zhang L., Huang W.J., Hou W., Fan C.F., Jin R.H., Feng Y.M., Wang Y.C., Yang J.K. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Tar. 2021;6 doi: 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.K., Feng F., Hu G.W., Wang Y.Y., Yu Y., Zhu Y.F., Xu W., Cai X., Sun Z.P., Han W.D., Ye R., Qu D., Ding Q., Huang X.X., Chen H.J., Xu W., Xie Y.H., Cai Q.L., Yuan Z.H., Zhang R. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]