Abstract
Oculocutaneous albinism type 1 (OCA1), resulting from pathogenic variants in the tyrosinase (TYR) gene, refers to a group of phenotypically heterogeneous autosomal recessive disorders characterized by a partial or a complete absence of pigment in the skin/hair and is also associated with common developmental eye defects. In this study, we identified two novel compound heterozygous TYR variants from a Chinese hypopigmentary patient by whole-exome sequencing. Specifically, the two variants were c.-89T>G, located at the core of the initiator E-box (Inr E-box) of the TYR promoter, and p.S16Y (c.47C>A), located within the signal sequence. We performed both in silico analysis and experimental validation and verified these mutations as OCA1 variants that caused either impaired or complete loss of function of TYR. Mechanistically, the Inr E-box variant dampened TYR binding to microphthalmia-associated transcription factor, a master transcriptional regulator of the melanocyte development, whereas the S16Y variant contributed to endoplasmic reticulum retention, a common and principal cause of impaired TYR activity. Interestingly, we found that the Inr E-box variant creates novel protospacer adjacent motif sites, recognized by nucleases SpCas9 and SaCas9-KKH, respectively, without compromising the functional TYR coding sequence. We further used allele-specific genomic editing by CRISPR activation to specifically target the variant promoter and successfully activated its downstream gene expression, which could lead to potential therapeutic benefits. In conclusion, this study expands the spectrum of TYR variants, especially those within the promoter and noncoding regions, which can facilitate genetic counseling and clinical diagnosis of OCA1.
Keywords: oculocutaneous albinism type 1, TYR, initiator E-box, signal peptide, allele-specific
Abbreviations: AAV, adeno-associated virus; CRISPRa, CRISPR activation; DOPA, 3,4-dihydroxyphenylalanine; Endo H, endoglycosidase H; ER, endoplasmic reticulum; Inr E-box, initiator E-box; L9P, leucine to proline at position 9; l-Tyr, l-tyrosine; MITF, microphthalmia-associated transcription factor; OCA1, oculocutaneous albinism type 1; sgRNA, single-guide RNA; TSS, transcription start site; TYR, tyrosinase; WES, whole-exome sequencing
Oculocutaneous albinism type 1 (OCA1) refers to a group of phenotypically heterogeneous autosomal recessive disorders that result from the impaired or complete loss of function of the tyrosinase (TYR) gene (1). OCA1 is characterized by a congenital absence of melanin or hypopigmentation in the skin and hair, accompanied by symptoms of the eyes, such as poor visual acuity, foveal hypoplasia, nystagmus, photophobia, strabismus, and iris transillumination (2, 3). The prevalence of OCA subtypes varies across different populations (4). For example, OCA2 has been reported to be the most predominant form of albinism in Africa and Japan, whereas in Chinese, European, and North American populations, OCA1 is the most common albinism (5, 6).
TYR is the rate-limiting enzyme involved in melanogenesis. During the conversion of tyrosine to melanin, the enzyme catalyzes the first two steps of the melanin biosynthesis pathway. It functions in the hydroxylation of tyrosine to 3,4-dihydroxyphenylalanine (DOPA) and then oxidation of DOPA to dopaquinone (7). Variants in TYR lead to the absence or decreased synthesis of melanin and thus cause hypopigmentary disease OCA1 (8). According to the Human Gene Mutation Database, last release (2021.4), 495 variants in TYR have been associated with OCA1. Most of the identified variants are located within the coding sequence, which generally give rise to dysfunctional TYR proteins, either immature, misfolded, or catalytically inactive, that fail to pass the quality control by endoplasmic reticulum (ER) and are degraded by subsequent proteolysis. Only properly folded melanosomal proteins can be transported from the ER to the cis-Golgi apparatus for complex glycosylation (9). As such, ER retention is believed to be a common and principal cause of TYR activity, irrespective of the variant sites (10, 11, 12).
Previous studies have shown that the human TYR promoter governs the tissue-specific transcriptional activation by recruiting specific transcription factors with the promoter cis-elements, such as TATA-box, CAAT-box, M-box, and E-box, among others (1, 13, 14, 15). Specifically, there exists an initiator E-box (Inr E-box), which is located upstream of the transcription start site (TSS), and also a distal E-box element (16). The E-box, containing a 6-nt core hexanucleotide sequence “CANNTG,” which is essential for the transcriptional activation of TYR by recruiting transcription factors, including microphthalmia-associated transcription factor (MITF), the master transcriptional regulator of the melanocyte lineage (17, 18). Variants inside the gene promoter have been tightly associated with Mendelian disorders or act to influence complex traits (19). However, very few genetic variants that could alter the activity of TYR promoter or noncoding regions have been identified in OCA1. And their correlations with patient genotypes and underlying molecular mechanisms are yet fully understood. Moreover, the pathogenicity of most TYR variants has not been further functionally validated (20).
In this study, we identified two novel TYR variants from a Chinese patient with hypopigmentation: Inr E-box variant (c.-89T>G) and p.S16Y variant (c.47C>A) located within the signal peptide. We then did both in silico analysis and experimental validation and finally verified them as OCA1 variants that caused either impaired or complete loss of function of TYR. We found that the Inr E-box variant greatly dampened its binding ability to MITF, whereas the S16Y variant contributed to ER retention probably by a novel yet undescribed mechanism. We also designed allele-specific genomic editing to specifically target the promoter harboring the Inr E-box variant and successfully activated the downstream gene expression, which might produce important therapeutic benefits, in terms of recovering partial, if not all, functional TYR expression.
Results
Clinical description and molecular analysis
At the time of this study, the proband (III:1 in the pedigree) was a 0.5-year-old Chinese boy diagnosed with OCA, whereas other family members have none of those phenotypes (Fig. 1). The proband was born with little melanin pigmentation manifested by white skin and hair but darkened a little bit at a later age (Fig. 1A). His irises were transparent and brownish with blurred optical disk boundary, which caused impaired visual acuity, and other symptoms include photophobia and nystagmus (Fig. 1B). To figure out any potential hereditary cause, the peripheral venous blood sample of the proband was therefore collected for whole-exome sequencing (WES). The analysis then gives rise to the identification of novel compound heterozygous variants in the TYR gene (c.-89T>G and c.47C>A), although one of which is surprisingly located on the proximal TYR promoter but not exons. The TYR sequence of other family members was later examined by targeted Sanger sequencing, showing as either heterozygous variant or unaffected healthy (Fig. 1C).
Figure 1.
Clinical description and molecular analysis.A and B, clinical manifestations of the proband. C, the family pedigree shows the proband (III1, arrow) in an autosomal recessive pattern over three generations. D and E, identification and cosegregation of novel compound heterozygous TYR variants by WES analysis and Sanger sequencing verification. The proband carries a novel signal variant, c.47C>A (p.S16Y), inherited from the mother and a novel c.-89T>G variant located at the core of Inr E-box element on the TYR promoter from the father. F, the evolutionary conservation of the N-terminal amino acids among TYR homologs across vertebrates. The N-terminal region with amino acids marked in black represents the core hydrophobic region. Inr E-box, initiator E-box; TYR, tyrosinase; WES, whole-exome sequencing.
