Abstract
High-grade (WHO grades III-IV) glioma remains one of the most lethal human cancers. Adoptive transfer of tumor-targeting chimeric antigen receptor (CAR)-redirected T cells for high-grade glioma has revealed promising indications of anti-tumor activity, but objective clinical responses remain elusive for most patients. A significant challenge to effective immunotherapy is the highly heterogeneous structure of these tumors, including large variations in the magnitudes and distributions of target antigen expression, observed both within individual tumors and between patients. To obtain a more detailed understanding of immunotherapy target antigens within patient tumors, we immunochemically mapped at single cell resolution three clinically-relevant targets, IL13Rα2, HER2 and EGFR, on tumor samples drawn from a 43-patient cohort. We observed that within individual tumor samples, expression of these antigens was neither random nor uniform, but rather that they mapped into local neighborhoods – phenotypically similar cells within regions of cellular tumor – reflecting not well understood properties of tumor cells and their milieu. Notably, tumor cell neighborhoods of high antigen expression were not arranged independently within regions. For example, in cellular tumor regions, neighborhoods of high IL13Rα2 and HER2 expression appeared to be reciprocal to those of EGFR, while in areas of pseudopalisading necrosis, expression of IL13Rα2 and HER2, but not EGFR, appeared to reflect the radial organization of tumor cells around hypoxic cores. Other structural features affecting expression of immunotherapy target antigens remain to be elucidated. This structured but heterogeneous organization of antigen expression in high grade glioma is highly permissive for antigen escape, and combinatorial antigen targeting is a commonly suggested potential mitigating strategy. Deeper understanding of antigen expression within and between patient tumors will enhance optimization of combination immunotherapies, the most immediate clinical application of the observations presented here being the importance of including (wild-type) EGFR as a target antigen.
Keywords: Glioblastoma, tumor heterogeneity, spatial organization of glioblastoma, antigen escape, immunotherapy, CAR T cells, IL13Rα2, HER2, EGFR
Introduction
The present standard of care for high-grade glioma (WHO grades III-IV) remains maximal surgical resection followed by a combination of radiation, chemotherapy [1], and electric field stimulation [2]. Unfortunately, despite considerable efforts, achieving effective and durable responses to treatment has remained elusive [3].
Immunotherapies in various forms [4], among them chimeric antigen receptor (CAR) T cell immunotherapy 5, 6, 7, 8, 9, 10, 11, 12, have provided tantalizing evidence of clinical responses [9,13] that, almost inevitably, have been followed by tumor recurrence. Failure to achieve satisfactory clinical responses has been attributed to multiple tumor responses initiated by the selection pressures imposed by immunotherapies, including, but not limited to, enhancement of the already highly disseminated nature of high grade glioma, activation of resistance mechanisms originating in the tumor microenvironment [9,14], and the potential for evasion of immune targeting (antigen escape) conferred by phenotypic heterogeneity within tumors 15, 16, 17, 18, 19, 20.
Work over the last several decades has revealed complex spatially heterogeneous distributions of genetic, epigenetic, metabolic and protein expression patterns that collectively define local neighborhoods within brain tumors [21]. We believe that fine-grained cell-level analyses across multiple patients will be required to achieve the level of understanding of high-grade glioma [22] required to design the most effective therapies.
Immunotherapies ideally target tumor-associated antigens with elevated expression on glioma cells with little or no confounding expression on normal cells in the brain or elsewhere. Several antigens fulfilling this requirement have been described, proposed, or are under evaluation, in ongoing clinical trials of chimeric antigen receptor (CAR) redirected T cells: IL13Rα2 [16,23]; HER2 24, 25, 26; EphA2 [27]; and wild-type EGFR as well as the EGFRvIII variant 28, 29, 30.
Our laboratories have focused on acquiring a deeper understanding of the organization of cell populations within the tumors of individual patients (intra-tumor heterogeneity), and of how these vary between different patients (inter-patient variability). In this study we consider spatial expression three immunotherapy target antigens across a cohort of patient samples: IL13Rα2, HER2 and EGFR. IL13Rα2 is a high-affinity IL13 receptor expressed by a high percentage of high-grade gliomas [23,31, 32, 33, 34, but not significantly in normal brain. The HER2/ErbB2 receptor tyrosine kinase is expressed by 15-30% of GBMs and is not found on normal postnatal neurons and glial cells [35,36]. EGFR is over-expressed in well over half (55-70%) of newly diagnosed glioblastomas [37], with appearance of the EGFRvIII mutation, when present, overlapping with wild-type EGFR 38, 39, 40. Immunological targeting of IL13Rα2, HER2 and EGFR individually has been shown to be safe in clinical practice [16,26,28].
We have here defined by immunochemistry the spatially-varying expression patterns of these three immunotherapy targets in formalin-fixed paraffin-embedded (FFPE) tissue samples from a cohort of 43 patients with high-grade glioma (WHO grades III-IV). We observed that when examined at cell-level resolution, target antigen expression was for the most part organized into millimeter-scale neighborhoods – phenotypically similar cells within regions of cellular tumor [21] –shaped by not well understood combinations of cell intrinsic processes and impinging microenvironmental cues. Strikingly, when considered across cellular tumor areas of many patient samples, the spatial extent of EGFR-dominant neighborhoods appeared be reciprocal to the distribution of IL13Rα2 and HER2. Areas of pseudopalisading tumor cells, however, showed tighter spatial organization in which expression of IL13Rα2 and HER2 could vary over a few cell diameters in cells arrayed around hypoxic centers [41]. Curiously, in these areas of pseudopalisading tumor cells, EGFR expression appeared independent of the hypoxic gradients presumed to shape IL13Rα2 and HER2 expression.
In future investigations of fundamental tumor biology we will consider the multiple overlapping mechanisms that together shape the organization of these antigen expression neighborhoods. Clinically, our data suggests that optimal multiantigen targeting strategies to “box in” high-grade gliomas and minimize antigen escape to yield more effective and durable therapeutic responses [42], 43, 44 should incorporate targeting of (wild-type) EGFR.
Materials and methods
Patient samples
A cohort of 43 deidentified high-grade glioma brain tumor samples (Supplemental Table 1), unselected except for the size of the tissue section, were drawn from archival material in the tumor bank maintained by the COH Department of Pathology: 25.6% (11/43) WHO grade III and 74.4% (32/43) WHO grade IV. Brain tumor samples were acquired at resection or biopsy from patients with either progressing or recurrent tumors (the patient populations available to us), and therefore from patients previously exposed to other therapies but who had not initiated immunotherapy. At the time that most of these tumor samples were collected, between 2004 and 2013, information on IDH status, EGFR amplification, TCGA subtype, and other tumor characteristics was not collected and therefore, while potentially informative, was not available. Some information on the tumors was incorporated in pathology reports, and is presented in Supplementary Table 1. As controls, five normal human brain samples were purchased from US Biomax (GL803), and four normal brain tissue samples (frontal cortex, hippocampus, medulla, and pons) were provided by the City of Hope Department of Pathology. All procedures and protocols were approved by the City of Hope Institutional Review Board (IRB).
Immunohistochemical staining
Serial 4 µm-thick sections were cut from formalin-fixed, paraffin-embedded (FFPE) blocks to generate sequential slides for multiple antigen staining. In each case, the first slide was processed for hematoxylin and eosin (H&E) staining, followed by (in sequence) EGFR, IL13Rα2, and HER2 immunohistochemistry (IHC), each with hematoxylin counterstain, using Envision+System-HRP 3,3′-diaminobenzidine (DAB) chemistry and AutostainerPlus hardware (both Dako) by the City of Hope Research Pathology Core shared resource. All patient sample and control slides were processed together to minimize batch effects. Antibodies and dilutions were: EGFR (1:100, Invitrogen 28-0005); IL13Rα2 (1:600, R&D Systems AF146); HER2 (1:200, Dako A0485). Cell populations identified by EGFR immunoreactivity likely included those displaying the EGFRvIII variant, as EGFRvIII expression is almost always associated with wild-type EGFR 38, 39, 40.
Visual evaluation of tumor grade and antigen expression
For each of the 43 patient tumor samples, the H&E slide was annotated by a neuropathologist (D'Apuzzo) to define four major histological regions: cellular tumor (defined as >60% tumor cells), pseudopalisading within cellular tumor areas, infiltrating tumor (<50% tumor cells), and normal brain (approximately 0% tumor cells). Intensity scales for each antigen were set based on positive and negative controls: testis and prostate tissue for IL13Rα2, and breast cancer and lung cancer for HER2 and EGFR, respectively. Visual scoring of antigen expression (H score = 0-300) used the system: intensity (1-3) × percent positive cells.
High resolution slide scanning
The stained tumor sections were imaged in their entirety at high resolution (0.46 × 0.46 µm pixels) using a Hamamatsu Nanozoomer 2.0 HT scanner (20 × source lens) (City of Hope Light Microscopy and Digital Imaging Core). Images were saved in NDPI format (Hamamatsu) and examined with NDPI.view v2 (Hamamatsu) or ImagePro Premier v9.1 (Media Cybernetics). For quantitative measurements, images were down-sampled 1:4 (to 1.84 × 1.84 µm pixels) and converted to TIFF format (Icy; http://icy.bioimageanalysis.org/), and then deconvolved into DAB and hematoxylin (nuclear counter stain) channels (Fiji/ImageJ; https://fiji.sc/) [45].
Quantitative considerations of target antigen expression
Images of serial sections were brought into registration using a two-step process. First, the images of deconvolved hematoxylin-stained nuclei were optimized for contrast and aligned [Fiji/ImageJ; Register Virtual Stack Slices (affine feature extraction, elastic registration by bUnwarpJ splines)]. Then the transformation parameters for the nuclear images were applied to the immunostained (DAB) sections (Fiji/ImageJ; Transform Virtual Stack Slices). The segmented immunostained images could then be pseudocolored and superimposed.
To examine histological regions of interest (ROIs) identified from H&E stained slides (cellular tumor, for example), ROIs were mapped onto sequential immunostained sections for region-specific analyses. Expression levels of EGFR, IL13Rα2 and HER2 in these ROIs were measured as regional integrated optical density (rIOD) using the DAB plug-in of ImagePro. Within each histologically-defined ROI, ImagePro identified features as areas of grouped immunostained pixels, and for each feature returned integrated optical density (IOD) and feature area. Values of rIOD, IOD scaled to the feature area within each ROI, were calculated as 255 – [(ΣIOD / Σarea) × (average area)]. These values of rIOD were used as metrics of antigen expression within histologically-defined regions. The threshold for positive expression of each antigen was two times the average rIOD determined for a cohort of normal brain controls (Figure 6B).
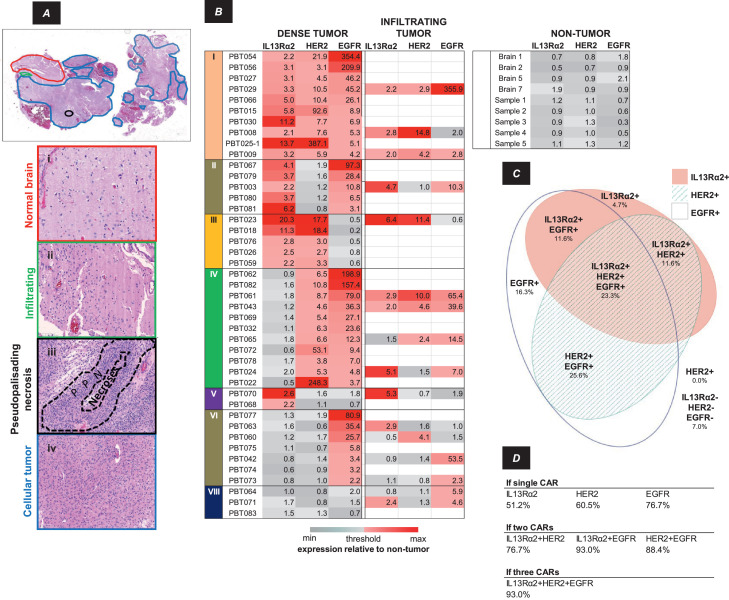
Figure 6.
