Abstract
A stable adhesion-deficient mutant of Burkholderia cepacia G4, a soil pseudomonad, was selected in a sand column assay. This mutant (ENV435) was compared to the wild-type strain by examining the adhesion of the organisms to silica sand and their transport through two aquifer sediments that differed in their sand, silt, and clay contents. We compared the longitudinal transport of the wild type and the adhesion mutant to the transport of a conservative chloride tracer in 25-cm-long glass columns. The transport of the wild-type strain was severely retarded compared to the transport of the conservative tracer in a variety of aquifer sediments, while the adhesion mutant and the conservative tracer traveled at similar rates. An intact sediment core study produced similar results; ENV435 was transported at a faster rate and in much greater numbers than G4. The results of hydrophobic interaction chromatography revealed that G4 was significantly more hydrophobic than ENV435, and polyacrylamide gel electrophoresis revealed significant differences in the lipopolysaccharide O-antigens of the adhesion mutant and the wild type. Differences in this cell surface polymer may explain the decreased adhesion of strain ENV435.
One approach for bioremediation of toxic chemicals by microorganisms is inoculation of contaminated sites with bacteria that are able to degrade or immobilize the relevant toxic compounds. After injection of biodegradative bacteria (i.e., bioaugmentation), the bacteria must be able to move throughout the surrounding porous media rather than collect at the injection site. Transport of bacteria through porous media can be prevented by filtration, the blocking of bacterial movement by pores that are smaller than the bacteria (26). It is assumed that binding of bacteria to particle surfaces also prevents movement and that bacteria must be nonadhesive for extensive bacterial transport through porous media (19, 32).
Studies of bacterial adhesive properties in marine and freshwater environments have indicated that a number of factors contribute to adhesiveness of the bacterial surface (11). These factors include cell surface charge, which affects electrostatic interactions between bacterial and substratum surfaces (33), bacterial hydrophobicity (3, 29), and the presence of extracellular polymers (1) and flagella (6). A wide range of approaches, including hydrophobic interaction chromatography (HIC) (21), electrostatic interaction chromatography (ESIC) (21), contact angle measurement (35), and electron microscopy (5), have been used to evaluate these parameters. The extent to which bacterial adhesion affects transport in the subsurface is not clear and depends on the organism and porous media conditions. It has been shown that the adhesiveness of Pseudomonas species and the transport of these organisms through porous media are both related to the presence and composition of flagella (6, 7) or lipopolysaccharide (LPS) (36). In contrast, in an evaluation of a wider range of organisms (Enterobacter, Pseudomonas, Bacillus, Achromobacter, Flavobacterium, and Arthrobacter species) Gannon et al. found no consistent relationship between transport and bacterial surface charge, hydrophobicity, or extracellular polymers (12).
Our previous studies focused on Pseudomonas strains obtained from freshwater (36) and soil (6, 7). LPS was involved in adhesion of the freshwater strain studied, whereas flagella played a role in attachment of the soil isolate. These findings raise the question of whether the same polymers or other polymers on the cell surface are involved in attachment of other pseudomonads. Thus, we extended our previous studies to include a bacterial species with bioremediation potential. In this study, we used a trichloroethylene (TCE)-degrading strain of Burkholderia cepacia, and our objectives were (i) to select an adhesion mutant that had decreased adhesion ability and thus increased transport potential compared with the wild type; and (ii) to determine the cell surface characteristics responsible for the altered transport properties. Once an adhesion mutant was obtained, we evaluated its cell surface characteristics by HIC and ESIC and performed outer membrane protein (OMP) and LPS analyses in order to determine the surface chemistry changes that were responsible for the alteration in adhesion ability. We also evaluated the relationship between adhesion and transport of the adhesion variant and of the wild type in repacked columns containing three types of porous media (homogeneous silica sand and two different aquifer sediments) and in intact sediment cores obtained from a borrow pit in Oyster, Va.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
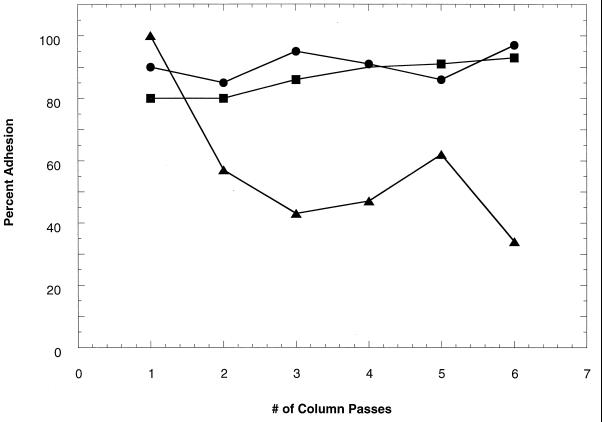
Wild-type B. cepacia G4 (formerly Pseudomonas cepacia G4) can degrade TCE and related compounds (24). B. cepacia PR1301 is a chemical mutant of G4; this mutant is constitutive for TCE degradation (23) and adhered like G4 in column tests (Fig. 1). ENV435 is a less adhesive chemical mutant of PR1301 and was produced during this study by exposing a culture to 99% methyl methanesulfonate (MMS) (Aldrich Chemical Co., Milwaukee, Wis.). The strains were grown in basal salts medium (BSM) (13) supplemented with either 0.2% (wt/vol) sodium glutamate or 0.2% (wt/vol) sodium lactate and incubated at 30°C.
FIG. 1.
Percent adhesion (retention) of bacteria from suspension after each passage (six repeated passages) through the sand column. Symbols: •, B. cepacia G4; ■, PR1301; ▴, mutagenized culture of PR1301, with which nonadhesive mutants were selected by their transport through the sand column. ENV435 was isolated from the effluent after the sixth passage.