Specifically, we confirmed that one variant (c.-89T>G), inherited from the father, was a T to G transversion at 7 nt before TSS, located at the Inr E-box element of TYR core promoter; whereas the other one, inherited from the mother, was a C to A transversion at 47 nt after the start codon ATG (c.47C>A), which is just located within the signal sequence (Fig. 1, D and E). To our best knowledge, the two variants were previously unreported after searching gnomAD, ExAC, Human Gene Mutation Database, and PubMed, among others (Tables S1 and S2). All this genetic information of families confirmed the inheritance and the allelic distribution of the two variants.
In silico analysis of variants
The TYR variant (c.47C>A) results in the substitution of serine by TYR at amino acid position 16 (p.S16Y). This position is highly conserved for serine in TYR homologs across vertebrates after the analysis of evolutionary conservation by WebLogo (http://weblogo.threeplusone.com) (Fig. 1F). Functional prediction of p.S16Y, performed by VarCards, which integrated SIFT (https://sift.bii.a-star.edu.sg), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2), MutationTaster (https://www.mutationtaster.org), FATHMM (http://fathmm.biocompute.org.uk), and several other bioinformatics algorithms, provided inconsistent scores (Table S1). Note that the S16 position is just located within the signal peptide of TYR, proximal to the core hydrophobic region (Fig. 1F); it is possible that this variant affects the cleavage of the signal peptide during the translation and translocation of the nascent TYR polypeptide into the ER.
As a control, we also analyzed another reported OCA1 variant (c.26T>C) on the peptide sequence (21), which causes the substitution of leucine to proline at position 9 (p.L9P). With the SignalP 5.0 Web server (http://www.cbs.dtu.dk/services/SignalP), the cleavage of TYR-WT, S16Y, and L9P was predicted with a probability of 0.7878, 0.9286, and 0.4702, respectively (Table S3). The results showed that the L9P variant could dramatically dampen the cleavage; however, the S16Y variant appears to have limited effect on, or even increase, its cleavage probability by serving as a signal sequence (Table S3). Given the similar properties between serine and tyrosine, it might partially account for this prediction (Fig. S1A). Since the central hydrophobic core region of signal peptide is indispensable for the translocation into ER, we further examined whether those substitutions affect the hydrophobicity using the Kyte and Doolittle method. As shown, L9P significantly changed the hydrophobicity of the signal peptide; however, S16Y performed almost consistently with WT and did not alter the hydrophobicity, which is also conserved among 12 vertebrates (Fig. S1, B and C).
Similar to the in silico analysis of the S16Y variant, we analyzed the pathogenicity of the other variant, TYR c.-89T>G, at the proximal promoter using GenomeCards Web server. The results also showed inconsistent pathogenicity among different algorithms (Table S2). Considering the predictive nature of in silico analysis, we thus performed experiments to validate their pathogenicity.
Abolished melanin synthesis and TYR activity caused by the S16Y variant
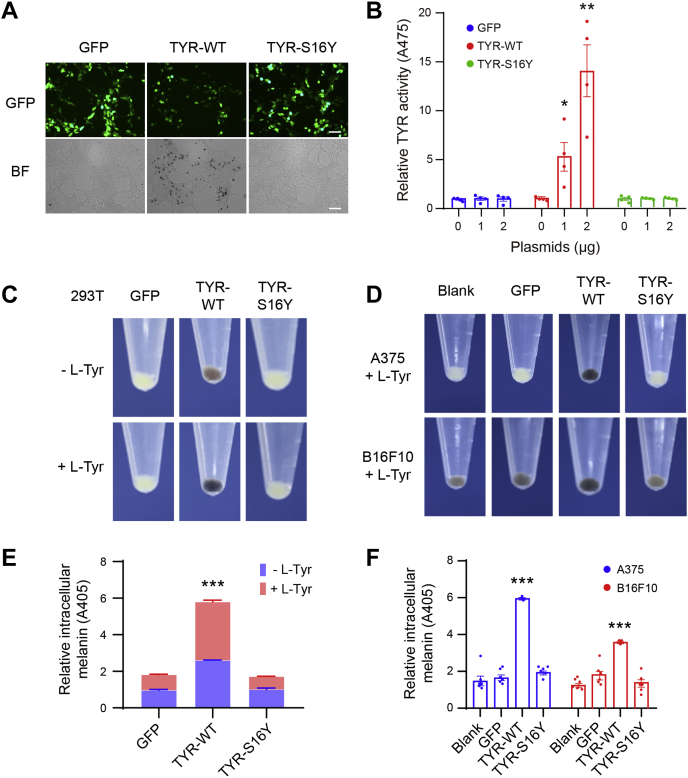
To examine the possible detrimental effect of the S16Y variant on melanin production, we constructed plasmids expressing full-length WT and S16Y TYR, respectively. The TYR-WT and TYR-S16Y plasmids, as well as an empty GFP control, were then transiently transfected into nonmelanocytic 293T cells and melanocytic human A375 and mouse B16F10 cells, with or without the treatment of TYR substrate, l-tyrosine (l-Tyr). After 48 h, a significant increase in TYR activity was observed in the TYR-WT transfected cells, in a dose-dependent manner, but not in TYR-S16Y and GFP control transfected 293T cells (Fig. 2, A and B). Cell pellets from both transfected nonmelanocytic and melanocytic cells were harvested to visualize their color and measure the relative intracellular melanin. Melanin production was apparently visible, shown as brown to black pellets, in both nonmelanocytic and melanocytic cells transfected by TYR-WT, rather than TYR-S16Y and GFP control, and was obviously increased by l-Tyr treatment (Fig. 2, C and D). Intracellular melanin contents were further detected, and the results were in line with the pellet observation (Fig. 2, E and F).
Figure 2.
Abolished melanin synthesis and tyrosinase (TYR) activity caused by the S16Y variant.A, for overexpression, the vectors containing GFP, TYR-WT-ires-GFP, and TYR-S16Y-ires-GFP variant were, respectively, transfected in 293T cells, without the treatment of TYR substrate, l-Tyr. Pictures were taken under the microscope. The scale bars represent 75 μm. B, dose-dependent evaluation of TYR activities. Different amounts (0, 1, and 2 μg) of TYR-WT and TYR-S16Y plasmids, as well as the empty GFP control, were respectively transfected in A375 cells. After 48 h, cells were harvested and lysed for assessing TYR activities. Values are means ± SEM (n = 4). C and D, the GFP, TYR-WT, and TYR-S16Y plasmids were transiently transfected into nonmelanocytic 293T cells and melanocytic human A375 and mouse B16F10 cells, with or without the l-Tyr treatment. After 48 h, cells were harvested in Eppendorf tubes, and the melanin production was then visualized by the color of cell pellets. E and F, the cell pellets of transfected cells were lysed for the measurement of intracellular melanin levels. Values are means ± SEM (n = 6). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 compared with control. l-Tyr, l-tyrosine.