Inter-patient heterogeneity of IL13Rα2, HER2 and EGFR expression within a cohort of 43 patients. A, H&E section annotated for four tissue characteristics as illustrated in the panels below: Normal brain (red), Infiltrating (green), Cellular (dense) tumor (blue), Pseudopalisading necrosis (black). B, Target antigen rIOD judged for the entirety of cellular tumor or infiltrating tumor (when present) regions in each patient sample relative to values in non-tumor tissues, colored low (gray) → high (red). Threshold was defined as 2 × the average rIOD in non-tumor samples. The eight possible combinations of three antigens are noted as I-VIII. Not all target antigens in each patient tumor sample showed expression above threshold. C, Euler diagram indicating the percentages of cellular tumor regions within the 43-patient cohort showing each of the eight possible expression patterns. The area of each ellipse is proportional to the percent of cellular tumor area expressing IL13Rα2, HER2 or EGFR, with overlap indicating co-expression within the tumor sample. D, Presumptive coverage of patients’ cellular tumor regions assuming delivery of CAR T cells targeting one, two or three antigens.
Alternatively, to evaluate IL13Rα2, HER2 and EGFR expression in regions of each tumor without prior selection of ROIs, a grid (5 × 5 pixels; 9.2 × 9.2 µm; total area 84.64 µm2) was imposed on the IL13Rα2/HER2/EGFR TIFF image stack, and OD was extracted for each antigen in each position. These were then plotted (down-sampled 10:1 for efficiency) using Origin (Origin Lab; Northampton, MA) with the presence of each antigen in each position indicated by color, and staining intensity (OD) by symbol size and transparency (Figure 2; Supplementary Figure 1) . Proportions of total cellular tumor area occupied by each of the eight possible combinations of three tumor antigens were computed for each position in the grid, and visualized in Euler diagrams (EulerAPE) [46] (Figures 2 and 6C; Supplementary Figure 1). Additionally, we determined the total percent of tumor area occupied by each antigen, singly or in combination (Figure 3).
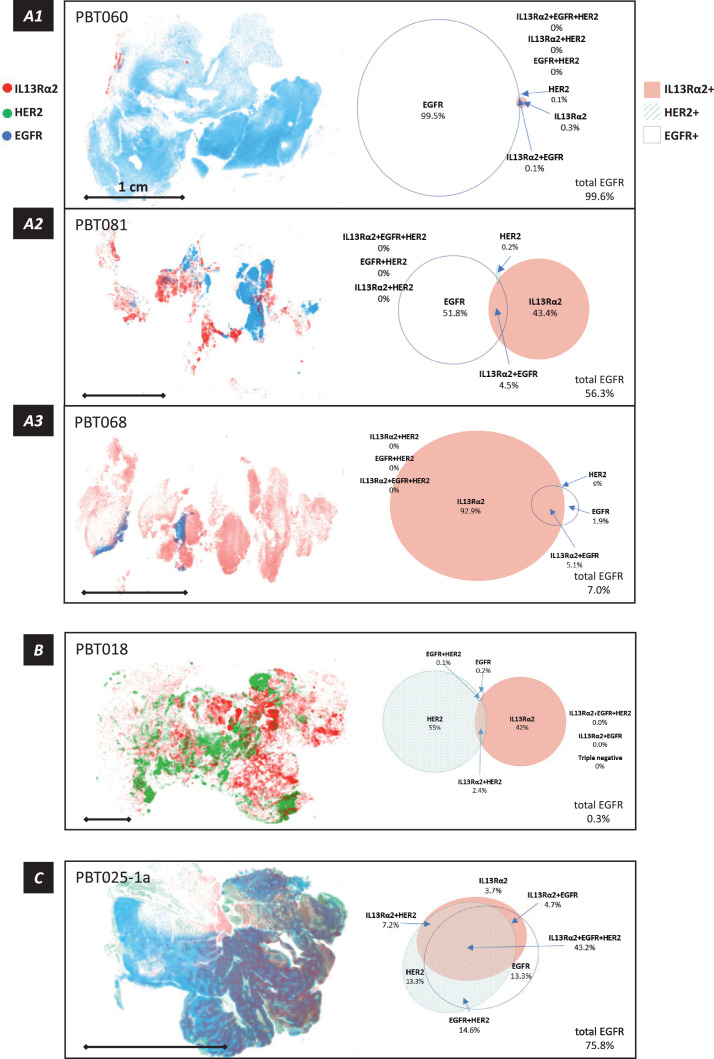
Figure 2.
Intra-tumoral organization of IL13Rα2, HER2 and EGFR antigen expression neighborhoods in cellular tumor regions, as determined from a grid superimposed on aligned serial sections. Within each grid element (9.2 × 9.2 µm), optical density (OD) for IL13Rα2 (red), HER2 (green) and EGFR (blue) was measured, scaled relative to the maximum value in that section, and plotted with OD determining symbol size [smaller (lower) → larger (higher)], and transparency [more transparent (lower) → more opaque (higher)]. For clarity of these plots, the threshold for display was 30% of maximum, and grid elements were binned 10:1. Associated with each map is an Euler diagram presenting the proportions of cellular tumor regions in each tumor section occupied by each of the eight possible antigen combinations. Scale bar = 1 cm in each panel. A1-A3, Three tumor samples (with relatively low HER2 expression) illustrating tumor-to-tumor progression from EGFR-dominant to IL13Rα2-dominant antigen expression. B, A tumor sample (PBT018) with very low EGFR expression, illustrating intermixing with relatively little high level co-expression in IL13Rα2- and HER2-dominant neighborhoods. C, A portion of tumor sample PBT025-1 with extensive pseudopalisading necrosis (PBT025-1a), showing the relation between the structure of the tumor cell palisades and target antigen expression (considered in more detail in Figure 4). The Euler diagram illustrates the extensive overlap of antigen expression within this subregion of pseudopalisading necrosis.
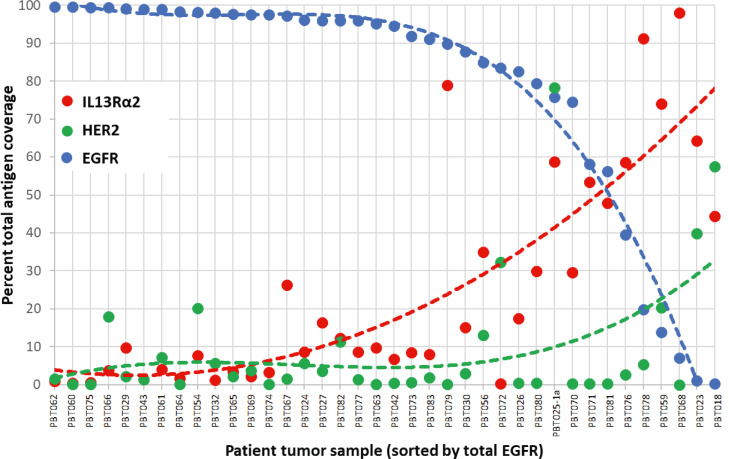
Figure 3.
Total percent of cellular tumor regions in each section occupied by each of the targeted antigens (IL13Rα2, HER2 and EGFR), ordered by total EGFR occupancy. Data are shown for the entirety of tumor samples, with the exception of PBT025a encompassing the area of pseudopalisading necrosis, and illustrate the overall reciprocal relationship between expression of EGFR and IL13Rα2 (and to some extent HER2). The complete set of antigen immunoreactivity maps from which these values were taken are shown in Supplemental Figure 1. The dotted lines highlight trends in the data.
Statistical considerations
Statistical significance was evaluated in Prism v8 (GraphPad Software) by Student's t-test; significance was assessed as p < 0.05.
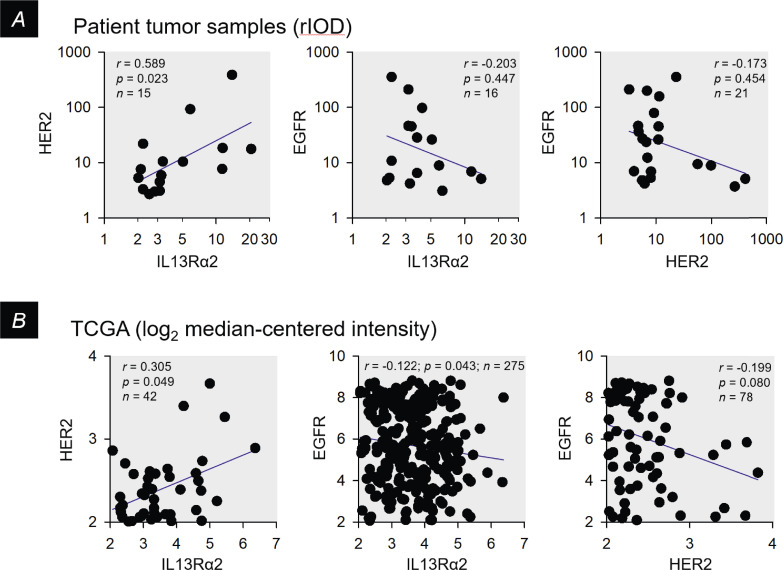
The Spearman correlation coefficient (r) for non-parametric data, calculated using Prism v8, was used to compare software-based quantification with conventional pathology scores (Figure 1C), to compare expression levels of IL13Rα2, HER2 and EGFR between tumor samples (Figure 7A), and to evaluate The Cancer Genome Atlas (TCGA) [47] expression data (Figure 7B). TCGA mRNA expression levels (Affymetrix U133A array) were determined using probes 206172_at for IL13Rα2, 216836_s_at for ERBB2, and 201983_s_at for EGFR, with threshold defined as 2-fold over-expression compared to normal brain.
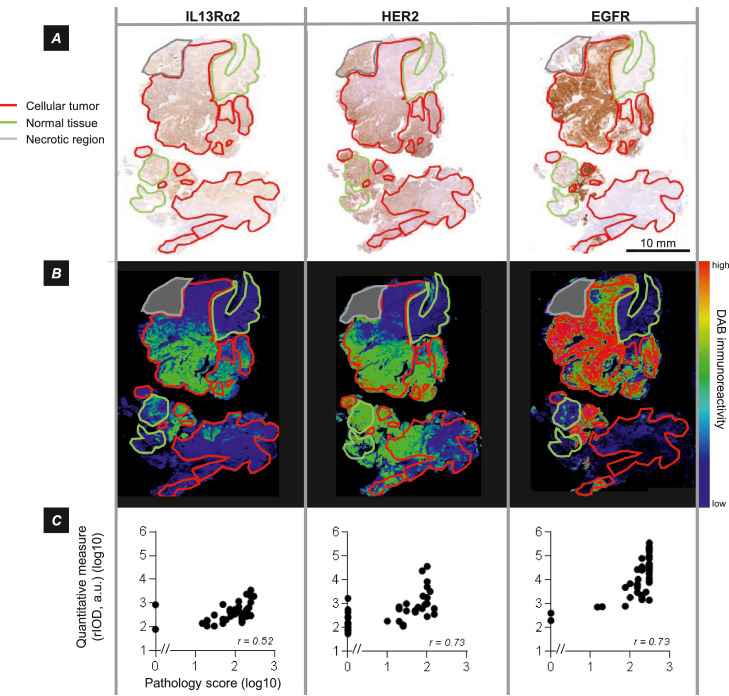
Figure 1.