Mutagenesis.
An overnight culture of strain PR1301 grown in BSM supplemented with 0.2% sodium lactate at 30°C was harvested by centrifugation, washed, and suspended in sterile BSM to an optical density at 550 nm (OD550) of 2.0. Aliquots (1.25 ml) of the washed cell suspension were placed into three microcentrifuge tubes. MMS (6.25 and 12.5 μl) was added to two of the tubes, and the tubes were mixed by vortexing and were incubated at 30°C for 1 h. After incubation, the cells were harvested by centrifugation and washed with 25 ml of BSM and then with 10 ml of BSM. The cells were then suspended in 10 ml of BSM supplemented with 0.2% sodium lactate and were incubated at 30°C for 1 h. After incubation, 100 μl of each preparation was diluted with phosphate-buffered saline (PBS) (30) and plated onto R2A (Difco Laboratories, Detroit, Mich.) plates. Colonies from these plates were inoculated into 100 ml of BSM containing 0.2% lactate and shaken at 200 rpm at 30°C overnight. These cultures were used as the inocula for selection of adhesion-deficient mutants as described below.
Selection of adhesion-deficient mutants.
Adhesion-deficient mutants were selected by repeated passage of effluent containing nonattached bacteria through sand columns by using the protocol of DeFlaun et al. (6). Briefly, columns were constructed from 20-ml syringe barrels that were filled with 12 g of dry, sterile sand (20/30 mesh [Ottawa Sand Standard]; Fisher Scientific Co., Pittsburgh, Pa.). Three cultures of mutants were grown to the log phase, washed, and suspended in PBS to densities of approximately 107 cells ml−1. The columns were inoculated with 3-ml portions of these suspensions, and after 5 min the excess liquid was drained off. After 1 h of incubation at room temperature, the columns were rinsed with 3 ml of PBS three times to remove nonattached bacteria. The adhesive bacteria were retained in the column, while the bacteria with reduced adhesion ability tended to pass through. Cells in the saline wash were then harvested, suspended in fresh medium, grown to the log phase, and passed through a second column. Cells were passed through several columns in this manner, so that adhesion-deficient organisms were enriched. Adhesion-deficient mutants were then tested individually with the adhesion assay to determine percentages of adhesion. The stability of the nonadhesive phenotype was also tested by growing strains for more than 100 generations in a nonselective medium and retesting to determine percentages of adhesion.
Transport column studies. (i) Repacked columns.
We compared the transport of ENV435 and the transport of wild-type G4 through porous media by using glass columns (diameter, 1.0 cm; length, 25 cm) that were packed with either sand (cleaned quartz play sand; particle diameter, ca. 160 μm) or sediment from a Savannah River site (site SR 10-20′; 86.7% sand, 9% silt, 4.3% clay; Savannah River Aquifer, Aiken, S.C.). The percentages of recovery and transport of adhesion-deficient mutant ENV435 in Roseland Aquifer sediment (73% sand, 10% silt, 17% clay; Roseland, N.J.) were also determined. Sediment samples were analyzed to determine the particle size distribution by using a commercially available kit (LaMotte Chemical, Chestertown, Md.). The sediment and sand were autoclaved twice at 15 lb/in2 and 121°C for 90 min, dried in a 95°C oven for 25 h, and then packed in the columns to a depth of 20 cm. Artificial groundwater (5 mM CaSO4 in deionized water) was pumped into the columns in an upflow mode at a rate of 0.25 ml h−1. At the start of each experiment, the columns were inoculated at the bottom with 0.2-ml portions of a ∼108-CFU ml−1 suspension of either ENV435 or wild-type strain G4. The cells were prepared for injection by washing them and suspending them in artificial groundwater. The columns were run for 24 h at room temperature. Samples were removed periodically from the top of each column, serially diluted, and plated onto R2A plates. After the last bacterial sample was removed, 0.2 ml of 4 M NaCl was injected into each column as a conservative tracer to determine the rate of water flow through the column. Chloride concentrations were measured with a chloride-specific electrode (Orion, Boston, Mass.).
(ii) Intact column studies.
The geology and mineralogy of the sediment in intact cores taken from the borrow pit at the U.S. Department of Energy (DOE) Oyster, Va., field site have been described elsewhere (8). The core used in this study was taken from a cross-bedded, shelly, gravelly sand facies in a formation that is predominantly quartz sand. The bottom of an aluminum Shelby tube (length, 70 cm; inside diameter, 7 cm) was cut off with a large pipe cutter, and the end piece was removed. Care was taken not to disturb the sediment remaining in the end piece and in the core. A bottom spacer consisting of a stainless steel mesh disc (wire diameter, 0.0075 in.; width opening, 0.0092 in.; 60 mesh per linear in.; McMaster-Carr, Dayton, N.J.) glued to a piece of acrylic pipe (height, 0.875 in.; outside diameter, 2.75 in.; Glasflex, Stirling, N.J.) was placed inside the bottom of the core with the mesh flush against the sediment. The top of the Shelby tube was cut off 53.5 cm from the bottom of the core, which left 2.5 cm for the bottom spacer, 50 cm for the sediment, and 1.0 cm for the headspace. A 4-in. square of 0.5-in.-thick acrylic that had a 0.125-in.-deep groove cut in the center and a center hole for inlet and outlet tubing was attached to the top of the core, and an identical place of acrylic was attached to the bottom of the core. The effluent tubing included a T-joint that was open to the atmosphere to prevent air bubbles from forming and to vent any pressure buildup that occurred. The core effluent was allowed to flow freely into a 100-ml plastic graduated cylinder. The effluent was then pumped out of the cylinder at the same flow rate as the influent flow rate and into a model 204 fraction collector (Gilson, Middleton, Wis.). This configuration prevented backpressure caused by the smaller tubing of the fraction collector from affecting flow through the column.
