Abstract
Advancements in the field of cancer research have enabled researchers and clinicians to access a massive amount of data to aid cancer patients and to add to the existing knowledge of research. However, despite the existence of reliable sources for extricating this data, it remains a challenge to accurately comprehend and draw conclusions based on the entirety of available information. Therefore, the current study aimed to design and develop a database for the identified variants of 5 different cancer types using 20 different cancer exomes. The exome data were retrieved from NCBI SRA and an NGS data clean-up protocol was implemented to obtain the best quality reads. The reads which passed the quality checks were then used for calling the variants which were then processed and filtered. This data was used to normalize and the normalized data generated was used for developing the database. MutaXome, which stands for mutations in cancer exome was designed in SQL, with the front end in bootstrap and HTML, and backend in PHP. The normalized data containing the variants inclusive of Single Nucleotide Polymorphisms (SNPs), were added into MutaXome, which contains detailed information regarding each type of identified variant. This database, available online via http://www.vidyalab.rf.gd/, serves as a knowledge base for cancer exome variations and holds much potential for enriching it by linking it to a decision support system as prospective studies.
Keywords: Cancer exomes, database, mutations, PHP, HTML and bootstrap, SQL
Introduction
In the present world, cancer is considered a disease associated with variations in the genes involved in regulation of cell cycle and repair mechanisms 1 and is popularly emphasized as challenging to treat if diagnosed at later stages. One major way of comprehending the cancer gene mutations and arriving at solutions for a treatment strategy is to thoroughly scrutinize the huge amount of existing data available on the cancer genomes. As whole-genome sequencing is the current preferred method for understanding mutations, both common and unique ones, at the genetic level, several studies are now exclusively dedicated to performing WES on cancer samples2-5 and making the data available to the public domain. Currently, there is a huge amount of data being generated by WES analyses, and maintaining these requires repositories that are also easily understandable and obtainable to the public.
Computerized database management systems (CDBMS) are now being widely implemented in hospitals and research centers to keep track of patient medical records for various cancer types. More recently, a study developed an intelligent CDM system for in silico diagnosis and detection of breast cancer using mammography, which retrieves, investigates, and analyzes images of a mammogram. 6 Another study introduced a computerized DBMS developed via MySQL to manage and store data from breast cancer patients. 7 With increasing importance being placed on using database management systems, developing digitized systems for storing, retrieving, using, and understanding cancer exome data for purposes of solving the existing dearth of handling a large amount of data is being greatly encouraged in the field of cancer research.
Ongoing research is concentrated on cancer etiology and its potential alternative strategies for therapy, and there has also been an exponential growth of cancer-associated data from sources like publications, protein-protein or gene-gene interaction studies, genome-wide association studies (GWAS), epigenetic and genetic experiments, etc., stored in related databases and repositories. Massive collaboration projects such as the Cancer Genome Atlas (TCGA), 4 International Cancer Genome Consortium (ICGC), 8 Clinical Proteomic Tumor Analysis Consortium (CPTAC), 9 and Cancer Genome Project (CGP), 10 primarily allow cancer data to be accessed, queried, scrutinized, and utilized for research. Likewise, specialized web-based tools for accessing and studying information of different cancer exomes/datasets are also available, including Cancer Genome Anatomy Project (CGAP), 11 Cancer Genome Workbench (CGW), 12 and other annotation databases like dbSNP, COSMIC, and 1000 Genomes. 10 Furthermore, several cancer-specific databases provide information particular to its cancer types such as the Dragon Database of Genes related to Prostate Cancer (DDPC) 13 and the Gene to systems Breast Cancer database (G2SBC). 14
Additionally, previous studies have examined cancer exomes at a large scale, generating valuable data pointing to various somatic mutations in cancer samples that could act as biomarkers for a specific cancer type.15,16 Moreover, some of the studies have also developed databases to deposit the amassed information in a tabulated, logical format for easy access to the same. 17 Although the evidence presented points toward the existence of several reliable resources for extricating data and performing analysis of the exomes of cancers, it remains challenging to precisely comprehend and draw solid conclusions based on the entirety of available data.
