Summary
Characterizing the interactions between RNAs and proteins in vivo is key to better understand how organisms regulate gene expression. Here, we describe a robust and quantitative protocol to measure specific RNA-protein interactions in a native context using RNA immunoprecipitation (RIP). We provide a comprehensive experimental framework to detect cotranslational interactions and detail the quantitative analysis of purified RNAs by PCR and high-throughput sequencing. Although we developed the protocol in fission yeast, it can be readily implemented in other yeast species.
For complete details on the use and execution of this protocol, please refer to Toullec et al. (2021).
Subject areas: Model Organisms, Molecular Biology, Protein Biochemistry
Graphical abstract

Highlights
-
•
Simple and robust detection of RNA-protein interactions in a native context
-
•
Ribonucleoprotein complex isolated by immunoprecipitation
-
•
Quantitative analysis of purified RNA by PCR or high-throughput sequencing
-
•
Detailed experimental pipeline to demonstrate cotranslational interactions
Characterizing the interactions between RNAs and proteins in vivo is key to better understand how organisms regulate gene expression. Here we describe a robust and quantitative protocol to measure specific RNA-protein interactions in a native context using RNA immunoprecipitation (RIP). We provide a comprehensive experimental framework to detect cotranslational interactions and detail the quantitative analysis of purified RNAs by PCR and high-throughput sequencing. Although we developed the protocol in fission yeast, it can be readily implemented in other yeast species.
Before you begin
Timing: 30 minto2 h
-
1.Make sure you have all reagents for buffer preparation, additives, and commercial kits, including:
-
a.Per sample, 100 μL of Pan mouse magnetic beads and 3 μL (3 μg) of your selected mouse monoclonal antibody.Note: This protocol has been set up using Pan mouse magnetic beads and anti-influenza virus hemagglutinin (anti-HA), anti-human c-Myc protein (anti-MYC) or anti-FLAG peptide (anti-FLAG) mouse monoclonal antibodies. Other antibodies should work but may require optimization by adjusting antibody and extract quantities.
-
b.50 mg/mL heparin stock solution.
-
c.40 units of RNase inhibitor per sample.
-
d.4.4 units of Turbo DNase per sample.
-
e.Acid-washed glass beads to be added up to the meniscus of the lysed extract (typically, 500 μL to 1 mL per sample).
-
f.1.5 mL tubes.
-
g.2 mL screw-cap tubes.
-
h.5 mL tubes.
-
i.50 mL tubes.
-
j.Needle (25G).
-
k.Bunsen burner.
-
l.PureLinkTM RNA Mini Kit (or equivalent).
-
m.RNase free water.
-
n.Ethanol.
-
o.Coomassie reagent for protein quantification.
-
p.Spectrophotometer.
-
q.Heat block at 70°C.
-
r.EDTA-free protease inhibitors cocktail, Pepstatin A, Bestatin, PMSF, (optional) phosphatase inhibitors.
-
s.Centrifuge at 4°C.
-
t.If assessing co-translational interactions, prepare stock solutions of puromycin (50 mg/mL) and GTP (200 mM). You can also use EDTA or ATG-mutated strains. Refer to Figures 1 and 2 for details about the effect of each perturbation on translation.Note: The PureLinkTM Micro Kit can also be used for the extraction of the IP RNA (see specific instructions below).Note: For further information about the mechanism of action and the use of puromycin see Aviner (2020).
-
a.
-
2.
Prepare all buffers and reagents, such as stock solutions of the protease inhibitors Bestatin (1 mg/mL; 1,000×), Pepstatin A (1 mg mL; 1,000×) and PMSF (100 mM; 100×). Keep all buffers at 4°C, except when containing detergents, such as Nonidet P-40 (NP40), in which case keep at ambient temperature.
-
3.
3–4 days before starting, streak your strains on appropriate media and let them grow at 32°C. Verify your markers. We recommend the use of epitope tagged strains.
Note: This protocol uses Schizosaccharomyces pombe as an experimental system. Adjust this step according to the growth conditions of your favorite organism.
Note: To increase the reproducibility of the procedure, we recommend to always keep similar amounts of inoculated biomass from plates and harvest the same number of yeast cells between samples and experiments.
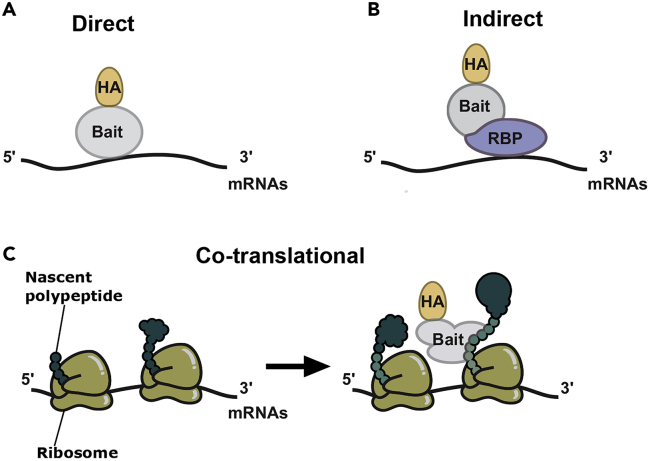
Figure 1.
Types of protein-RNA interactions detected by this RIP protocol
(A) Direct interaction between the protein bait tagged with an epitope (HA in this example) and an RNA.
(B) Indirect interaction between an epitope-tagged bait and its target RNAs through an RNA-binding protein (RBP).
(C) Cotranslational interaction between an epitope-tagged protein bait and a nascent polypeptide detected through indirect binding to the translated mRNA.
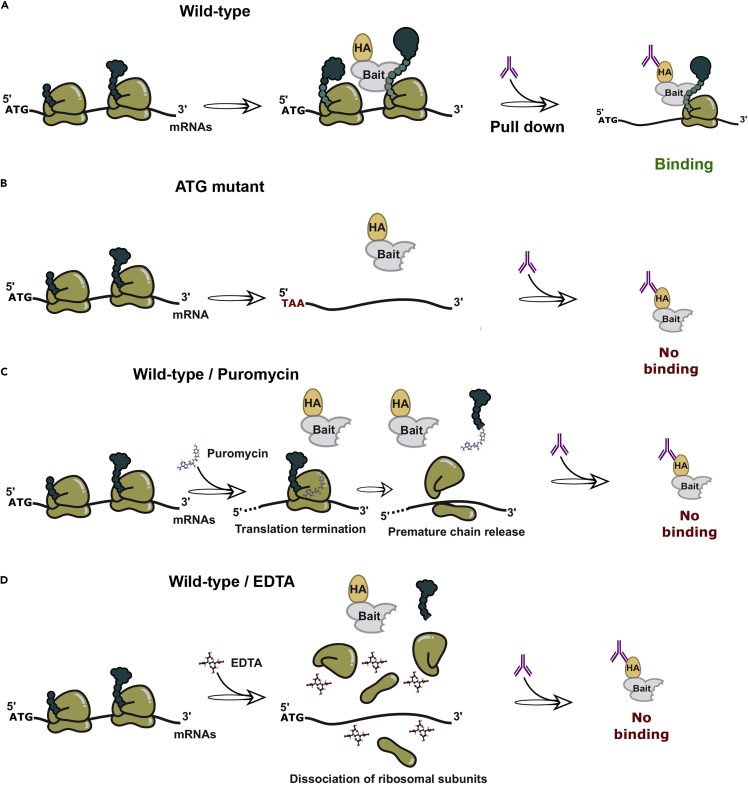
Figure 2.
Experimental strategies to test whether protein-mRNA interactions detected by RIP are dependent on translation
(A) Cotranslational interaction between a protein bait and an mRNA in a wild-type strain in the absence of any treatment (control condition).
(B) Impact of mutating the ATG of the targeted mRNA on the co-translational binding of the bait.