(iii) Pore volume determination.
The pore volume of the core was determined by using the intact end pieces that were cut off the main body of the core. The end pieces were placed onto a 150-mm glass petri dish, and the sediment was left to dry at room temperature for 4 days. The weight of the dried sediment was then recorded. The sediment was then saturated by pipetting distilled water into the bottom of the piece of Shelby tube and allowing the water to rise up into the sediment by capillary action. The weight of the saturated sediment was recorded. The sediment was then dried overnight in a 105°C oven, and the procedure was repeated. The mean value of the two measurements obtained was used in the following calculations. The dry weight was subtracted from the saturated weight, and this value was divided by the volume of the piece of Shelby tube to determine the volume of water (in milliliters) per cubic centimeter of sediment. This value was multiplied by the total volume of the core to obtain the pore volume of the core.
(iv) Determination of water velocity.
The composition of Oyster artificial groundwater (OAGW) is based on the Oyster groundwater chemistry; OAGW contains (per liter of distilled water) 60 mg of MgSO4 · 7H2O, 20 mg of KNO3, 36 mg of NaHCO3, 36 mg of CaCl2, 35 mg of Ca(NO3)2, 25 mg of CaSO4 · 2H2O, 28 mg of NaH2PO4, and 0.35 ml of 1.0 N HCl (pH 5.99). Once 1 pore volume of OAGW had passed completely through the core, 10 ml of 5 M NaCl was injected into the influent tubing. The flow velocity (in meters per day) of the chloride tracer was measured with a combination chloride probe that was inserted through a section of effluent tubing.
(v) Radiolabeling of cells.
Bacteria were radiolabeled by using a modification of the method of McEldowney and Fletcher (21). Cells grown on R2A streak plates for 5 days were scraped off the plates and suspended in 2.5 ml of BSM. The resulting suspensions were adjusted to an OD550 of 0.05 with BSM. To the wild-type strain G4 culture 1.35 μCi of d-[U-14C]glucose (specific activity, 250 mCi/mmol; ICN Pharmaceuticals Inc., Irvine, Calif.) per ml was added, and the ENV435 culture was supplemented with 1 μCi of d-[6-3H]glucose (specific activity, 26 Ci/mmol) per ml. These cultures were incubated with shaking at room temperature (22°C) for 8 h, after which 0.05% unlabeled glucose was added. The cultures were returned to the shaker for 48 h, after which the cells were harvested, suspended in OAGW, and incubated with shaking for an additional 48 h.
(vi) Core injection and sampling procedure.
After being starved for 48 h, the cultures were washed once and suspended in OAGW to an OD550 of 0.03 (∼3 × 107 cells/ml). The specific activities of the cells were determined by calibrating serial dilutions of radiolabeled cells versus CFU (14C-labeled G4, 618 CFU/dpm; 3H-labeled ENV435, 1,312 CFU/dpm). Then 370 ml (0.5 core pore volume) of each cell suspension was combined in a 1-liter container and mixed well. The cells were pumped into the column via the influent tubing. Once the injection was complete, the inflow tubing was moved to a container containing Oyster site groundwater. Oyster site groundwater was continuously pumped into the core for 6 days (corresponding to 12 pore volumes). The stability of the radiolabel was checked by determining the specific activity of cells throughout the experiment. There was no decrease in the specific activity over an 11-day period (data not shown). Core effluent fractions were analyzed at 15-min intervals. One milliliter of Liquiscint scintillation cocktail (National Diagnostics, Atlanta, Ga.) was added to a 4-ml sample, and the preparation was then vortexed for 30 s before the radioactivity was counted with a model 1209 Rackbeta scintillation counter (Pharmacia LKB Nuclear, Gaithersburg, Md.). The percentage of recovery was calculated from the total disintegrations per minute collected in the fractions and the total disintegrations per minute of the cells injected into the core.
(vii) Bacterial distribution within the core material.
The distribution of the cells that remained in the core material (i.e., the cells that adhered to the sediment and were not recovered in the effluent) was determined by sampling the sediment. Once the transport experiment was completed, the core was frozen at −80°C for 1 week and then fractured in half and allowed to thaw. A grid constructed out of polyethylene hardware cloth (opening size, 0.78 in.) was laid across the core to insure that sampling was consistent. Samples were taken through this grid by using Teflon tubing (outside diameter, 0.5 in.; inside diameter, 0.375 in.) which was inserted through a hole in the grid until it reached the aluminum casing. The sediment in the tubing was extruded into a 50-ml disposable plastic centrifuge tube. The samples were mixed well, and duplicate 1.2-g aliquots of each sample (equivalent to 1 g [dry weight]) were placed into 20-ml scintillation vials. Five milliliters of OptiPhase HiSafe 3 scintillation cocktail (Pharmacia LKB Nuclear) was added to each vial, and the vials were vortexed at the highest speed for 1 min before the radioactivity was counted with a scintillation counter.
Characterization of OMP and LPS.
For the OMP analysis, 30-ml cultures were grown as described above. Cells were harvested and washed in 0.1 M potassium phosphate buffer (pH 7.4). Each pellet was suspended in 4 ml of a Tris-EDTA solution (50 mM Tris, 2 mM EDTA; pH 7) and passed twice through a French press, and 1-ml aliquots were transferred to two microcentrifuge tubes, each containing 1.5 ml of lysate. Nonlysed cells were removed by centrifugation, and outer membrane fractions were prepared (20). For each culture, one of the two prepared samples was solubilized, heated as described by McCarter and Silverman (20), and used for a protein electrophoretic analysis. The second sample was used for protein quantification by the BCA protein assay (Pierce, Rockford, Ill.), in which bovine serum albumin was used as the standard. OMP profiles were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 14% polyacrylamide gel (20 cm by 1 mm) by using the Tris-glycine Laemmli buffer system (16). Approximately 25 μg of each sample was electrophoresed at a constant voltage (200 V). The gels were stained with Coomassie brilliant blue R-250 (Sigma Chemical Co., St. Louis, Mo.).