Spatially varying expression of immunotherapy target antigens IL13Rα2, HER2 and EGFR. A, Immunohistochemical (DAB) visualization of target antigens in aligned serial sections, annotated for cellular tumor regions as well as for normal and necrotic regions, illustrating variation in their expression across the tumor section. Note in these sections the absence of immunostaining for CAR T cell target antigens in regions of normal brain. Tumor PBT025-1 B, DAB optical density for images in part A, pseudocolored blue (low) → red (high). The same color scale was used for each image. C, Overall agreement of visual immunostaining score by a neuropathologist [H score = intensity (1-3) × percent positive cells], and quantitative measures of regional integrated optical density (rIOD) for the same tumor sections. Spearman coefficients are indicated; p = <0.001 for all comparisons.
Figure 7.
Examination of pair-wise correlations of IL13Rα2, HER2 and EGFR antigen expression within cellular regions of patient tumors. A, For patient tumor samples, correlation of IL13Rα2 and HER2 rIOD values, and anti-correlation of IL13Rα2 or HER2 with EGFR rIOD values, within cellular tumor regions for the subset of tumors showing above-threshold expression of both antigens (from Figure 6B). B, Similar presentation of mRNA expression profiles from the TCGA data set for glioblastoma [47], with threshold defined as 2-fold overexpression compared to control brain. In all graphs, blue lines are linear regressions to illustrate overall trends in the data.
The Shannon diversity index (H) is a metric, viewed in an ecological context, of species diversity within a mixed population of many species. It incorporates consideration of the number of species in the population (richness) and their relative abundance (evenness). Here, for cellular tumor regions, N is the number of antigen species, in this case three (IL13Rα2, HER2, and EGFR), and pi (calculated from relative rIOD) is the proportional expression of each antigen within a given region or neighborhood. It is evaluated as ln(pi). Thus an H value of 0 indicates no diversity (one antigen present), and increasing values of H reflect the presence of both multiple antigens and greater differences in their relative expression.
Results
Expression of potential immunotherapy target antigens was evaluated in individual surgical resections from a 43-patient cohort (Supplemental Table 1). For each tumor sample, consecutive FFPE sections were H&E stained, or immunostained for the immunotherapy target antigens of interest: IL13Rα2, HER2, and EGFR. From the H&E sections, a neuropathologist outlined histologically-defined regions of normal brain, infiltrating tumor, cellular tumor, pseudopalisading necrosis (a subset of cellular tumor), and necrotic tissue that was omitted from any further analyses. In the example of immunostained sections shown in Figure 1A (PBT025-1) and presented in pseudocolor in Figure 1B, it is clear that within cellular tumor regions, expression of IL13Rα2, HER2 and EGFR could be intermixed and not spatially uniform.
In this study we evaluated antigen expression by pixel-level quantitative measures of DAB-visualized immunoreactivity, expressed as regional integrated optical density (rIOD) in tumor regions, or as OD within elements of grids superimposed on regions of cellular tumor. The comparisons presented in Figure 1C confirm that rIOD measurements and visual scoring (H scoring) show similar trends and significant correlation (p = <0.001 for each of the three antigens). Note that quantitative measures of antigen expression revealed fine distinctions within the cellular tumor regions visually assigned a score of zero.
Intra-tumoral heterogeneity
We examined variations in antigen expression within cellular tumor regions of patient samples using three approaches: an arbitrarily imposed grid, a structural morphology highly characteristic of glioblastoma (WHO grade IV), and subregions identified by image analysis software.
(a) Antigen expression within elements of an imposed grid
We mapped antigen expression across cellular tumor regions by arbitrarily superimposing a grid onto the aligned sections of each of the 38 patient tumors for which this analysis was possible, with each element a 5 × 5 square of 1.84 × 1.84 µm pixels (9.2 × 9.2 µm; total area of 84.6 µm2). The optical density (OD) for each antigen averaged across each grid element was then determined and mapped. Five illustrative examples of alternative expression patterns are presented in Figure 2; the entire series of patient samples is presented in Supplemental Figure 1. In these maps, for visual clarity grid elements were binned 10:1 (92 × 92 µm; 8,464 µm2), and average OD in each binned element is represented in diameter and opacity. For each antigen, the threshold for display was 30% of maximum OD, emphasizing neighborhoods of higher antigen expression. Adjacent Euler diagrams plot the percentages of the total cellular tumor area occupied by each of the eight combinations of the three antigens.
These expression maps demonstrate non-random and non-independent antigen expression in neighborhoods within cellular tumor regions, with for the most part little spatial overlap in areas with the highest antigen expression levels. In one common pattern, illustrated in Figure 2A1-A3, there is a progression from high EGFR to high IL13Rα2 spatial occupancy, with relatively few neighborhoods of high HER2 expression. In the cellular tumor regions with the lowest EGFR occupancy, PBT018 (Figure 2B, Supplemental Figure 1) and PBT023 (Supplemental Figure 1), neighborhoods of high HER2 and IL13Rα2 immunostaining were intermixed; this is a pattern never seen when EGFR immunostaining was prominent except in a single case, PBT079 (Supplemental Figure 1). Pseudopalisading necrosis (Figure 2C), however, imposed a different antigen expression pattern that closely followed tumor morphology (considered in more detail below).
When examined across all 38 tumor samples, a progression from high to low percentage of total EGFR occupancy (determined from the Euler diagrams for each tumor) was accompanied by an inverse and reciprocal progression for IL13Rα2. This can be seen in Supplemental Figure 1, where expression maps and Euler diagrams for cellular regions of the 38 tumor samples are ordered according to the total percentage area occupied by EGFR in each sample (high → low). Figure 3 presents these data graphically: in most cellular tumor regions, lower percentages of EGFR occupancy are associated with higher percentage coverage by IL13Rα2, and, at the lowest EGFR occupancies, HER2.
(b) Granularity of target antigen expression in an area of pseudopalisading necrosis
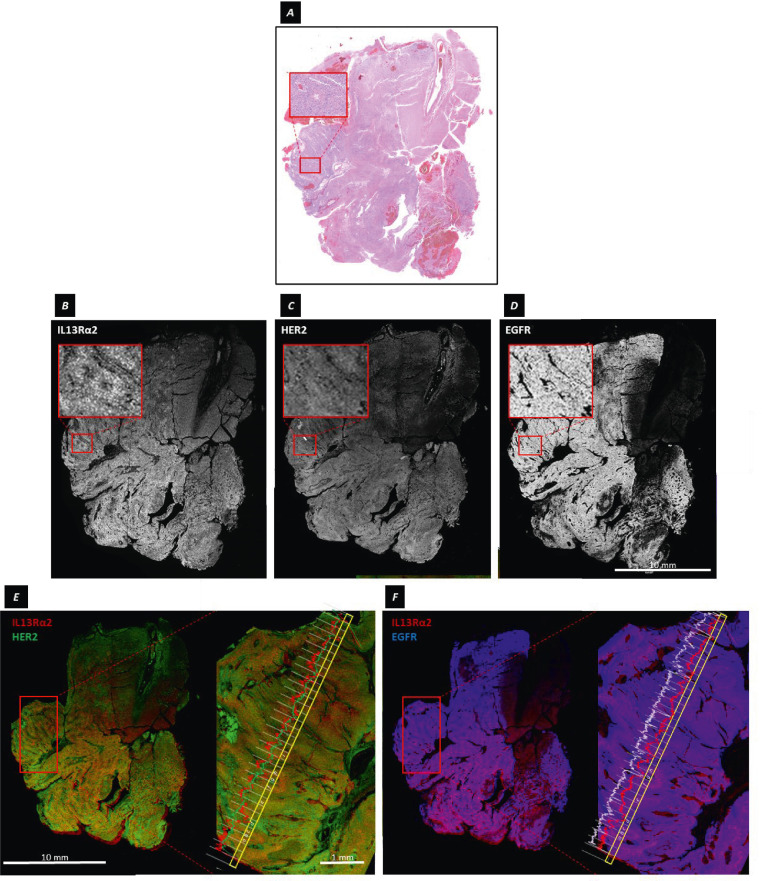
As noted above, the cellular tumor subregion of pseudopalisading necrosis (a cell architecture characteristic of GBM) in PBT025-1a (Figure 4A) presented a pattern in which IL13Rα2 and HER2 expression varied according to tumor architecture (evident in Figure 2C), and were organized around the central area of presumed hypoxia [41]. Within this space, IL13Rα2 immuoreactivity closely followed the radially-arranged contours of tightly packed tumor palisades (Figure 4B), with the distribution of HER2 immunoreactivity being somewhat more diffuse (Figure 4C) but still reflecting the underlying pseudopalisading structure. In contrast, EGFR immunoreactivity was more uniformly distributed (Figure 4D). As illustrated in Figures 4E and 4F, where lines plot the ODs of IL13Rα2 and HER2, or IL13Rα2 and EGFR, immunoreactivities across a transect of pseudopalisading cells, changes in IL13Rα2 expression (red) could occur over 3-5 cell diameters, often but not always in parallel with HER2 immunoreactivity (green) (Figure 4E). In contrast, over the same transect, EGFR immunoreactivity (blue-white) was relatively uniform as compared to IL13Rα2 (red), decreasing only over areas devoid of cells (Figure 4F).
Figure 4.
Differing microgranularity of IL13Rα2, HER2 and EGFR expression at the resolution of single pixels. A, H&E stained section of tumor sample PBT025-1a. The inset shows a subregion of pseudopalisading necrosis within the region of cellular tumor. B-D, Gray-scale images of aligned serial sections immunostained for IL13Rα2,HER2, and EGFR. The insets show the subregion of pseudopalisading necrosis, and illustrate in B rapid variation of IL13Rα2 immunoreactivity over a few cell diameters around the presumably hypoxic core, (C) more dispersed HER2 immunoreactivity, and (D) relatively uniform EGFR immunoreactivity. E-F, Superimposed aligned tumor sections showing (E) IL13Rα2 and HER2, and (F) IL13Rα2 and EGFR. The enlarged images on the right of E and F are the subregion of pseudopalisading necrosis, with a transect (yellow) and plot of OD across each pixel-length portion of the transect averaged across its width. Along the transect in E, the ODs of IL13Rα2 (red) and HER2 (green) immunostaining varied with the architecture of the pseudopalisades, although not necessarily in parallel (compare c at the peak of the palisade with d in the trough, or g and h). Along the same transect in F, the OD of EGFR (blue-white) immunostaining remained essentially constant except when the transect crossed areas of low cellularity (d, g).