(C) Effect of puromycin addition on translation and on the detection of cotranslational interactions. Puromycin, which partly resembles the 3′ end of an amino-acylated tRNA, enters the A site of the ribosome and transfer to the growing polypeptide, leading to the formation of a puromycylated chain and causing premature chain release.
(D) Effect of EDTA on ribosome integrity and on the detection of cotranslational interactions. EDTA, which is added in vitro to the extraction buffers used in the protocol, causes dissociation of the small and large ribosomal subunits.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Myc antibody, clone 9E11 | Abcam | Cat#ab56; RRID: AB_304976 |
| Mouse monoclonal anti-FLAG antibody, clone M2 | Sigma | Cat#F1804; RRID: AB_262044 |
| Mouse monoclonal anti-HA antibody, clone 16B12 |
BioLegend | Cat#901514; RRID: AB_2565336 |
| Chemicals, peptides, and recombinant proteins | ||
| Bestatin | Sigma | Cat#B8385 |
| Pepstatin A | Sigma | Cat#P5318 |
| Phenylmethanesulfonyl fluoride (PMSF) | Sigma | Cat#P7626 |
| β-Glycerol phosphate disodium salt hydrate | Sigma | Cat#G9422 |
| Sodium orthovanadate (Na3VO4) | Sigma | Cat#450243 |
| Sodium fluoride (NaF) | Sigma | Cat#201154 |
| Dynabeads Pan-Mouse IgG | Thermo Fisher Scientific | Cat#11041 |
| Promega Recombinant RNasin™ Ribonuclease Inhibitor | Thermo Fisher Scientific | Cat#PR-N2511 |
| Igepal CA-630 | Sigma | Cat#56741 |
| Heparin sodium salt from porcine intestinal mucosa | Sigma | Cat#H3393 |
| cOmplete EDTA-free cocktails tablets | Roche | Cat#04693132001 |
| Puromycin hydrochloride (optional) | Santa Cruz Biotechnology | Cat#sc-108071B |
| Adenine sulphate | Formedium | Cat#DOC0229, |
| Uracil | Formedium | Cat#DOC0213, |
| L-Lysine | Formedium | Cat#DOC0160 |
| L-Histidine monohydrochloride monohydrate | Sigma | Cat#H8125 |
| L-Leucine | Formedium | Cat#DOC0156 |
| Bacto yeast extract | Thermo Fisher Scientific-Gibco | Cat# 212750 |
| D-(+)-Glucose | Sigma | Cat# G7021 |
| Tris base | EUROMEDEX | Cat#26-128-3094-B |
| Hydrochloric acid (HCl) | Sigma | Cat#320331 |
| Potassium chloride (KCl) | Sigma | Cat#P9541 |
| Magnesium chloride (MgCl2) | Sigma | Cat# M8266 |
| Glycerol | Sigma | Cat#G6279 |
| Igepal CA630 | Sigma | Cat#I8896 |
| Bromophenol blue sodium salt | Sigma | Cat#B5525 |
| Polyoxyethylene ether W-1 (for qPCR master mix) | Sigma | Cat#P7516 |
| Bovine Serum Albumin (BSA) (for qPCR master mix) | Sigma | Cat#A4378 |
| dATP (for qPCR master mix) | Thermo Fisher Scientific-Invitrogen | Cat#10216018 |
| dCTP (for qPCR master mix) | Thermo Fisher Scientific-Invitrogen | Cat#10217016 |
| dGTP (for qPCR master mix) | Thermo Fisher Scientific-Invitrogen | Cat#10218014 |
| dTTP (for qPCR master mix) | Thermo Fisher Scientific-Invitrogen | Cat#10219012 |
| Potassium chloride (KCl) (for qPCR master mix) | Merck-Sigma-Supelco | Cat#1.04936.1000 |
| Magnesium chloride (MgCl2) (for qPCR master mix) | Merck-Sigma-Supelco | Cat#1058331000 |
| SYBR Green (for qPCR master mix) | FMC BioProducts / Cambrex | Cat#50513 |
| Glycerol (for qPCR master mix) | Thermo Fisher Scientific-MP Biomedicals | Cat#ICN800687 |
| 2-Amino-2-methyl-1,3-propanediol for qPCR master mix | Sigma | Cat#A9074 |
| Hydrochloric acid fuming 37% for qPCR master mix | Sigma-Merck | Cat#1003171000 |
| Platinum Taq DNA polymerase for qPCR master mix | Thermo Fisher Scientific-Invitrogen | Cat#10966-034 |
| Critical commercial assays | ||
| SuperScript III First-Strand System | Thermo Fisher Scientific | Cat#18080051 |
| Purelink RNA Mini kit | Thermo Fisher Scientific | Cat#12183018A |
| Purelink RNA Micro Scale kit | Thermo Fisher Scientific | Cat#12183016 |
| TURBO DNA-free kit | Invitrogen | Cat#AM1907 |
| Experimental models: Organisms/strains | ||
| Schizosaccharomyces pombe strains | See Table S4 at Toullec et al., 2021. | Derivatives from h- 972 strain (ATCC 24843) |
| Oligonucleotides | ||
| Oligonucleotides | See Table S5 at Toullec et al., 2021. | N/A |
| Software and algorithms | ||
| GraphPad Prism | version 9.2.0 https://www.graphpad.com/ |
|
Note: Any other chemical or media additive were bought from Sigma-Merk or Formedium, respectively.
Materials and equipment
Liquid Yeast Extract with Supplements (YES) media
| Reagent | Final concentration | Amount |
|---|---|---|
| Bacto yeast extract | 0.5% w/v | 5 g |
| Glucose | 3% w/v | 30 g |
| Supplements∗ | 0.225 g/L each | 1.125 g |
| Adenine | 0.225 g/L | 0.225 g |
| Uracil | 0.225 g/L | 0.225 g |
| Lysine | 0.225 g/L | 0.225 g |
| Histidine | 0.225 g/L | 0.225 g |
| Leucine | 0.225 g/L | 0.225 g |
| ddH2O | n/a | Up to 1 L |
| Total | n/a | 1 L |
It is possible to use either a powder mix containing all supplements (adenine, uracil, lysine, histidine and leucine) or add each of them individually.
After preparing the media, autoclave it for 10 min at 113°C and let it cool down to room temperature before using it.
To prepare solid media, add 2% w/v of bacto agar before autoclaving.
Immunoprecipitation buffer (IP buffer)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 8.0 (1 M) | 20 mM | 1 mL |
| KCl (1 M) | 140 mM | 7 mL |
| MgCl2 (1 M) | 1.8 mM | 0.1 mL |
| NP40 (10%) | 0.1% | 0.5 mL |
| Heparin (50 mg/mL) | 0.2 mg/mL | 0.2 mL |
| ddH2O | n/a | 41.2 mL |
| Total | n/a | 50 mL |
Keep heparin stock solution aliquoted at −20°C; avoid repeated cycles of thawing and freezing. All other stock solutions can be kept at room temperature. Nonetheless, to be able to prepare a cold IP buffer at will, we recommend keeping stock solutions at 4°C, with the exception of NP40. Once prepared, keep IP buffer at 4°C until use.
CRITICAL: NP40 (or its alternative Igepal CA-630) is highly toxic through oral, skin, or eye contact. Manipulate with extreme care, using gloves and protection glasses and following the instructions of the manufacturer.
Alternatives: NP40 can be replaced by Igepal CA-630 (same concentration of stock and working solutions).
Complete immunoprecipitation buffer (complete IP buffer)
| Reagent | Volume per sample (without phosphatase inhibitors) | Volume per sample (with phosphatase inhibitors)∗ |
|---|---|---|
| IP buffer | 1039.5 μL | 962.5 μL |
| Heparin (50 mg/mL) | 1.1 μL | 1.1 μL |
| Turbo DNase | 2.2 μL | 2.2 μL |
| EDTA-free protease inhibitors cocktail (25× stock solution; 1 pill into 2 μL sterile distilled water) | 44 μL | 44 μL |
| Bestatin 1,000× | 1.1 μL | 1.1 μL |
| Pepstatin 1,000× | 1.1 μL | 1.1 μL |
| PMSF 100× | 11 μL | 11 μL |
| Beta-glycerol phosphate 20× | – | 55 μL |
| Na3VO4 1,000× | – | 11 μL |
| NaF 1,000× | – | 11 μL |
| Total | 1,100 μL | 1,100 μL |
If needed.