For LPS determinations, pellets were collected from 1-ml aliquots of cultures (3.5 ml of BSM) by centrifugation. The cells were washed once in 0.1 M potassium phosphate buffer (pH 7.4) and then treated by using the procedure of Hitchcock and Brown (15) for whole-cell lysates and proteinase K digestion. Samples (6 μl) of LPS preparations were electrophoresed on sodium dodecyl sulfate–15% polyacrylamide mini gels as described above. The LPS gels were silver stained by using the Hitchcock-Brown (15) protocol with the following two modification: (i) the gels were briefly rinsed in Millipore water prior to the oxidation step; and (ii) following the oxidation step, the gels were subjected to eight 15- to 20-min washes.
HIC and ESIC.
HIC was performed by using the protocol described by Dahlback et al. (4) and Octyl-Sepharose CL-4B (Pharmacia Biotech, Uppsala, Sweden) diluted 1:1 in OAGW and packed into Pasteur pipettes. Cultures were grown to the mid-log phase as described above, except that they were incubated with 0.27 μCi of [14C]glucose (final glucose concentration, 0.97 μg/ml; Sigma Chemical Co.) per ml for 8 to 10 h at room temperature before 0.2% sodium lactate was added. This method resulted in the highest percent incorporation of label, and the sodium lactate added provided sufficient carbon for growth. The cultures were adjusted to densities of approximately 109 cells/ml and washed three times with 1 ml of OAGW per ml of cells to remove any exogenous radioactivity. One milliliter of each washed cell suspension was applied to a gel bed and eluted with four 3-ml OAGW washes into separate vials. One milliliter of Liquiscint scintillation cocktail was added to each 3-ml wash. The gel was then transferred from the pipette into a scintillation vial by using one 5-ml OAGW wash. Once chromatography was complete, a test for leakage of radiolabel from the cells was performed by centrifuging (16,000 × g, 5 min) duplicate 1-ml samples of the washed cell suspension and counting the radioactivity in the supernatant. Hydrophobicity was expressed as the ratio of the counts in the gel to the counts in the eluant (g/e ratio). Higher g/e ratios indicated that there was a greater tendency toward hydrophobic interactions. A negative log g/e ratio implied hydrophilicity.
ESIC was performed in essentially the same manner as HIC (25), except that the pipettes were packed with either the anion-exchange resin Dowex 1x8 (a strongly basic anion exchanger whose ionic form is chloride) or the cation-exchange resin Dowex 50Wx8 (a strongly acidic cation exchanger whose ionic form is hydrogen; Sigma Chemical Co.). Columns were constructed with 1 g of resin suspended in 1 ml of OAGW and packed into a Pasteur pipette. Bacterial affinity for the ion-exchange resins was expressed as the ratio of the radioactivity remaining in the resin fraction to the radioactivity in the eluant from the same column (r/e ratio). Higher r/e ratios for the anion-exchange resin indicated that there was a higher negative surface charge, while higher r/e ratios for the cation-exchange resin indicated that there was a higher positive surface charge.
RESULTS
After bacterial suspensions had been passed through sand columns six times, the percentage of the mutagenized cells retained was reduced from 100 to 34%, whereas the percentages of the parent strain PR1301 and G4 cells retained ranged from 80 to 93% (Fig. 1). A number of colonies obtained from the effluent of the sixth pass of the mutagenized cells were tested with the adhesion assay, and the adhesion values obtained were as low as 13%. ENV435, an adhesion-deficient mutant and constitutive TCE degrader, was selected from these cultures on the basis of its low adhesion ability. The stability of the adhesion phenotype of this strain was confirmed by culturing the organism for more than 100 generations and retesting adhesion with the sand column assay three times (data not shown).
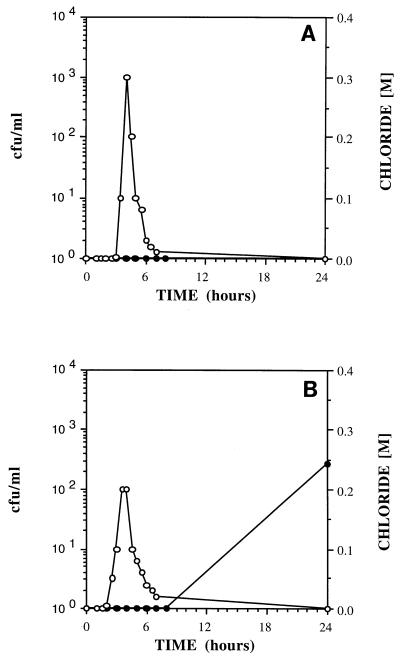
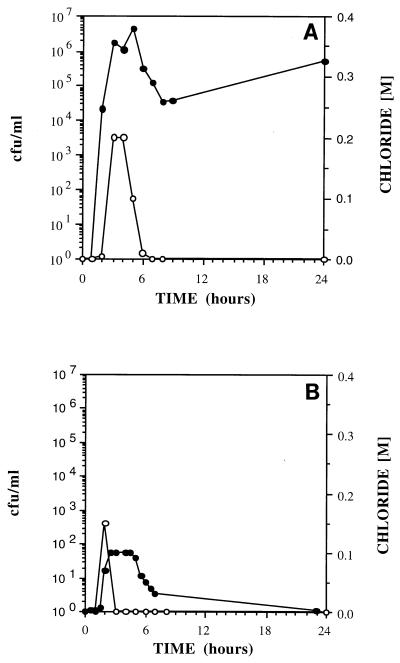
In the transport experiments, adhesion-deficient mutant ENV435 was compared to strain G4. This comparison was valid, even though ENV435 was derived from strain PR1301, because PR1301 was derived from G4 and these strains exhibited similar adhesion characteristics in the sand column assay (Fig. 1). Wild-type B. cepacia G4 was always retained and never passed through 25-cm-long glass columns packed with fine commercial sand (Fig. 2A); in addition, transport of this strain was severely retarded in Savannah River sediment, a more complex medium containing 9% silt and 4.3% clay, during 24-h transport studies (Fig. 2B). In contrast, the adhesion-deficient mutant, ENV435, moved through the glass columns containing the sand matrix (Fig. 3A), Savannah River sediment (Fig. 3B), and Roseland Aquifer sediment (Fig. 4A). The transport rate of ENV435 through the sand and sediment was approximately the same as the transport rate of the chloride tracer.