(c) Heterogeneity within subregions of cellular tumor
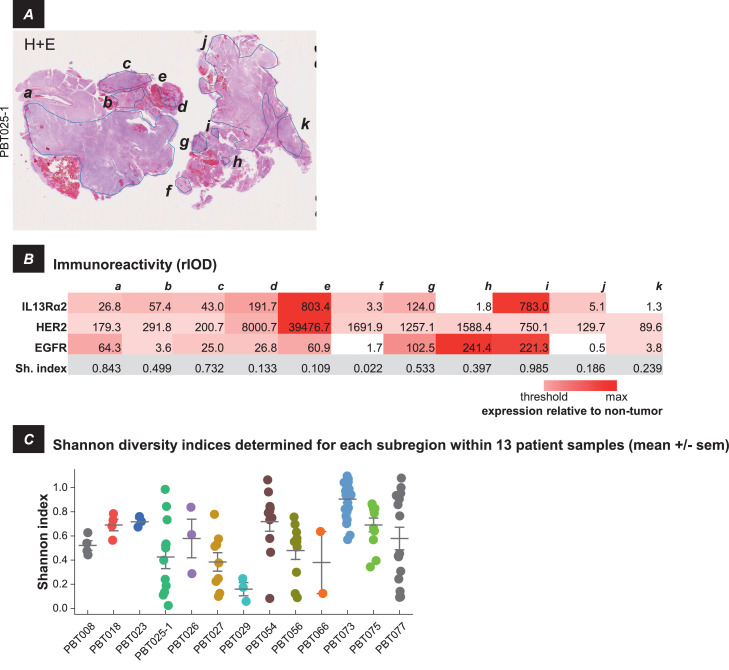
When multiple subregions of DAB immunostaining were identified by ImagePro within cellular tumor regions, antigen expression in these subregions could vary widely. This is illustrated by patient sample PBT025-1, in which all three target antigens were expressed in spatially disparate subregions. In this tumor sample, 11 subregions were identified, a-k in Figure 5A. Of these areas (Figure 5B; intensity coded light red → dark red), seven (a, b, c, d, e, g, and i) showed elevated levels of IL13Rα2, two (f and j) showed low IL13Rα2 levels, and two (h and k) were IL13Rα2 negative.
Figure 5.
Intra-tumoral heterogeneity of IL13Rα2, HER2 and EGFR antigen expression within separate subregions of cellular tumor. A, H&E section of tumor sample PBT025-1, with 11 separate cellular tumor subregions identified by the DAB plug-in of ImagePro, outlined and labeled a-k. B, Immunoreactivity (rIOD) within each of these subregions, colored in the table white (below threshold) → red (maximum). The bottom row shows the Shannon diversity index (Sh. Index) calculated for each of the subregions, with higher numbers indicating greater diversity. C, Shannon diversity indices for separate subregions within cellular tumor regions of each of 13 patient samples for which these could be identified. Data are mean +/- s.e.m.
Overall, there were 13 tumor samples (of 43 total) within which multiple subregions of antigen expression were detected by the DAB plug-in to ImagePro and could be considered individually (in the remainder, antigen expression in cellular regions was more spatially uniform). This variability was quantified by calculating the Shannon diversity index (H) for antigen expression each of these areas. Here, H is an index increasing in value with the numbers of antigens expressed and with greater differences in their relative abundance. In the majority of tumor samples (9 of 13), values of H were highly variable, indicating that levels of antigen expression across cellular tumor regions could be highly divergent. Only in tumor samples with the smallest number of subregions (PBT008, PBT018, PBT023, PBT029) were the values of H tightly clustered.
Inter-patient heterogeneity: variations of antigen expression between patients
We next assessed how tumor antigens were expressed in tumors considered as a whole across multiple patients. Our analysis is summarized in Figure 6B for regions of cellular tumor/pseudopalisading necrosis, and regions of infiltrating tumor when these were present (examples shown in Figure 6A). Observations were grouped according to the eight possible patterns of IL13Rα2, HER2, and EGFR expression, considered individually, and in combinations (designated I-VIII). Values of rIOD, presented as normalized to average values in non-tumor brain, are presented numerically and as color scale (gray → red). The immunostaining threshold for positive expression was defined as an rIOD twice the average in non-tumor brain.
While vagaries of antigen accessibility and antibody avidity confound quantitative comparisons between antigens, we take these measures of antigen expression relative to control tissue to be appropriate for semiquantitative comparisons of each antigen.
Within regions of cellular tumor, and infiltrating tumor when present, antigen expression varied markedly between patient samples, and, as expected, expression of multiple antigens was common. Within regions of infiltrating tumor, antigen expression patterns could be similar (50%; 8/16) or divergent (50%; 8/16) from those of cellular tumor regions. As infiltrating tumor was present in only a minority of our samples (37%; 16/43), we consider here only the cellular tumor regions present in all samples. For these regions of cellular tumor, the combinatorics of antigen expression are illustrated by the Euler diagram in Figure 6C. Except for EGFR, relatively few of the 43 tumor samples were scored as expressing single antigens (16.3% for EGFR, versus 4.7% for IL13Rα2 and none for HER2). Relatively more tumors were scored as double positive: 11.6% for EGFR and IL13Rα2; 11.6% for IL13Rα2 and HER2; 25.6% for EGFR and HER2). The percentage of triple positive tumors (IL13Rα2, HER2 and EGFR was 23.3%. Only 7.0% of samples were negative for all three antigens.
Overall, cellular tumor areas in 93.0% of patient samples expressed at least one of the three potential target antigens, and greater than 72.1% of tumors expressed two or three of these antigens. Based on these data, Figure 6D presents the percentages of patient tumors potentially targeted by single, dual, or triple CAR T cell therapies. Depending on the particular CAR T cell targets, multiple CAR T cell targeting increased potential patient coverage from as low as 51.2% for a single CAR to as high as 93.0% for all three CAR species.
Examination of mRNA expression in the TCGA database [47] yielded a similar pattern. Overall, when considered alone or in combination with other antigens, 54.0% (293/543) of tumors would be accessible to an IL13Rα2-CAR T cell, 15.1% (82/543) to a HER2-CAR T cell, and 92.1% (500/543) to an EGFR-CAR T cell.
Non-random, non-independent expression of target antigens across patient tumor samples
For regions of cellular tumor, we next assessed how the eight possible combinations of antigen immunoreactivity differed from patient to patient, using the compilation presented in Figure 6B. Here, our results were highly divergent from expectations assuming that the probability of expression of each antigen was random and equal, with no interdependency. As summarized in Table 1, across the entire 43-patient cohort, EGFR appeared with significantly higher than expected frequency when considered as a single antigen, when paired with HER2, and when in combination with IL13Rα2 and HER2. Of note, this dominance of EGFR expression was also evident within multiple subregions of cellular tumor regions (Figure 5, Table 1). This pattern was confirmed for the 543 patient tumors in the TCGA database [47], which also yielded patterns of antigen transcript expression that differed significantly from expectations of random and independent expression, with the largest divergence also seen for EGFR (Table 1).
Table 1.
Combinatorics of CAR T cell target antigen expression compared to predictions assuming random and independent expression of each antigen.
 |
Eight possible combinations of three antigens: IL13Rα2, HER2, EGFR. Data are by protein immunochemistry for patient samples and patient sample regions, and RNA expression profiling for TCGA. P-values are the result of the binomial test comparing the predicted and observed percentages with the null hypothesis that the samples are from the same distribution (one-tailed). RED = expression significantly above expectation; BLUE = expression significantly below expectation
We also examined the expression data in Figure 6B for potential associations of antigen expression (Figure 7A). Here, IL13Rα2 and HER2 expression showed preferential pairing, with expression of these antigens trending to be negatively associated with EGFR. A similar pattern was observed for TCGA mRNA expression data [47] (Figure 7B).
Discussion and conclusions
Summary of findings
Here, we have used immunohistochemistry to map cellular tumor regions of formalin-fixed paraffin-embedded (FFPE) tissues from patients with high-grade glioma (WHO grades III-IV) to evaluate spatially varying expression of clinically-relevant immunotherapy target antigens. We compared our measurements within the tumors of individual patients, and across multiple patients. To overcome the limitations of manual semi-quantitative scoring, as well as the subjectivity of visually evaluating of IHC staining, we employed digital imaging methods to more quantitatively examine expression of multiple antigens within tumor regions defined according to conventional neuropathology criteria.
We focused on three immunotherapy target antigens that are currently being utilized in clinical trials for recurrent high-grade glioma: IL13Rα2, HER2, and EGFR [7]. We observed that expression of these antigens was highly-variable, non-homogeneous, non-random, and non-independent. We observed this at all resolutions of our measurements: over the individual elements of 9.2 µm grids superimposed on cellular tumor areas, over 10 cm-scale regions of cellular tumor in tumor sections evaluated as a whole, and in subregions of pseudopalisading necrosis characteristic of glioblastoma.
Distributions of target antigen expression
Across entire cellular tumor regions in tumor sections, neighborhoods of EGFR expression could predominate over millimeter to centimeter-sized areas. In these tumors, neighborhoods of IL13Rα2 and HER2 expression were quite compact, an example being PBT060 (Figure 2A1). In other tumors where EGFR expression was more spatially restricted, IL13Rα2 expression was progressively more expansive, as in PBT081 (Figure 2A2) and PBT068 (Figure 2A3). Finally, in tumors in which EGFR expression was minimal, such as PBT018, intermixed neighborhoods of IL13Rα2 and HER2 expression were seen (Figure 2B). When all of the tumor maps were arrayed in descending order of total percent of EGFR coverage (Supplemental Figure 1), a reciprocal relationship between spatial dominance of EGFR versus IL13Rα2/HER2 became apparent (Figure 3). This reciprocal pattern of EGFR and IL13Rα2/HER2 expression was preserved when tumor sections were evaluated as a whole (Figure 6B) and when plotted pair-wise (Figure 7A). TCGA mRNA expression profiles [47] showed a similar pattern (Figure 7B), suggesting that this relationship was not limited to our patient cohort.
Nonuniform spatially varying expression patterns have been seen for other cell-cell signaling elements in brain tumors: growth factors and their receptors including PDGF and PDGFR [48]; c-Met [49]; wild type EGFR and the EGFRvIII variant [38,39,50, 51, 52; pro-angiogenic angiopoietin-2 [53]; and integrins αvβ3 and αvβ5 [54]. While the origins of these neighborhoods are not well understood, they impose a structure on tumor heterogeneity.
Interplay of hierarchical lineage and microenvironment shape glioma phenotypic variation
Overall, high-grade glioma may be viewed as highly complex microenvironment overlaid on developing tumor lineages to give rise to heterogeneous neighborhood organizations not unique to these target antigens 55, 56, 57, [58].
Observations from single cell expression and genomic profiling [52,59, 60, 61, 62 suggest that variegated glioma cell phenotypes derive in part from the cell-intrinsic variation found in diverging lineages [63,64] differentiating within a stem cell hierarchy [65], and recapitulating that of normal brain development [66], including appearance of cancer stem-like cells resembling normal stem cells [67]. That heterogeneity of target antigen expression is maintained down to the finest granularity available to us is consistent with a role of EGFR (and other receptor tyrosine kinases) amplification in shaping of individual glioma cell phenotypes.
Microenvironment also appears to be critical. Hypoxic niches are proposed to be dynamically regulated organizers of glioblastoma [68], including pseudopalisading necrosis [41]. Thus, the aligned tumor cells and their highly spatially restricted pattern of IL13Rα2 expression in neighborhoods of pseudopalisading necrosis may reflect a common association with hypoxia, as hypoxia is reported to up-regulate IL13Rα2 expression [69], and the distribution of IL13Rα2 was coincident with the layer of cells adjacent to the presumably hypoxic core [70]. A surrounding region of less severe hypoxia was characterized by HER2 expression and the cell migration assembling tumor palisades [41].