Keep all stock solutions at −20°C and IP buffer at 4°C. Once prepared, keep complete IP buffer at 4°C until use.
CRITICAL: PMSF is a highly toxic neurotoxin. Prepare the stock solution (100 mM in DMSO) in a fume hood. To add it to your buffer, take the stock out of the −20°C and warm the tube in your hands, wearing gloves, until no PMSF crystals are visible. Alternatively, warm the tube in a table-top incubator preset to RT. Solubilized PMSF should be added last, as it rapidly degrades in aqueous solution. Add PMSF and mix gently immediately to prevent precipitation.
CRITICAL: NaVO4 and NaF are highly toxic via oral and can cause skin and eye irritation. Prepare 1 mg/mL (1,000×) stock solutions in a fume hood, wearing gloves and glasses. Keep stock solutions at −20°C. To add it to your buffer, take the stock solutions out of the −20°C and warm the tube in your hands, wearing gloves. Keep solutions on ice until added to the buffer.
Immunoprecipitation wash buffer (W buffer)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 8.0 (1 M) | 20 mM | 1 mL |
| KCl (1 M) | 140 mM | 7 mL |
| MgCl2 (1 M) | 1.8 mM | 0.1 mL |
| Glycerol (50%) | 10% | 10 mL |
| NP40 (10%) | 0.01% | 0.1 mL |
| ddH2O | n/a | 31.8 mL |
| Total | n/a | 50 mL |
All stock solutions can be kept at room temperature. Nonetheless, to be able to prepare a cold W buffer at will, we recommend keeping stock solutions at 4°C, with the exception of NP40 and optionally glycerol 50%.
Immunoprecipitation wash buffer with heparin and PMSF (W1)
Add 3 μL of heparin (50 mg/mL) and 30 μL of PMSF (100×) to 3 mL of W buffer.
Immunoprecipitation wash buffer with heparin (W2)
Add 1.5 μL heparin (50 mg/mL) to 1.5 mL of W buffer.
10× qPCR master mix
| Reagent | Reference | Final concentration |
|---|---|---|
| Polyoxyethylene ether W-1 | Sigma P7516 | 0.24% |
| Bovine Serum Albumin (BSA) | Sigma A4378 | 500 μg/mL |
| dNTPs | Invitrogen 10216, 10217, 10218, 10219 | 300 μM |
| KCl | Merck proanalysis 1.04936.1000 | 50 mM |
| MgCl2 | Merck proanalysis 1058331000 | 30 mM |
| SYBR Green | FMC BioProducts 50513 | 1/3000 |
| Glycerol | ICN 800687 | 16.24% |
| 2-Amino-2-methyl-1,3-propanediol buffer at pH 8.3 using HCl (Merck proanalysis 1.00317.1000) | Sigma A9074 | 400 mM |
| Platinum Taq DNA polymerase | Invitrogen 10966-034 | 0.4 U/μL |
Note: Polyoxyethylene ether W-1 can be replaced by a combination of two detergents: Brij 56 (0.09%, Sigma P5759) and Brij 58P (0.15%, Sigma P5884).
Step-by-step method details
Cell growth and beads preparation—Day 1 and day 2
Timing: 5–10 min (Day 1) and 1 h (Day 2)
Note: This protocol is an adaptation of previous protocols (Gerber et al., 2004; Duncan and Mata, 2011).
Note: This protocol will detect specific protein-RNA interactions occurring directly, indirectly or co-translationally (Figure 1).
Note: Upon harvesting yeast cells, keep your samples and buffers cold (0°C–4°C), unless otherwise indicated.
-
1.
Inoculate one single colony into 5 mL of YES rich media (or your favorite/required media). Let it grow at 32°C for 24 h to reach saturation.
Note: Timing should be adjusted depending on media, strain and growth rate, in order to obtain a saturated culture. Ideally, a concentration of 1 × 108 cells/mL should be reached.
Note: Use colonies of similar size for each strain and between biological replicates. Maintaining the same timing from streaking out your strains from the −80°C to inoculation helps. If mutants form bigger or smaller colonies as compared to control/wild-type strain, adjust the amount of biomass for each inoculate.
CRITICAL: Always include at least one negative control. Either a non-tagged strain or a strain containing another, unrelated protein tagged with the same epitope can be used. We recommend using both when performing RIP for the first time or when testing a bait for the first time. The former gives an idea of the non-specific binding of RNAs to antibody-coated beads. The latter serves as a control for non-specific binding due to stickiness of nucleic acids to proteins and epitopes. If using antibodies against an endogenous protein, a deletion mutant can be used as a negative control.
-
2.
Measure OD at 595 nm and dilute the culture in 60 mL of fresh media to reach a cell concentration of 1.5 × 107 cells/mL at the time of harvesting the culture (step 10), aiming for at least 4–5 doubling. Incubate at 32°C.
Note: Beware that if additional conditions are required, such as testing cotranslational interactions by using EDTA and/or puromycin (Figure 2), the volume should be adjusted accordingly. For example, to check the cotranslational dependency of the interaction using puromycin, inoculate 120 mL of media per strain.
Note: To allow proper oxygenation of the culture, its volume should be less than about 20% of the flask.
CRITICAL: If using slow- or fast-growing mutants, adjust the cell number to guarantee that all cultures reach the concentration of 1.5 × 107 cells/mL at the same time. Performing prior proliferation assays is recommended to optimize this step.
-
3.
Prepare IP buffer. Keep on ice.
-
4.For each sample: Wash 100 μL of Pan mouse IgG magnetic beads 3 times with 500 μL IP buffer each time:
-
a.Take 100 μL slurry of beads into a 1.5 mL tube.
-
b.Remove the supernatant using the magnetic stand.
-
c.Add 500 μL ice-cold IP buffer.
-
d.Mix by constantly inverting the tube for 1 min.
-
e.Remove IP buffer using the magnetic stand.
-
f.Repeat steps c-e twice.
-
a.
-
5.
Resuspend the beads in 500 μL ice-cold IP buffer.
-
6.
Add 3 μL (3 μg) of your favorite mouse monoclonal primary antibody.
-
7.
Incubate overnight at 4°C rotating.
Cell harvesting, lysis, and immunoprecipitation (part 1)—Day 3
Timing: 3–4 h
Goal: To obtain sufficient material for subsequent RNA isolation and immunoprecipitation.
-
8.
Prepare complete IP buffer. Keep on ice.
Note: If assessing cotranslational interactions using EDTA (Figure 2), also prepare complete IP buffer containing 25 mM EDTA (from a 0.5 M pH 8.0 stock in distilled water; filter sterilized). Always use this buffer (complete IP buffer+EDTA) instead of complete IP buffer for EDTA-treated samples during the rest of the protocol. For a more rigorous assessment of the cotranslational nature of protein-RNA interaction, we recommend using either puromycin or an ATG mutant version of a known target gene, if possible.
-
9.
Count and annotate the concentration of the cultures. It should be about 1.5 × 107 cells/mL.
-
10.
Transfer 50 mL of culture to a 50 mL tube.
Note: Adjust small differences in the required volume for each strain. Use the less concentrated culture at first and adjust the other cultures accordingly. To minimize variability and/or allow comparisons, it is recommended to use a similar number of cells for each sample and subsequent replications of the experiment.
Optional: When using puromycin (Figure 2), transfer 2 × 50 mL of culture to two different flasks. Then add 1 mL of puromycin hydrochloride stock solution (50 mg/mL in sterile distilled water) to one of them and 1 mL of sterile distilled water to the other one. Incubate for 15 minutes at 32°C before harvesting them as indicated in step 10.