Notably, we also constructed TYR-(BC027179)-WT and TYR-(BC027179)-S16Y plasmids, in which TYR coding sequences were from National Center for Biotechnology Information RefSeq BC027179 but lack exons 3 to 5 (22). However, both truncated TYR did not give rise to any melanin synthesis after transfection into 293T cells, no matter l-Tyr treatment (Fig. S2), reminiscent of the competence of full-length TYR. Similarly, a 340.5 kb deletion encompassing exons 3 to 5 of TYR was reported in an OCA1 patient (23). Together, these results indicated that the S16Y variant could abolish the TYR activity and melanin production.
The Inr E-box variant attenuated the transcription ability of the TYR promoter
Basically, the E-box contains a 6-nt consensus sequence “CANNTG,” which is recognized by a family of basic helix–loop–helix leucine zipper transcription factors (1). Of those, MITF is a master regulator of melanocyte development and is mechanistically recruited to bind with E-box, proximal to TSS, for the transcriptional activation of TYR (18, 24). Notably, a series of MITF variants were also identified that are tightly associated with pigment deficiency syndromes, by affecting its DNA-binding ability to either E-box or M-box (17). For instance, MITF with R217G variant cannot efficiently bind with the E-box DNA sequence (CACGTG), albeit with ∼25% binding activity, to activate expression from the TYR promoter (17, 25). Interestingly, specificity analysis revealed that MITF favors a 5′ T and/or 3′ A residue, adjacent to the core 6-nt CACGTG or CATGTG motif for binding with E-box (26). Moreover, compared with the CATGTG motif, the CACGTG motif appears to be with higher affinity for MITF binding, without flanking residue requirement (26, 27).
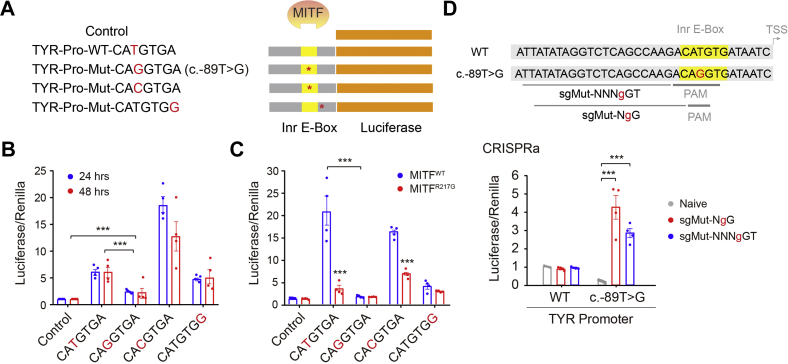
To investigate the possible effect of the Inr E-box variant (c.-89T>G:CAgGTG versus WT:CATGTG) on TYR expression, we thus performed dual-luciferase reporter assays to examine whether the variants affect the TYR promoter activity. The pGL3-Basic vector was employed and cloned with the 1000 bp length of TYR promoter with different variants located before the luciferase transcript (Fig. 3A). Specifically, we also introduced CAcGTGA (may serve as a positive control) and CATGTGg (with a flanking residue mutation from A to G). All constructs were then transfected into A375 cells for luciferase detection after 24 or 48 h. The Inr E-box variant (CAgGTGA) caused a significantly decreased luciferase activity by ∼60%, compared with the WT (CATGTGA) (Fig. 3B). As expected, the CAcGTG motif performed far better than WT in activating luciferase expression, whereas the variant of flanking residue (CATGTGg) reduced ∼23% activation capability (Fig. 3B).
Figure 3.
The Inr E-box variant attenuated the transcription ability of the TYR promoter.A, diagram of pGL3-Basic vectors cloned with the 1000 bp length of TYR promoter harboring different variants. B, A375 cells were cultured in a 24-well plate till 70% confluency. Each firefly luciferase reporter plasmid: pGL3-Control, pGL3-Basic-TYR-WT-Pro, or multiple pGL3-Basic-TYR-mut-Pro (200 ng) were respectively cotransfected with 10 ng renilla luciferase–expressing vector phRL-TK. After 24 h or 48 h, cells were harvested and lysed to perform dual-luciferase reporter assays. The relative reporter activities were presented by the normalization of firefly activities to renilla activities. Values are means ± SEM (n = 4). C, 293T cells in the 24-well plate were cotransfected with 200 ng pGL3 constructs, 200 ng pCSC-MITFs, and 10 ng phRL-TK plasmid. After 48 h, the transfected cells were collected and lysed to perform dual-luciferase reporter assays for assessing relative reporter activities. Values are means ± SEM (n = 4). D, allele-specific CRISPRa was applied to specifically target the promoter with the Inr E-box variant. 293T cells in the 24-well plate were cotransfected with 200 ng pGL3 plasmids, 200 ng Cas9/sgRNA plasmids, and 10 ng phRL-TK plasmids. After 48 h, the transfected cells were collected and lysed to perform dual-luciferase reporter assays. The relative promoter activities were presented by the normalization of firefly activities to renilla activities. Values are means ± SEM (n = 4). ∗∗∗p < 0.001. CRISPRa, CRISPR activation; Inr E-box, initiator E-box; TYR, tyrosinase.
As 293T cells do not express MITF endogenously, we thus cotransfected with the aforementioned promoter constructs and MITF expression plasmids (WT or R217G), respectively, in 293T cells. The results showed that MITF effectively activated the TYR promoter with both WT (CATGTGA) and variant (CAcGTGA) motif but failed to activate the TYR promoter with the Inr E-box variant (CAgGTGA) and 3′ residue mutation (CATGTGg) (Fig. 3C). Unsurprisingly, the MITFR217G variant greatly attenuated its transcriptional activity by approximately fivefold because of the dampened binding with E-box (Fig. 3C). Collectively, our results confirmed that the Inr E-box variant could impair the transcription ability of the TYR promoter by impeding MITF binding.
Allele-specific CRISPR activation of the Inr E-box variant produced therapeutic benefits
Inactivation of the TYR expression leads to a blockage of melanosome biogenesis and causes hypopigmentation in melanocytes and retinal pigment epithelium cells. The most significant consequence of melanin deficiency is the impairment of the eye and visual system. Encouragingly, the strategy of gene therapy by the administration of adeno-associated virus (AAV)–based vectors, encoding the human TYR gene, has been applied for treating OCAs. In adult Tyrc-2j mice, an OCA1 mouse model, subretinal injection of AAV2/1-cytomegalovirus-hTYR resulted in melanosome biogenesis and prevented progressive photoreceptor degeneration that led to long-term retinal functional amelioration (28). Although the replacement strategy is promising, CRISPR/Cas9mediated genome targeting has also produced tremendous therapeutic benefits in recent years, especially for treating eye diseases such as retinitis pigmentosa, because of the unique advantage of ocular immune privilege for gene therapy (29, 30). Of those, the CRISPR activation (CRISPRa) is one of the promising strategies that only targets endogenous regulatory elements, without breaking genomic DNAs, to enhance the expression of functional genes (31).