Other microenvironmental variations related to the physical position of neighborhoods within tumor masses also appear to contribute to establishing neighborhood phenotypes. Border niches, sites of progression and recurrence, can contain variegated populations of oligodendrocyte progenitor cells (OPCs), macrophages/microglia, and glioma stem cells [71]. Here, abnormal secretion of EGF by glioma-associated microglia is proposed to initiate a feedback loop promoting GBM invasion and functional disruption of brain tissue [72]. EGF secreting tumor cells counter this self-reinforcing process, as increased EGF is reported to result in loss of EGFR gene amplification [73].
Other aspects of metabolism may also be reflected in multiple local metabolic neighborhoods [74].
Between neighborhoods, systems of reciprocal interactions [75] may be operating [56,76, 77, 78, [79]. The striking observation was that EGFR expression appeared anti-correlated with IL13Rα2 and HER2 at larger spatial scales, as suggested by both by our immunochemistry (Figure 7A) and by TCGA mRNA analyses (Figure 7B), may well indicate more widespread systems of mutual interactions.
Considerations for further development of glioma-directed immunotherapies
From a translational and clinical perspective, understanding patterns of tumor-related antigen expression in the context of high-grade glioma is an essential step towards developing effective immunotherapies.
One implication of our observations concerns the limited spatiotemporal sampling that can inform precision oncology for high-grade glioma [80]. Phenotypic and genetic aberrations [47,81,82] may be used to stratify patients and guide therapy choices [83]. We suggest the possibility, however, that the presence of highly variable phenotypic neighborhoods as reflected by regions of varying TCGA subtypes within individual tumors [63], by local and intermixed amplification of EGFR and other receptor tyrosine kinases [52], and by distortions of molecular heterogeneity in recurrent verses naïve tumors [84], may thwart application of narrowly targeted therapies [30]
Rather, as it is widely held that tumor recurrence has its origins in multiple preëxisting cell populations incorporating target antigen-deficient populations coming forward to fill the space (literally and in the evolutionary sense) vacated by killed tumor cells 15, 16, 17,19,20,85], examinations of antigen escape have led to considerations of potentially more effective immunotherapies targeting multiple antigens simultaneously.
One recently explored strategy involves designing dual or triple targeting CAR T cell therapies to substantially increase the proportion of tumor cells in primary patient samples potentially targetable by immunotherapies, thereby reducing opportunities for antigen escape [42], 43, 44. We suggest that the antigen expression patterns presented here indicate that multitarget immunotherapy targeting strategies incorporating EGFR along with IL13Rα2, HER2, or other targets, could be more clinically effective than antigen combinations omitting EGFR.
Alternatively, efforts are underway to design CARs recognizing immunotherapy targets broadly distributed within individual tumors and across patients. Examples include chondroitin sulfate proteoglycan 4 (CSPG4) [86] and the molecular complex bound by the scorpion-derived peptide chlorotoxin (CLTX) [87].
The evolutionary challenge of phenotypic heterogeneity in high-grade glioma
When considering the evolution of a species, theory posits that fitness in one environment does not necessarily guarantee fitness in a successor [88,89], and that genetic and phenotypic heterogeneity provides the raw material for adaptation under selection pressures [90,91]. This principle can be applied to consideration of malignancy, with the conclusion [92] that “… acquisition of phenotypic heterogeneity by populations of tumor cells imposes a degree of stability on the tumor as a whole”. Thus, clinically, heterogeneity within tumors confers resilience to the challenges imposed by radiation, chemotherapies, electric fields, and immune-based therapies, resilience that ultimately leads to treatment failure and recurrence [14,93]. Or, from a patient's perspective, more extensive and dynamic heterogeneity of antigen expression correlates with less favorable prognoses [59,94]. Thus, evolutionary theory suggests that surmounting the consequences of the extensive heterogeneity of high-grade glioma will remain a major challenge to effective and durable therapies.
Financial Support
This work was supported in part by California Institute for Regenerative Medicine (CIRM) grants TR3-05641 and CLIN2-10248, and the Ben and Catherine Ivy Foundation.
The results shown here are in part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga).
Research reported in this publication included work performed in the City of Hope Pathology Research Services Core, supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CRediT authorship contribution statement
Michael E. Barish: Conceptualization, Methodology, Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization, Supervision, Funding acquisition. Lihong Weng: Conceptualization, Methodology, Formal analysis, Investigation, Visualization. Dina Awabdeh: Formal analysis, Visualization. Yubo Zhai: Methodology, Formal analysis, Investigation. Renate Starr: . Massimo D'Apuzzo: Resources, Data curation. Russell C. Rockne: Formal analysis. Haiqing Li: Formal analysis. Behnam Badie: Resources. Stephen J. Forman: Supervision, Funding acquisition. Christine E. Brown: Conceptualization, Methodology, Data curation, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
S.J.F. and C.E.B. receive royalty payments from Mustang Bio; S.J.F, C.E.B. and M.E.B. receive royalty payments from Chimeric Therapeutics; C.E.B. and M.E.B. have equity interests in Chimeric Therapeutics; all in areas outside of the work presented here. All other authors declare no competing interests.
Acknowledgments
We thank Michael Nelson and Drs. James O'Hearn and Ian Talisman for their comments on the manuscript, Blake Brewster for assistance with the analyses, and Dr. Brian Armstrong and the Light Microscopy and Digital Imaging Core for assistance with image acquisition.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100801.
Contributor Information
Michael E. Barish, Email: mbarish@coh.org.
Christine E. Brown, Email: cbrown@coh.org.
Appendix. Supplementary materials
Supplementary Figure 1. Maps of IL13Rα2, HER2 and EGFR immunoreactivity and associated Euler diagrams.
Shown are data from 38 tumor samples with sufficiently large regions of cellular tumor for this mapping. Details of the construction of these maps are in the legend to Figure 2. Tumor sections are ordered by total area occupied by EGFR (left → right, top → bottom). In some tumor sections, the total percentage area occupied by HER2 was below the presentation threshold of 30% of maximum, and therefore not evident in all maps. Scale bar = 1 cm in each panel.
References
- 1.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, National Cancer Institute of Canada Clinical Trials Group, Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R., Taillibert S., Kanner A., Read W., Steinberg D.M., Lhermitte B., Toms S., Idbaih A., Ahluwalia M.S., Fink K., Di Meco F., Lieberman F., Zhu J.J., Stragliotto G., Tran D.D., Brem S., Hottinger A.F., Kirson E.D., Lavy-Shahaf G., Weinberg U., Kim C.Y., Paek S.H., Nicholas G., Burna J., Hirte H., Weller M., Palti Y., Hegi M.E., Ram Z. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma a randomized clinical trial. JAMA - Journal of the American Medical Association. 2017;318:2306–2316. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R., Hegi M.E. Targeting brain-tumor stem cells. Nat Biotechnol. 2007;25:193–194. doi: 10.1038/nbt0207-193. [DOI] [PubMed] [Google Scholar]
- 4.Lim M., Xia Y., Bettegowda C., Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 5.Irving M., de Silly R.V., Scholten K., Dilek N., Coukos G. Engineering chimeric antigen receptor T-cells for racing in solid tumors: Don’t forget the fuel. Front Immunol. 2017;8:267. doi: 10.3389/fimmu.2017.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirzaei H.R., Rodriguez A., Shepphird J., Brown C.E., Badie B. Chimeric antigen receptors T cell therapy in solid tumor: Challenges and clinical applications. Front Immunol. 2017;8:1850. doi: 10.3389/fimmu.2017.01850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bagley S.J., Desai A.S., Linette G.P., June C.H., O’Rourke D.M. CAR T-cell therapy for glioblastoma: recent clinical advances and future challenges. Neuro-oncol. 2018;20:1429–1438. doi: 10.1093/neuonc/noy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe K., Kuramitsu S., Posey A.D., June C.H. Expanding the therapeutic window for CAR T cell therapy in solid tumors: The knowns and unknowns of CAR T cell biology. Front Immunol. 2018;9:2486. doi: 10.3389/fimmu.2018.02486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akhavan D., Alizadeh D., Wang D., Weist M.R., Shepphird J.K., Brown C.E. CAR T cells for brain tumors: Lessons learned and road ahead. Immunol Rev. 2019;290:60–84. doi: 10.1111/imr.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen C.T., Krenciute G. Next generation CAR T cells for the immunotherapy of high-grade glioma. Front Oncol. 2019;9:69. doi: 10.3389/fonc.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuntova P., Downey K.M., Hegde B., Almeida N.D., Okada H. Genetically engineered T-cells for malignant glioma: Overcoming the barriers to effective immunotherapy. Front Immunol. 2019;9:3062. doi: 10.3389/fimmu.2018.03062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwok D., Okada H. T-Cell based therapies for overcoming neuroanatomical and immunosuppressive challenges within the glioma microenvironment. J Neurooncol. 2020;147:281–295. doi: 10.1007/s11060-020-03450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurz S.C., Wen P.Y. Quo Vadis—Do immunotherapies have a role in glioblastoma? Current Treatment Options in Neurology. 2018;20 doi: 10.1007/s11940-018-0499-0. [DOI] [PubMed] [Google Scholar]
- 14.Qazi M.A., Vora P., Venugopal C., Sidhu S.S., Moffat J., Swanton C., Singh S.K. Intratumoral heterogeneity: Pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol. 2017;28:1448–1456. doi: 10.1093/annonc/mdx169. [DOI] [PubMed] [Google Scholar]