-
11.
Centrifuge for 3 min at 2,500 g, 4°C. Discard supernatant.
-
12.
Wash: add 5 mL cold (4°C) distilled water, resuspend and centrifuge for 3 min at 2,500 g, 4°C.
-
13.
Remove supernatant, resuspend into 500 μL of ice-cold complete IP buffer and transfer to 2 mL screw-cap tubes.
-
14.
Centrifuge 1 min at 2,500 g at 4°C. Discard supernatant. Keep pellets on ice.
-
15.
Add 100 μL of ice-cold complete IP buffer and 10 μL RNasin and resuspend by pipetting.
-
16.
Add acid washed glass beads (stored at 4°C) up to the meniscus of your liquid sample.
-
17.
Lyse cells in a fast prep (MP Biomedicals) set at 6.5 power setting, for 3 cycles consisting of 40 s ON and 3 min OFF. Chill the tube holder using grinded dry ice.
Note: Cell lysis can be performed differently depending on strain background requirements, equipment, or yeast model. At least 90% of lysed cells are recommended to proceed with the protocol. See troubleshooting problem 1 for further details.
-
18.
Add 500 μL of ice-cold complete IP buffer and vortex.
-
19.
Heat up a needle under the Bunsen lighter (Figure 3A) and poke a hole within the cap and the bottom of the tube (Figure 3B). Place it immediately into a 5 mL tube, on ice (Figure 3C).
-
20.
Centrifuge for 1 min at 300 g at 4°C (Figure 3C).
Note: For centrifugation in step 20, use 15–50 mL tube centrifuge buckets to accommodate the 2 mL screw-cap tubes inserted into the 5 mL tubes.
-
21.
Transfer each sample (supernatant and any pellet formed) to a 1.5 mL tube (Figure 3C).
-
22.
Centrifuge 5 min at 7,500 g at 4°C.
-
23.
Transfer supernatant into a new 1.5 mL tube. Discard the pellet.
-
24.
Centrifuge 5 min at 13,300 g at 4°C.
-
25.
Transfer supernatant into a new 1.5 mL tube. Keep on ice. Discard the pellet.
Optional: If performing puromycin treatment, add 12 μL of puromycin stock solution (50 mg/mL) and 6 μL of GTP stock solution (200 mM) to puromycin treated samples. Mix and incubate 15 minutes on ice before proceeding to step 26.
CRITICAL: Save 10 μL for Western blot (input). Add 10 μL of 2× Laemmli buffer (0.15 M Tris-HCl pH 6.8, 4% Sodium dodecyl sulfate (SDS), 20% glycerol, 0.02% bromophenol blue).
Note: If the protein bait contains a N-terminal tag or a long C-terminal tag (such as HA3-TAP2 and you use anti-HA antibodies for IP), you will be able to check if your IP worked by testing binding of the bait to its own mRNA. In these cases, Western blot is optional (see second “Note” after step 88).
-
26.
Measure protein concentration using the Bradford method.
Note: This protocol was optimized using 4 mg of total protein for RNA immunoprecipitation and 400 μg of total protein for total RNA extraction (10% of the amount used for IP). Thus, you will need at least 4.4 mg of total protein for each sample to continue the experiment. See troubleshooting problem 1 for further details.
CRITICAL: It is essential to use the same amount of total protein for IP and total RNA extractions between samples. If one or more of the samples contain less than 4.4 mg of total protein but nevertheless decide to proceed with the procedure, adjust the concentration of the other samples accordingly.
-
27.For each sample, wash 100 μL of antibody-coated Pan mouse IgG magnetic beads 3 times with 500 μL ice-cold IP buffer:
-
a.Transfer 100 μL of beads to a 1.5 mL tube.
-
b.Add 500 μL of ice-cold IP buffer.
-
c.Constantly invert the tube for 1 min at room temperature.
-
d.Remove supernatant using a magnetic stand.
-
e.Repeat b-d twice.
-
a.
-
28.
Add 4 mg of total protein extract to the beads.
-
29.
If needed, fill the volume up to 500 μL using ice-cold complete IP buffer.
-
30.
Add 5 μL RNasin (40 U/μL) and 5 μL PMSF (100 mM).
-
31.
Incubate 2 h at 4°C, rotating.
Note: During step 31, you can proceed with the “Total RNA extraction” step.
Figure 3.
Scheme of steps 19–21 for sample recovery after cell lysis
(A) Heat up a needle (G25) under the Bunsen burner until incandescence.
(B) Using the ablaze needle, make a hole in the cap of 2 mL screw-cap tube containing the sample. Then repeat the process to make a hole in the bottom of the tube.
(C) Immediately place the 2 mL screw-cap tube into a 5 mL tube on ice. Centrifuge for 1 min at 300 g and 4°C to recover the sample. Put back the 5 mL tube on ice and remove the 2 mL screw-cap tube containing glass beads. Transfer the supernatant into a 1.5 mL tube and continue with step 22. Original images were obtained and modified with permission from smart.servier.com.
Total RNA extraction—Day 3
Timing: 1 h
Note: If using other RNA extraction kits, please refer to manufacturer’s instructions.
Goal: To recover total RNA that will be used as input in the final qPCR assay.
Note: If instead of qPCR, a transcriptome analysis is performed, such as microarrays or high-throughput sequencing, please see the “limitations” section below and supplemental information.
-
32.
Prepare lysis buffer (provided in the kit) by addition of β-mercaptoethanol (not provided in the kit). Add 10 μL of β-mercaptoethanol 13.4 M per 1 mL of lysis buffer.
-
33.
Take the appropriate volume of sample to get 400 μg of total protein extract and complete with complete IP buffer up to 50 μL final volume.
-
34.
Add 250 μL of lysis buffer containing β-mercaptoethanol.
-
35.
Add 250 μL of ethanol 100%.
-
36.
Load onto a column.
-
37.
Centrifuge 30 s at 16,100 g.
-
38.
Discard flow through.
-
39.
Add 700 μL of wash buffer I.
-
40.
Centrifuge 30 s at 16,100 g.
-
41.
Discard flow through.
-
42.
Add 500 μL of wash buffer II.
-
43.
Centrifuge 30 s at 16,100 g.
-
44.
Discard flow through.
-
45.
Add 500 μL of wash buffer II.
-
46.
Centrifuge 2 min at 16,100 g.
-
47.
Transfer the column to a new tube.
-
48.
Centrifuge for 1 min at 16,100 g.
-
49.
Transfer the column to a collection tube.
-
50.
Add 30 μL of RNase-free water.
-
51.
Incubate for 1 min at 20°C–30°C.
-
52.
Centrifuge for 1 min at 16,100 g.
-
53.
Reload the eluate into the column.
-
54.
Incubate for 1 min at 20°C–30°C.
-
55.
Centrifuge for 1 min at 16,100 g.
-
56.
Measure concentration.
-
57.
Keep the samples on ice until proceeding with DNase treatment and reverse transcription.
Pause point: Total RNA samples can be stored at −80°C before proceeding with DNase treatment and reverse transcription.
Immunoprecipitation (part 2-washes)—Day 3
Timing: 2–3 h
Goal: To immunoprecipitate the bait and all associated RNAs.
Note: Per sample, you will need 3,033 μL W1 buffer [To 3,000 μL of W buffer, add 3 μL heparin (50 mg/mL) and 30 μL PMSF (100 mM)], 1501.5 μL W2 buffer [To 1,500 μL of W buffer, add 1.5 μL heparin (50 mg/mL)] and 1,500 μL W buffer.
Note: To prepare sequencing libraries from the IP RNA do not add heparin to the washing buffers since it can impact downstream steps during library preparation. We use heparin in washing buffers as a cheaper alternative to commercial RNase inhibitors and we have systematically and successfully used it when performing RIP followed by qPCR (Toullec et al., 2021).
-
58.