Therefore, the current study aims to overcome this challenge by incorporating previously analyzed data, identified as SNPs existing in 20 different cancer exomes in a logical format as a database. The developed database allows for querying, submitting, and displaying information of mutations discovered in different cancer types that could point toward valuable information for clinicians, researchers, patients, and the general public. The database was developed by initially analyzing the mutational profiles and normalizing the data. The normalized data were then developed into a database that contains extensive cancer-related information on gene aberrations, which can act as a vital knowledge base for prospective future studies.
Materials and Methods
Sequence retrievals and pre-processing of raw data for calling variants
Twenty NGS sequenced cancer exomes belonging to 5 different cancer types were downloaded from NCBI SRA 18 along with the human reference genome from the FTP location ftp.broadinstitute.org/bundle. The cancer types intrahepatic cholangiocarcinoma, human diffuse-type gastric cancer, non-BRCA1/BRCA2 familial breast cancer, high-grade serous ovarian cancer, and pancreatic adenocarcinoma were selected for the study based on the most common ones that affect the Indian population.19-23 These included Human Diffuse Type Gastric Cancer (NCBI SRA ID: SRR941051, SRR941052, SRR941053, SRR941054), High-grade serous ovarian cancer (NCBI SRA ID: ERR035487, ERR035488, ERR035489), Non BRCA1/BRCA2 familial breast cancer (NCBI SRA ID: ERR166303, ERR166304, ERR166307, ERR166310, ERR166312, ERR166335, ERR166336), Intrahepatic cholangiocarcinoma (NCBI SRA ID: SRR894452, SRR900123, SRR900099), and Pancreatic adenocarcinoma (NCBI SRA ID: ERR232253, ERR232254, ERR232255). Each of the 20 sample IDs refer to single patient samples that are available in NCBI-SRA. The pipeline for variant identification was implemented previously. 24 In brief, sequence reads were initially pre-processed via the general NGS pipeline via quality checks. FastQC and MultiQC25,26 were used for preliminary quality checks followed by adaptor trimming via CutAdapt,27,28 gapped alignment with the reference genome through Burrows-Wheeler Aligner (BWA) 29 using Bowtie2, 30 converting from SAM to BAM format using SAMtools 31 and variant processing. Once the sequence reads cleared all these quality tests, variants were called using PICARD and GATK 32 and processed via snpSIFT algorithms.24,33
Normalization of data for identified variants
The uncovered data containing identified, sorted, and filtered variants were then normalized, and random columns of numerics were transformed into standard scales of understandable format. Raw data frequently contains missing or non-standardized values, preventing the appropriate development of a database. Therefore, in the current study, data normalization was carried out by writing specific codes in Python for each sample depending on their requirements. In each case, the preliminary data was imported, loaded, created, and read. Codes were then written in Python and executed for all 20 retrieved exome samples depending on their requirements. The normalized data obtained were scrutinized and appropriately used to design and develop the database. The number of variants present in each cancer exome sample was also examined.
Design of the database
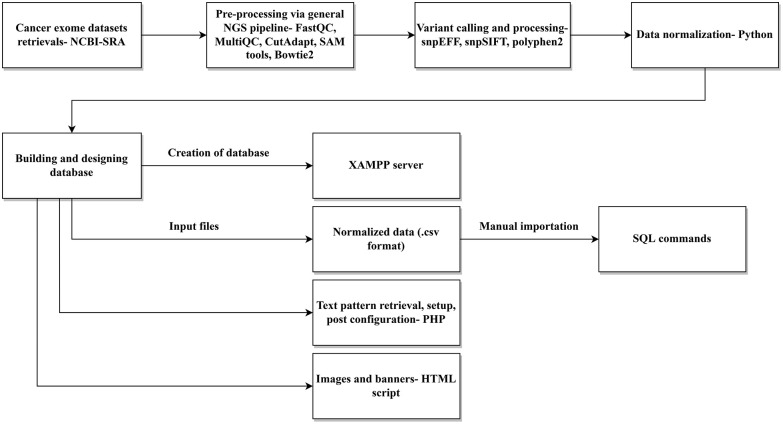
Our novel MutaXome database allows users to access all processed variants identified from the previous steps. The database was created using the XAMPP server, a multi-platform combination of Apache, MySQL, PHP, FileZilla and other essential servers. 34 The identified variants post normalization were present in .csv format which was taken as input files for creating tables for the database. Manual import of the input files was carried out using SQL commands. PHP was utilized for text pattern retrieval, set-up, and post configuration. Images and banners for the database were inserted using HTML scripts. The final designed database could then be installed and run. The pipeline used to design and develop the database is shown in Figure 1.
Figure 1.
Schematic representation of the MutaXome workflow. An initial experimental analysis of retrieved cancer exomes, including preprocessing, variant calling, and data normalization, revealed the different mutations in .csv format, which was used as the input for developing the database. MutaXome was created using the XAMPP server. The retrieval of text patterns, setup, and post configuration was carried out using PHP and the images and banners were added via HTML scripts. The final designed database can be installed and run as stated in the article.
Installation and running the database on Windows platform
To run the database on Windows, a WAMP server needs to be installed. WAMP (Windows, Apache, MySQL, PHP) is a web development package intended for Windows OS. 35 For installation in Linux OS, XAMP can be installed and used to run the developed database. Thus, WAMP was initially downloaded and installed from https://sourceforge.net/projects/wampserver/ 36 and was run after acceptance of the agreement. Once the preferred browser and text editor was set at default, the WAMP server was run and a green WAMP icon observed on the system indicated successful installation. The MySQL database was then set up after logging in with appropriate credentials.
Once MySQL was installed and set up, the phpMyAdmin option was selected and the desired name of the database, MutaXome, was provided. The new database was then created from the drop-down menu options. An automatic server provided a window to create tables for which the identified variant file comprising of the information that was to be incorporated into the database was imported in .sql format from the system. The addition of the data was followed by running the program with appropriate parameters and saving the required .php files in WAMP before opening the local host and running the program. Alternatively, MutaXome was also hosted in Infinity free domain (https://infinityfree.net/) for easy access to users and is available at http://www.vidyalab.rf.gd/. A simple case study was also run to demonstrate the use of the database and the results it displays. This protocol was followed to develop the resource.
Results and Discussion
Identification of variants and normalization of data
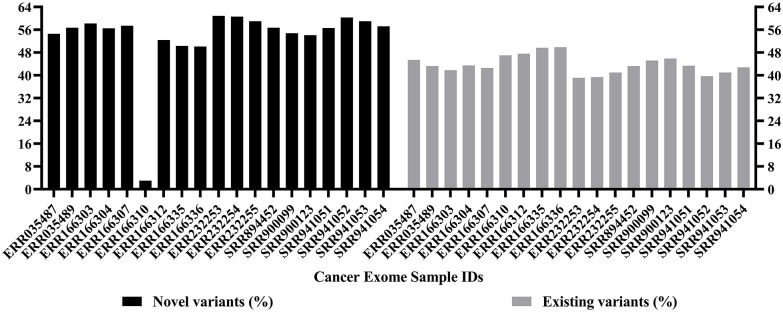
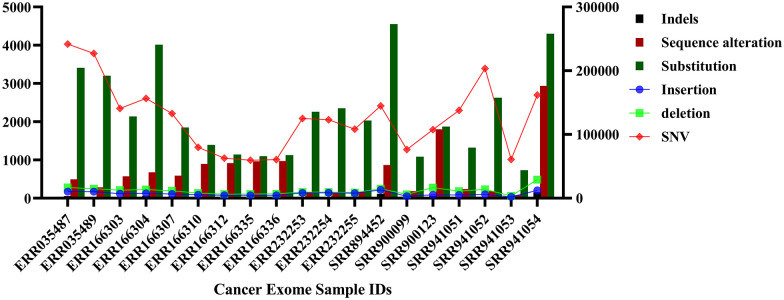
Mutational analysis showed that 60.3% of novel variants were found in the SRR941052 sample and 59% of novel variants were observed in the sample IDs SRR941053 and ERR232255. It was also observed that 49.9% of existing variants were found in ERR166336, 49.7% in ERR166335, and 47.6% in ERR166312 (Figure 2). An initial mutational analysis of the identified variants also revealed the presence of indels, SNPs, SNVs, sequence alterations, deletions, insertions, and substitutions. A total of 13 127 sequence alterations were observed, among which the sample ID SRR941054 showed the highest, that is, 2937 alterations and 1052 indels were observed overall, with 158 indels in SRR941054. Likewise, 42 524 substitutions were observed among which, 4554 were found in SRR894452. 133 302 insertions were found in all 20 exome samples, with 13 104 in SRR894452. Further, a total of 221 544 deletions were observed, with 29 175 deletions in ID SRR941054 and 241 1869 SNVs were found with 241 816 observed in ERR035487. The number of variants, types, and data sets they were observed in is displayed comprehensively in Figure 3. It was also noted that various types of mutations were responsible for these variants such as frameshift mutations, missense mutations, stop gained, splice region variants, in-frame deletions, splice acceptors, and splice donor variants.
Figure 2.
Bar plot showing the percentage of novel and existing variants identified by variant calling. X-axis shows the sample IDs while y-axis shows the percentage of novel and existing variants. The sample ID SRR941052 was found to have 60.3% of novel variants and 59% of novel variants were observed in SRR941053 and ERR232255. Further, 49.9% of existing variants were observed in ERR166336, 49.7% in ERR166335, and 47.6% in ERR166312. Several novels and existing variants were called; however, organized and meaningful data were obtained only after normalization.
Figure 3.
Plots showing the number of insertions, deletions, substitutions, sequence alterations, indels, and single nucleotide variations. x-Axis shows the sample IDs and y-axis shows the number of mutations. 13 127 sequence alterations were observed, among which the highest number, 2937 alterations were found in SRR941054. A total of 1052 indels were observed, and the sample SRR941054 was found to have 158 of them. Likewise, 42 524 substitutions were observed among which 4554 were found in SRR894452. 133 302 insertions were found in all 20 exome samples, with 13 104 in SRR894452. Moreover, a total of 221 544 deletions were observed, with 29 175 deletions in SRR941054 and 241 1869 SNVs were found with 241 816 observed in ERR035487.
Normalization and filtering of the data revealed a total of 4181 variants for 20 cancer exomes. It was noted that 934 variants, the highest among all, were identified in the sample ID SRR941054 belonging to human diffuse-type gastric cancer, followed by 544 variants in SRR900123 (intrahepatic cholangiocarcinoma) and 320 in ERR166304 (non-BRCA1/BRCA2 familial breast cancer) (Table 1). Furthermore, the normalized data indicated information regarding each of the 4181 variations, such as its gene symbols, HGNC code (HUGO Gene Nomenclature Committee), the position of the variant, its chromosome number, amino acid change, nucleotide change with reference to the human genomes hg19 and hg 38, the type of consequence, etc., (Figure 3). The normalized data were obtained in the form of understandable rows and columns as a result of which an appropriate database was developed.
Table 1.
The number of variants for each of the 20 cancer exome datasets totaling up to 4181 mutations analyzed after data normalization.
| Sl. No | Cancer Exome Dataset ID (NCBI SRA) | Number of variants identified after normalization |
|---|---|---|
| 1 | ERR035487 | 164 |
| 2 | ERR035488 | 182 |
| 3 | ERR035489 | 198 |
| 4 | ERR166303 | 75 |
| 5 | ERR166304 | 320 |
| 6 | ERR166307 | 92 |
| 7 | ERR166310 | 60 |
| 8 | ERR166312 | 73 |
| 9 | ERR166335 | 46 |
| 10 | ERR166336 | 57 |
| 11 | ERR232253 | 203 |
| 12 | ERR232254 | 182 |
| 13 | ERR232255 | 192 |
| 14 | SRR894452 | 252 |
| 15 | SRR900099 | 37 |
| 16 | SRR900123 | 544 |
| 17 | SRR941051 | 266 |
| 18 | SRR941052 | 198 |
| 19 | SRR941053 | 106 |
| 20 | SRR941054 | 934 |
| Total 4181 | ||
Previous studies have used similar NGS pipelines to call and process variants.37,38 However, this study incorporates the data obtained from the identified somatic variants, as a continuation of our previous work, to design a database so that the information can be easily available for prospective use. Additionally, another study designed a database called SomaMutDB. This database comprises somatic mutations in normal tissues in humans. The database has 0.12 million indels and 2.42 million SNVs that were identified in 19 tissues. SomaMutDB also has a user-friendly interactive web browsers that allows to search for mutations of interest. 39 Unlike SomaMutDB, MutaXome presents all somatic mutations, comprising of all identified indels, sequence alterations, substitutions, insertions, deletions and SNVs, identified from 20 cancer exome samples retrieved from NCBI and has an interactive webpage, which allows the users to search for mutations of interest, depending on the cancer type. Furthermore, another study discussed the importance of identification of mutations such as point mutations, insertions, deletions, SNVs, and CNVs for precision medicine in cancer. The study validated and developed a whole-exome sequencing test for identification of such somatic variations that can aid in advanced cancer care. 40 This corroborates the outcomes obtained in the present study since the primary aim of the current work was to identify such mutations and develop a comprehensive, easy-to-use database that can help advanced cancer research. The current database contains a large amount of data generated from 20 cancer exomes available to researchers and clinicians to explore, with future developments to focus on expanding the data content of the database further.