- 15.Sampson J.H., Heimberger A.B., Archer G.E., Aldape K.D., Friedman A.H., Friedman H.S., Gilbert M.R., Herndon J.E., McLendon R.E., Mitchell D.A., Reardon D.A., Sawaya R., Schmittling R.J., Shi W., Vredenburgh J.J., Bigner D.D. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28:4722–4729. doi: 10.1200/JCO.2010.28.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown C.E., Badie B., Barish M.E., Weng L., Ostberg J.R., Chang W.C., Naranjo A., Starr R., Wagner J., Wright C., Zhai Y., Bading J.R., Ressler J.A., Portnow J., D’Apuzzo M., Forman S.J., Jensen M.C. Bioactivity and safety of IL13Ra2-redirected chimeric antigen receptor CD8+ T cells in patients with recurrent glioblastoma. Clin Cancer Res. 2015;21:4062–4072. doi: 10.1158/1078-0432.CCR-15-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rapoport A.P., Stadtmauer E.A., Binder-Scholl G.K., Goloubeva O., Vogl D.T., Lacey S.F., Badros A.Z., Garfall A., Weiss B., Finklestein J., Kulikovskaya I., Sinha S.K., Kronsberg S., Gupta M., Bond S., Melchiori L., Brewer J.E., Bennett A.D., Gerry A.B., Pumphrey N.J., Williams D., Tayton-Martin H.K., Ribeiro L., Holdich T., Yanovich S., Hardy N., Yared J., Kerr N., Philip S., Westphal S., Siegel D.L., Levine B.L., Jakobsen B.K., Kalos M., June C.H. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med. 2015;21:914–921. doi: 10.1038/nm.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown C.E., Alizadeh D., Starr R., Weng L., Wagner J.R., Naranjo A., Ostberg J.R., Blanchard M.S., Kilpatrick J., Simpson J., Kurien A., Priceman S.J., Wang X., Harshbarger T.L., D’Apuzzo M., Ressler J.A., Jensen M.C., Barish M.E., Chen M., Portnow J., Forman S.J., Badie B., D’Apuzzo M., Ressler J.A., Jensen M.C., Barish M.E., Chen M., Portnow J., Forman S.J., Badie B., D’Apuzzo M., Ressler J.A., Jensen M.C., Barish M.E., Chen M., Portnow J., Forman S.J., Badie B. Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N Engl J Med. 2016;375:2561–2569. doi: 10.1056/NEJMoa1610497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krenciute G., Prinzing B.L., Yi Z., Wu M.F., Liu H., Dotti G., Balyasnikova I.V., Gottschalk S. Transgenic expression of IL15 improves antiglioma activity of IL13Rα2-CAR T cells but results in antigen loss variants. Cancer Immunol Res. 2017;5:571–581. doi: 10.1158/2326-6066.CIR-16-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Rourke D.M., Nasrallah M.P., Desai A., Melenhorst J.J., Mansfield K., Morrissette J.J.D.D., Martinez-Lage M., Brem S., Maloney E., Shen A., Isaacs R., Mohan S., Plesa G., Lacey S.F., Navenot J.-M.M., Zheng Z., Levine B.L., Okada H., June C.H., Brogdon J.L., Maus M.V., O’Rourke D.M., Nasrallah M.P., Desai A., Melenhorst J.J., Mansfield K., Morrissette J.J.D.D., Martinez-Lage M., Brem S., Maloney E., Shen A., Isaacs R., Mohan S., Plesa G., Lacey S.F., Navenot J.-M.M., Zheng Z., Levine B.L., Okada H., June C.H., Brogdon J.L., Maus M.V. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017;9:eaaa0984. doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhate S.S., Barlow G.L., Schürch C.M., Nolan G.P. Tissue schematics map the specialization of immune tissue motifs and their appropriation by tumors. Cell Syst. 2022;13:109–130. doi: 10.1016/J.CELS.2021.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Park J.H., de Lomana A.L.G., Marzese D.M., Juarez T., Feroze A., Hothi P., Cobbs C., Patel A.P., Kesari S., Huang S., Baliga N.S. A Systems Approach to Brain Tumor Treatment. Cancers. 2021;13:3152. doi: 10.3390/CANCERS13133152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debinski W., Gibo D.M., Hulet S.W., Connor J.R., Gillespie G.Y. Receptor for interleukin 13 is a marker and therapeutic target for human high-grade gliomas. Clin Cancer Res. 1999;5:985–990. [PubMed] [Google Scholar]
- 24.Ahmed N., Salsman V.S., Kew Y., Shaffer D., Powell S., Zhang Y.J., Grossman R.G., Heslop H.E., Gottschalk S. HER2-specific T cells target primary glioblastoma stem cells and induce regression of autologous experimental tumors. Clin Cancer Res. 2010;16:474–485. doi: 10.1158/1078-0432.CCR-09-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed N., Brawley V., Hegde M., Bielamowicz K., Wakefield A., Ghazi A., Ashoori A., Diouf O., Gerken C., Landi D., Kalra M., Yi Z., Rooney C., Dotti G., Gee A., Heslop H., Gottschalk S., Powell S., Grossman R., Wels W., Kew Y., Baskin D., Zhang J., New P., Hicks J. Autologous HER2 CMV bispecific CAR T cells are safe and demonstrate clinical benefit for glioblastoma in a Phase I trial. J Immunother Cancer. 2015;3:O11. doi: 10.1186/2051-1426-3-S2-O11. [DOI] [Google Scholar]
- 26.Ahmed N., Brawley V., Hegde M., Bielamowicz K., Kalra M., Landi D., Robertson C., Gray T.L., Diouf O., Wakefield A., Ghazi A., Gerken C., Yi Z., Ashoori A., Wu M.F., Liu H., Rooney C., Dotti G., Gee A., Su J., Kew Y., Baskin D., Zhang Y.J., New P., Grilley B., Stojakovic M., Hicks J., Powell S.Z., Brenner M.K., Heslop H.E., Grossman R., Wels W.S., Gottschalk S. HER2-specific chimeric antigen receptor–modified virus-specific T cells for progressive glioblastoma: A phase 1 dose-escalation trial. JAMA Oncol. 2017;3:1094–1101. doi: 10.1001/jamaoncol.2017.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow K.K., Naik S., Kakarla S., Brawley V.S., Shaffer D.R., Yi Z., Rainusso N., Wu M.-F., Liu H., Kew Y., Grossman R.G., Powell S., Lee D., Ahmed N., Gottschalk S. T cells redirected to EphA2 for the immunotherapy of glioblastoma. Mol Ther. 2012;21:629–637. doi: 10.1038/mt.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan R.A., Johnson L.A., Davis J.L., Zheng Z., Woolard K.D., Reap E., Feldman S., Chinnasamy N., Kuan C.-T., Song H., Zhang W., Fine H.A., Rosenberg S.A. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Hum Gene Ther. 2012;23:1043–1053. doi: 10.1089/hum.2012.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J., Chu J., Keung Chan W., Zhang J., Wang Y., Cohen J.B., Victor A., Meisen W.H., Kim S.H., Grandi P., Wang Q.E., He X., Nakano I., Chiocca E.A., Glorioso J.C., Kaur B., Caligiuri M.A., Yu J. CAR-engineered NK cells targeting wild-type EGFR and EGFRvIII enhance killing of glioblastoma and patient-derived glioblastoma stem cells. Sci Rep. 2015;5:1–13. doi: 10.1038/srep11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson L.A., Scholler J., Ohkuri T., Kosaka A., Patel P.R., McGettigan S.E., Nace A.K., Dentchev T., Thekkat P., Loew A., Boesteanu A.C., Cogdill A.P., Chen T., Fraietta J.A., Kloss C.C., Posey A.D., Engels B., Singh R., Ezell T., Idamakanti N., Ramones M.H., Li N., Zhou L., Plesa G., Seykora J.T., Okada H., June C.H., Brogdon J.L., Maus M.V. Rational development and characterization of humanized anti–EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aaa4963. 275ra22-275ra22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarboe J.S., Johnson K.R., Choi Y., Lonser R.R., Park J.K. Expression of interleukin-13 receptor α2 in glioblastoma multiforme: Implications for targeted therapies. Cancer Res. 2007;67:7983–7986. doi: 10.1158/0008-5472.CAN-07-1493. [DOI] [PubMed] [Google Scholar]
- 32.Joshi B.H., Puri R.A., Leland P., Varricchio F., Gupta G., Kocak M., Gilbertson R.J., Puri R.K. Identification of interleukin-13 receptor α2 chain overexpression in situ in high-grade diffusely infiltrative pediatric brainstem glioma. Neuro-oncol. 2008;10:265–274. doi: 10.1215/15228517-2007-066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawakami M., Kawakami K., Takahashi S., Abe M., Puri R.K. Analysis of interleukin-13 receptor α2 expression in human pediatric brain tumors. Cancer. 2004;101:1036–1042. doi: 10.1002/cncr.20470. [DOI] [PubMed] [Google Scholar]
- 34.Brown C.E., Warden C.D., Starr R., Deng X., Badie B., Yuan Y.C., Forman S.J., Barish M.E. Glioma IL13Rα2 is associated with mesenchymal signature gene expression and poor patient prognosis. PLoS One. 2013;8:e77769. doi: 10.1371/journal.pone.0077769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koka V., Potti A., Forseen S.E., Pervez H., Fraiman G.N., Koch M., Levitt R. Role of Her-2/neu overexpression and clinical determinants of early mortality in glioblastoma multiforme. Am J Clin Oncol. 2003;26:332–335. doi: 10.1097/01.coc.0000020922.66984.e7. [DOI] [PubMed] [Google Scholar]
- 36.Potti A., Forseen S.E., Koka V.K., Pervez H., Koch M., Fraiman G., Mehdi S.A., Levitt R. Determination of HER-2/neu overexpression and clinical predictors of survival in a cohort of 347 patients with primary malignant brain tumors. Cancer Invest. 2004;22:537–544. doi: 10.1081/CNV-200026523. [DOI] [PubMed] [Google Scholar]
- 37.Faulkner C., Palmer A., Williams H., Wragg C., Haynes H.R., White P., DeSouza R.M., Williams M., Hopkins K., Kurian K.M. EGFR and EGFRvIII analysis in glioblastoma as therapeutic biomarkers. Br J Neurosurg. 2015;29:23–29. doi: 10.3109/02688697.2014.950631. [DOI] [PubMed] [Google Scholar]
- 38.Biernat W., Huang H., Yokoo H., Kleihues P., Ohgaki H. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol. 2004;14:131–136. doi: 10.1111/j.1750-3639.2004.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishikawa R., Sugiyama T., Narita Y., Furnari F., Cavenee W.K., Matsutani M. Immunohistochemical analysis of the mutant epidermal growth factor, ΔEGFR, in glioblastoma. Brain Tumor Pathol. 2004;21:53–56. doi: 10.1007/BF02484510. [DOI] [PubMed] [Google Scholar]