After the 2 h incubation at 4°C (step 31), place the IP on a magnetic stand and remove the supernatant.
CRITICAL: Keep 10 μL for Western blot (flow through). Add 10 μL of 2× Laemmli buffer.
Note: Remember that Western blot is optional if the bait contains N-terminal or long C-terminal tags (See second “Note” after step 88).
Note: For washing the beads (steps 59–61), please follow these instructions. Each wash step involves addition of buffer, either long or short incubation, buffer removal. Short washes are performed by constant inversion of the tubes for 1 minute at room temperature. Long washes are performed by rotating the tubes for 10 minutes at 4°C.
-
59.
Wash 4 times with 750 μL of W1. 2 short washes, followed by 1 long wash and 1 short wash.
-
60.
Wash 2 times with 750 μL of W2. Long washes.
-
61.
Wash 2 times with 750 μL of W buffer. 1 long wash, then 1 short wash.
-
62.
Briefly spin down and remove the supernatant.
-
63.
Resuspend the beads in 110 μL W buffer.
CRITICAL: Take 10 μL (from the 110 μL in step 63) for Western blot, to which add 10 μL of 2× Laemmli buffer, heat up at 90°C for 5 minutes and centrifuge 5 minutes at 16,100 g. Store supernatant at −20°C (IP sample) or immediately proceed with SDS-PAGE and Western blotting. When possible, we recommend probing the blot with a different antibody from the one used for the IP, or TrueBlot secondary antibodies, to avoid detection of heavy and light chains.
Note: As stated above, Western blot could be optional if the bait contains a N-terminal or a long C-terminal tag (see second “Note” after step 88). In such case, if Western blot is not performed, resuspend the beads of step 63 in 100 μL of W buffer.
Note: If using the PureLinkTM RNA Micro Kit for the following steps (see next heading), resuspend in 55 μL W buffer in step 63 (or 50 μL if Western blot is not performed) and use 5 μL for Western blot, to which add 5 μL of 2× Laemmli buffer, heat up at 90°C for 5 minutes and centrifuge 5 minutes at 16,100 g. Store supernatant at −20°C (IP sample) or immediately proceed with SDS-PAGE and Western blot.
Isolation of immunoprecipitated RNA—Day 3
Timing: 30 minto1 h
Goal: To recover RNAs bound to the bait.
Note: We have successfully used both the PureLinkTM RNA Mini and Micro kits. If using other RNA extraction kits, please refer to manufacturer instructions. The following instructions (from step 64) are for the Mini kit. If using the Micro kit, adjust the volumes as follows. In step 63, resuspend in 55 μL W buffer (save 5 μL for Western blot). In step 79, use 15 μL for elution. In step 86.a, take 13 μL of the IP RNA (adjust to 13 μL with RNase-free water if needed). In step 86.a.i, add 2 μL of 10× reaction buffer and 4 μL of RNase-free water. In step 86.a.vii, transfer 15 μL of the supernatant to a PCR tube. In step 86.b.i, add 1.25 μL of 10 mM dNTPs. In step 86.b.ii, add 0.21 μL of random primers and 0.54 μL of RNase-free water. In step 86.b.vi, add 5 μL of 5× superscript buffer, 1 μL of DTT (0.1 M), 1 μL of RNasin and 1 μL of Superscript III.
-
64.
To the 100 μL beads-sample from step 63, add a mix containing 350 μL of lysis buffer (provided in the kit), 3.5 μL of β-mercaptoethanol (13.4 M) and 5 μL of diluted carrier RNA. Mix well.
CRITICAL: When diluting the carrier RNA, aliquot it such as to avoid more than 2 thaw-freeze cycles.
-
65.
Incubate at room temperature for 20 min, rotating.
-
66.
Remove beads with a magnet and transfer supernatant to a new tube.
-
67.
Add 350 μL of ethanol 100%. Mix well.
-
68.
Load sample into a column and centrifuge 30 s at 16,100 g.
Note: Use a maximum of 700 μL of sample from step 68. If needed, re-load the column with the rest of the sample.
-
69.
Add 700 μL of wash buffer 1.
-
70.
Centrifuge 30 s at 16,100 g.
-
71.
Discard flow through.
-
72.
Add 500 μL of wash buffer 2.
-
73.
Centrifuge 30 s at 16,100 g.
-
74.
Discard flow through.
-
75.
Repeat steps 72 and 73.
-
76.
Transfer the column to a new tube.
-
77.
Spin for 1 min at 16,100 g.
-
78.
Transfer the column into a collection tube.
-
79.
Add 40 μL of RNase-free water.
-
80.
Incubate 1 min at room temperature.
-
81.
Centrifuge 1 min at 16,100 g.
-
82.
Reload the eluate into the column.
-
83.
Incubate 1 min at room temperature.
-
84.
Centrifuge 1 min at 16,100 g.
Pause point: Immunoprecipitated RNAs can be now stored at −80°C until used for DNase treatment and reverse transcription.
DNase treatment and reverse transcription—Day 3 or day 4
Timing: 2–3 h
-
85.For total RNA samples:
-
a.Take 10 μg of total RNA (input) from step 57 for DNase treatment adjusted to a final volume of 26 μL with water:
-
i.Add 3 μL of 10× reaction buffer.
-
ii.Add 1 μL Turbo DNase.
-
iii.Incubate 30 min at 37°C.
-
iv.Add 3 μL of nuclease inhibitor reagent.
-
v.Incubate for 2 min at room temperature flicking the tubes constantly.Note: If room temperature is below 22°C, use a heat block to control the temperature; otherwise, DNase inactivation might be incomplete.
-
vi.Centrifuge 2 min at 16,100 g.
-
vii.Transfer 15 μL of the supernatant to a new tube.
 CRITICAL: Do not transfer any of the pellet (containing DNase inhibitor and DNase) to the new tube.
CRITICAL: Do not transfer any of the pellet (containing DNase inhibitor and DNase) to the new tube.
-
i.
-
b.Transfer 3 μL (1 μg) of the DNase-treated total RNA for reverse transcription into a PCR tube:Note: Use another 3 μL (1 μg) of the DNase-treated total RNA samples to be used as control for genomic DNA contamination in the qPCR (see next heading). Proceed these samples exactly the same way in the reverse transcription reaction, but without adding reverse transcriptase in step 85.b.vii).
-
i.Add 8.83 μL of RNase-free water.
-
ii.Add 0.17 μL of random primers.
-
iii.Add 1 μL of 10 mM dNTPs.
-
iv.Pre-set the following program in the thermocycler:65°C/5 min.4°C/90 s.50°C/60 min.4°C/infinite hold.
-
v.Put the tubes in the thermocycler, start the program and let it run for the 65°C/5 min step and just 30 s from the 4°C/90 s step. Pause the thermocycler and proceed with the following steps.
-
vi.Take out your PCR tubes and keep them on ice while adding and mixing the components indicated in the next step.
-
vii.Per sample, add a mix containing: 4 μL of 5× superscript buffer, 1 μL of DTT (0.1 M), 1 μL of RNasin and 1 μL of Superscript III.
-
viii.Mix by pipetting.
-
ix.Put back the tubes in the thermocycler and re-start the program from the 4°C/90 s step (of which there should be 60 s left).
-
i.
-
a.
-
86.For immunoprecipitated (IP) RNA samples:
-
a.Take 35 μL of IP-RNA from step 84:
-
i.Add 4 μL of 10× reaction buffer.
-
ii.Add 1 μL of Turbo DNase.
-
iii.Incubate 30 min at 37°C.
-
iv.Add 3 μL of nuclease inhibitor reagent.
-
v.Incubate for 2 min at room temperature flicking the tubes constantly.
-
vi.Centrifuge 2 min at 16,100 g.
-
vii.Transfer 33.5 μL of the supernatant to a PCR tube.
-
i.
-
b.Use all your DNase-treated IP-RNA for reverse transcription:
-
i.Add 3 μL of 10 mM dNTPs.