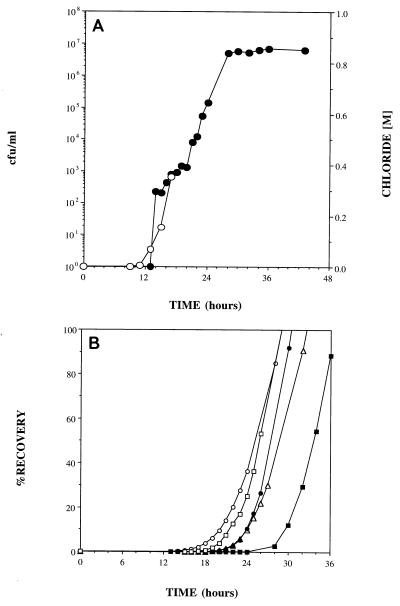
FIG. 2.
Effluent concentrations of B. cepacia G4 (•) and chloride (○) in 25-cm columns packed with sand (A) and Savannah River sediment (B).
FIG. 3.
Effluent concentrations of ENV435 (•) and chloride (○) in 25-cm columns packed with sand (A) and Savannah River sediment (B).
FIG. 4.
(A) Effluent concentrations of ENV435 (•) and chloride (○) in 25-cm columns packed with Roseland Aquifer sediment. Chloride data were collected only until breakthrough occurred. (B) Recovery of ENV435 in the effluent of 25-cm columns packed with Roseland Aquifer sediment. The different symbols show the results of replicate transport experiments.
In addition to influencing the rate of transport, the sediment type also influenced recovery of the organisms. The low CFU values for the effluent curve in Fig. 3B indicate that a higher percentage of ENV435 cells was retained by the Savannah River sediment than by the homogeneous sand (Fig. 3A).
For the Roseland Aquifer sediment the transport rate and the recovery of the adhesion-deficient strain, ENV435, were determined by using five replicate columns (Fig. 4). Figure 4A shows the results obtained when the bacterial transport rate in one of these columns was compared to the chloride tracer transport rate. The transport rate of ENV435 was similar to the transport rate of the chloride tracer up to 18 h, at which time 1 pore volume of chloride had passed through the column and chloride measurements were terminated. Similar rates of chloride and bacterial transport were observed in all five replicate columns. The recovery data obtained for all five columns are shown in Fig. 4B. More than 50% of the inoculum was recovered in 32 h and more than 80% of the inoculum was recovered in 36 h for all five columns. The average retention time for water in these columns, as determined by the conservative tracer tests, was 18 h.
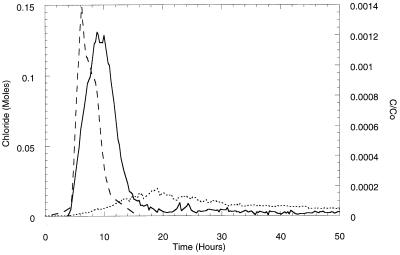
Figure 5 shows the results obtained when G4 and ENV435 were injected simultaneously into an intact sediment core. Although the peak concentration of ENV435 in the effluent lagged behind the conservative chloride tracer peak, initial detection of ENV435 and initial detection of chloride occurred at approximately the same time. The peak concentration of wild-type strain G4, however, took approximately twice as long (19 versus 9 h) to appear in the effluent. Instead of occurring in the effluent as a definitive peak, G4 was transported through the core at low numbers throughout the 50-h experiment. The overall level of recovery of G4 in the effluent was 13.2%, while the level of recovery of adhesion mutant ENV435 was 26.3%.
FIG. 5.
Effluent concentrations of chloride (---), ENV435 (—), and B. cepacia G4 (⋯) transported simultaneously through an intact core column containing Oyster site sediment. Bacterial concentrations are expressed as the concentration in the effluent (C) divided by the initial concentration injected (Co).
The distributions of the cells retained in the columns, however, were very similar. Approximately 94% of the G4 cells and approximately 95% of the ENV435 cells were retained in the first 16 cm of the core, and the remaining cells were distributed fairly evenly throughout the top 34 cm of the core. The number of ENV435 cells detected in the sediment plus the number of ENV435 cells detected in the effluent accounted for 102% of the cells injected. For G4, however, the number of cells in the effluent and plus the number of cells in the sediment accounted for only 50% of the cells added. The reason for the low level of recovery of the 14C-labeled cells is not known, although it is likely that respiratory loss as 14CO2 is responsible at least in part. Some of the loss can be attributed to quenching of the radioactive signal by the sediment. Samples radiolabeled with 14C and counted in the presence of Oyster sediment had an average of 11.9% fewer counts than samples without sediment. The loss was slightly greater for tritium; the average loss was 25.5%. Correcting the percent recovery values for quenching effects, however, did not account for the greater loss in 14C counts because the quenching of 14C is less than the quenching of 3H.
There were no detectable differences in OMP between wild-type strain G4 and the two chemical mutants. In contrast, the LPS structure of ENV435 differed from the LPS structures of the more adhesive strains; G4 and PR1301 produced a ladderlike pattern consistent with the presence of the O-antigen, whereas ENV435 did not (Fig. 6).
FIG. 6.
Polyacrylamide gel electrophoresis of LPS of strains G4, PR1301, and ENV435.
HIC of wild-type strain G4, PR1301, and ENV435 revealed large differences in hydrophobicity among these strains. G4 clearly had the strongest tendency toward hydrophobic interactions, with a mean g/e ratio of 16.6; PR1301 had a mean g/e ratio of 1.5 (Table 1), while the mean g/e ratio of ENV435 was 0.3 (Table 1).
TABLE 1.
Affinities of B. cepacia G4, PR1301, and ENV435 for hydrophobic, cationic, and anionic matrices
| Strain | HIC g/e ratioa | ESIC affinity for the cation exchangerb | ESIC affinity for the anion exchangerc |
|---|---|---|---|
| B. cepacia G4 | 16.6 (0.8)d | 0.1 (0.1) | 8.2 (0.3) |
| PR1301 | 1.5 (0.1) | 0.1 (0.0) | 0.6 (0.2) |
| ENV435 | 0.3 (0.3) | 0.1 (0.1) | 13.0 (3.2) |
A higher g/e ratio indicates a greater tendency for hydrophobic interactions.
A higher value indicates a higher positive cell surface charge.
A higher value indicates a higher negative cell surface charge.
The values in parentheses are standard deviations (n = 3).
There was no difference among the affinities of the three strains for the cation exchanger (Table 1). There were, however, differences among the affinities of the three strains for the anion exchanger. The anion exchanger affinity value for G4 was 8.2, while the value for ENV435 was higher, 13.0. PR1301, however, had an anion exchanger affinity value (0.6) that was 13 times lower than that of G4 and 21 times lower than that of ENV435. The data were confirmed by performing each experiment at least three times (Table 1).
DISCUSSION
Adhesion to inanimate materials is the net result of a variety of attractive and repulsive forces (e.g., electrostatic interactions, hydrophobic interactions, van der Waals interactions, hydrogen bonding, and steric interactions). The balance of these forces and the net adhesiveness of a given cell can vary from organism to organism, depending on variations in complex mixtures of surface polymers and structures. Therefore, adhesiveness is not an all-or-none property that can be described by well-defined affinity constants. For any given bacterial population, some cells attach, while others remain suspended, and there can be appreciable variation in adhesion even among clones of a single organism (36). Variations in adhesiveness are also fostered by (i) phenotypic modifications resulting from differences in life cycle stages or physiological conditions, and (ii) spontaneous mutations resulting in altered cell surface polymers or structures. By selecting for mutants with altered surface characteristics, we can obtain strains that have adhesive properties that are significantly different from those of the parent. Chemical mutagenesis was used in this study; however, transposon mutagenesis has also been used to generate cells with altered attachment properties (6, 7, 36).
Bacterial transport rates can be affected by differences in porosity (2) and mineralogy (22) in various sediments. The porosity of sediments or the fissures in rocks may be small enough to prevent movement of bacteria with the groundwater flow. Alternatively, pores and fissures may be large enough to collect transporting cells, while dissolved solutes move more quickly through channels and preferred flow paths (9). Presumably, the tendency for bacteria to collect on sediment and rock surfaces is enhanced by adhesive polymers or mechanisms (22), but there is little direct evidence demonstrating that the adhesive properties of cells are a significant factor in determining transport of the cells in complex porous media.
Although the adhesion variant ENV435 was transported through sand and sediment at approximately the same rate as the conservative tracer, the peak concentrations of cells in the column effluent were lower for the Savannah River sediment than for the sand. Apparently, adhesion and hence retention are influenced by the mineralogy, as well as the size distribution, of the sediment. Pure sand is relatively homogeneous; it is composed solely of quartz and does not contain clay or silt fractions.
Recovery assessed in the Roseland Aquifer sediment demonstrated that although the transport rate of ENV435 was similar to the transport rate of the conservative tracer, it took approximately twice as long to transport all of the cells as it took to transport the tracer. Retardation of the cells was presumably due to significant silt and clay fractions in the sediment, which decreased porosity, resulting in filtration of cells and tortuous flow paths. If adhesion was responsible for retardation of cells, then it was reversible, as more than 85 to 100% of the bacteria were eventually recovered. The differences in the results obtained with the five columns can be attributed to the variability associated with obtaining five subsamples of the same sediment and packing of the columns. The level of recovery of ENV435 from the intact core was much lower than the level of recovery from the repacked cores (26.3%) after 12 pore volumes of groundwater was used. This low level of recovery may demonstrate the effect on bacterial transport of physical heterogeneities which block or bind cells and are lost when a sediment is repacked into columns.
The significant differences in adhesion and transport of wild-type strain G4 and adhesion mutant ENV435 were presumed to be due to differences in cell surface properties. We attempted to identify the cell surface polymer(s) involved in adhesion of B. cepacia G4, PR1301, and ENV435 by comparing the OMP and LPS profiles of each isolate. Flagella (6) and pili (34) have been identified with adhesion of Pseudomonas fluorescens in the environment, but we were not able to detect any differences in cell surface proteins that correlated with differences in adhesiveness.
The polyacrylamide gel electrophoresis analysis of ENV435 demonstrated that the LPS of this adhesion-deficient strain lacked the ladderlike pattern that is consistent with the presence of O-antigen, which was present in G4 and PR1301. G4 and PR1301 were also more hydrophobic than ENV435, as determined by HIC (G4 > PR1301 > ENV435), whereas ENV435 had the highest negative charge (ENV435 > G4 ≫ PR1301). These results are consistent with the results of studies performed with Pseudomonas aeruginosa that demonstrated that the presence of A-band LPS resulted in a more hydrophobic surface and greater adhesion to polystyrene than was observed with bacteria lacking O-polysaccharide chains (18). Makin and Beveridge (18) suggested that their results could be explained by the presence of a relatively hydrophobic LPS O-polysaccharide that shielded charged groups near the membrane, such as phosphate groups in the core-lipid A region. These results contrast with the results of other studies that demonstrated that an increase in adhesion (31, 36) or hydrophobicity (14, 17, 31) accompanied a loss or attenuation of O-polysaccharide, presumably due to greater exposure of the lipid moiety. Similarly, P. aeruginosa cells with B-band O-polysaccharide were less hydrophobic (10), indicating that the specific chemistries of the components of the LPS (i.e., the O-polysaccharide, the oligosaccharide core, and the lipid moiety) are involved in determining the overall adhesiveness of the cells.
The fact that a positive relationship between hydrophobicity and adhesion to sand or minerals was found in this and other studies (31, 36) indicates that hydrophobic interactions may control attachment to these types of surfaces. Numerous studies have demonstrated that hydrophobic interactions can play a significant role in bacterial attachment phenomena (11, 27, 29). This is somewhat surprising, as adhesion to silica sand might be expected to be similar to adhesion to glass, which does not appear to be favored by hydrophobic interactions (18); we cannot explain this inconsistency at this time.
The results of this study illustrate the validity of evaluating the relationship between the adhesiveness of bacteria and transport of the bacteria through porous media. Adhesion is a relatively simple phenotype to measure and can be used to evaluate bacterial strains for bioaugmentation applications. Information concerning the surface characteristics that determine adhesion can potentially be used to alter these properties and therefore influence the transport of organisms. Enhanced transport of ENV435 has been demonstrated in a system larger than the intact cores which we used. Using a pilot-scale in situ bioremediation trench (length, 4 ft), Reardon et al. (28) found that the wild-type strain B. cepacia PR1301 could not be detected more than a few inches from the addition zone, while ENV435 was rapidly transported through the soils used. ENV435 is currently being field tested in a sand aquifer contaminated with TCE. In previous studies workers determined that the degradative ability of the adhesion-deficient, constitutive isolate ENV435 was similar to that of the wild type (8a). Bioaugmentation with ENV435 could have considerable potential for remediating the most common groundwater contaminant in the United States, TCE.
ACKNOWLEDGMENTS
This work was supported in part by National Science Foundation-SBIR Phase II award II-931077; by DOE ORNL-Martin Marietta subcontract 1GX-SR004; and by DOE Subsurface Science Program grant DF-FG02-94ER61814 under the direction of Frank Wobber.
We are grateful to T. C. Onstott and Doug Johnson of the Department of Geological Sciences, Princeton University, for assistance with core fracturing. We also thank the EPA Gulf Breeze Laboratory, Gulf Breeze, Fla., and Malcolm Shields for providing B. cepacia G4 and PR1301, respectively.
REFERENCES
- 1.Allison D G, Sutherland I W. The role of exopolysaccharides in adhesion of freshwater bacteria. J Gen Microbiol. 1987;133:1319–1327. [Google Scholar]
- 2.Barton J W, Ford R M. Determination of effective transport coefficients for bacterial migration in sand columns. Appl Environ Microbiol. 1995;61:3329–3333. doi: 10.1128/aem.61.9.3329-3335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busscher H J, Sjollema J, van der Mei H C. Relative importance of surface free energy as a measure of hydrophobicity in bacterial adhesion to solid surfaces. In: Doyle R J, Rosenberg M, editors. Microbial cell surface hydrophobicity. Washington, D.C: American Society for Microbiology; 1990. pp. 335–359. [Google Scholar]
- 4.Dahlback B, Hermansson M, Kjelleberg S, Norkrans B. The hydrophobicity of bacteria—an important factor in their initial adhesion at the air-water interface. Arch Microbiol. 1981;128:267–270. doi: 10.1007/BF00422527. [DOI] [PubMed] [Google Scholar]
- 5.DeFlaun M F, Mayer L M. Relationships between bacteria and grain surfaces in intertidal sediments. Limnol Oceanogr. 1983;28:873–881. [Google Scholar]
- 6.DeFlaun M F, Tanzer A S, McAteer A L, Marshall B, Levy S B. Development of an adhesion assay and characterization of an adhesion-deficient mutant of Pseudomonas fluorescens. Appl Environ Microbiol. 1990;56:112–119. doi: 10.1128/aem.56.1.112-119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeFlaun M F, Marshall B M, Kulle E-P, Levy S B. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl Environ Microbiol. 1994;60:2637–2642. doi: 10.1128/aem.60.7.2637-2642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeFlaun M F, Murray C J, Holben W, Scheibe T, Mills A, Ginn T, Griffin T, Majer E, Wilson J L. Preliminary observations on bacterial transport in a coastal plan aquifer. FEMS Microbiol Rev. 1997;20:473–487. [Google Scholar]
- 8a.DeFlaun, M. F., and R. J. Steffan. Unpublished data.
- 9.DeMarsily G. Quantitative hydrogeology. New York, N.Y: Academic Press; 1986. [Google Scholar]
- 10.deWeger L A, Van Loosdrecht M C M, Klaassen H E, Lugtenberg B. Mutational changes in physicochemical cell surface properties of plant-growth-stimulating Pseudomonas spp. do not influence the attachment properties of the cells. J Bacteriol. 1989;171:2756–2761. doi: 10.1128/jb.171.5.2756-2761.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher M. Bacterial attachment in aquatic environments: a diversity of surfaces and adhesion strategies. In: Fletcher M, editor. Bacterial adhesion: molecular and ecological diversity. New York, N.Y: Wiley-Liss; 1996. pp. 1–24. [Google Scholar]
- 12.Gannon J T, Manilal V B, Alexander M. Relationship between cell surface properties and transport of bacteria through soil. Appl Environ Microbiol. 1991;57:190–193. doi: 10.1128/aem.57.1.190-193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hareland W A, Crawford R L, Chapman P J, Dagley S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol. 1975;121:272–285. doi: 10.1128/jb.121.1.272-285.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermansson M, Kjelleberg S, Korhonen T K, Stenstrom T-A. Hydrophobic and electrostatic characterization of surface structures of bacteria and its relationship to adhesion to an air-water interface. Arch Microbiol. 1982;131:308–312. [Google Scholar]
- 15.Hitchcock P J, Brown T M. Morphological heterogeneity among salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Magnusson K-E, Johansson G. Probing the surface of Salmonella typhimurium and Salmonella minnesota SR and R bacteria by aqueous biphasic partitioning in systems containing hydrophobic and charged polymers. FEMS Microbiol Lett. 1977;2:225–228. [Google Scholar]
- 18.Makin S A, Beveridge T J. The influence of A-band and B-band lipopolysaccharide on the surface characteristics and adhesion of Pseudomonas aeruginosa to surfaces. Microbiology. 1996;142:299–307. doi: 10.1099/13500872-142-2-299. [DOI] [PubMed] [Google Scholar]
- 19.Martin R E, Bouwer E J, Hanna L M. Application of clean-bed filtration theory to bacterial deposition in porous media. Environ Sci Technol. 1992;26:1053–1058. [Google Scholar]
- 20.McCarter L L, Silverman M. Phosphate regulation of gene expression in Vibrio parahaemolyticus. J Bacteriol. 1987;169:3441–3449. doi: 10.1128/jb.169.8.3441-3449.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McEldowney S, Fletcher M. Effect of growth conditions and surface characteristics of aquatic bacteria on their attachment to solid surfaces. J Gen Microbiol. 1986;132:513–523. [Google Scholar]
- 22.Mills A, Powelson D K. Bacterial interactions with surfaces in soils. In: Fletcher M, editor. Bacterial adhesion: molecular and ecological diversity. New York, N.Y: Wiley-Liss; 1996. pp. 25–57. [Google Scholar]
- 23.Munakata-Marr J, McCarty P L, Shields M S, Reagin M, Francesconi S C. Enhancement of trichloroethylene degradation in aquifer microcosms bioaugmented with wild-type and genetically altered Bukholderia (Pseudomonas) cepacia G4 and PR1. Environ Sci Technol. 1996;30:2045–2052. [Google Scholar]
- 24.Nelson M J K, Montgomery S O, O’Neill E I, Pritchard P H. Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl Environ Microbiol. 1986;52:383–384. doi: 10.1128/aem.52.2.383-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen K. Electrostatic interaction chromatography: a method for assaying the relative surface charge of bacteria. FEMS Microbiol Lett. 1980;12:365–367. [Google Scholar]
- 26.Peterson T C, Ward R C. Development of a bacterial transport model for coarse soils. Water Resour Bull. 1989;20:349–357. [Google Scholar]
- 27.Pringle J H, Fletcher M, Ellwood D C. Selection of attachment mutants during the continuous culture of Pseudomonas fluorescens and relationship between attachment ability and surface composition. J Gen Microbiol. 1983;129:2557–2569. [Google Scholar]
- 28.Reardon K F, Malusis M A, Adams D J, Shackelford C D, Mosteller D C, Bourquin A W. In situ and on-site bioremediation. 4. Fourth International In Situ and On-Site Bioremediation Symposium. Columbus, Ohio: Battelle Press; 1997. Microbial transport in a pilot-scale biological treatment zone; pp. 559–564. [Google Scholar]
- 29.Rosenberg M, Kjelleberg S. Hydrophobic interactions: role in bacterial adhesion. Adv Microb Ecol. 1986;9:353–393. [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Stenström T A. Bacterial hydrophobicity, an overall parameter for the measurement of adhesion potential to soil particles. Appl Environ Microbiol. 1989;55:142–147. doi: 10.1128/aem.55.1.142-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor S W, Jaffe P R. Substrate and biomass transport in a porous medium. Water Resour Res. 1990;26:2181–2194. [Google Scholar]
- 33.van Loosdrecht M C M, Lyklema J, Norde W, Zehnder A J B. Bacterial adhesion: a physicochemical approach. Microb Ecol. 1989;17:1–16. doi: 10.1007/BF02025589. [DOI] [PubMed] [Google Scholar]
- 34.Vesper S J. Production of pili (fimbriae) by Pseudomonas fluorescens and correlation with attachment to corn roots. Appl Environ Microbiol. 1987;53:1397–1405. doi: 10.1128/aem.53.7.1397-1405.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan J, Wilson J L, Kieft T L. Influence of the gas-water interface on transport of microorganisms through unsaturated porous media. Appl Environ Microbiol. 1994;60:509–516. doi: 10.1128/aem.60.2.509-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams V, Fletcher M. Pseudomonas fluorescens adhesion and transport through porous media are affected by lipopolysaccharide composition. Appl Environ Microbiol. 1996;62:100–104. doi: 10.1128/aem.62.1.100-104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]