- 40.Brennan C.W., Verhaak R.G.W., McKenna A., Campos B., Noushmehr H., Salama S.R.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., Beroukhim R., Bernard B., Wu C.-J.J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S.A., Ciriello G., Yung W.K.K.A., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D.D., Van Meir E.G., Prados M., Sloan A.E., Black K.L.L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D.W., Guha A., Iacocca M., O’Neill B.P., Foltz G., Myers J., Weisenberger D.J., Penny R., Kucherlapati R., Perou C.M., Hayes D.N., Gibbs R., Marra M., Mills G.B., Lander E.S., Spellman P., Wilson R.K., Sander C., Weinstein J.N., Meyerson M., Gabriel S.B., Laird P.W., Haussler D., Getz G., Chin L., Van Meir E.G., Prados M., Sloan A.E., Black K.L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D.W., Guha A., Iacocca M., O’Neill B.P., Foltz G., Myers J., Weisenberger D.J., Penny R., Kucherlapati R., Perou C.M., Hayes D.N., Gibbs R., Marra M., Mills G.B., Lander E.S., Spellman P., Wilson R.K., Sander C., Weinstein J.N., Meyerson M., Gabriel S.B., Laird P.W., Haussler D., Getz G., Chin L., Benz C., Barrett W., Ostrom Q., Wolinsky Y., Bose B., Boulos P.T., Boulos M., Brown J., Czerinski C., Eppley M., Kempista T., Kitko T., Koyfman Y., Rabeno B., Rastogi P., Sugarman M., Swanson P., Yalamanchii K., Otey I.P., Liu Y.S.Y., Xiao Y., Auman J.T., Chen P.C., Hadjipanayis A., Lee E., Lee S., Park P.J., Seidman J., Yang L.L., Kalkanis S., Poisson L.M., Raghunathan A., Scarpace L., Bressler R., Eakin A., Iype L., Kreisberg R.B., Leinonen K., Reynolds S., Rovira H., Thorsson V., Annala M.J., Paulauskis J., Curley E., Hatfield M., Mallery D., Morris S., Shelton T., Shelton C., Sherman M., Yena P., Cuppini L., DiMeco F., Eoli M., Maderna E., Pollo B., Saini M., Balu S., Hoadley K.A., Li L., Miller C.R., Shi Y., Topal M.D., Wu J., Dunn G., Giannini C., Aksoy B.A., Antipin Y., Borsu L., Cerami E., Gao J., Gross B., Jacobsen A., Ladanyi M., Lash A., Liang Y., Reva B., Schultz N., Shen R., Socci N.D., Viale A., Ferguson M.L., Chen Q.R., Demchok J.A., Dillon L.A.L., Mills Shaw K.R., Sheth M., Tarnuzzer R., Wang Z., Yang L.L., Davidsen T., Guyer M.S., Ozenberger B.A., Sofia H.J., Bergsten J., Eckman J., Harr J., Smith C., Tucker K., Winemiller C., Zach L.A., Ljubimova J.Y., Eley G., Ayala B., Jensen M.A., Kahn A., Pihl T.D., Pot D.A., Wan Y., Hansen N., Hothi P., Lin B., Shah N., Yoon J.G., Lau C., Berens M., Ardlie K., Carter S.L., Cherniack A.D., Noble M., Cho J., Cibulskis K., DiCara D., Frazer S., Gabriel S.B., Gehlenborg N., Gentry J., Heiman D., Kim J., Jing R., Lawrence M., Lin P., Mallard W., Onofrio R.C., Saksena G., Schumacher S., Stojanov P., Tabak B., Voet D., Zhang H., Dees N.N., Ding L., Fulton L.L., Fulton R.S., Kanchi K.L., Mardis E.R., Wilson R.K., Baylin S.B., Harshyne L., Cohen M.L., Devine K., Sloan A.E., Van Den Berg S.R., Berger M.S., Carlin D., Craft B., Ellrott K., Goldman M., Goldstein T., Grifford M., Ma S., Ng S., Stuart J., Swatloski T., Waltman P., Zhu J., Foss R., Frentzen B., McTiernan R., Yachnis A., Mao Y., Akbani R., Bogler O., Fuller G.N., Liu W., Liu Y.S.Y., Lu Y., Protopopov A., Ren X., Sun Y., Yung W.K.K.A., Zhang J., Chen K., Weinstein J.N., Bootwalla M.S., Lai P.H., Triche T.J., Van Den Berg D.J., Gutmann D.H., Lehman N.L., Brat D., Olson J.J., Mastrogianakis G.M., Devi N.S., Zhang Z., Lipp E., McLendon R. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brat D.J., Castellano-Sanchez A.A., Hunter S.B., Pecot M., Cohen C., Hammond E.H., Devi S.N., Kaur B., Van Meir E.G. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 2004;64:920–927. doi: 10.1158/0008-5472.CAN-03-2073. [DOI] [PubMed] [Google Scholar]
- 42.Hegde M., Corder A., Chow K.K.H., Mukherjee M., Ashoori A., Kew Y., Zhang Y.J., Baskin D.S., Merchant F.A., Brawley V.S., Byrd T.T., Krebs S., Wu M.F., Liu H., Heslop H.E., Gottschalk S., Gottachalk S., Yvon E., Ahmed N. Combinational targeting offsets antigen escape and enhances effector functions of adoptively transferred T cells in glioblastoma. Molecular Theraspy. 2013;21:2087–2101. doi: 10.1038/mt.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegde M., Mukherjee M., Grada Z., Pignata A., Landi D., Navai S.A., Wakefield A., Fousek K., Bielamowicz K., Chow K.K.H., Brawley V.S., Byrd T.T., Krebs S., Gottschalk S., Wels W.S., Baker M.L., Dotti G., Mamonkin M., Brenner M.K., Orange J.S., Ahmed N. Tandem CAR T cells targeting HER2 and IL13Rα2 mitigate tumor antigen escape. J Clin Invest. 2016;126:3036–3052. doi: 10.1172/JCI83416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bielamowicz K., Fousek K., Byrd T.T., Samaha H., Mukherjee M., Aware N., Wu M.-F.F., Orange J.S., Sumazin P., Man T.-K.K., Joseph S.K., Hegde M., Ahmed N. Trivalent CAR T cells overcome interpatient antigenic variability in glioblastoma. Neuro-oncol. 2018;20:506–518. doi: 10.1093/neuonc/nox182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Micallef L., Rodgers P. eulerAPE: Drawing area-proportional 3-Venn diagrams using ellipses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLendon R., Friedman A., Bigner D., Van Meir E.G., Brat D.J., Mastrogianakis G.M., Olson J.J., Mikkelsen T., Lehman N., Aldape K., Yung W.K.A., Bogler O., Weinstein J.N., VandenBerg S., Berger M., Prados M., Muzny D., Morgan M., Scherer S., Sabo A., Nazareth L., Lewis L., Hall O., Zhu Y., Ren Y., Alvi O., Yao J., Hawes A., Jhangiani S., Fowler G., San Lucas A., Kovar C., Cree A., Dinh H., Santibanez J., Joshi V., Gonzalez-Garay M.L., Miller C.A., Milosavljevic A., Donehower L., Wheeler D.A., Gibbs R.A., Cibulskis K., Sougnez C., Fennell T., Mahan S., Wilkinson J., Ziaugra L., Onofrio R., Bloom T., Nicol R., Ardlie K., Baldwin J., Gabriel S., Lander E.S., Ding L., Fulton R.S., McLellan M.D., Wallis J., Larson D.E., Shi X., Abbott R., Fulton L., Chen K., Koboldt D.C., Wendl M.C., Meyer R., Tang Y., Lin L., Osborne J.R., Dunford-Shore B.H., Miner T.L., Delehaunty K., Markovic C., Swift G., Courtney W., Pohl C., Abbott S., Hawkins A., Leong S., Haipek C., Schmidt H., Wiechert M., Vickery T., Scott S., Dooling D.J., Chinwalla A., Weinstock G.M., Mardis E.R., Wilson R.K., Getz G., Winckler W., Verhaak R.G.W., Lawrence M.S., O’Kelly M., Robinson J., Alexe G., Beroukhim R., Carter S., Chiang D., Gould J., Gupta S., Korn J., Mermel C., Mesirov J., Monti S., Nguyen H., Parkin M., Reich M., Stransky N., Weir B.A., Garraway L., Golub T., Meyerson M., Chin L., Protopopov A., Zhang J., Perna I., Aronson S., Sathiamoorthy N., Ren G., Yao J., Wiedemeyer W.R., Kim H., Sek W.K., Xiao Y., Kohane I.S., Seidman J., Park P.J., Kucherlapati R., Laird P.W., Cope L., Herman J.G., Weisenberger D.J., Pan F., Van Den Berg D., Van Neste L., Joo M.Y., Schuebel K.E., Baylin S.B., Absher D.M., Li J.Z., Southwick A., Brady S., Aggarwal A., Chung T., Sherlock G., Brooks J.D., Myers R.M., Spellman P.T., Purdom E., Jakkula L.R., Lapuk A.V., Marr H., Dorton S., Yoon G.C., Han J., Ray A., Wang V., Durinck S., Robinson M., Wang N.J., Vranizan K., Peng V., Van Name E., Fontenay G.V., Ngai J., Conboy J.G., Parvin B., Feiler H.S., Speed T.P., Gray J.W., Brennan C., Socci N.D., Olshen A., Taylor B.S., Lash A., Schultz N., Reva B., Antipin Y., Stukalov A., Gross B., Cerami E., Wei Q.W., Qin L.X., Seshan V.E., Villafania L., Cavatore M., Borsu L., Viale A., Gerald W., Sander C., Ladanyi M., Perou C.M., Hayes D.N., Topal M.D., Hoadley K.A., Qi Y., Balu S., Shi Y., Wu J., Penny R., Bittner M., Shelton T., Lenkiewicz E., Morris S., Beasley D., Sanders S., Kahn A., Sfeir R., Chen J., Nassau D., Feng L., Hickey E., Barker A., Gerhard D.S., Vockley J., Compton C., Vaught J., Fielding P., Ferguson M.L., Schaefer C., Zhang J., Madhavan S., Buetow K.H., Collins F., Good P., Guyer M., Ozenberger B., Peterson J., Thomson E. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermanson M., Funa K., Hartman M., Claesson-Welsh L., Heldin C.-H., Westermark B., Nistér M. Platelet-derived growth factor and its receptors in human glioma tissue: Expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52:3213–3219. [PubMed] [Google Scholar]
- 49.Nabeshima K., Shimao Y., Sato S., Kataoka H., Moriyama T., Kawano H., Wakisaka S., Koono M. Expression of c-Met correlates with grade of malignancy in human astrocytic tumours: an immunohistochemical study. Histopathology. 1997;31:436–443. doi: 10.1046/j.1365-2559.1997.3010889.x. [DOI] [PubMed] [Google Scholar]
- 50.Su Huang H.J., Nagane M., Klingbeil C.K., Lin H., Nishikawa R., Ji X.D., Huang C.M., Gill G.N., Wiley H.S., Cavenee W.K. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 51.Inda M.D.M., Bonavia R., Mukasa A., Narita Y., Sah D.W.Y.Y., Vandenberg S., Brennan C., Johns T.G., Bachoo R., Hadwiger P., Tan P., DePinho R.A., Cavenee W., Furnari F. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snuderl M., Fazlollahi L., Le L.P., Nitta M., Zhelyazkova B.H., Davidson C.J., Akhavanfard S., Cahill D.P., Aldape K.D., Betensky R.A., Louis D.N., Iafrate A.J. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 53.K. Koga, T. Todaka, M. Morioka, J.-I. Hamada, Y. Kai, S. Yano, A. Okamura, N. Takakura, T. Suda, Y. Ushio, Expression of Angiopoietin-2 in Human Glioma Cells and Its Role for Angiogenesis 1, 2001. [PubMed]
- 54.Bello L., Francolini M., Marthyn P., Zhang J., Carroll R.S., Nikas D.C., Strasser J.F., Villani R., Cheresh D.A., McL P. Black, αvβ3 and αvβ5 integrin expression in glioma periphery. Neurosurgery. 2001;49:380–390. doi: 10.1097/00006123-200108000-00022. [DOI] [PubMed] [Google Scholar]
- 55.Yuan Y. Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Broekman M.L., Maas S.L.N., Abels E.R., Mempel T.R., Krichevsky A.M., Breakefield X.O. Multidimensional communication in the microenvirons of glioblastoma. Nat Rev Neurol. 2018;14:1–14. doi: 10.1038/s41582-018-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dirkse A., Golebiewska A., Buder T., Nazarov P.V., Muller A., Poovathingal S., Brons N.H.C., Leite S., Sauvageot N., Sarkisjan D., Seyfrid M., Fritah S., Stieber D., Michelucci A., Hertel F., Herold-Mende C., Azuaje F., Skupin A., Bjerkvig R., Deutsch A., Voss-Böhme A., Niclou S.P. Stem cell-associated heterogeneity in glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10:1–16. doi: 10.1038/s41467-019-09853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chumakova A.P., Hitomi M., Sulman E.P., Lathia J.D. High-throughput automated single-cell imaging analysis reveals dynamics of glioblastoma stem cell population during state transition. Cytometry Part A. 2019;95:290–301. doi: 10.1002/cyto.a.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., Louis D.N., Rozenblatt-Rosen O., Suvà M.L., Regev A., Bernstein B.E. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Francis J.M., Zhang C.Z., Maire C.L., Jung J., Manzo V.E., Adalsteinsson V.A., Homer H., Haidar S., Blumenstiel B., Pedamallu C.S., Ligon A.H., Love J.C., Meyerson M., Ligon K.L. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4:956–971. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer M., Reimand J., Lan X., Head R., Zhu X., Kushida M., Bayani J., Pressey J.C., Lionel A.C., Clarke I.D., Cusimano M., Squire J.A., Scherer S.W., Bernstein M., Woodin M.A., Bader G.D., Dirks P.B. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Nat Acad Sci USA. 2015;112:851–856. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eskilsson E., Røsland G.V., Solecki G., Wang Q., Harter P.N., Graziani G., Verhaak R.G.W., Winkler F., Bjerkvig R., Miletic H. EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro-oncol. 