-
ii.Add 0.5 μL of random primers.
-
iii.Pre-set the following program in the thermocycler:65°C/5 min.4°C/90 s.50°C/60 s.4°C/infinite hold.
-
iv.Put the tubes in the thermocycler, start the program and let it run for the 65°C/5 min step and just 30 s from the 4°C/90 s step. Pause the thermocycler and proceed with the following steps.
-
v.Take out your PCR tubes and keep them on ice while adding and mixing the components indicated in the next step.
-
vi.Per sample, add a mix containing: 10 μL of 5× superscript buffer, 1 μL of DTT (0.1 M), 1 μL of RNasin and 1 μL of Superscript III.
-
vii.Mix by pipetting.
-
viii.Put back the tubes in the thermocycler and re-start the program from the 4°C/90 s step (of which there should be 60 s left).
 Pause point: The resulting cDNA samples can be now stored at −20°C until proceeding with qPCR, microarray or high-throughput sequencing.
Pause point: The resulting cDNA samples can be now stored at −20°C until proceeding with qPCR, microarray or high-throughput sequencing.
-
i.
-
a.
Quantification of RNA binding to the bait by qPCR—Day 4 or day 5
Binding levels of the protein bait to the RNAs of interest are quantified using qPCR. The relative binding frequency is estimated by calculating the fraction of input cDNA recovered in the IP.
Note: This protocol was designed to test for interaction between a specific bait and candidate RNAs. For an unbiased screen, use a RIP-chip or RIP-seq protocol, in which cDNAs are analyzed either through microarray hybridization or high throughput sequencing, respectively (see Costello et al., 2015). Here, we also provide a quick guide on how to proceed with a global approach in the “limitations” section below. Regardless, whenever possible, we recommend performing an initial check by qPCR using a positive control RNA.
Note: For qPCR, we typically use a 50-fold dilution of total cDNA and a 3-fold dilution of IP cDNA samples, but higher dilutions of IP cDNA up to 10-fold work equally well. We used 10× home-made SYBR Green Master Mix (see materials and equipment), 96 well plates and Stratagene MX3000P and MX3005P qPCR machines. Our amplicons are typically below 200 bp in size. Optimization might be required when using other dilutions, qPCR reagents, machine or amplicon length.
-
87.
For each primer pair, prepare dilutions of one of the input cDNAs in sterile distilled water to make a standard curve and calculate primer pair efficiency.
Note: To prepare the standards, we use input cDNA from a non-tagged strain which has grown and has been processed in standard conditions. For an initial standard of a first-time used pair of priming oligonucleotides, we recommend making 4, 20, 100, 500, 2,500 and 12,500-fold dilutions of one of the input cDNAs. Once the primers are validated, we reduce the number of dilutions for the standard down at least three.
Alternatives: We strongly recommend using the same type of material for the standard as for the samples that are analyzed. However, if this is not possible, dilutions of genomic DNA can be used to prepare the standard for all primer pairs. 5, 0.5, 0.05, 0.005 and 0.0005 ng/μL concentrated genomic DNA samples could be used in a first try to prepare the standards. Always use clean, high-quality genomic DNA preparations.
Note: We include a standard curve for each primer pair in the same plate than the samples.
-
88.
Prepare a master mix for each primer pair that you want to test in your qPCR experiment, by mixing the reagents indicated in the table below.
Note: Primers specific for a negative control should be included. For example, a gene which has no functional relationship with the bait and does not interact with it. If there is no previous information, include several potential negative controls for your first assay.
Note: As a positive control, primers targeting the mRNA of the bait itself can be used. We have successfully used this type of control for proteins with N-terminal epitope tag used for immunoprecipitation, as well as with long C-terminal tags, such as HA3-TAP2 using an HA antibody for IP. Beware that if using a short C-terminal tag in the bait, this positive control might not work because the native polypeptide quickly detaches from polysomes, hence its own mRNA, upon synthesis of the C-terminal epitope tag. If using an antibody against an endogenous protein, the success of this strategy depends on where the epitope is in the sequence.
Note: To calculate the volume needed for each primer pair master mix consider the number of samples you have, 3 technical replicates and the standard (also performed in triplicates).
PCR reaction master mix
| Reagent | Amount per well | Amount per sample (3 technical replicates) |
|---|---|---|
| 10× SYBR Green PCR Master Mix | 2.5 μL | 7.5 μL |
| Primer 1 (10 μM) | 0.5 μL | 1.5 μL |
| Primer 2 (10 μM) | 0.5 μL | 1.5 μL |
| ddH2O | 16.5 μL | 49.5 μL |
-
89.
Prepare a 50-fold dilution of the input cDNAs and a 3-fold dilution of your IP cDNA into sterile distilled water.
-
90.
Prepare an equivalent dilution of the control DNase-treated total RNA samples in which reverse transcriptase was not added during the reverse transcription reaction (see Note in step 85.b) to be used as control for genomic DNA contamination. We will name these samples No-RT samples.
-
91.
Distribute 20 μL of the master mix prepared in step 88 per well. Mix well before dispensing.
-
92.
Distribute 5 μL of the diluted cDNA or No-RT samples in each of the corresponding wells. Mix well before dispensing.
-
93.
Cover the PCR plate with compatible tube caps or a transparent film as required by the qPCR machine instrument and mix by hand.
-
94.
Briefly spin down the qPCR plate in a centrifuge at 200 g.
-
95.
Run the plate into the qPCR machine using the appropriate program according to the primer pairs used and machine instructions. We used the program below for our assays described in Toullec et al. (2021):
PCR cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Initial Denaturation | 95°C | 10 min | 1 |
| Denaturation | 95°C | 30 s | 40 cycles |
| Annealing | 60°C | 1 min | |
| Extension | 72°C | 1 min | |
| Melting curve | 70°C to 95°C, increment 0.5°C, measure fluorescence. | ||
| Hold | 12°C | forever | |
Note: It is possible to keep the plate at 4°C in the dark. We have had successful runs after up to 2 hours.
Note: A melting curve at the end of the program is important to obtain the dissociation curve of the PCR product. Only a single specific product should form.
-
96.
Calculate the efficiency of the primers. Most qPCR machine software do this automatically. The efficiency should be between 90%–110% (−3.6 ≥ slope ≥ −3.1) to proceed with quantification.
Note: To manually calculate the efficiency of your primers, first calculate the equation of their standard curve to obtain the slope. The slope is derived from a graph that plots the cycle threshold (Ct) values against the Log10 of a given amount of DNA template. Efficiency (%) = [10ˆ(-1/slope) – 1] × 100. A slope of -3.32 indicates an amplification efficiency of 100%.
-
97.
After exporting the data in the selected format, calculate the starting quantities by relying on the standard curve of each primer pair. Most qPCR machine software do this automatically and allow to directly export these data.
-
98.
Calculate the mean of the starting quantities of each sample by averaging the three (or more) technical replicates.
-
99.
Correct each input and IP value for the corresponding dilutions and calculate a “percent IP” value, which is a proxy of the RNA IP efficiency relative to the amount of RNA in the input and calculated by dividing each IP value by its corresponding input value (see tables below).