In our case, the TYRS16Y variant destroyed its coding sequences. However, the allele with the Inr E-box variant retained a functional coding region, and more excitingly, brought about novel protospacer adjacent motif sites, including “NgG” and “NNNgGT,” recognized by SpCas9 and SaCas9-KKH, respectively (Fig. 3D). This leads us to apply allele-specific genomic targeting by CRISPR/Cas9, which has been increasingly exploited for treating human genetic diseases and other versatile applications, to maximally avoid the potential off-targets (32, 33). Specifically, we used the AsCRISPR Web server to design potential allele-specific single-guide RNAs (sgRNAs) that may selectively target only one allele, the Inr E-box variant allele, without affecting the other allele, based on the stringent selectivity of DNA sequences that are recognized by Cas9. By doing this, we figured out two candidate sgRNAs (Table S4; marked in red) that are adjacent to novel protospacer adjacent motif sites created by the Inr E-box variant.
We thus cloned candidate sgRNAs into SpCas9 and SaCas9-KKH vectors and cotransfected with pGL3 promoter vectors in 293T cells. The luciferase assay showed that allele-specific sgRNAs indeed specifically elevated the transcriptional activity of the TYR promoter harboring the variant motif but not the WT one (Fig. 3D). Compared with control, the luciferase activity was increased by ∼19.7-fold and ∼13.0-fold, respectively, after CRISPRa on the variant promoter (Fig. 3D). Therefore, the results demonstrated that allele-specific CRISPRa of the Inr E-box variant might produce potential therapeutic benefits. Although we applied lentiviral vectors to deliver CRISPRa and sgRNAs in this study, further AAV-based delivery, especially involving the smaller size of SaCas9-KKH, might be promising for the disease treatment.
ER retention caused by the S16Y variant
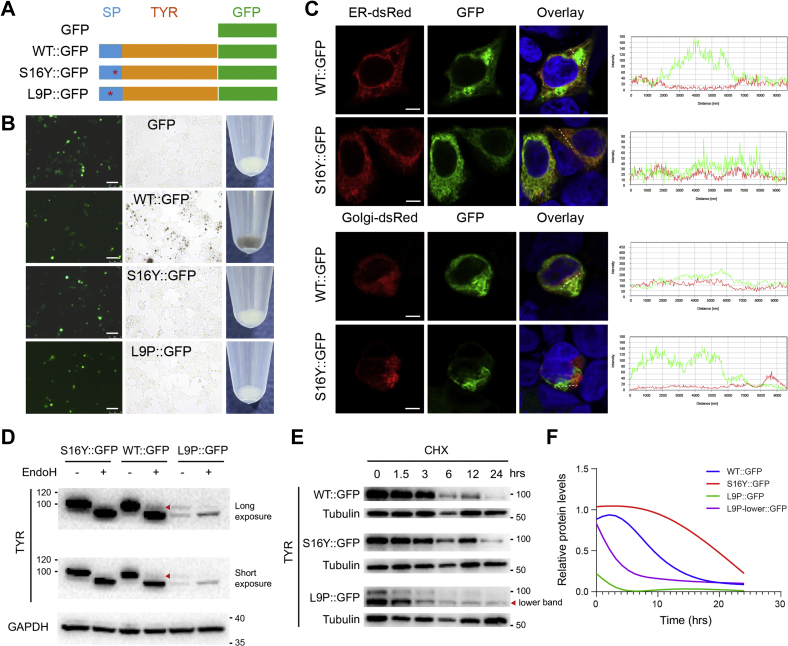
As demonstrated earlier, we confirmed that the S16Y variant abolished melanin synthesis and TYR activity. However, we would like to further dissect its potential mechanism. We construct the expression of TYR fusion proteins with GFP at the C terminus for further visualization and other assays (Fig. 4A). Those constructs recapitulated their phenotypes on melanin production (Fig. 4B). As OCA1 is recognized as an ER retention disorder, we thus investigated the subcellular localization of translated TYR among organelles. We cotransfected TYR::GFP constructs and organelle reporter plasmids into cells. After 48 h, we observed that TYRWT::GFP was localized as granules in a reticular pattern. However, TYRS16Y::GFP exhibited only the reticular pattern identical to ER-dsRed, an ER reporter, without any prominent granular structures (Fig. 4C). Moreover, TYRS16Y::GFP did not overlap with Golgi-dsRed, a Golgi reporter, suggesting that the variant TYR might not be translocated to the Golgi apparatus (Fig. 4C). As TYR overexpression in nonpigment cells might also be sorted to lysosomes, which share a common biogenesis pathway with melanosomes (34), we thus examined the location of TYR inside lysosomes. Similarly, TYRWT::GFP was mostly colocalized with lysosome-associated membrane protein 1-mCherry, a lysosome reporter, whereas TYRS16Y::GFP or TYRL9P::GFP did not show obvious overlap with lysosomes (Fig. S3).
Figure 4.
ER retention of variant TYR as the cause of functional loss.A, diagram of TYR::GFP fusion proteins. B, the melanin production of fusion proteins. The fusion vectors were respectively transfected in 293T cells, without l-Tyr treatment. The melanin production was visualized by the color of cell pellets. The scale bars represent 75 μm. C, subcellular localization of translated fusion TYR among organelles. The fusion vectors were cotransfected with ER-dsRed and Golgi-dsRed reporter vectors, respectively, in 293T cells. After 48 h, cells were fixed and observed under the confocal microscopy. The scale bars represent 10 μm. D, 293T cells were transfected with 500 ng fusion vectors, respectively, in a 6-well plate. After 48 h, cells were harvested for Endo H digestion. The digested proteins were separated by Western blot. E and F, 293T cells were transfected with 1 μg fusion vectors, respectively. After 24 h, cells were treated with 100 μg/ml CHX and collected respectively at desired time points (0, 1.5, 3, 6, 12, and 24 h). The half-lives of fusion proteins were measured by Western blot. CHX, cycloheximide; Endo H, endoglycosidase H; ER, endoplasmic reticulum; l-Tyr, l-tyrosine; SP, signal peptide; TYR, tyrosinase.
Endoglycosidase H (Endo H) specifically removes oligosaccharides of the high mannose in ER and hybrid forms of N-linked carbohydrates in pre-Golgi but is resistant to complex carbohydrate structures in the trans-Golgi network. We thus further employed Endo H digestion to assess the delivery of TYR from ER to Golgi apparatus. The results showed that WT::GFP was detected as two distinct bands after Endo H treatment, a faster migrating immature form (∼90 kDa) and a slower migrating mature form that is Endo H resistant. On the contrary, the glycosylated mature band was completely invisible in both S16Y::GFP and L9P::GFP groups, suggesting of OCA1 phenotypes characterized by absolute lack of mature TYR (Fig. 4D). Together, those results demonstrated that the S16Y variant might also cause ER retention that results in ER-associated degradation and thus stall the TYR delivery into Golgi apparatus for further complex glycosylation and maturation.
Although both the S16Y and L9P variants lead to ER retention, we still observed differences between these two variants. Apart from the differential effects on the TYR expression per se (Fig. 4D), they also contribute to the different half-lives of TYR proteins. After cycloheximide treatment, we could observe that the TYRL9P degraded more rapidly than TYRWT; however and unexpectedly, the TYRS16Y survived for a longer duration (Fig. 4, E and F). This difference between S16Y and L9P, converging with their distinct effects on the TYR expression and probably the signal peptide cleavage, might indicate a novel yet undescribed mechanism for the biology of signal sequences.