2018;20:743–752. doi: 10.1093/neuonc/nox191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sottoriva A., Spiteri I., Piccirillo S.G.M.M., Touloumis A., Collins V.P., Marioni J.C., Curtis C., Watts C., Tavaré S., Tavare S., Tavaré S., Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Nat Acad Sci USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim H., Zheng S., Amini S.S., Virk S.M., Mikkelsen T., Brat D.J., Grimsby J., Sougnez C., Muller F., Hu J., Sloan A.E., Cohen M.L., Van Meir E.G., Scarpace L., Laird P.W., Weinstein J.N., Lander E.S., Gabriel S., Getz G., Meyerson M., Chin L., Barnholtz-Sloan J.S., Verhaak R.G.W. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25:316–327. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lan X., Jörg D.J., Cavalli F.M.G., Richards L.M., Nguyen L.V., Vanner R.J., Guilhamon P., Lee L., Kushida M.M., Pellacani D., Park N.I., Coutinho F.J., Whetstone H., Selvadurai H.J., Che C., Luu B., Carles A., Moksa M., Rastegar N., Head R., Dolma S., Prinos P., Cusimano M.D., Das S., Bernstein M., Arrowsmith C.H., Mungall A.J., Moore R.A., Ma Y., Gallo M., Lupien M., Pugh T.J., Taylor M.D., Hirst M., Eaves C.J., Simons B.D., Dirks P.B. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549:227–232. doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Couturier C.P., Ayyadhury S., Le P.U., Nadaf J., Monlong J., Riva G., Allache R., Baig S., Yan X., Bourgey M., Lee C., Wang Y.C.D., Wee Yong V., Guiot M.-C., Najafabadi H., Misic B., Antel J., Bourque G., Ragoussis J., Petrecca K. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun. 2020;11:3406. doi: 10.1038/s41467-020-17186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhaduri A., Di Lullo E., Jung D., Müller S., Crouch E.E., Espinosa C.S., Ozawa T., Alvarado B., Spatazza J., Cadwell C.R., Wilkins G., Velmeshev D., Liu S.J., Malatesta M., Andrews M.G., Mostajo-Radji M.A., Huang E.J., Nowakowski T.J., Lim D.A., Diaz A., Raleigh D.R., Kriegstein A.R. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell. 2020;26:48–63. doi: 10.1016/j.stem.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evans S.M., Judy K.D., Dunphy I., Jenkins W.Timothy, Hwang W.T., Nelson P.T., Lustig R.A., Jenkins K., Magarelli D.P., Hahn S.M., Collins R.A., Grady S., Koch C.J. Hypoxia is important in the biology and aggression of human glial brain tumors. Clin Cancer Res. 2004;10:8177–8184. doi: 10.1158/1078-0432.CCR-04-1081. [DOI] [PubMed] [Google Scholar]
- 69.Minchenko O.H., Tsymbal D.O., Minchenko D.O., Riabovol O.O., Ratushna O.O., Karbovskyi L.L. Hypoxic regulation of the expression of cell proliferation related genes in U87 glioma cells upon inhibition of ire1 signaling enzyme. Ukrainian Biochemical Journal. 2016;88:11–21. doi: 10.15407/ubj88.01.011. [DOI] [PubMed] [Google Scholar]
- 70.Sobhanifar S., Aquino-Parsons C., Stanbridge E.J., Olive P. Reduced expression of hypoxia-inducible factor-1α in perinecrotic regions of solid tumors. Cancer Res. 2005;65:7259–7266. doi: 10.1158/0008-5472.CAN-04-4480. [DOI] [PubMed] [Google Scholar]
- 71.Hide T., Komohara Y., Miyasato Y., Nakamura H., Makino K., Takeya M., ichi Kuratsu J., Mukasa A., Yano S. Oligodendrocyte progenitor cells and macrophages/microglia produce glioma stem cell niches at the tumor border. EBioMedicine. 2018;30:94–104. doi: 10.1016/j.ebiom.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pudełek M., Król K., Catapano J., Wróbel T., Czyż J., Ryszawy D. Epidermal growth factor (EGF) augments the invasive potential of human glioblastoma multiforme cells via the activation of collaborative egfr/ros-dependent signaling. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21103605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.William D., Mokri P., Lamp N., Linnebacher M., Classen C.F., Erbersdobler A., Schneider B. Amplification of the EGFR gene can be maintained and modulated by variation of EGF concentrations in in vitro models of glioblastoma multiforme. PLoS One. 2017;12:1–13. doi: 10.1371/journal.pone.0185208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vartanian A., Singh S.K., Agnihotri S., Jalali S., Burrell K., Aldape K.D., Zadeh G. GBM’s multifaceted landscape: Highlighting regional and microenvironmental heterogeneity. Neuro-oncol. 2014;16:1167–1175. doi: 10.1093/neuonc/nou035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonavia R., Inda M.D.M., Cavenee W.K., Furnari F.B. Heterogeneity maintenance in glioblastoma: A social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jung E., Alfonso J., Osswald M., Monyer H., Wick W., Winkler F. Emerging intersections between neuroscience and glioma biology. Nat Neurosci. 2019;22:1951–1960. doi: 10.1038/s41593-019-0540-y. [DOI] [PubMed] [Google Scholar]
- 77.Pinto G., Brou C., Zurzolo C. Tunneling nanotubes: The fuel of tumor progression? Trends Cancer. 2020;6:874–888. doi: 10.1016/j.trecan.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 78.Montgomery M.K., Kim S.H., Dovas A., Zhao H.T., Goldberg A.R., Xu W., Yagielski A.J., Cambareri M.K., Patel K.B., Mela A., Humala N., Thibodeaux D.N., Shaik M.A., Ma Y., Grinband J., Chow D.S., Schevon C., Canoll P., Hillman E.M.C. Glioma-induced alterations in neuronal activity and neurovascular coupling during disease progression. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S. Jamous, A. Comba, P.R. Lowenstein, S. Motsch, Self-organization in brain tumors: How cell morphology and cell density influence glioma pattern formation, 16 (2020) e1007611. https://doi.org/10.1371/journal.pcbi.1007611. [DOI] [PMC free article] [PubMed]
- 80.Lee J.K., Wang J., Sa J.K., Ladewig E., Lee H.O., Lee I.H., Kang H.J., Rosenbloom D.S., Camara P.G., Liu Z., Van Nieuwenhuizen P., Jung S.W., Choi S.W., Kim J., Chen A., Kim K.T., Shin S., Seo Y.J., Oh J.M., Shin Y.J., Park C.K., Kong D.S., Seol H.J., Blumberg A., Il Lee J., Iavarone A., Park W.Y., Rabadan R., Nam D.H. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49:594–599. doi: 10.1038/ng.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L., Williams P.M., Modrusan Z., Feuerstein B.G., Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 82.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., Alexe G., Lawrence M., O’Kelly M., Tamayo P., Weir B.A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H.S., Hodgson J.G., James C.D., Sarkaria J.N., Brennan C., Kahn A., Spellman P.T., Wilson R.K., Speed T.P., Gray J.W., Meyerson M., Getz G., Perou C.M., Hayes D.N., N. Cancer Genome Atlas Research Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eckel-Passow J.E., Lachance D.H., Molinaro A.M., Walsh K.M., Decker P.A., Sicotte H., Pekmezci M., Rice T., Kosel M.L., Smirnov I.V., Sarkar G., Caron A.A., Kollmeyer T.M., Praska C.E., Chada A.R., Halder C., Hansen H.M., McCoy L.S., Bracci P.M., Marshall R., Zheng S., Reis G.F., Pico A.R., O’Neill B.P., Buckner J.C., Giannini C., Huse J.T., Perry A., Tihan T., Berger M.S., Chang S.M., Prados M.D., Wiemels J., Wiencke J.K., Wrensch M.R., Jenkins R.B. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schäfer N., Gielen G.H., Rauschenbach L., Kebir S., Till A., Reinartz R., Simon M., Niehusmann P., Kleinschnitz C., Herrlinger U., Pietsch T., Scheffler B., Glas M. Longitudinal heterogeneity in glioblastoma: Moving targets in recurrent versus primary tumors. J Transl Med. 2019;17:96. doi: 10.1186/s12967-019-1846-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J., Cazzato E., Ladewig E., Frattini V., Rosenbloom D.I.S., Zairis S., Abate F., Liu Z., Elliott O., Shin Y.J., Lee J.K., Lee I.H., Park W.Y., Eoli M., Blumberg A.J., Lasorella A., Nam D.H., Finocchiaro G., Iavarone A., Rabadan R. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48:768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pellegatta S., Savoldo B., Di Ianni N., Corbetta C., Chen Y., Patané M., Sun C., Pollo B., Ferrone S., DiMeco F., Finocchiaro G., Dotti G. Constitutive and TNFα-inducible expression of chondroitin sulfate proteoglycan 4 in glioblastoma and neurospheres: Implications for CAR-T cell therapy. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aao2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang D., Starr R., Chang W.C., Aguilar B., Alizadeh D., Wright S.L., Yang X., Brito A., Sarkissian A., Ostberg J.R., Li L., Shi Y., Gutova M., Aboody K., Badie B., Forman S.J., Barish M.E., Brown C.E. Chlorotoxin-directed CAR T cells for specific and effective targeting of glioblastoma. Sci Transl Med. 2020;12:eaaw2672. doi: 10.1126/scitranslmed.aaw2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gould S.J., Vrba E.S. Exaptation—A missing term in the science of form. Paleobiology. 1982;8:4–15. [Google Scholar]
- 89.Gould S.J. W. W. Norton & Co.; 1989. Wonderful Life: The Burgess Shale and the Nature of History. [Google Scholar]
- 90.Gould S.J., Hildreth J.E.K., Booth A.M. The evolution of alloimmunity and the genesis of adaptive immunity. Q Rev Biol. 2004;79:359–382. doi: 10.1086/426088. [DOI] [PubMed] [Google Scholar]
- 91.Sánchez-Romero M.A., Casadesús J. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc Nat Acad Sci USA. 2014;111:355–360. doi: 10.1073/pnas.1316084111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fidler I.J. The Ernst W. Bertner Memorial Award lecture: the evolution of biological heterogeneity in metastatic neoplasms. Symp Fundam Cancer Res. 1983;36:5–26. [PubMed] [Google Scholar]
- 93.Kleppe M., Levine R.L. Tumor heterogeneity confounds and illuminates: Assessing the implications. Nat Med. 2014;20:342–344. doi: 10.1038/nm.3522. [DOI] [PubMed] [Google Scholar]
- 94.Rajapakse V.N., Herrada S., Lavi O. Phenotype stability under dynamic brain-tumor environment stimuli maps glioblastoma progression in patients. Sci Adv. 2020;6:eaaz4125. doi: 10.1126/sciadv.aaz4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Maps of IL13Rα2, HER2 and EGFR immunoreactivity and associated Euler diagrams.
Shown are data from 38 tumor samples with sufficiently large regions of cellular tumor for this mapping. Details of the construction of these maps are in the legend to Figure 2. Tumor sections are ordered by total area occupied by EGFR (left → right, top → bottom). In some tumor sections, the total percentage area occupied by HER2 was below the presentation threshold of 30% of maximum, and therefore not evident in all maps. Scale bar = 1 cm in each panel.