Input and IP quantities or volumes at each step
| Step | Input | IP | IN/IP |
|---|---|---|---|
| Starting material for RNA extraction | 400 μg protein extract | 4,000 μg protein extract | 0.1 |
| RNA extraction final volume | 33 μL | 40 μL | 1.212 |
| DNase | 10 μg (max. 86.67% of total volume of RNA sample) | 87.5% of total volume of RNA samples | TBD (max. 0.991) |
| RT | 1 μg = 3 μL of DNase-treated RNA) | 33.5 μL (95,71% of DNase) | 0.105 |
| qPCR | 50-fold dilution of cDNA | 3-fold dilution of cDNA | 0.06 |
| plate | 5 μL/25 μL total, per well | 5 μL/25 μL total, per well | 1 |
| Total | – | – | 7.6 × 10-4 |
Input and IP dilution factors
| Input dilution factor relative to IP |
Input and IP individual dilution factors at each step |
|||
|---|---|---|---|---|
| Step | Input | IP | Input | IP |
| RNA extraction starting material | ×0.1 | 1 | ×0.1 | ×1 |
| RNA extraction final volume | ×1.212 | 1 | ×1.212 | ×1 |
| DNase | TBD, maximum is ×0.991 (max = 26 μL of RNA samples = 86.67% of total volume) | 1 (87.5% of total volume of RNA sample) | Max × 0.867 | ×0.875 |
| RT | ×0.105 (10% of DNase; 3 μL) | 1 (95.71% of DNase) | ×0.1 | ×0.957 |
| qPCR | ×0.06 (50-fold) | 1 (3-fold) | ×0.02 | ×0.33 |
| plate | 1 (5 μL/25 μL total, per well) | 1 (5 μL/25 μL total, per well) | ×1 | ×1 |
| “Total dilution factor” | ×0.00076 (7.6 × 10-4) | 1 | ×0.00021 | ×0.276 |
| IN/IP relative dilution factor∗ | ×0.00076 (7.6 × 10-4) | ×0.00076052219 (7.6 × 10-4) | ||
For each IP, we use 0.00076-times less input (0.076%). Thus, the IP/IN ratio must be multiplied by 0.00076 ([IP value] × 1)/([input value] × 7.6 × 10-4)]
Expected outcomes
A successful RIP-qPCR should give a robust and reproducible enrichment of the targeted RNA over background. “Percent IP” values can be, however, variable and close to background. Therefore, including negative and positive controls and a sufficient number of biological replicates is essential to carefully interpret these values. The level of enrichment will depend on several parameters:
-
•
The frequency of bait binding to its target RNAs (dynamics). In other words, the percentage of cells in the population where the protein bait is bound to a specific RNA at a given time. Each specific bait will have a different binding frequency to its target RNAs. Baits that share RNA targets might, therefore, give rise to different enrichment results by RIP-qPCR.
-
•
The epitope used. IP efficiency depends, among other parameters, on the strength of the epitope-antibody interaction. Thus, a RIP-qPCR assay using the same bait but tagged with different epitopes could result in significant differences in the final enrichment results.
-
•
The nature of the bait. Depending on whether using an RNA binding protein or a protein that indirectly binds RNA through intermediate factors, the dynamics and efficiency of the RIP-qPCR could change.
Comparing “percent IP” values between test and negative control samples, we consider that enrichment below 5-fold correspond to background signal, based on the background levels that we have typically observed in RIP-qPCR using an unrelated tagged protein as bait (Toullec et al., 2021).
The identification of co-translational RNA-protein interactions can be used to predict protein-protein interactions and to identify new components of protein complexes (Duncan and Mata, 2014). Nonetheless, several aspects limit the use of this protocol with this aim (see below).
Quantification and statistical analysis
Statistical analysis of RIP-qPCR data is performed using either a t-test when comparing two means or analysis of variance (ANOVA) when comparing more than two means, across one (one-way ANOVA) or two and more variables (two-way ANOVA and up). Post-hoc tests are used for pairwise comparison. In this protocol, we typically used a significance level of 0.01 and a minimum of four biological replicates, which correspond to four independent experiments. We recommend representing quantitative percent IP values as individual data points overlaid with the mean and the standard deviation.
Limitations
RIP followed by qPCR, microarray hybridization or high-throughput sequencing has become a standard method for studying protein-RNA interactions in different organisms (Jayaseelan et al., 2014; Gagliardi and Matarazzo, 2016; Kachaev et al., 2017; Bierhoff, 2018; Seo and Chua, 2019). Nonetheless, RIP has the following limitations.
Because the RIP-qPCR protocol presented here does not rely on crosslinking to stabilize protein-RNA interactions, its success will depend on the strength and dynamics of the interaction. We implemented this protocol with the goal of detecting cotranslational interactions between interacting proteins (Toullec et al., 2021), which are transient and engage only a small fraction of the bait present in the cell. RIP-qPCR is therefore a sensitive method but we do not know what range of affinities is detectable with this methodology. Indeed, we do not have affinity measurements for such cotranslational interactions, which are challenging to reconstitute in vitro. Other direct or indirect interactions, weak and/or transient, may be missed by this method and might therefore require some form of stabilization to be captured, such as crosslinking.
The use of puromycin or EDTA makes this protocol suitable for studying co-translational protein-RNA interactions (Figure 2). However, it does not allow to distinguish between direct binding of the protein bait to RNA and indirect binding, for example, through an RNA-binding protein (Figure 1).
Although the identification of co-translational protein-RNA binding can predict protein-protein interactions, biochemical validation is needed using, for instance, co-immunoprecipitation or colocalization of the proteins of interest (Duncan and Mata, 2014). Also, because the interactions revealed by RIP might not be direct (for example, a preformed protein complex might interact with a nascent peptide), complementing RIP with mass spectrometry or imaging-based techniques is crucial when studying new partners of a protein bait, new components of a protein complex, or protein complex assembly mechanisms, as shown in (Duncan and Mata, 2014; Kassem et al., 2017; Kamenova et al., 2019; Elías-Villalobos et al., 2019; Toullec et al., 2021; Lautier et al., 2021).
This protocol has been used with epitope-tagged proteins only, for which highly efficient antibodies are commercially available. Using specific antibodies directed against the protein of interest or home-made antibodies might impact the quality of the results and require optimization.
Strong RNA-binding proteins frequently co-elute with non-specific RNAs during purifications. Individual qPCRs can quickly become cumbersome to verify the specificity of protein-RNA interactions, unless using several control mRNAs clearly indicate that only a specific type of mRNA binds to the bait. Alternatively, a genome-wide analysis will be required to estimate how many RNAs bind to the protein of interest and address this issue. The several controls proposed along this protocol, including a non-tagged strain, a strain tagged in an unrelated bait and including in the qPCR analysis mRNAs to which the bait should not bind, would also help determining the specificity of the detected interactions and the requirement or not of alternative approaches.
Considerations for genome-wide analyses of RIP: In its present form, this protocol is not directly suitable for high-throughput analyses since it is conceived to check for the binding capacity of a bait to specific RNAs of interest. To identify unknown RNA binding targets of an RBP there are currently two genome-wide derivatives of this protocol, RIP-chip and RIP-seq. RIP-chip requires specialized equipment and carefully designed microarrays. Due to these limitations, RIP-seq has become the dominant technique. While the objective of this protocol is not to describe either technique in detail, we describe in a supplemental file the main modifications and considerations for combining this protocol with established high-throughput sequencing pipelines (Methods S1).
Troubleshooting
Problem 1
Low protein yield (step 26).
Potential solution
Low protein yield is typically caused by inefficient cell lysis (see problem 2). Alternatively, it can be caused by protein degradation during sample processing. Make sure that the samples remain at low temperature, below 4°C, throughout the procedure, by prechilling all tubes and buffers before use and by adding protease inhibitors at the indicated steps. Note that when using media lacking nutrients, for example for quiescent cells, protein and RNA yields will drop (see problem 5).
Problem 2
Inefficient cell lysis (step 17).
Potential solution
In S. pombe and other yeasts, cell lysis can be monitored by direct observation under a light microscope. It is important to verify the integrity of yeast cells before and after bead-beating by monitoring cells under a light microscope, using 5 μL of the extract spread on a glass slide. Efficient cell lysis corresponds to about 90% of lysed cells, which appear as black and refringent. Alternatively, if there are only cellular debris, estimate the number of intact cells before and after lysis. Lysis efficiency should reach close to 90%. Otherwise, repeat the bead-beating procedure until reaching this threshold. In this case, to avoid heating of the samples and protein degradation, place samples on ice between each lysis cycle. Alternative lysis methods, such as cryogrinding can be used, but larger culture volumes are typically required to end up with sufficient material for subsequent steps and to avoid variability between samples.