Using the database: A case study
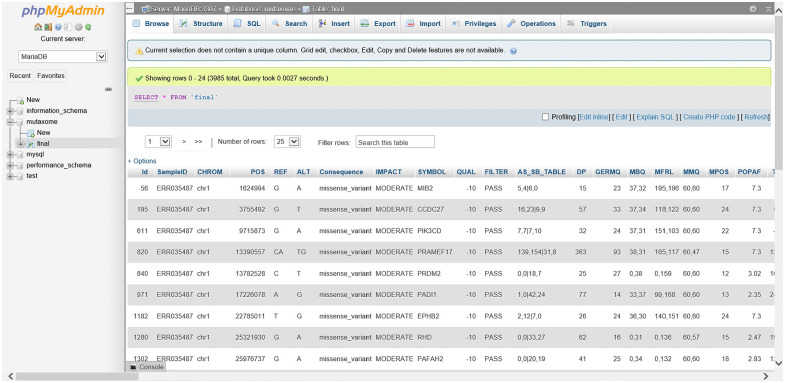
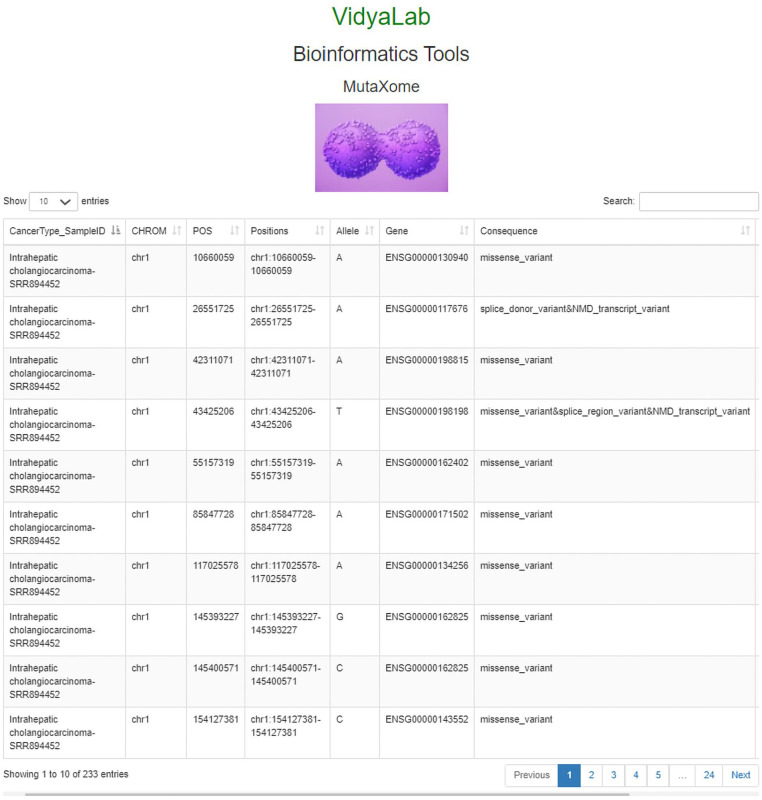
Successful designing and development of database in SQL format with the front end in bootstrap and HTML, and backend in PHP also ensured easy GUI development for the same. The developed database when opened in WAMP or XAMP displays all tabs as necessary for it to function appropriately. Alternatively, when accessed through http://www.vidyalab.rf.gd/, it shows the 20 different cancer exome sample IDs from the dropdown, based on which the cancer type can be selected accordingly. The database displays the different mutations in rows and columns identified after variant calling. The view of the database in backend in PHP, with the original file is shown in Figure 4. As a case study, a cancer exome ID SRR894452 was entered in the search bar, and the results obtained were demonstrated (Figure 5). Comprehensive information such as the sample ID number, chromosome on which the mutation is present, its position, the location range with respect to the chromosomes, the allele change, gene ensemble ID, consequence (type of mutation), etc., was displayed. MutaXome allows searches to be made based on the sample ID (provided as drop-down list), gene symbol, or by providing one of the 5 cancer types. Minimal clinical information such as the type of cancer that the sample ID belongs to is also provided to aid the users to select their query. When typed in the search bar, the gene names return only those results that are existing in the database, that is, only specific gene names belonging to specific cancer types and those identified in our analysis. The list of genes can be obtained from “List of genes” tab from the database. The database GUI also displays 2 search bars, 1 for querying the mutational information of the sample IDs and another 1 for searching the patient information of the sample IDs. Since the database will be updated and enriched as and when further analysis is carried out (as part of prospective work) and adding more columns to the database will cause redundancy of the patient information for each sample ID, 2 search bars are provided to simplify the use of the database. Users can search for mutational information under “Mutational information” search bar, and patient details under “clinical information” search bar. Additionally, there are also options to display 10, 25, 50, or 100 entries are also present, which can be set according to the user’s preferences. Figure 5 shows the top 10 entries for sample SRR894452. Likewise, any of the 20 cancer exome NCBI SRA IDs that are provided in the paper can be used to obtain mutation information for the same.
Figure 4.
Screenshot of MutaXome database on backend, when viewed on the WAMP server using phpMyAdmin, when the database was installed locally on the system using the original variant file. The designed database displays information on the variants organized as rows and columns. The sample ID, chromosome number, position, reference nucleotide, altered nucleotide, the type of consequence, impact of the variation, gene symbol, quality, filter, etc. are initially observed.
Figure 5.
Search results for a single sample ID (SRR894452), as viewed on the online resource http://www.vidyalab.rf.gd/ presented as a case study. The top 10 entries are shown in the figure, revealing the chromosome number, position, allele, Ensembl gene number, and type of mutation as a consequence.
A web-based tool called VarStack interprets the variants somatic variants in cancer, where data retrieval is carried out using several publicly available databases such as ClinVar, COSMIC, OncoKB, etc. 41 This tool comprises of several components such as MySQL, Python, and R. The present study however, has designed and developed a comprehensive database from MySQL, PHP, and HTML. In addition, another database COSMIC (Catalogue of Somatic Mutations in Cancer) stores all somatic mutations related to different human cancers, specifically on mutations of BRAF, KRAS2, HRAS, and NRAS genes. 42 This database reports about 10 647 mutations, while MutaXome currently has about 4181 mutations. Despite this, MutaXome offers information on several 1000 genes that are possibly related to different cancer types. Data on the impact of the type and impact of mutation serve as value added components of MutaXome. Moreover, MutaXome is user-friendly, easy to maneuver through the pages and displays only relevant information when user searches a query. Since it houses only the data identified from current study and previous work, search by gene names provides only the variations that have been detected from specific cancer types in the study. The search returns zero records and “no data available in table” if the user queries something outside the scope of the specific cancer types.
Other cancer somatic variant databases include SomamiR 2.0, for detecting mutations in microRNAs 43 and DSMNC that shows alterations in the normal tissues that might trigger various cancers. 44 Despite there being other related databases, the information obtained from MutaXome, can be used to gather more data on the variants present in 20 different cancer exomes that can be implicated in the prognosis or diagnosis of the disease. The major contribution of this work is the design of a database that houses 4181 mutations identified from analysis of 20 cancer exomes. This database serves as a knowledge base for all vital information related to the 5 specific cancer types (intrahepatic cholangiocarcinoma, pancreatic adenocarcinoma, non-BRCA1/BRCA2 familial breast cancer, human diffuse type gastric cancer, and high grade serous ovarian cancer). Since this work identified a huge number of somatic mutations as a continuation of our previous work, the outcomes pave the way for similar prospective studies. The database created can be accessible via the link provided and thus, offers valuable information on each of the variant, such as the position of occurrence of the SNP and the altered nucleotide base, making this research work novel. Further, an in-depth analysis of these mutations may provide additional insights and possibilities into potential biomarker identification that can be used for specific cancer detection quickly and easily. Thus, the current study is the first of its kind to develop a specialized database for mutations that were identified through the NGS pipeline. The database has immense relevance, as it encompasses 20 cancer exomes belonging to 5 cancer types that contribute a wide array of information for advancement in cancer research.
Conclusions
The vast amount of information associated with mutations in vital cancer exomes makes it challenging to access them all in 1 place and use it for further research and analysis. Based on the entirety of the available data, the type of cancers significant in the current Indian population was considered to develop the database. The database was developed for the identified variants with the front end in bootstrap and HTML and backend in PHP, for all 20 cancer exomes encompassing 5 types of cancers. The database developed hosts information regarding 4181 variants detected, filtered, and normalized from 20 cancer exomes. Each variant has information related to its chromosome position, chromosome number, sample ID it was detected in, reference nucleotide, altered nucleotide in the cancer type, type of consequence, impact, filter, HGNC number, amino acid change, etc. There are several ways in which this database can be refined to add more scope to its applications. First, an attempt to include more cancer exome data will be looked at by analyzing more exome samples of various cancer types to make the database more comprehensive. The NGS pipeline will be followed, and variants that are called will be incorporated into the existing information. Secondly, designing and developing a decision support system (DSS) model that can be used to provide a preliminary prognosis for cancer patients is another future prospective. The DSS model will be developed using several machine learning algorithms to improve the accuracy of the model so that the best result can be predicted. Therefore, this project will be carried forward to establish a solid DSS system to enrich and provide value addition to the developed database.
Acknowledgments
The authors acknowledge the staff and non-staff members of the Department of Biotechnology and Centre of Excellence at R V College of Engineering, Bangalore, India for providing the motivation and skills to carry out this research work. Special thanks to Mr. Akshay Uttarkar for reviewing the manuscript, Mr. Vasanth Kumar Desai for helping in the preliminary database development process and Vanishree G for helping in database modifications. We would also like to acknowledge Mr. Aravind Ganessin, Managing Director, Intergene Biosciences Pvt. Ltd, Bangalore, for providing insights on the methodology.
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Padmavathi P was involved in implementation of methodology, data collection and analysis. Chandrashekar K and Anagha S Setlur were involved in manuscript writing, implementation of methodology and analysis of data. Vidya Niranjan conceptualized the idea, analysed the data and supervised the project.
ORCID iD: Vidya Niranjan  https://orcid.org/0000-0001-9187-7753
https://orcid.org/0000-0001-9187-7753
References
- 1. Weeden C, Liesse M, Labat A. Mechanisms of DNA damage repair in adult stem cells and implications for cancer formation. Biochimica et Biophysica Acta (BBA) – Mol Basis Dis. 2018;1864:89-101. [DOI] [PubMed] [Google Scholar]
- 2. Hodis E, Watson I, Kryukov G, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research Network; Weinstein JN, Collisson EA, Mills CB, et al. The Cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Agrawal N, Akbani R, Aksoy BA, et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adusei I, Kuljaca O, Agyepong K. Intelligent mammography database management system for a computer aided breast cancer detection and diagnosis. Int J Manag Inf Technol. 2010;2:1-13. [Google Scholar]
- 7. Sim KS, Chong SS, Tso CP, Nia ME, Chong AK, Abbas SF. Computerized database management system for breast cancer patients. Springerplus. 2014;3:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. International Cancer Genome Consortium; Hudson TJ, Anderson W, Artez A. International network of cancer genome projects. Nature. 2010;464:993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellis MJ, Gillette M, Carr SA, et al. Connecting genomic alterations to cancer biology with proteomics: the NCI clinical proteomic tumor analysis consortium. Cancer Discov. 2013;3:1108-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pavlopoulou A, Spandidos DA, Michalopoulos I. Human cancer databases. Oncol Rep. 2015;33:3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hess JL. The cancer genome anatomy project: power tools for cancer biologists. Cancer Invest. 2003;21:325-326. [DOI] [PubMed] [Google Scholar]
- 12. Zhang J, Finney RP, Rowe W, et al. Systematic analysis of genetic alterations in tumors using cancer genome workBench (CGWB). Genome Res. 2007;17:1111-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maqungo M, Kaur M, Kwofie SK, et al. DDPC: dragon database of genes associated with prostate cancer. Nucleic Acids Res. 2010;39:D980-D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mosca E, Alfieri R, Merelli I, Viti F, Calabria A, Milanesi L. A multilevel data integration resource for breast cancer study. BMC Syst Biol. 2010;4:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poulos RC, Wong YT, Ryan R, Pang H, Wong JWH. Analysis of 7,815 cancer exomes reveals associations between mutational processes and somatic driver mutations. PLOS Genet. 2018;14:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leinonen R, Sugawara H, Shumway M; on behalf of the International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2010;39(suppl 1):D19-D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ibrahim M, Gilbert K. Management of gastric cancer in Indian population. Transl Gastroenterol Hepatol. 2017;2:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maurya AP, Brahmachari S. Current status of breast cancer management in India. Indian J Surg. 2020;83: 1-6. [Google Scholar]
- 21. Kammar PS, Shrikhande SV, Goel M. The reality of cholangiocarcinoma in India: experience of over 500 patients from tata memorial Centre, Mumbai. J Cancer Res Ther. 2017;13:S128-S128. [Google Scholar]
- 22. Shrikhande SV, Barreto S, Sirohi B, et al. Indian council of medical research consensus document for the management of pancreatic cancer. Indian J Med Paediatr Oncol. 2019;40:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaur S, Singh R. Patterns of care for ovarian cancer. Cancer Res Stat Treat. 2019;2:217-220. [Google Scholar]
- 24. Padmavathi P, Setlur AS, Chandrashekar K, Niranjan V. A comprehensive in-silico computational analysis of twenty cancer exome datasets and identification of associated somatic variants reveals potential molecular markers for detection of varied cancer types. Inf Med Unlocked. 2021;26:100762. [Google Scholar]
- 25. Andrews S. Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. Published online 2010. Accessed 30 August 2021. https//wwwbioinformaticsbabrahamacuk/projects/fastqc.
- 26. Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10-12. [Google Scholar]
- 28. Trim galore Krueger F. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. 2015;516:517. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore [Google Scholar]
- 29. Li H, Durbin R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinf. 2009;25:1754-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinf. 2009;25:2078-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruden DM, Cingolani P, Patel VM, et al. Using drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, snpsift. Front Genet. 2012;3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dvorski DD. Installing, configuring, and developing with Xampp. Ski Canada. Published online 2007. [Google Scholar]
- 35. WampServer Bourdon R. Comput software WampServer-Apache, PHP, MySQL Wind Version. 2012;2. https://www.wampserver.com/en/ [Google Scholar]
- 36. Sourceforge. WampServer. Accessed September 5, 2021. https://sourceforge.net/projects/wampserver/
- 37. Singh RR. Next-generation sequencing in high-sensitive detection of mutations in tumors: challenges, advances, and applications. J Mol Diagn. 2020;22:994-1007. [DOI] [PubMed] [Google Scholar]
- 38. Bailey MH, Meyerson WU, Dursi LJ, et al. Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples. Nat Commun. 2020;11:1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun S, Wang Y, Maslov AY, Dong X, Vijg J. SomaMutDB: a database of somatic mutations in normal human tissues. Nucleic Acids Res. Published online 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rennert H, Eng K, Zhang T, et al. Development and validation of a whole-exome sequencing test for simultaneous detection of point mutations, indels and copy-number alterations for precision cancer care. NPJ Genomic Med. 2016;1:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Howard M, Kane B, Lepry M, Stey P, Ragavendran A, Gamsiz Uzun ED. VarStack: a web tool for data retrieval to interpret somatic variants in cancer. Database. 2020; 2020. doi:10.1093/database/baaa092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhattacharya A, Cui Y.SomamiR 2.0: a database of cancer somatic mutations altering microRNA—ceRNA interactions. Nucleic Acids Res. 2016;44:D1005-D1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miao X, Li X, Wang L, Zheng C, Cai J. DSMNC: a database of somatic mutations in normal cells. Nucleic Acids Res. 2019;47:D971-D975. [DOI] [PMC free article] [PubMed] [Google Scholar]