Discussion
TYR variants: coding and noncoding
In this study, we identified two novel TYR compound heterozygous noncoding variants by WES in a Chinese patient with hypopigmentation. However, most previous studies have been focused on the in silico analysis of identified TYR variants, and few have further functionally validated the pathogenicity. We thus experimentally confirmed that the Inr E-box variant (c.-89T>G) significantly impaired the transcription ability of TYR promoter and the S16Y variant (c.47C>A) abolished the TYR activity and melanin synthesis.
To our best knowledge, it is the first time to report a variant inside Inr E-box and also among those few cases of variants located at noncoding regions of TYR. Besides the promoter, several variants were identified within the introns that may produce cryptic splicing sites (21). It has to be noted that most of the previous studies have applied WES to identify pathogenic variants, which partially account for the fact that almost all TYR variants are located within coding regions. However, there are still numerous variants within the noncoding regions that may cause OCAs, which have been ignored for years, and in considerable amounts of OCA patients, molecular causes have been missing (6, 35). In this study, the Inr E-box variant was nearly undetected, only that this variant is proximal to TSS. As such, whole-genome sequencing is encouraging for the identification of more noncoding variants. It thus underlies the importance of both WES and whole-genome sequencing in precision medicine and disease diagnosis as well as prenatal genetic testing.
In silico analysis and functional validation of TYR variants
As we know, in silico algorithms cannot always give consistent pathogenicity scores of given variants, which still awaits experimental validation. It is the same situation for the variants in this study, especially for the S16Y variant. Even with the help of most updated algorithms, empowered by machine/deep learning or artificial intelligence, such as VARITY (http://varity.varianteffect.org) (36) and EVE (https://evemodel.org) (37), the S16Y variant was all recognized as not pathogenic, which might also hint at a potentially novel mechanism that is distinct from existing ones, such as L9P, in the literature (Fig. S4, A and B).
To examine whether S16Y and L9P might both abolish the cleavage capability of the signal peptide, a small tag, FLAG (DYKDDDDK; ∼1 kDa), was cloned just after the ATG start codon yet preceding the signal sequence (with WT, S16Y, and L9P variant, respectively) and TYR::GFP sequence. After cell transfection, we can detect the TYR production by Western blot. As we predicted, FLAG::WT::TYR shall be translated into functional TYR (FLAG was removed together with cleaved signal peptide) that produces melanin, whereas FLAG::S16Y::TYR and FLAG::L9P::TYR may produce dysfunctional TYR proteins (still fused with FLAG; can be detected by Western blot) if the signal cleavage is abolished. Unexpectedly, we can hardly see melanin production even in FLAG::WT::TYR transfected cells. This means that the signal cleavage has already disrupted by the incorporation of FLAG peptide located in front of WT::TYR, so that we cannot, at least in this experiment, examine whether the S16Y and L9P variants could affect the cleavage (Fig. S5). Nevertheless, considering the multiple differences between L9P and S16Y variants (Fig. 4), it definitely merits further in-depth investigations, especially into the novel mechanisms of signal peptide cleavage.
The extra challenge for probing functional effects by TYR variants is that no crystal structure of human TYR is available until now, because of its transmembrane and glycosylation nature. The recently launched AlphaFold algorithm (https://alphafold.ebi.ac.uk) (38), excitingly, may help predict the TYR protein structure with variable reliability. The AlphaFold prediction showed that S16 might form a hydrogen bond with F167, which is also a highly conserved site (Fig. S4, C and D). This might be reasonable, since the partial cleavage of the signal peptide is just observed when the translated TYR polypeptide was 166 amino acids in length (39). The N-terminal hydrophobic region inside the translocon Sec61, orchestrating with the S16–F167 hydrogen bond, might help anchor nascent polypeptide to the translocon, thereby facilitating the cleavage of the signal peptide. However, the S16Y variant might abolish this hydrogen bond, which will increase the dwell time inside the translocon and further stall the cleavage. Yet, another explanation might be that Y16 forward the signal peptide’s cleavage position, since tyrosine (Y) rather than serine (S) is more prone to be within a signal peptide, as predicted by the SignalP 5.0 server.
Specificity of the Inr E-box motif for TYR transcription
The Inr E-box variant adheres to the consensus “CANNTG” motif for E-box but surprisingly gives rise to detrimental effects. Nevertheless, this is in line with the notion that the E-box motif works specifically within different gene promoters (26). In our case, the binding activity of the WT motif (CATGTG) with MITF is greatly dampened by the Inr E-box variant (CAGGTG), reminiscent of the motif specificity for the TYR gene. Interestingly, other than the master MITF, the variant might impair the binding with other transcription factors such as upstream transcription factor 1, which shows similar binding specificity to MITF (26). A recent study has demonstrated that upstream transcription factor 11 cannot bind the E-box variant (CAGGTG) on the promoter of the splicing gene, epithelial splicing regulatory protein 1 (ESRP1), which hinders its expression in both MCF-7 and MDA-MB-231 breast cancer cells (40). This further reinforces the functional specificity of E-box motifs under different gene contexts.
Experimental procedures
Subjects
One Chinese family, the proband with OCA and all available family members, was enrolled in the Xiangya Hospital of Central South University, Hunan, China. Peripheral venous blood samples were collected in EDTA containing vacutainer tubes from all family members. All participants provided written informed consent in this genetic study, which is approved by the Institutional Review Board of the Xiangya Hospital of Central South University and abides by the Declaration of Helsinki principles.
WES and in silico analysis
Genomic DNA was extracted from peripheral leukocytes of the proband with the standard phenol–chloroform extraction method. WES was performed on the proband (III:1) by the Novogene Bioinformatics Institute. The SureSelect Human All Exon V6 kit (Agilent Technologies) was used for exome capture, and the Illumina HiSeq 2000 platform was used for sequencing, following the manufacturer's protocol. A total of 300 ng genomic DNA was used to construct the exome library, and the genomic DNA was sheared into 150 to 200 bp for enrichment using a Covaris instrument (Covaris, Inc). Enrichment libraries for target regions were sequenced by the HiSeq 2000 platform, which generated 101 bp pair-end reads. The candidate variants were further examined by PCR and targeted Sanger sequencing from all other family members. Primer sequences for PCR are listed in Table S5.
In silico analysis of candidate variants was assessed with VarCards (41) and GenomeCards (http://genemed.tech/genomecards/), respectively. Furthermore, in silico prediction of the TYR p.S16Y variant was analyzed by the recently developed algorithms, such as EVE (37) and VARITY (36). The conservation of amino acids was performed by WebLogo3 (42). Hydrophobicity plots of the signal peptides from WT TYR, among TYR homologs across vertebrates, as well as TYR variants were carried out by the Kyte and Doolittle method (43). Properties of the signal sequences were predicted and analyzed with the SignalP 5.0 server (44). The protein structure of TYR was predicted and deposited by AlphaFold (45).
Plasmid constructions
For the construction of luciferase reporter plasmids, a promoter fragment of 1000 nt (just before the start codon of TYR) was amplified from the patient’s genomic DNA, which contains the WT T allele or variant G allele and were subcloned into the KpnI and XhoI restriction sites of the pGL3-Basic vector (Promega). Other vectors carrying promoter variants were constructed by site-directed mutagenesis of the generated pGL3-Basic-TYR-WT-Pro construct. To investigate the effect of signal peptide variants during TYR synthesis, the full-length coding sequence of human TYR was amplified from 293T cells and cloned into the pCSC-IRES-GFP vector; and other vectors with signal peptide missense variants, including p.S16Y (c.47C>T) and p.L9P (c.26T>C) were generated by site-directed mutagenesis, respectively. Similarly, the TYR ORF clone (BC027179) and its variant form were constructed. Human MITF was amplified from A375 melanocytic cells and further ligated into the pCSC-IRES-GFP vector, which provided as the donor plasmid for constructing MITFR217G variant. The multiple TYR::GFP or FLAG::TYR::GFP fusion constructs were generated based on the pEGFP-TYR vector (addgene; plasmid #32781), either by site-directed mutagenesis or PCR amplification. All final constructs were verified by bidirectional Sanger sequencing.
Cell culture
Immortalized human embryonic kidney 293T cells, human malignant melanoma A375 cells, as well as murine melanoma B16F10 cells were obtained from Procell. Cells were cultured in Dulbecco’s modified Eagle's medium (Procell) supplemented with 10% fetal bovine serum (Biological Industries), containing 1% penicillin, 1% streptomycin, and 1% amphotericin B (Beyotime) in a humidified incubator in which 5% CO2 was supplied and maintained at 37 °C.
Luciferase reporter assays
To evaluate the TYR promoter activities, A375 cells were seeded at a density of 6 × 104 in a 24-well plate and cultured overnight. When cells were grown till 70% confluency, 200 ng of each firefly luciferase reporter plasmid:pGL3-Control, pGL3-Basic-TYR-WT-Pro, or multiple pGL3-Basic-TYR-mut-Pro were, respectively, cotransfected with 10 ng renilla luciferase–expressing vector phRL-TK using Lipofectamine 3000 (Invitrogen). After 24 h or 48 h, cells were harvested and lysed using the dual-luciferase reporter assay system (Promega; catalog no.: E1910), according to the manufacturer’s protocol. The fluorescence intensities were finally measured with a tube luminometer (Berthold Sirius). To elaborate the role of Inr E-box element and its possible regulation by MITF, 293T cells in the 24-well plate were cotransfected with 200 ng pGL3 constructs, 200 ng pCSC-MITFs, and 10 ng phRL-TK plasmid. After 48 h, the transfected cells were collected and lysed to perform dual-luciferase reporter assays. The relative reporter activities were presented by the normalization of firefly activities to renilla activities.
CRISPR-mediated activation of the TYR promoter
The sgRNAs specifically targeting c.-89T>G (sgMut-NgG; sgMut-NNNgGT) allele were designed by the AsCRISPR Web server (46) (http://www.genemed.tech/ascrispr). To generate sgRNA expression plasmids, the dCas9-VP64 and dSaCas9-KKH-VP64 plasmids were digested with BsmBI, respectively, and cloned with a pair of annealed oligonucleotides sgMut-NgG or sgMut-NNNgGT (Table S5). 293T cells in the 24-well plate were cotransfected with 200 ng pGL3 plasmids, 200 ng Cas9/sgRNA plasmids, and 10 ng phRL-TK plasmid using Lipofectamine 3000. After 48 h, the transfected cells were collected and lysed to perform dual-luciferase reporter assays. The relative promoter activities were presented by the normalization of firefly activities to renilla activities.
Cellular TYR activity
The TYR activities were evaluated independently for each TYR (WT and variants). A375 cells were transfected with 0, 1, and 2 μg plasmids (pCSC-TYR-IRES-GFP), respectively, in a 6-well plate. After 48 h, cells were harvested and washed three times with PBS. The cell pellets were lysed with 300 μl lysis buffer (0.1 M PBS containing 5% Triton X-100). After incubation on ice for 30 min, transfer out 200 μl of the mixture in a 96-well plate followed by adding 50 μl l-DOPA (2 mg/ml). The mixture was immediately monitored at 475 nm using a multimode plate reader (Cytation 5; BioTek). The mixtures were further incubated for 30 min at 37 °C and measured again at 475 nm. The TYR relative activity was calculated as: TYR activity (%) = (A′30–A′0)/(A30–A0) × 100, in which A′ represents absorbance values of samples at 0 min and 30 min, and A represent absorbance values of controls.
Intracellular melanin measurement
The measurements of melanin contents were performed as described previously (47). A375 and B16F10 cells were seeded at a density of 4 × 105 in a 6-well plate and cultured overnight. The next day, cells were transfected with 2 μg plasmids (pCSC-TYR-IRES-GFP), respectively, with or without 2 mM l-Tyr treatment. Forty-eight hours later, the cell pellets were collected in Eppendorf tubes by spinning down at 800g for 4 min to examine their color and further measure intracellular melanin levels. The pellets were washed three times with PBS, followed by adding 500 μl of 1 N NaOH containing 10% dimethyl sulfoxide. The reaction was heated at 80 °C for 20 min, and the relative intracellular melanin was then measured at 405 nm absorbance using a microplate spectrophotometer (Cytation5; BioTek).
Endo H digestion and Western blot
293T cells were transfected with 500 ng plasmids expressing WT::GFP, S16Y::GFP, and L9P::GFP fusion proteins, respectively, in a 6-well plate. After 48 h, cells were digested with 150 μl radioimmunoprecipitation assay buffer. For Endo H digestion, each supernatant containing 20 μg of total proteins was denatured in 1× glycoprotein denaturation solution for 10 min at 100 °C and then cooled to room temperature. The denatured proteins were then digested with 1 μl Endo H (New England Biolabs) at 37 °C for 10 min. After the digestion, each sample was supplemented with 4× sample loading buffer and 2-mercaptoethanol and heated at 100 °C for 10 min.
A total of 60 μg proteins were separated by SDS-PAGE and transferred to the polyvinylidene difluoride membrane (Millipore). The blots were incubated with primary antibodies, including TYR (Affinity Biosciences; catalog no.: AF5491; 1:1000 dilution), GAPDH (Abclonal; catalog no.: AC033; 1:20,000 dilution), and tubulin (Sigma; catalog no.: T5168; 1:10,000 dilution), at 4 °C for overnight. The blots were washed with Tris-buffered saline with Tween-20 and incubated with secondary antibodies conjugated with horseradish peroxidase at room temperature for 1 h. After the repeated washing by PBS with Tween-20, protein blots were detected by using ECL substrates and finally visualized under the ChemiDoc XRS Imaging System (Bio-Rad).
Degradation kinetics of TYR proteins
To examine the degradation kinetics or half-lives of both WT and variant TYR proteins, 1 × 105 cells were seeded in a 12-well plate and transfected with 1 μg plasmids (pEGFP-TYR-WT, pEGFP-TYR-S16Y, and pEGFP-TYR-L9P). After 24 h, 100 μg/ml cycloheximide (Selleck; catalog no.: S7418) was added into each well, and cells were collected, respectively, at desired time points (0, 1.5, 3, 6, 12, and 24 h). The degradation kinetics were measured by Western blot, and TYR densities were quantified using ImageJ software (NIH).
Statistical analysis
The statistical analysis was conducted with GraphPad Prism software (version 7; GraphPad Software, Inc), which was also used for graphics. Statistical analysis was performed using the Student’s t test. A 95% confidence level was considered the significance of differences between groups, with p < 0.05 indicating statistical significance.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge the participation of the patient and his family members in this study. We also thank members of the Tang laboratory as well as Dr Jian Qiu, Zhengmao Hu, and Jieqiong Tan of Central South University for insightful discussions.
Author contributions
Y. T. conceptualization; Y. T. methodology; J. L. formal analysis; Y. T., C. L. and Q. C. investigation; J. W. resources; Q. C., C. L. and Y. T. writing–original draft.
Funding and additional information
This study was funded by the National Natural Sciences Foundation of China (grant no.: 81801200; to Y. T.), Hunan Provincial Natural Science Foundation of China (grant no.: 2019JJ40476; to Y. T.), Talents Startup Fund (grant no.: 2209090550; to Y. T.), and Youth Science Foundation (grant no.: 2021Q04; to J. J. W.) of Xiangya Hospital, Central South University, Changsha, China.
Edited by Craig Cameron
Supporting information
References
- 1.Ray K., Chaki M., Sengupta M. Tyrosinase and ocular diseases: Some novel thoughts on the molecular basis of oculocutaneous albinism type 1. Prog. Retin. Eye Res. 2007;26:323–358. doi: 10.1016/j.preteyeres.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Gronskov K., Ek J., Brondum-Nielsen K. Oculocutaneous albinism. Orphanet J. Rare Dis. 2007;2:43. doi: 10.1186/1750-1172-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruijt C.C., de Wit G.C., Bergen A.A., Florijn R.J., Schalij-Delfos N.E., van Genderen M.M. The phenotypic spectrum of albinism. Ophthalmology. 2018;125:1953–1960. doi: 10.1016/j.ophtha.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q., Yi S., Li M., Xie B., Luo J., Wang J., Rong X., Zhang Q., Qin Z., Hang L., Feng S., Fan X. Genetic analyses of oculocutaneous albinism types 1 and 2 with four novel mutations. BMC Med. Genet. 2019;20:106. doi: 10.1186/s12881-019-0842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norman C.S., O'Gorman L., Gibson J., Pengelly R.J., Baralle D., Ratnayaka J.A., Griffiths H., Rose-Zerilli M., Ranger M., Bunyan D., Lee H., Page R., Newall T., Shawkat F., Mattocks C., et al. Identification of a functionally significant tri-allelic genotype in the Tyrosinase gene (TYR) causing hypomorphic oculocutaneous albinism (OCA1B) Sci. Rep. 2017;7:4415. doi: 10.1038/s41598-017-04401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei A., Wang Y., Long Y., Wang Y., Guo X., Zhou Z., Zhu W., Liu J., Bian X., Lian S., Li W. A comprehensive analysis reveals mutational spectra and common alleles in Chinese patients with oculocutaneous albinism. J. Invest. Dermatol. 2010;130:716–724. doi: 10.1038/jid.2009.339. [DOI] [PubMed] [Google Scholar]
- 7.Lai X., Wichers H.J., Soler-Lopez M., Dijkstra B.W. Structure and function of human tyrosinase and tyrosinase-related proteins. Chemistry. 2018;24:47–55. doi: 10.1002/chem.201704410. [DOI] [PubMed] [Google Scholar]
- 8.Dolinska M.B., Kus N.J., Farney S.K., Wingfield P.T., Brooks B.P., Sergeev Y.V. Oculocutaneous albinism type 1: Link between mutations, tyrosinase conformational stability, and enzymatic activity. Pigment Cell Melanoma Res. 2017;30:41–52. doi: 10.1111/pcmr.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang N., Hebert D.N. Tyrosinase maturation through the mammalian secretory pathway: Bringing color to life. Pigment Cell Res. 2006;19:3–18. doi: 10.1111/j.1600-0749.2005.00288.x. [DOI] [PubMed] [Google Scholar]
- 10.Berson J.F., Frank D.W., Calvo P.A., Bieler B.M., Marks M.S. A common temperature-sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J. Biol. Chem. 2000;275:12281–12289. doi: 10.1074/jbc.275.16.12281. [DOI] [PubMed] [Google Scholar]
- 11.Chaki M., Sengupta M., Mondal M., Bhattacharya A., Mallick S., Bhadra R., Ray K. Molecular and functional studies of tyrosinase variants among Indian oculocutaneous albinism type 1 patients. J. Invest. Dermatol. 2011;131:260–262. doi: 10.1038/jid.2010.274. [DOI] [PubMed] [Google Scholar]
- 12.Mondal M., Sengupta M., Ray K. Functional assessment of tyrosinase variants identified in individuals with albinism is essential for unequivocal determination of genotype-to-phenotype correlation. Br. J. Dermatol. 2016;175:1232–1242. doi: 10.1111/bjd.14977. [DOI] [PubMed] [Google Scholar]
- 13.Fang D., Tsuji Y., Setaluri V. Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF. Nucleic Acids Res. 2002;30:3096–3106. doi: 10.1093/nar/gkf424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarkar D., Leung E.Y., Baguley B.C., Finlay G.J., Askarian-Amiri M.E. Epigenetic regulation in human melanoma: Past and future. Epigenetics. 2015;10:103–121. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawakami A., Fisher D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Invest. 2017;97:649–656. doi: 10.1038/labinvest.2017.9. [DOI] [PubMed] [Google Scholar]
- 16.Um J.M., Kim H.J., Lee Y., Choi C.H., Hoang Nguyen D., Lee H.B., Shin J.H., Tai No K., Kim E.K. A small molecule inhibitor of Mitf-E-box DNA binding and its depigmenting effect in melan-a cells. J. Eur. Acad. Dermatol. Venereol. 2012;26:1291–1297. doi: 10.1111/j.1468-3083.2011.04286.x. [DOI] [PubMed] [Google Scholar]
- 17.Grill C., Bergsteinsdóttir K., Ogmundsdóttir M.H., Pogenberg V., Schepsky A., Wilmanns M., Pingault V., Steingrímsson E. MITF mutations associated with pigment deficiency syndromes and melanoma have different effects on protein function. Hum. Mol. Genet. 2013;22:4357–4367. doi: 10.1093/hmg/ddt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goding C.R., Arnheiter H. MITF-the first 25 years. Genes Dev. 2019;33:983–1007. doi: 10.1101/gad.324657.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French J.D., Edwards S.L. The role of noncoding variants in heritable disease. Trends Genet. 2020;36:880–891. doi: 10.1016/j.tig.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Wei A.H., Zang D.J., Zhang Z., Yang X.M., Li W. Prenatal genotyping of four common oculocutaneous albinism genes in 51 Chinese families. J. Genet. Genomics. 2015;42:279–286. doi: 10.1016/j.jgg.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 21.King R.A., Pietsch J., Fryer J.P., Savage S., Brott M.J., Russell-Eggitt I., Summers C.G., Oetting W.S. Tyrosinase gene mutations in oculocutaneous albinism 1 (OCA1): Definition of the phenotype. Hum. Genet. 2003;113:502–513. doi: 10.1007/s00439-003-0998-1. [DOI] [PubMed] [Google Scholar]
- 22.Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Zeeberg B., Buetow K.H., Schaefer C.F., Bhat N.K., Hopkins R.F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morice-Picard F., Lasseaux E., Cailley D., Gros A., Toutain J., Plaisant C., Simon D., François S., Gilbert-Dussardier B., Kaplan J., Rooryck C., Lacombe D., Arveiler B. High-resolution array-CGH in patients with oculocutaneous albinism identifies new deletions of the TYR, OCA2, and SLC45A2 genes and a complex rearrangement of the OCA2 gene. Pigment Cell Melanoma Res. 2014;27:59–71. doi: 10.1111/pcmr.12173. [DOI] [PubMed] [Google Scholar]
- 24.Vetrini F., Auricchio A., Du J., Angeletti B., Fisher D.E., Ballabio A., Marigo V. The microphthalmia transcription factor (Mitf) controls expression of the ocular albinism type 1 gene: Link between melanin synthesis and melanosome biogenesis. Mol. Cell Biol. 2004;24:6550–6559. doi: 10.1128/MCB.24.15.6550-6559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H., Jiang L., Xie Z., Mei L., He C., Hu Z., Xia K., Feng Y. Novel mutations of PAX3, MITF, and SOX10 genes in Chinese patients with type I or type II Waardenburg syndrome. Biochem. Biophys. Res. Commun. 2010;397:70–74. doi: 10.1016/j.bbrc.2010.05.066. [DOI] [PubMed] [Google Scholar]
- 26.Aksan I., Goding C.R. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell Biol. 1998;18:6930–6938. doi: 10.1128/mcb.18.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strub T., Giuliano S., Ye T., Bonet C., Keime C., Kobi D., Le Gras S., Cormont M., Ballotti R., Bertolotto C., Davidson I. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011;30:2319–2332. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- 28.Gargiulo A., Bonetti C., Montefusco S., Neglia S., Di Vicino U., Marrocco E., Corte M.D., Domenici L., Auricchio A., Surace E.M. AAV-mediated tyrosinase gene transfer restores melanogenesis and retinal function in a model of oculo-cutaneous albinism type I (OCA1) Mol. Ther. 2009;17:1347–1354. doi: 10.1038/mt.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannelli S.G., Luoni M., Castoldi V., Massimino L., Cabassi T., Angeloni D., Demontis G.C., Leocani L., Andreazzoli M., Broccoli V. Cas9/sgRNA selective targeting of the P23H Rhodopsin mutant allele for treating retinitis pigmentosa by intravitreal AAV9.PHP.B-based delivery. Hum. Mol. Genet. 2018;27:761–779. doi: 10.1093/hmg/ddx438. [DOI] [PubMed] [Google Scholar]
- 30.Li P., Kleinstiver B.P., Leon M.Y., Prew M.S., Navarro-Gomez D., Greenwald S.H., Pierce E.A., Joung J.K., Liu Q. Allele-specific CRISPR-Cas9 genome editing of the single-base P23H mutation for rhodopsin-associated dominant retinitis pigmentosa. CRISPR J. 2018;1:55–64. doi: 10.1089/crispr.2017.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., Lim W.A., Weissman J.S., Qi L.S. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J., Tang B., Tang Y. Allele-specific genome targeting in the development of precision medicine. Theranostics. 2020;10:3118–3137. doi: 10.7150/thno.43298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu X., Li C., Chen Y., Ou R., Cao B., Wei Q., Hou Y., Zhang L., Song W., Zhao B., Wu Y., Shang H. Mutation screening and burden analysis of VPS13C in Chinese patients with early-onset Parkinson's disease. Neurobiol. Aging. 2020;94:311.e1–311.e4. doi: 10.1016/j.neurobiolaging.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Toyofuku K., Wada I., Spritz R.A., Hearing V.J. The molecular basis of oculocutaneous albinism type 1 (OCA1): Sorting failure and degradation of mutant tyrosinases results in a lack of pigmentation. Biochem. J. 2001;355:259–269. doi: 10.1042/0264-6021:3550259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lasseaux E., Plaisant C., Michaud V., Pennamen P., Trimouille A., Gaston L., Monfermé S., Lacombe D., Rooryck C., Morice-Picard F., Arveiler B. Molecular characterization of a series of 990 index patients with albinism. Pigment Cell Melanoma Res. 2018;31:466–474. doi: 10.1111/pcmr.12688. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y., Li R., Sun S., Weile J., Roth F.P. Improved pathogenicity prediction for rare human missense variants. Am. J. Hum. Genet. 2021;108:1891–1906. doi: 10.1016/j.ajhg.2021.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frazer J., Notin P., Dias M., Gomez A., Min J.K., Brock K., Gal Y., Marks D.S. Disease variant prediction with deep generative models of evolutionary data. Nature. 2021;599:91–95. doi: 10.1038/s41586-021-04043-8. [DOI] [PubMed] [Google Scholar]
- 38.Jumper J., Evans R., Pritzel A., Green T., urnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang N., Daniels R., Hebert D.N. The cotranslational maturation of the type I membrane glycoprotein tyrosinase: The heat shock protein 70 system hands off to the lectin-based chaperone system. Mol. Biol. Cell. 2005;16:3740–3752. doi: 10.1091/mbc.E05-05-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng K., He B., Yang B.B., Xu T., Chen X., Xu M., Liu X., Sun H., Pan Y., Wang S. The pro-metastasis effect of circANKS1B in breast cancer. Mol. Cancer. 2018;17:160. doi: 10.1186/s12943-018-0914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Shi L., Zhang K., Zhang Y., Hu S., Zhao T., Teng H., Li X., Jiang Y., Ji L., Sun Z. VarCards: An integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2018;46:D1039–D1048. doi: 10.1093/nar/gkx1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 44.Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 45.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., Velankar S., Kleywegt G.J., Bateman A., Evans R., Pritzel A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao G., Li J., Tang Y. AsCRISPR: A web server for allele-specific single guide RNA design in precision medicine. CRISPR J. 2020;3:512–522. doi: 10.1089/crispr.2020.0071. [DOI] [PubMed] [Google Scholar]
- 47.Zheng H., Zhou L., Shi Y., Tian J., Wang F. Tyrosinase-based reporter gene for photoacoustic imaging of MicroRNA-9 regulated by DNA methylation in living subjects. Mol. Ther. Nucl. Acids. 2018;11:34–40. doi: 10.1016/j.omtn.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.