Problem 3
Low RNA yield (steps 57 and 84).
Potential solution
RNA degradation is frequently caused by inappropriate manipulation of RNA-containing samples or absence of RNA inhibitors. This can be minimized by wearing gloves when manipulating your samples, using nuclease-free filter-tips and making sure that a sufficient amount of RNase inhibitors is added at the indicated steps. Using media lacking nutrients can impact protein and RNA yields - see problem 5.
Problem 4
High variability in RIP signal (step 99).
Potential solution
This RIP protocol involves many steps and differences in any of them can lead to significant changes in RIP signal. It is thus crucial to be precise and rigorous to avoid variations across samples and experiments.
Protein and RNA degradation is another frequent source of variability (see problems 1, 3, and 5). Therefore, it is important to make sure that the samples remain at low temperature throughout the procedure, to prechill all tubes and buffers before use and to add protease and RNase inhibitors at the indicated steps.
Problem 5
Low protein or RNA yields when using media lacking nutrients (steps 1, 26, 57, and 84).
Potential solution
The physiological state of the yeast culture can significantly impact the synthesis and stability of RNAs and proteins. For example, nutrient starvation is typically accompanied by an increased expression and activation of proteases and nucleases as well as global changes in gene expression (Nakashima et al., 2002, 2006; Kohda et al., 2007; Marguerat et al., 2012). In this case, Western blotting and RT-qPCR should be used to control for variations in the levels of the protein bait and the RNA targets, respectively. In our experience, with S. pombe, protein degradation becomes detectable following a 60 min shift to starvation medium. In this case, adding higher amounts of protease and RNase inhibitors can help. In addition, scaling up the amount of starting material by increasing the number of cells grown can also help. Finally, using the RNA micro kit instead of the RNA mini kit allows increasing the concentration of the RNA samples (see first “Note” under the heading “isolation of immunoprecipitated RNA – day 3”).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dominique Helmlinger (dhelmlinger@crbm.cnrs.fr).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank all members of the laboratory for helpful discussions. A.E.-V. was a recipient of a postdoctoral fellowship from the Fondation pour la Recherche Médicale (SPF20130526854) at the CRBM and of a Maria Zambrano grant, funded by the European Union – NextGenerationEU, at the Universidad de Sevilla. J.M. and C.D. were supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC BB/G011869/1 and BB/S015833/1). This work was supported by funds from the Agence Nationale de la Recherche (ANR-15-CE12-0009-01 and ANR-15-CE11-0022-03) and the Fondation ARC (PJA 20181208277) to D.H.
Author contributions
Conceptualization: A.E.-V and C.D.; investigation: A.E.-V. and C.D.; writing – original draft, A.E.-V.; writing – review and editing, A.E.-V., C.D., J.M., and D.H.; funding acquisition, J.M. and D.H.; supervision, J.M. and D.H.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101373.
Contributor Information
Alberto Elías-Villalobos, Email: aelias1@us.es.
Dominique Helmlinger, Email: dhelmlinger@crbm.cnrs.fr.
Supplemental information
Data and code availability
This study did not generate/analyze datasets.
References
- Aviner R. The science of puromycin: from studies of ribosome function to applications in biotechnology. Comput. Struct. Biotechnol. J. 2020;18:1074–1083. doi: 10.1016/j.csbj.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H. Analysis of lncRNA-protein interactions by RNA-protein pull-down assays and RNA immunoprecipitation (RIP) Methods Mol. Biol. 2018;1686:241–250. doi: 10.1007/978-1-4939-7371-2_17. [DOI] [PubMed] [Google Scholar]
- Costello J., Castelli L.M., Rowe W., Kershaw C.J., Talavera D., Mohammad-Qureshi S.S., Sims P.F., Grant C.M., Pavitt G.D., Hubbard S.J., Ashe M.P. Global mRNA selection mechanisms for translation initiation. Genome Biol. 2015;16:10. doi: 10.1186/s13059-014-0559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C.D., Mata J. Widespread cotranslational formation of protein complexes. PLoS Genet. 2011;7:e1002398. doi: 10.1371/journal.pgen.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C.D., Mata J. Cotranslational protein-RNA associations predict protein-protein interactions. BMC Genom. 2014;15:298. doi: 10.1186/1471-2164-15-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elías-Villalobos A., Toullec D., Faux C., Séveno M., Helmlinger D. Chaperone-mediated ordered assembly of the SAGA and NuA4 transcription coactivator complexes in yeast. Nat. Commun. 2019;10:5237. doi: 10.1038/s41467-019-13243-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi M., Matarazzo M.R. RIP: RNA immunoprecipitation. Methods Mol. Biol. 2016;1480:73–86. doi: 10.1007/978-1-4939-6380-5_7. [DOI] [PubMed] [Google Scholar]
- Gerber A.P., Herschlag D., Brown P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaseelan S., Doyle F., Tenenbaum S.A. Profiling post-transcriptionally networked mRNA subsets using RIP-Chip and RIP-seq. Methods. 2014;67:13–19. doi: 10.1016/j.ymeth.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachaev Z.M., Gilmutdinov R.A., Kopytova D.V., Zheludkevich A.A., Shidlovskii Y.V., Kurbidaeva A.S. [RNA immunoprecipitation technique for Drosophila melanogaster S2 cells] Mol. Biol. 2017;51:85–93. doi: 10.1134/s002689331606008x. [DOI] [PubMed] [Google Scholar]
- Kamenova I., Mukherjee P., Conic S., Mueller F., El-Saafin F., Bardot P., Garnier J.M., Dembele D., Capponi S., Timmers H.T.M., et al. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 2019;10:1740. doi: 10.1038/s41467-019-09749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem S., Villanyi Z., Collart M.A. Not5-dependent co-translational assembly of Ada2 and Spt20 is essential for functional integrity of SAGA. Nucleic Acids Res. 2017;45:1186–1199. doi: 10.1093/nar/gkw1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda T.A., Tanaka K., Konomi M., Sato M., Osumi M., Yamamoto M. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells. 2007;12:155–170. doi: 10.1111/j.1365-2443.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- Lautier O., Penzo A., Rouvière J.O., Chevreux G., Collet L., Loïodice I., Taddei A., Devaux F., Collart M.A., Palancade B. Co-translational assembly and localized translation of nucleoporins in nuclear pore complex biogenesis. Mol. Cell. 2021;81:2417–2427. doi: 10.1016/j.molcel.2021.03.030. [DOI] [PubMed] [Google Scholar]
- Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., Bähler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Yoshida M., Nakayama K., Kato-Furuno A., Ueno M., Ushimaru T., Uritani M. Genes for a nuclease and a protease are involved in the drastic decrease in cellular RNA amount in fission yeast cells during nitrogen starvation. J. Biochem. 2002;131:391–398. doi: 10.1093/oxfordjournals.jbchem.a003114. [DOI] [PubMed] [Google Scholar]
- Nakashima A., Hasegawa T., Mori S., Ueno M., Tanaka S., Ushimaru T., Sato S., Uritani M. A starvation-specific serine protease gene, isp6+, is involved in both autophagy and sexual development in Schizosaccharomyces pombe. Curr. Genet. 2006;49:403–413. doi: 10.1007/s00294-006-0067-0. [DOI] [PubMed] [Google Scholar]
- Seo S.J., Chua N.H. Analysis of interaction between long noncoding RNAs and protein by RNA immunoprecipitation in Arabidopsis. Methods Mol. Biol. 2019;1933:289–295. doi: 10.1007/978-1-4939-9045-0_18. [DOI] [PubMed] [Google Scholar]
- Toullec D., Elías-Villalobos A., Faux C., Noly A., Lledo G., Séveno M., Helmlinger D. The Hsp90 cochaperone TTT promotes cotranslational maturation of PIKKs prior to complex assembly. Cell Rep. 2021;37:109867. doi: 10.1016/j.celrep.2021.109867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets.