Introduction
The generation of hematopoietic stem cells (HSCs) from human pluripotent stem cells (hPSCs) is a major goal for regenerative medicine. During embryonic development, HSCs derive from hemogenic endothelium (HE) in a NOTCH- and retinoic acid (RA)-dependent manner. While a WNT-dependent (WNTd) patterning of nascent hPSC mesoderm specifies clonally multipotent intra-embryonic-like HOXA+ definitive HE, this HE is functionally unresponsive to RA. Here we show that WNTd mesoderm, prior to HE specification, is actually comprised of two distinct KDR+CD34neg populations. CXCR4negCYP26A1+ mesoderm gives rise to HOXA+ multilineage definitive HE in an RA-independent manner, while CXCR4+ALDH1A2+ mesoderm gives rise to HOXA+ multilineage definitive HE in a stage-specific, RA-dependent manner. Further, both RA-independent (RAi) and -dependent (RAd) HE harbor transcriptional similarity to distinct populations found in the early human embryo, including HSC-competent HE. This revised model of human hematopoietic development provides essential resolution to the regulation and origins of the multiple waves of hematopoiesis. These insights provide the basis for the generation of specific hematopoietic populations, including the de novo specification of HSCs.
Main
Hematopoietic development during embryogenesis is comprised of multiple spatio-temporally orchestrated hematopoietic programs, each regulated by BMP, WNT, NOTCH, and RA signaling1. Of these, the specific role of RA in the specification of hPSC-derived HE has not been extensively characterized. Therefore, we sought to define the dependence of RA on known hPSC-derived HE populations2. Briefly, through a WNT-independent (WNTi) process, hPSCs differentiate rapidly towards either a CD43+ population of primitive hematopoietic progenitor cells (HPCs), as well as a CD34+ HE2–4, consistent with extra-embryonic hematopoiesis. Conversely, in a WNT-dependent (WNTd) process, hPSCs give rise to HOXA+ HE with definitive erythroid-myeloid-lymphoid potential, indicative of intra-embryonic-like definitive hematopoiesis (Extended Data Fig. 1A–C)2,3,5–7.
RA is essential for definitive hematopoietic development during embryogenesis1,8,9. Therefore, we sought to identify an RA-dependent hPSC-derived hematopoietic program. Both WNTi and WNTd hPSC-derived populations are obtained in an RA-independent manner, as these are chemically-defined conditions with no exogenous sources of RA2,3,5. Similarly, manipulation of RA signaling on hPSC differentiation cultures have failed to yield functional improvements10–12. As precise mesodermal patterning is critical for specifying ontogenically-distinct hematopoietic programs2, we performed single cell (sc)RNA-seq on day 3 of differentiation cultures to better understand the mesodermal population(s) obtained during early differentiation. Unsupervised clustering (Extended Data Fig. 2A–C) revealed 19 transcriptionally distinct clusters (Fig. 1A,B). CDX1/2/4, which correlates with the development of intra-embryonic-like HE6,13, was uniquely expressed across multiple clusters. Conversely, cluster 6 was highly enriched for WNTi cells that expressed GYPA/GYPB, which is a unique subset of mesoderm that harbors extra-embryonic-like hemogenic potential2 (Fig. 1B,C, Extended Data Table 1). This indicated that it is possible to identify functionally distinct hemogenic mesodermal populations. We next searched for cells expressing ALDH1A2, which governs the enzymatic conversion of retinol (ROH) to all-trans retinoic acid (ATRA) during embryogenesis14, and is essential for intra-embryonic HE development1. KDR+GYPA+ WNTi cells had virtually no ALDH1A2 expression, but instead robustly expressed CYP26A1, an enzyme that degrades RA (Extended Data Fig. 2D). In contrast, there was a significant population of ALDH1A2+ cells within WNTd KDR+ cells (Extended Data Fig. 2D).
Figure 1: scRNAseq reveals distinct mesoderm populations.
A-B, UMAP plots of (A) sample origin or (B) transcriptionally distinct clusters within WNTi or WNTd day 3 of differentiation cultures. C, Expression of KDR, GYPA, or CDX genes within each differentiation culture. Scale bar: relative expression scaled for KDR and GYPA, and calculated module score for expression of CDX1/2/4. D, Clusters of WNTd KDR+ cells. UMAP plots visualizing (i) clustering of KDR+ WNTd cells, (ii) ALDH1A2 and (iii) CXCR4 expression. Scale bar: scaled expression within KDR+ mesodermal cells. E, Mesodermal CXCR4 expression under WNTd or WNTi differentiation conditions. (i) Representative flow cytometric analysis of KDR and CXCR4 expression on day 3 of differentiation, following control, WNTi, or WNTd differentiation conditions. (ii) Quantification of CXCR4+ cells within each KDR+ fraction, on day 3 of differentiation, across various hPSC lines. Two-way ANOVA with Tukey’s test comparing all biological replicates: H1 (n=12; Control vs. WNTi, p=0.0112; Control vs. WNTd, WNTi vs. WNTd, p<0.0001), H9 (n=5; Control vs. WNTi, p=0.1388; Control vs. WNTd, p=0.0008; WNTi vs. WNTd, p<0.0001), iPSC-1 (Control n=5, WNTi/WNTd n=7; Control vs. WNTi, p=0.0003; Control vs. WNTd, p=0.1109; WNTi vs. WNTd, p<0.0001). F, Expression of CYP26A1, ALDH1A1, and ALDH1A2 in day 3 KDR+ cells, as in (E). SEM, Two-way ANOVA with Tukey’s test comparing all biological replicates (n=3), CD235a+ vs. CXCR4+ (CYP26A1 p=0.152225, ALDH1A1 p=0.068067, ALDH1A2 p=0.003529), CD235a+ vs. CXCR4neg (CYP26A1 p=0.140911, ALDH1A1 p=0.103219, ALDH1A2 p=0.010088), CXCR4+ vs CXCR4neg (CYP26A1 p=0.000833, ALDH1A1 p=0.429912, ALDH1A2 p=0.035653). G, WNTd KDR+ cells with ALDEFLUOR (AF) activity have enriched expression of ALDH1A2. (i) Representative ALDEFLUOR (AF) flow cytometric analysis. (ii) ALDH1A2 and CYP26A1 expression within CXCR4+/negALDF+/neg KDR+ cells. One-way ANOVA with Tukey’s test comparing all biological replicates (n=3). SEM, ns=not significant, ALDH1A2 (CXCR4–ALDF– vs. CXCR4–ALDF+, p=0.0904; CXCR4–ALDF– vs. CXCR4+ALDF–, p=0.0818; CXCR4–ALDF– vs. CXCR4+ALDF+, p<0.0001; CXCR4–ALDF+ vs. CXCR4+ALDF–, p=0.9999; CXCR4–ALDF+ vs. CXCR4+ALDF+, p=0.0006; CXCR4+ALDF– vs. CXCR4+ALDF+, p=0.0006), CYP26A1 (CXCR4–ALDF– vs. CXCR4–ALDF+, p=0.0128; CXCR4–ALDF– vs. CXCR4+ALDF–, p=0.0033; CXCR4–ALDF– vs. CXCR4+ALDF+, p=0.0025; CXCR4–ALDF+ vs. CXCR4+ALDF–, p=0.7111; CXCR4–ALDF+ vs. CXCR4+ALDF+, p=0.5751; CXCR4+ALDF– vs. CXCR4+ALDF+, p=0.9945).
As approximately 40% of the WNTd cells harbored some degree of KDR expression (Fig. 1C), we first sought to discriminate mesodermal cells from non-mesodermal cell types. Independent clustering of the WNTd population revealed KDR expression within most clusters (Extended Data Fig. 2E). However, there was separation of germ layer-like populations within many of these clusters (Extended Data Fig. 2Fi, ii), indicating that there is significant heterogeneity within these cultures. The population identified as mesoderm exclusively harbored KDR expressing cells, although only 58% of them had detectable KDR transcripts (Extended Data Fig. 2Fiii). As we have previously demonstrated that all WNTd hematopoiesis is derived from a KDR+ population2, we conservatively focused on the cells with detectable KDR expression, which consisted of 9 transcriptionally distinct clusters (Fig. 1Di). While multiple clusters harbored ALDH1A2+ cells, no single cluster was uniquely enriched in ALDH1A2 expression (Fig. 1Di–ii, Extended Data Table 2A,B). However, when we performed differential gene expression analysis between all of the KDR+ALDH1A2+ vs KDR+ALDH1A2neg populations, we identified CXCR4 as a candidate cell surface marker (Extended Data Table 2C).
Flow cytometry confirmed that, across hESC and iPSC lines, CXCR4 was expressed within day 3 of differentiation KDR+ cells, and that its expression is significantly upregulated under WNTd conditions in comparison to WNTi (Fig. 1E). We next performed whole-transcriptome analyses on FACS-isolated WNTd KDR+CXCR4+/neg populations, WNTi KDR+CD235a+ cells13, and WNTi and WNTd CD34+CD43negCD73negCD184neg HE cells4. In comparison to later-stage HE, each KDR+ population had higher expression of mesodermal genes, but substantially lower expression of canonical hemato-endothelial markers (Extended Data Fig. 3A, Extended Data Table 3A). This was further reflected in flow cytometric analyses, which confirmed that WNTd KDR+ cells did not express CD34, CD144/VE-Cadherin (CDH5), or TIE2 (TEK) on their surface (Extended Data Fig. 3B), establishing these cells as an early stage mesodermal cell-state that precedes hemato-endothelial specification.
In comparison to WNTi CD235a+ mesoderm, both WNTd KDR+ populations exhibited substantially higher expression of primitive streak markers such as T (TBXT), CDX1/2/4, and members of the HOXA cluster6,13,15 (Extended Data Fig. 3A,C, Extended Data Table 3A), indicating that these populations comprise gastrulation and/or early mesodermal cell-states. Hierarchical clustering of KDR+ and HE populations revealed that the WNTd KDR+ populations were more similar to each other than WNTi KDR+CD235a+ cells (Extended Data Fig. 3A). Despite these similarities, each WNTd mesodermal population was transcriptionally distinct from one another (Extended Data Fig. 3D,E, Extended Data Table 3B). Critically, these populations harbored differences in the gene expression of two key enzymes regulating RA signaling (Fig. 1F). CYP26A1 was highly expressed within both WNTi CD235a+ and WNTd CXCR4neg mesoderm, while ALDH1A2 was highly expressed in CXCR4+ mesoderm (Fig. 1F, Extended Data Table 3A,B). ALDEFLUOR (“AF”) analysis confirmed that overall ALDH activity, which includes that of ALDH1A2, is enriched within this CXCR4+ mesoderm (Fig. 1Gi). Consistent with this, ALDH1A2 expression was significantly enriched within KDR+CXCR4+AF+ cells, while CYP26A1 was enriched within KDR+CXCR4negAFneg cells (Fig. 1Gii). Collectively, these observations reveal that at least two mesodermal populations exist following WNTd differentiation conditions, with a subset of CXCR4+ cells being enriched in ALDH1A2, and are poised to respond to RA.
To assess which WNTd KDR+ subset(s) could give rise to HOXA+ definitive HE6, we isolated each KDR+CXCR4+/neg population by FACS, and then cultured them for an additional 5 days to allow for HE specification (Fig. 2Ai)2,5. Both populations gave rise to a CD34+CD43neg population (Fig. 2Aii). However, only the CD34+ cells derived from CXCR4neg mesoderm harbored HE, as they formed a monolayer of endothelial cells that gave rise to non-adherent hematopoietic progenitors (Extended Data Fig. 4Ai). This HE harbored multilineage definitive hematopoietic potential, evidenced by its ability to give rise to definitive erythro-myeloid and T-lymphoid cells (P1; Fig. 2Aiii,B). In contrast, CD34+ cells derived from the KDR+CXCR4+ population (“P2”) formed an endothelial monolayer (Extended Data Fig. 4Ai), but did not efficiently give rise to multilineage lympho-myeloid potential (Fig. 2Aiii,B). This strongly suggested that definitive hematopoietic specification from hPSCs2,5,6 originates from a KDR+CXCR4negCD34negCDX4+ mesodermal population. Further, consistent with its CYP26A1 expression (Fig. 1F,G), HE can be specified from CXCR4neg mesoderm in the presence of the pan-ALDH inhibitor DEAB (Extended Data Fig. 4B), indicating that that this is an RA-independent (RAi) hematopoietic program.
Figure 2: CXCR4neg and CXCR4+ mesoderm gives rise to hemogenic endothelium in a RA-independent and RA-dependent manner, respectively.
A, Separation of mesodermal progenitors of HE, based on CXCR4 cell surface expression. (i) Representative FACS gating scheme of KDR+ mesoderm for CXCR4 expression, within day 3 WNTd differentiation cultures. (ii) Representative FACS gating scheme of CD34 and CD43 expression, following 5 days of culture after mesoderm isolation. (iii) Representative flow cytometric analyses of CD4+CD8+T-lymphoid potential of CD34+CD43neg populations. B, Quantification of erythro-myeloid CFC potential from different HE populations, as in (Aii). Two-way ANOVA with Tukey’s test for all biological replicates (n=3), SEM, statistics shown for BFU-E (CXCR4– Control vs. CXCR4+ Control, p=0.0008; CXCR4+ Control vs. CXCR4+ ROH, p=0.009, ns=not significant), remaining statistics included in Source Data Fig. 2. Bar: mean count by colony type across all biological replicates, 〇: total CFCs for each biological replicate. C, Quantification of erythro-myeloid CFC potential of CD34+CD43neg populations, as in (Ai,ii), following ROH treatment initiating on either day 3, 4, or 5. Two-way ANOVA with Tukey’s test for all biological replicates (n=3), SEM, statistics shown for BFU-E (CXCR4+ Control v. D3 p=0.0005, D3 vs. D4 and D3 v. D5 p<0.0001, ns=not significant), remaining statistics included in Source Data Fig. 2. Bar: mean count by colony type across all biological replicates, 〇: total CFCs for each biological replicate. D, Quantification of erythro-myeloid CFC potential of CD34+CD43neg cells, following ATRA treatment on day 3, from isolated (i) WNTd KDR+CXCR4+, (ii) WNTd KDR+CXCR4neg, or (iii) WNTi KDR+CD235a+ cells, as in (A). Two-way ANOVA compared to DMSO with Dunnett’s test for all biological replicates, statistics shown for RAi/RAd BFU-E and WNTi Ery-P, remaining statistics included in Source Data Fig. 2. Bar: mean count by colony type across all biological replicates, 〇: total CFCs for each biological replicate. SEM, **p<0.01, ***p<0.001, ****p<0.0001. WNTi CD235a+: DMSO control, 0.1nM, 1nM (n=6); 0.01nM, 100nM, 1000nM (n=3); 10nM (n=5); WNTd CXCR4neg: DMSO control, 1nM (n=5); 0.01nM, 0.1nM, 10nM (n=2); 100nM (n=3). WNTd CXCR4+: DMSO control (n=5); 0.01nM, 0.1nM, 10nM (n=3); 1nM, 100nM (n=4).
Given that the non-hemogenic CXCR4+ population was enriched in ALDH1A2 expression, we hypothesized that this population may instead require RA signaling for HE specification. We therefore cultured the freshly isolated KDR+ populations with ROH (Fig. 2Ai). Critically, this treatment resulted in the specification of CD34+ HE that harbored definitive erythroid, myeloid, and lymphoid hematopoietic potential (P2’; Fig. 2Aii,iii,B, Extended Data Fig. 4Aii). Interestingly, this RA-mediated response was temporally-restricted, as only treatment of freshly isolated CXCR4+ mesoderm on day 3 of differentiation, but not thereafter, resulted in the robust specification of HE (Fig. 2C). Therefore, a KDR+CD34negCXCR4+ mesodermal population harbors stage-specific, RA-dependent (RAd) definitive hematopoietic potential.
ATRA has been identified as being required for definitive hematopoiesis, but repressive of extra-embryonic hematopoiesis8,15. So, we next asked whether ATRA would also specify functional HE from WNTd CXCR4+ mesoderm. Titration of ATRA on isolated KDR+CXCR4+ mesoderm revealed that 1 nM was capable of eliciting robust specification of definitive HE, while concentrations lower than 1 nM and higher than 10nM did not (Fig. 2Di). This indicated that a narrow range of RA signaling is required to establish an RAd hematopoietic program. Indeed, 1 nM ATRA elicited an RA-dependent response within CXCR4+ mesoderm across 3 hPSC lines (Extended Data Fig. 4C), indicating this is a conserved, reproducible population that can be obtained from hPSCs. In contrast, 1–10 nM ATRA exhibited a reduction of overall definitive hematopoietic development from WNTd KDR+CXCR4neg mesoderm, while ≥100 nM was repressive to HE specification (Fig. 2Dii). Similarly, ≥1 nM ATRA was repressive to extra-embryonic-like HE specification from WNTi CD235a+ mesoderm (Fig. 2Diii), consistent with a repressive role of RA signaling on extra-embryonic hematopoiesis8,15.
As these cells represent distinct hemogenic mesodermal populations, we next asked whether correlates exist for each within the early human embryo16 (Fig. 3A). We first sought to identify mesodermal populations that either simultaneously expressed both CXCR4 and ALDH1A2, or expressed CDX genes but lacks CXCR4 expression, similar to our RNA-seq analyses (Fig. 1F, Extended Data Fig. 3E). Following Seurat label transfer (Fig. 3B), we observed that most embryonic cell types directly correspond with similarly-labeled hPSCs, and cells with matched labels shared many marker genes (Extended Data Table 4A), but emergent mesoderm and the yolk sac-derived lineages (yolk sac mesoderm, HE progenitors) had very few corresponding hPSC populations, while axial mesoderm and erythrocytes had very few cells in either dataset (Fig. 3B).
Figure 3: In vivo correlates of hPSC-derived populations within the early human embryo.
A-B, Cells from hPSC WNTd differentiation cultures and early gastrulating human embryos cluster together following integration of the datasets. A, UMAP visualizing the contribution of embryonic and hPSCs to the integrated dataset (1 biological replicate each). B, UMAP visualizing (i) cell types in the human embryo, as defined by Tyser et al16, and (ii) the cell type labels transferred from embryonic cells to hPSCs. C, UMAPs visualizing the calculated module score for the expression of CDX1/2/4, CXCR4, or ALDH1A2 within each dataset. Scale bar: expression scaled to each dataset and gene. D, UMAP of primitive streak and nascent mesoderm CS7 cells visualizing the (i) clusters and (ii) cell labels. E, Violin plot for scaled expression of ALDH1A2 and CXCR4 across clusters, as in D. F, Simultaneous expression of CDX1/2/4 or ALDH1A2/CXCR4 across the subset dataset, as in D. Scale bar: module score calculated for each gene combination. G, GSEA for enrichment of WNTd mesodermal gene signatures and RA-related processes within each subset cluster. (i) Normalized enrichment scores (NES) for each cluster using genes upregulated in KDR+CXCR4+ and KDR+CXCR4neg cells, based on all cells within each cluster in Di. Gene signatures were defined as in Extended Data Table 4D. *FDR<0.25, ns=not significant. Clusters 1, 2 and 3 were enriched for the hPSC-derived KDR+CXCR4neg transcriptional signature (Cluster 1: NES=1.689, FDR=0; Cluster 2: NES=1.541, FDR=0.001; Cluster 3 CXCR4neg: NES=1.530, FDR=0), while only cluster 4 harbored a statistically significant KDR+CXCR4+ gene signature (NES=1.972, FDR = 0). Additionally, cluster 2 was negatively correlated with the gene signature from KDR+CXCR4+ cells (NES=−1.234, FDR=0.051), while cluster 4 negatively correlated with the KDR+CXCR4neg gene signature (NES=−1.350, FDR=0.017). Other clusters include: Cluster 1 CXCR4+ (NES=1.511, FDR=0); Cluster 3 CXCR4+ (NES=1.530, FDR=0); Cluster 5 CXCR4+ (NES=0.800, FDR=0.880) and CXCR4neg (NES=−1.675, FDR=0); Cluster 6 CXCR4+ (NES=−0.716, FDR=0.980) and CXCR4neg (NES=−1.241, FDR=0.123); Cluster 7 CXCR4+ (NES=0.676, FDR=0.983) and CXCR4neg (NES=1.081, FDR=0.603). (ii) Enrichment plots for RA-related metabolic processing GO terms with opposite trends in cluster 2 (NES=−1.60, p=0.007, FDR=0.2147) and cluster 4 (NES=1.45, p=0.047, FDR=0.2498).
We next focused on identifying mesodermal cell types present in both data sets that preceded hemato-endothelial specification (Extended Data Fig. 3A), but were not expressing markers characteristic of commitment to a non-hematopoietic fate (Extended Data Table 4B). This excluded epiblast, ectoderm, endoderm, erythrocytes and HE progenitors as candidate populations. Axial mesoderm was similarly excluded due to its enrichment of somitic genes, while both yolk sac mesoderm and advanced mesoderm were excluded for their enrichment of genes associated with cardiac development. Conversely, the poorly-correlated emergent mesoderm-like cells were enriched in genes associated with kidney development. Therefore, if correlates to WNTd hemogenic mesoderm exist, they would be found within the remaining cell types of primitive streak and/or nascent mesoderm. Indeed, both these populations within the embryo had enrichment for CDX1/2/4 expression, and nascent mesoderm was exclusively enriched in the expression of both ALDH1A2 and CXCR4 (Fig. 3C, Extended Data Table 4B).
Independent clustering of these two embryonic populations revealed 7 transcriptionally distinct clusters (Fig. 3D). Of those lacking significant CXCR4 expression (Fig. 3E, Extended Data Table 4C), only cluster 3 had enrichment of CDX1/2 gene expression (Fig. 3F, Extended Data Table 4C). Gene set enrichment analyses (GSEA) using gene signatures derived from our whole-transcriptome analyses on hPSC-derived KDR+CXCR4+/neg mesodermal populations (Extended Data Fig. 3A, Extended Data Table 4D) revealed that clusters 1, 2 and 3 were enriched for an hPSC-derived KDR+CXCR4neg transcriptional signature (Fig. 3Gi, Extended Data Table 4E). Importantly, cluster 2 was also negatively correlated with the gene signature from KDR+CXCR4+ cells, and had a negative association with an RA-related biological process GO term (Fig. 3Gi,ii). Collectively, these comparisons suggest that the primitive streak cells within cluster 2 may be enriched for functionally equivalent RAi hemogenic mesoderm. However, in the absence of a positive-identifying or cell-autonomous marker for RAi hemogenic mesoderm, the discrete identification of its in vivo correlate within clusters 1, 2, and/or 3 remains unclear.
In sharp contrast, ALDH1A2 and CXCR4 were exclusively enriched in the nascent mesodermal cluster 4, (Fig. 3E, Extended Data Table 4C). Further, GSEA revealed that only cluster 4 harbored a statistically significant KDR+CXCR4+ gene signature and simultaneously negatively correlated with the KDR+CXCR4neg gene signature (Fig. 3Gi, Extended Data Table 4E). Finally, GO term analysis found that only cluster 4 exhibited a positive enrichment for the biological process relating to retinol metabolism (Fig. 3Gii). Collectively, these analyses strongly suggest that the cells within cluster 4 are the in vivo correlates to hPSC-derived RAd mesoderm. Thus, an ALDH1A2+CXCR4+ RA-responsive hemogenic mesoderm found in hPSC differentiation cultures is recapitulating the emergence of a similar population found within CS7 nascent mesoderm in the human embryo.
To better understand each hPSC-derived HE, we treated unfractionated WNTd differentiation cultures with either DEAB or ROH on day 3 of differentiation, to induce RAi or RAd definitive hematopoiesis, respectively (Extended Data Fig. 5A). Each culture gave rise to a CD34+CD43neg population, which could be subset by CD73 and CXCR4 expression (Extended Data Fig. 5B). Critically, multilineage hematopoietic potential of both RAi and RAd HE was found within a NOTCH-dependent CD34+CD43negCD73negCXCR4neg population (Extended Data Fig. 5Ci,ii). Notably, compared to RAi HE, RAd HE gave rise to significantly more CD34+CD45+ progenitors after 9 days of EHT culture, had higher erythro-myeloid CFC potential, and their resultant BFU-E exhibited higher expression of both fetal (HBG) globin and BCL11A (Extended Data Fig. 5Cii,iii,D).
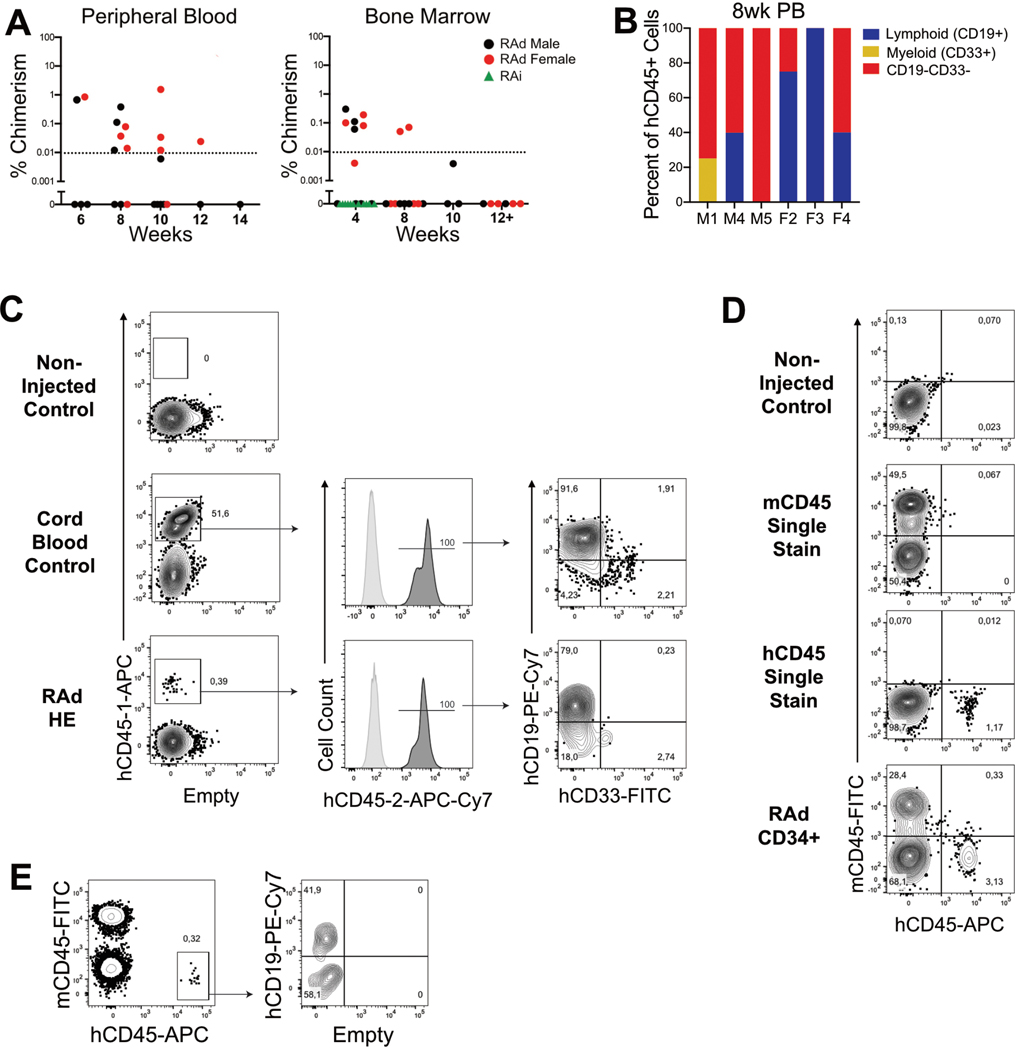
As a final functional assay, we isolated each CD34+ population and assessed their ability to engraft in a murine neonatal recipient. As expected17, RAi CD34+ cells completely failed to persist in murine hosts (Extended Data Fig. 6A). However, the RAd CD34+ cells gave rise to a transiently engrafting human CD45+ population, detectable within both the marrow of the femurs, as well as peripheral blood (Extended Data Fig. 6B–E). The frequency of the observed human chimerism remained low, becoming undetectable in the marrow after 8–10 weeks post-transplant while persisting in the blood until 10–12 weeks post-injection. While we observed CD45+CD19-CD33- hematopoietic cells of unspecified lineage, the presence of both myeloid CD33+ and B-lymphoid CD19+ lineages, the latter of which was not interrogated in vitro, confirmed the multi-lineage potential of RAd CD34+ cells. Altogether, this indicates that RA signaling results in the generation of a unique population of CD34+ cells that can transiently give rise to multilineage hematopoiesis in vivo.
We next performed whole-transcriptome analyses on each hPSC-derived population. Overall, while RAi and RAd HE were more similar to each other than to either WNTi HE or their derivative CD34+CD45+ HPCs, they did exhibit clear segregation from one another, with >300 differentially expressed genes (Extended Data Fig. 7Ai,ii, Extended Data Table 5A,B). Genes enriched in RAi HE were involved in a diverse array of transcriptional pathways, including endothelium development, cellular adhesion, the epithelial-to-mesenchymal transition, and response to mechanical stimuli (Extended Data Table 5C). In contrast, RAd HE was uniquely enriched in RA signaling, as expected, but also several histone modification pathways, suggesting there may be unexplored regulation at the chromatin level within these cells. Collectively, the transcriptional differences between each population are subtle, highlighting the need for functional assays when contrasting these definitive hematopoietic populations.
We next asked if either HE population had transcriptional similarity to that found in the developing human embryo. We first compared hPSC-derived RAi and RAd HE to 5th week of gestation CD34+CD90+CD43neg aorta-gonad-mesonephros (AGM) cells6, where the first HSC-competent HE is detectable18. RAi and RAd HE both expressed hemato-endothelial genes similar to that of primary embryonic tissue (Extended Data Fig. 7Bi), but had vastly different enrichment of broad metabolic processes (Extended Data Table 5D-F), which could be reflective of differences between primary in vivo cells and their in vitro correlates. Both RAi and RAd HE exhibited enriched transcriptional signatures for hematopoietic stem and progenitor differentiation (Extended Data Fig. 7Bii), consistent with their multilineage in vitro potential. AGM cells, in contrast, harbored pathway enrichment for aorta and vascular development, and BMP and VEGF signaling pathways (Extended Data Fig.7Bii, Extended Data Table 5E,F), consistent with ongoing vasculogenesis at this developmental stage. Curiously, gene expression of the HOXA cluster varied between each population. RAi HE had higher expression of posterior HOXA genes, while RAd HE exhibited generally higher expression of anterior and medial HOXA genes, with overall higher similarity to that observed in AGM CD34+ cells (Fig. 4A, Extended Data Table 5A). However, this HOXA expression was not retained in either population, as hPSC-derived HPCs had lower HOXA expression in comparison to the embryonic stem and progenitor population (“HSPC”, CD34+CD90+CD43+; Fig. 4A), consistent with their limited engraftment potential.
Figure 4: Distinct HOXA+ intra-embryonic-like HE from different ontogenic origins can be specified from hPSCs.
A, Heatmap visualizing the relative mean expression of HOXA genes within indicated hPSC and embryonic populations, averaged across all biological replicates (WNTi HE, RAi HE, RAd HE, RAi HPC, and RAd HPC: n=4; AGM CD34+, AGM HSPC, and AGM PR: n=1). Left: hPSC-derived CD34+CD43negCD73negCXCR4neg HE cells and primary embryonic CD34+CD90+CD43- cells from 5th week of gestation AGM (“AGM CD34+”). Right: hPSC-derived CD34+CD45+ cells (“HPC”) and primary embryonic CD34+CD90+CD43+ hematopoietic stem/progenitor (“HSPC”) or CD34+CD90negCD43+ committed progenitor (“AGM PR”) from 5th week of gestation AGM. Scale bar: robust z-score B, RAi and RAd HE exhibit transcriptional similarity to distinct subsets of intra-embryonic HECs in the human embryo. (i) Heatmap visualizing the similarity of individual early (CS10–11) and late (CS12–14) human embryonic arterial endothelial cells (“AEC”, CDH5+CXCR4+GJA5+DLL4+HEY2+SPNneg PTPRCneg) or HE cells (“HEC”, CDH5+RUNX1+HOXA+ITGA2BnegSPNnegPTPRCneg) compared to hPSC-derived CD34+CXCR4+ arterial endothelium (“CD34+ EC”), WNTi, WNTd RAi or RAd HE. Scale bar: relative Spearman coefficients. Each column is representative of a single embryonic cell scored across each hPSC-derived population indicated by the row name. (ii) Average similarity scores for each group of human embryonic cells. Two-way ANOVA with Tukey’s multiple comparison test comparing all single cells within each group illustrated in Bi (group 1: n=117, group 2: n=44, group 3: n=12, group 4: n=93, group 5: n=45, group 6: n=54), SEM, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant. C, Heatmap of relative mean expression for arterial genes within hPSC-derived RAi HE, RAd HE, and CD34+CXCR4+ arterial endothelial cells, averaged across all biological replicates (RAi HE, RAd HE, CD34+ EC: n=3). Scale bar: robust z-score D, Schematic of hPSC-derived HE from different ontogenic origins. KDR+CD235a+CYP26A1+ mesoderm (red) is obtained in a WNT-independent (WNTi) manner, and this population subsequently gives rise to HOXAlow/neg extra-embryonic-like HE and HPCs. Conversely, two distinct KDR+ populations are obtained under WNTd differentiation conditions, each of which gives rise to HOXA+ intra-embryonic-like HE. KDR+CXCR4negCYP26A1+ mesoderm (green) gives rise to multilineage, definitive HE in an RA-independent manner, while KDR+CXCR4+ALDH1A2+ mesoderm (blue) gives rise to multilineage, definitive HE, in a stage-specific RA-dependent manner.
As a final comparison, we next contrasted these hPSC-derived CD34+ populations with a human embryonic HE scRNA-seq dataset19. This comprised intra-embryonic populations of arterial endothelial cells (AEC) and HE cells (HEC)6,19 (Extended Data Fig. 8A–C), segregated as CS10–11 and CS12–14 (Fig. 4Bi,ii), as these populations possess temporally distinct HE subsets, of which the latter is presumed to be HSC-competent19. As expected, there was relatively poor similarity between the intra-embryonic cell types with hPSC-derived WNTi HE (Fig. 4B), as the latter is extra-embryonic-like4. The embryonic AEC (groups 1 and 4) exhibited strong similarity to hPSC-derived CD34+CXCR4+ arterial-like endothelial cells, regardless of developmental stage (Fig. 4B). Curiously though, many transcriptionally-defined embryonic HECs also harbored high similarity to hPSC-derived endothelial cells (groups 2 and 5; Fig. 4B). However, as RUNX1 can also be expressed within non-hemogenic arterial endothelium20,21, the strong similarity to hPSC-derived ECs suggests groups 2 and 5 likely comprise non-hemogenic endothelial cells. Critically, embryonic HECs in groups 3/6 exhibited stage-restricted correlation to either WNTd HE, with CS10–11 (“early”) HECs harboring high transcriptional similarity to RAi HE (group 3) and CS12–14 (“late”) HECs exclusively having strong similarity to RAd HE (group 6; Fig. 4B). Interestingly, HECs in group 3 were characterized by only 8 marker genes, while group 6 HECs were characterized by 194 marker genes, including several genes relating to HSC specification such as MYB and ANGPT1 (Extended Data Table 6A)19.
The high similarity of group 6 HSC-competent HECs with RAd HE suggested that these cells may be a precursor to HSC-competent HE. HSC-competent HE harbors an arterial gene signature comprising CXCR4, GJA4, GJA5, HEY2 and DLL419,22. However, CXCR4 expression is higher in AECs than in HECs19. Indeed, groups 3 and 6 exhibited the lowest CXCR4 expression (Extended Data Fig. 8D, Extended Data Table 6B). This expression pattern was also recapitulated when comparing hPSC-derived CD34+ populations, as RAd HE exhibited higher expression of GJA4, GJA5, HEY2 and DLL4 in comparison to RAi HE, but lower CXCR4 expression in comparison to AECs (Fig. 4C). Collectively, these analyses have identified that both HOXA+ HE populations derived from hPSCs have transcriptionally similar populations within the early human embryo. RAi HE is more similar to a non-arterial “early” HE, while RAd HE exhibits high transcriptional similarity to embryonic HSC-competent HECs.
We sought to identify a unique signal pathway-dependence that defines definitive HE development from hPSCs. Both NOTCH-dependency and HOXA expression have been reported as distinguishing characteristics of WNTd CD34+ HE derived from hPSCs5. However, here we demonstrate the existence of 2 ontogenically distinct HOXA+ NOTCH-dependent HE populations. Therefore, neither criteria can be solely used to distinguish between various hPSC-derived HE populations. In contrast, here we present evidence of a population that positively responds to RA in a stage-specific manner, giving rise to embryonic-like HE. As this population responds to a narrow concentration range of ATRA, but is inhibited at higher concentrations, our observations indicate that physiologically-relevant concentrations are required for this process23. Thus, ATRA may not be inhibitory to extra-embryonic-like hPSC hematopoiesis, but is a positive regulator of intra-embryonic like hematopoiesis.
Notably, these studies demonstrate that hPSC-derived hematopoietic potential is restricted to distinct KDR+CD34neg mesodermal subpopulations, which are specified very rapidly within differentiation cultures and recapitulate populations found in the early embryo. This is reminiscent of a similar developmental trajectory for cardiomyocyte specification from hPSCs24, suggesting that major cell fates are specified immediately following gastrulation. There are several lines of evidence that similarly suggest that hemogenic specification is a very early event in the murine conceptus25,26. Each hPSC-derived mesodermal population gives rise to an immunophenotypically similar HE population, but each respective population harbors subtle but significant transcriptional and functional differences.
Finally, these studies provide additional insight into the multiple, distinct hematopoietic progenitors that can be obtained from hPSCs (Fig. 4D). hPSC-derived WNTd HE expresses medial HOXA genes6, which we also observe in RAi and RAd HE. However, RAi HE has posterior enrichment of HOXA expression, whereas RAd HE has more anterior and medial HOXA expression, giving it a higher similarity to primary HSC-competent HE. Our observations that hPSCs can give rise to functionally distinct HOXA+ HE populations is inconsistent with the canonical 3-wave model of embryonic hematopoietic development27, wherein all intra-embryonic HE is assumed to be HSC-competent. Instead, these observations are consistent with the identification of heterogeneity in HE within the AGM 19,20,28,29.
The RAd HE derived under these conditions did not give rise to an HSC-like population. Determining whether RAd HE represent bona fide HSC-competent HE precursors, or represent an additional HSC-independent wave of hematopoietic development, is a remaining challenge. Nevertheless, given its functional and transcriptional similarity to a primary intra-embryonic population that harbors HSC-competent HE, we anticipate that this methodology will provide a useful platform for developmental hematopoiesis and disease modeling studies, and in the pursuit of the in vitro generation of therapeutically relevant hematopoietic cells.
Methods
Maintenance and differentiation of human ES and iPS cells
The previously established hESC lines H130 and H96, and iPSC131 were maintained on irradiated mouse embryonic fibroblasts in hESC media as described previously2,32. For differentiation, hPSC were cultured on Matrigel-coated plasticware (Corning Life Sciences) for 24 hours, followed by embryoid body (EB) generation, as described previously3,33,34. Briefly, hPSCs were dissociated with brief trypsin-EDTA (0.05%) treatment, followed by scraping. Embryoid body (EB) aggregates were resuspended in SFD media35 supplemented with L-glutamine (2 mM), ascorbic acid (1 mM), monothioglycerol (MTG, 4×10−4 M; Sigma), transferrin (150 μg/mL), and BMP4 (10 ng/mL). 24 hours later, bFGF (5 ng/mL) was added. On the second day of differentiation, ACTIVIN A (1 ng/mL), SB-431542 (6 μM), CHIR99021 (3 μM), and/or IWP2 (3 μM) were added, as indicated. On the third day, EBs were changed to StemPro-34 media supplemented with L-glutamine, ascorbic acid, MTG and transferrin, as above, with additional bFGF (5 ng/mL) and VEGF (15 ng/mL). On day 6, IL-6 (10 ng/mL), IGF-1 (25 ng/mL), IL-11 (5 ng/mL), SCF (50 ng/mL), EPO (2 U/mL final). All differentiation cultures were maintained at 37°C. All embryoid bodies and mesodermal aggregates were cultured in a 5% CO2/5% O2/90% N2 environment. All recombinant factors are human and were obtained from Biotechne. Analysis of hematopoietic colony potential via Methocult (H4034; Stem Cell Technologies) was performed as described previously3,5.
For RA manipulation within bulk EB differentiation cultures: Cells were treated exactly as above, except that on day 3 of differentiation, cells were treated with either 10 μM of the pan-ALDH inhibitor DEAB (4-Diethylaminobenzaldehyde, Sigma #D86256; “RA-independent”) or 5 μM retinol (ROH, Sigma #R7632; “RA-dependent”). DEAB or ROH were added again on day 6 of differentiation. HE was FACS-isolated for terminal assays on day 8 (DEAB) or day 10 (ROH).
Stem cell studies have been approved by the Washington University Embryonic Stem Cell Research Oversight Committee, ESCRO #14–001, the Ospedale San Raffaele Ethical Committee (TIGET-HPCT), and the Icahn School of Medicine Embryonic Stem Cell Research Oversight Committee, ESCRO #20–06.
Flow Cytometry and Cell Sorting.
Cultures were dissociated to single cells, as previously described2. All cell sorting was performed in the absence of fetal bovine serum. Cells were washed, labeled, sorted and collected in StemPro-34 media. The antibodies used include those previously described2,3,5. For hPSC studies, antibodies include KDR-Biotin (clone 89106, Biotechne #MAB3572, 15:100), Streptavidin-PE (BD #554061, 1:200), CD4-PerCP-Cy5.5 (clone RPA-T4, BD #560650, 1:100), CD8-PE (clone RPA-T8, BD #561950, 5:100), CD34-APC (clone 8G12, BD #340441, 1:100), CD34-PE-Cy7 (clone 8G12, BD #348791, 1:100), CD43-FITC (clone 1G10, BD #555475, 10:100), CD45-APC-Cy7 (clone 2D1, BD #557833, 3:100), CD45-BV421 (clone 2D1, BD #642275, 3:100), CD56-APC (clone B159, BD #555518, 4:100), CD73-PE (clone AD2, BD #550257, 2:100), CXCR4-APC (clone 12G5, BD #555976, 2:100), CXCR4-BV421 (clone 12G5, BD #562448, 2:100), and CD235a-APC (clone HIR-2, BD #551336, 1:100). For murine xenograft analyses, antibodies used were CD19-PerCP-Cy5.5 (clone HIB19, BD #561295, 1:100), CD33-BV605 (clone HIM3–4, BD #744352, 1:100), mCD45-Alexa Fluor 488 (clone 30-F11, BioLegend #103122, 1:100), and hCD45-APC (clone 2D1, BD #557833, 1:100). Cells were sorted with a FACSAria™II (BD) cell sorter and analyzed on a LSRFortessa (BD) cytometer. For murine xenograft analyses, antibodies used were CD19-PerCP-Cy5.5 (clone HIB19) or -PE-Cy7 (clone SJ25C1), CD33-BV605 or -FITC (clone HIM3–4), mCD45-FITC (clone 30-F11, BioLegend), and hCD45-APC or -APC-Cy7 (clone 2D1). Representative gating strategies are demonstrated in Extended Data Fig. 9.
Mesoderm isolation.
For isolation of mesodermal populations, day 3 of differentiation WNTd KDR+CD235anegCXCR4+/neg were FACS-isolated and reaggregated at 250,000 cells/mL in day 3 media, as above. Cultures were plated in 250 μL volumes in a 24 well low-adherence culture plate, and grown overnight in a 37°C incubator, with a 5% CO2/5% O2/90% N2 environment. As specified, RA was manipulated with either 5 μM ROH or ATRA (Sigma #R2625), or 10 μM DEAB. On day 4, an additional 1 mL of RA-supplemented day 3 media was added to reaggregates. On day 6 of differentiation, CD34+ and CD43+ cells from WNTi cultures were FACS-isolated for terminal assays. WNTd cultures were fed as normally, but without additional RA manipulation. CD34+ cells were sorted from all WNTd populations on day 8 of differentiation.
Hemato-endothelial growth conditions of hPSC-derived hemogenic endothelium.
CD34+CD43- hemogenic endothelium was isolated by FACS and allowed to undergo the endothelial-to-hematopoietic transition (EHT) as described previously5,34. Briefly, cells (CD34+CD43neg or CD34+CD43negCD73negCXCR4neg cells) were aggregated overnight at a density of 2×105 cells/mL in StemPro-34 media supplemented with L-glutamine (2 mM), ascorbic acid (1 mM), monothioglycerol (MTG, 4 × 10−4 M; Sigma-Aldrich), holo-transferrin (150 μg/mL), TPO (30 ng/mL), IL- 3 (30 ng/mL), SCF (100 ng/mL), IL-6 (10 ng/mL), IL-11 (5 ng/mL), IGF-1 (25 ng/mL), EPO (2 U/mL), VEGF (5 ng/mL), bFGF (5 ng/mL), BMP4 (10 ng/mL), FLT3L (10 ng/mL), and SHH (20 ng/mL). Aggregates were spotted onto Matrigel-coated plasticware and were cultured for additional 3 or 9 days for WNTi and WNTd cultures, respectively. Cultures were maintained in a 37°C incubator, in a 5% CO2/5% O2/90% N2 environment. All resultant cells within the hemato-endothelial cultures were subsequently harvested by trypsinization, and assessed for hematopoietic potential by Methocult in a 37°C incubator, in a 5% CO2/air environment. In some cases, NOTCH-dependency was assessed by culturing cells, for the entire duration, in the presence or absence of the NOTCH inhibitor, gamma secretase inhibitor (“gSI”, L-685,458 Sigma #L1790).
OP9-DL4 co-culture for T-lineage differentiation.
OP9 cells expressing Delta-like 4 (OP9-DL4) were a gift from Juan-Carlos Zúñiga-Pflücker, and were generated and described previously36,37. 1–10 ×104 isolated CD34+CD43neg cells were added to individual wells of a 6-well plate containing OP9-DL4 cells, and cultured with rhFlt-3L (5 ng/mL) and rhIL-7 (5 ng/mL). rhSCF (30 ng/mL) was added for the first 5 days. Cultures were maintained at 37°C, in a 5% CO2/air environment. Every five days co-cultures were transferred onto fresh OP9-DL4 cells by vigorous pipetting and passaging through a 40μm cell strainer. Following 21–28 days of co-culture, cells were analyzed using a LSRFortessa flow cytometer (BD).
Gene expression analyses.
Total RNA was prepared for whole-transcriptome sequencing using the Clontech SMARTer kit and was sequenced using an Illumina HiSeq 2500 with 1×50 single reads. Reads were aligned to GENCODE GRCh38 version 23 using STAR (version 2.7.1a), gene counts were obtained using Subread (version 1.6.5), and batch correction was achieved using Limma. Differential gene expression analysis, FPKM calculation, and principal component analysis was performed using DESeq2 (version 1.24.0), as described on https://github.com/sturgeonlab/Luff-etal-2022/tree/master/Bulk-analyses. Expression values were filtered to only include protein coding genes (GRCh38) using Ensembl BioMart38. Genes were considered significantly enriched with an adjusted p value < 0.05. Euclidean distance was calculated using the generic R function for generating distance matrices, and the accompanying heatmap with hierarchical clustering (Extended Data Figure 8Aii) was plotted using the R package pheatmap. Preranked Gene Set Enrichment Analysis (Broad Institute GSEA, version 4.1.0) using C5 biological processes as the gene set database was performed including gene sets containing between 5 and 500 genes, with default parameters. All genes were ranked using the product of the signed log2 fold change and inverse log of the p-value39. GO terms were considered significantly enriched with a p value < 0.05 and FDR < 0.25. Morpheus (https://software.broadinstitute.org/morpheus) was used to create all gene heatmaps of FPKM values, perform the accompanying hierarchical clustering with gene heatmaps (one minus the Pearson correlation with average linkage), and transform data to z-scores. Bulk RNA-seq comparison to scRNA-seq was performed using the SingleR package (version 1.0.1)40 implemented in R (version 3.6.2), as described at https://github.com/sturgeonlab/Luff-etal-2022/tree/master/SingleR. Embryonic populations from Zeng, et al.19 were selected based on the expression of known markers for arterial endothelial cells (“AEC”, CDH5+CXCR4+GJA5+DLL4+HEY2+SPNnegPTPRCneg) and HE cells (“HEC”, CDH5+RUNX1+HOXA+ITGA2BnegSPNnegPTPRCneg)6. Genes driving the similarities scores are included in Extended Data Table 6C.
qRT-PCR was performed as previously described2. Briefly, total RNA was isolated with the RNAqueous RNA Isolation Kit (Ambion), followed by reverse transcription using random hexamers and Oligo (dT) with Superscript III Reverse Transcriptase (Invitrogen). Real-time quantitative PCR was performed on a StepOnePlus thermocycle (Applied Biosystems), using Power Green SYBR mix (Invitrogen). Primers used include: ALDH1A2 (5’-TTGCATTCACAGGGTCTACTG-3’ and 5’-GCCTCCAAGTTCCAGAGTTAC-3’), CYP26A1 (5’-CTGGACATGCAGGCACTAAA-3’ and 5’-TCTGGAGAACATGTGGGTAGA-3’), and BCL11A (5’-GCCAGAGGATGACGATTGTTTA-3’ and 5’-CCCTCCAGTGCAGAAGTTTATC-3’). Gene expression was evaluated as DeltaCt relative to control (ACTB). For globin analysis, the following TaqMan assays (Applied Biosystems) were used: HBB (Hs00747223_g1), HBE1 (Hs00362215_g1), HBG1/2 (Hs00361131_g1), and GAPDH (Hs02786624_g1).
scRNA-seq analyses.
Cells from each day 3 differentiation culture condition were methanol-fixed as previously described41. Libraries were prepared following the manufacturer’s instruction using the 10X Genomics Chromium Single Cell 3′ Library and Gel Bead Kit v2 (PN-120237), Chromium Single Cell 3′ Chip kit v2 (PN-120236), and Chromium i7 Multiplex Kit (PN-120262). 17,000 cells were loaded per lane of the chip, capturing >6000 cells per transcriptome. cDNA libraries were sequenced on an Illumina HiSeq 3000. Sequencing reads were processed and aligned to GRCh38 using the Cell Ranger software pipeline (version 3.1.0). A detailed workflow for these scRNA-seq analyses is available at https://github.com/sturgeonlab/Luff-etal-2022/tree/master/Seurat. Briefly, using Seurat (version 3.2.2) implemented in R (version 3.6.2), the dataset was filtered by removing genes expressed in fewer than 3 cells and cells where greater than 10% of the UMIs are mitochondrial genes, and to retain cells with unique gene counts between 200 and 6000. The remaining UMI counts were log-normalized with a scale factor of 10,000 and variable features were calculated using the vst selection method. The data were then scaled by regressing out mitochondrial gene percentage and UMI count. Principal component analysis utilizing 50 dimensions was performed and a JackStraw plot was used to determine the dimensionality of the dataset. To finalize the initial processing, uniform manifold approximation and project (UMAP) and shared nearest neighbors were calculated utilizing 50 dimensions, with optimal Louvain clustering resolution determined by maximizing cluster (SC3) stability, as calculated using the clustree package (version 0.4.3). All gene thresholds were determined using density plots. WNTi and WNTd cells were integrated to account for different sequencing runs using the maximum number of dimensions (50) at all applicable steps. For integration of WNTd mesodermal cultures with the human embryo gastrulation dataset16, the standard workflow for integration between two datasets was performed in a similar manner to the previous dataset, described in detail in the online repository (https://github.com/sturgeonlab/Luff-etal-2022/tree/master/Seurat).
Murine xenografts.
0.5–4 × 105 CD34+ cells from either anonymous cord blood, or DEAB- or ROH-treated hPSC bulk differentiation cultures, was isolated by CD34+ MACS (Milenyi) and immediately transplanted into neonatal NBSGW or NSG mice of both sexes via intrahepatic injection. To assess xenograft persistence, peripheral blood and bone marrow aspirates were obtained at 2-to-4 week intervals. Peripheral blood was obtained from tail veins, followed by successive washes with red blood cell lysis buffer. Flow cytometry was performed using a BD Fortessa or BD Canto cytometer. Bone marrow aspirates were obtained from right femurs under deep anesthesia, by inserting a 29G needle into the distal femur shaft and aspirating the content into a syringe containing 100μl PBS, followed by red blood cell lysis. Or, following sacrifice, bone marrow were harvested from femurs by aspirating each bone with IMDM supplemented with 5% FBS, followed by red blood cell lysis. The animals were housed in standard BSL2 level animal facility, in dedicated room for immunocompromised animals, according to institutional guidelines.
This study is compliant with all relevant ethical regulations regarding animal research and has been approved by the Washington University Institutional Animal Care and Use Committee, IACUC #19–0959, the Animal Care and Use Committee of Ospedale San Raffaele (IACUC no. 841) and authorized by the Italian Ministry of Health, and the Icahn School of Medicine Institutional Animal Care and Use Committee, IACUC #PROTO202000171.
Data Availability.
All gene expression analysis datasets are available in the Gene Expression Omnibus (GEO) under the accession numbers GSE139853 (scRNA-seq in Fig. 1, Fig. 3, and Extended Data Fig. 2; CXCR4+/− mesoderm RNA-seq in Fig. 1 and Extended Data Fig. 3; RAi/RAd HE RNA-seq in Fig. 4, Extended Data Fig. 1, Extended Data Fig. 3, and Extended Data Fig. 7; RAi/RAd HPC RNA-seq in Fig. 4, Extended Data Fig. 7), or BioProjects #PRJNA352442 (WNTi mesoderm RNA-seq in Fig. 1, Extended Data Fig. 1, and Extended Data Fig. 3; WNTd mesoderm RNA-seq in Extended Data Fig. 1) and #PRJNA525404 (WNTi HE RNA-seq in Fig. 4, Extended Data Fig. 1, Extended Data Fig. 3, and Extended Data Fig. 8). Publicly-available datasets used include: GEO SuperSeries GSE81102, “AGM CD34+” (GSM2142333), “AGM PR” (GSM2142334), “AGM HSPC” (GSM2142332) for Human 5 week AGM (Fig. 4 and Extended Data Fig. 7); GEO GSE135202 for Human CS10-CS15 embryo (Fig. 4 and Extended Data Fig. 8); ArrayExpress E-MTAB-9388 for Human CS7 embryo (Fig. 3). Genome alignments were performed with GENCODE GRCh38.p3 version 23. All source data has been deposited within the San Raffaele Open Research Data Repository and are available at doi: 10.17632/pbbn55mhy9.1
Code Availability.
All code used to analyze and visualize datasets has been shared on a repository and can be found at: https://github.com/sturgeonlab/Luff-etal-2022.
Statistics & Reproducibility.
For all multivariate statistical analyses, ANOVA were performed with the appropriate corrections for multiple comparisons. Two-way ANOVA with Tukey’s multiple comparison test were used when comparing ≥2 metrics with >2 sorted populations, while one-way ANOVA with Tukey’s multiple comparison test were chosen for single metrics with >2 populations. For comparing ≥2 metrics with exactly 2 populations, two-way ANOVA with Bonferroni’s multiple comparison test was used. For Figure 2D, two-way ANOVA with Dunnett’s multiple comparison test was used to analyze the difference in BFU-E between each concentration as compared to the vehicle control. Data distribution was not formally tested but assumed to be normally distributed with equal variance. Sample size and replication was determined by historical controls2,5. In general, biological replicates were only excluded if internal controls failed and technical replicates (e.g. methylcellulose colony forming assays) were not excluded. Specific to the CXCR4 quantification in the H9 cell line, experiments with poor mesodermal specification (<50% KDR+) were excluded. Experimental conditions were not randomized but covariates were controlled by equal distribution of sorted cells across controls and experimental conditions. Blinding of experimental conditions was not relevant as our studies do not require grading of the results.
Extended Data
Extended Data Fig. 1. Specification of HOXA+ HE from hPSCs in a WNT-dependent manner.
A, Schematic of hPSC directed differentiation towards hemogenic endothelium, as described in Sturgeon et. al2. Definitive intra-embryonic-like hematopoietic potential is specified in a WNT-dependent (“WNTd”) manner, while extra-embryonic-like hematopoietic potential is WNT-independent (“WNTi”). B, Representative flow cytometric analyses of the T-lymphoid potential of WNTd CD34+ cells and WNTi CD34+CD43neg and CD43+ populations. T cell potential is positively identified by the presence of a CD4+CD8+ population following 21+ days of OP9-DL4 co-culture, while an absence of potential is identified by an absence of CD45+ lymphocytes3,30. C, Heatmaps visualizing the mean expression of HOXA genes across all biological replicates in mesoderm and hemogenic endothelium populations, as in (A). Grey indicates undetected gene. Scale bar: log10 FPKM. Biological replicates: WNTi/WNTd KDR+ (n=4); WNTi/WNTd HE (n=3). The expression of HOXA genes within WNTd-derived populations is suggestive of an intra-embryonic-like population6, while a lack of HOXA expression in WNTi-derived populations is suggestive of an extra-embryonic-like population7.
Extended Data Fig. 2. scRNA-seq analyses of day 3 WNTi and WNTd differentiation cultures.
A, Violin plots visualizing the number of genes per cell (left), the number of unique molecular identifiers (“UMIs”, middle), and the percent of expressed genes that are mitochondrial (right) following filtering of low quality cells from both WNTi and WNTd datasets combined (1 biological replicate each). B, UMAP visualizing before and after integration of WNTi (red) and WNTd (blue) datasets to account for batch effects between sequencing runs. C, UMAP visualizing quality control metrics, as in A, for the dataset following integration. Scale bar: values range as indicated in (A). D, Violin plots for CYP26A1 and ALDH1A2 expression within WNTi KDR+GYPA+ and WNTd KDR+GYPAneg cells, as indicated. E,F, Day 3 WNTd cultures are comprised of all germ layers. E, UMAP visualizing (i) clustering and (ii) KDR expression within WNTd differentiation cultures. Scale bar: gene expression scaled to WNTd subset. F, (i) UMAP plot with the projection of predicted germ layer type, where each label includes cells expressing the following genes: Pluripotent (SOX2, NANOS3, DND1, POU5F1, or TBXT), Ectoderm (TFAP2A, DLX5, or GATA3), Endoderm (FOXA2, APOA1, or APOA2), and Mesoderm (KDR, MEST, MESP1, TEK, or FLT1). 44 (0.64%) remaining cells were labeled based on clustering. (ii) Dot plot visualizing expression of germ layer-specific genes within each identified cell type, as in (i). Scale
Extended Data Fig. 3. Day 3 of differentiation WNTd KDR+CXCR4neg and KDR+CXCR4+ cells are transcriptionally distinct mesodermal subsets.
A, Heatmaps visualizing the mean expression across all biological replicates (log10 FPKM) of mesodermal, endothelial, hematopoietic, and RA-related genes within hPSC-derived day 3 WNTi KDR+CD235a+ cells and WNTd KDR+CXCR4+/neg cells, day 6 WNTi HE, and day 8 WNTd HE. Grey indicates undetected gene. Hierarchical clustering based on the expression of genes shown. Scale bar: log10 FPKM. Biological replicates: WNTi KDR+ (n=4); CXCR4+/neg KDR+, WNTi HE, WNTd HE (n=3). B, Representative flow cytometric analysis for endothelial markers CD34, CD144 (VE-Cadherin/CDH5), and TIE2 (TEK) within WNTd KDR+ cells. C, Heatmaps visualizing the mean expression of HOXA genes across all biological replicates in day 3 WNTi KDR+CD235a+ cells, or WNTd KDR+CXCR4+ or KDR+CXCR4neg cells, as in A. Scale bar: log10 FPKM. Biological replicates: WNTi KDR+ (n=4); CXCR4+/neg KDR+ (n=3). D, PCA plot for batch-corrected KDR+CXCR4+ and KDR+CXCR4neg replicates, as in A. E, Expression of CDX genes within WNTi and WNTd mesodermal populations, as in A, Two-way ANOVA with Tukey’s multiple comparison test comparing all biological replicates: WNTd CXCR4+/neg (n=3), WNTi CD235a+ (n=4). SEM, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant.
Extended Data Fig. 4. CXCR4neg mesoderm gives rise to HE in an RA-independent manner, while specification of HE from CXCR4+ mesoderm is RA-dependent.
A, Representative phase contrast microscopy of CD34+ cells from DMSO-treated CXCR4+/neg mesoderm (i) and retinol-treated CXCR4+ mesoderm (ii) following 1, 3, and 5 days after FACS isolation. 100X magnification, scale bar: 50um. B, Erythro-myeloid colony forming potential of HE specified
Extended Data Fig. 5. RA-dependent HE, similar to RAi HE, is a CD34+CD43negCD73negCXCR4neg population and undergoes the endothelial-to-hematopoietic transition (EHT) in a NOTCH-dependent manner.
A, Schematic for the differentiation of WNTd cultures towards either RAi and RAd CD34+ cells and their respective hematopoietic progenitor cells (HPCs) in the presence or absence of NOTCH inhibitor L-685458. B, Representative flow cytometric analyses and FACS isolation strategies within DEAB-treated (RAi) or retinol (“ROH”)-treated (RAd) cultures. Isolated populations were then assessed for hematopoietic potential. C(i), Representative flow cytometric analyses of T-lymphoid potential of populations, as in (B). (ii) Quantification of the erythro-myeloid CFC potential from the populations in (B), averaged across all biological replicates. Two-way ANOVA with Tukey’s multiple comparison test comparing all biological replicates: RAd HE plus NOTCH inhibitor (n=3), remaining samples (n=4), SEM, statistics shown for BFU-E (RAi HE vs. RAi HE+gSI (p=0.0059), RAi HE vs. RAi CXCR4+ (p=0.008), RAi HE vs. RAi CD73+ (p=0.0044), RAi HE vs. RAd CXCR4+ (p=0.0128), RAi HE vs. RAd CD73+ (p=0.0033), p<0.0001 for RAi HE vs. RAd HE, RAi HE+gSI vs. RAd HE, RAi CXCR4+ vs. RAd HE, RAi CD73+ vs. RAd HE, RAd HE vs. RAd HE+gSI, RAd HE vs. RAd CXCR4+, RAd HE vs. RAd CD73+), all comparisons not shown are not significant. All colony counts and statistical analyses are included in Source Extended Data Fig. 5. (iii) Representative flow cytometric analysis of CD34 and CD45 at 9 days after HE FACS isolation and percentage of CD34+CD45+ cells from each culture, averaged across all biological replicates (n=4). Two-tailed paired t-test, p=0.0458, SEM. D, Average expression across all biological replicates of embryonic (HBE), fetal (HBG), and adult (HBB) globin genes and BCL11A in BFU-E derived from WNTi CD34+ cells (purple), WNTd RAi HE (green), and RAd HE (blue). Ordinary one-way ANOVA comparing all biological replicates: HBE (n=3; WNTi vs. RAi, p=0.4636, WNTi vs. RAd, p=0.8811; RAi vs. RAd, p=0.3841), HBG (n=3; WNTi vs. RAi, p=0.234; WNTi vs. RAd, p=0.0018; RAi vs. RAd, p=0.007), HBB (n=3, WNTi vs. RAi, p=0.3253; WNTi vs. RAd, p=0.0662; RAi vs. RAd, p=0.286), BCL11A (n=6, p=0.002552), SEM, ns=not significant.
Extended Data Fig. 6. Xenograft analyses of hPSC-derived HE populations.
A, Transient xenograft persistence of RAd hematopoietic progenitors following injection in neonatal mice. Percent chimerism observed of hCD45 cells present in either the peripheral blood and bone marrow following injection with hPSC-derived CD34+ cells. B, Lineage distribution of hCD45+ cells present in the peripheral blood (PB) 8 weeks post-transplant. C-E, Detailed analysis of human engraftment in the bone marrow or peripheral blood of mice transplanted with RAd HE cells. C, Two independent CD45 antibodies (“CD45–1” and “CD45–2”) were used to label human cells. Representative flow cytometric 4-week analysis of the bone marrow of a non-injected control mouse (top), a recipient of 105 CD34+ cord blood cells (middle) and a recipient of 5×104 RAd CD34+ cells. D-E, Alternative strategy for human chimerism analysis based on mCD45 and hCD45 exclusive staining. D, Representative flow cytometric analysis for single stains of both mCD45 and hCD45, from the bone marrow of an non-injected control mouse or a recipient of 2×105 RAd CD34+ cells. E, Representative flow cytometric strategy for the detection of human chimerism in the peripheral blood, from a recipient of 105 RAd CD34+ cells.
Extended Data Fig. 7. Whole-transcriptome analysis of hPSC and human embryonic CD34+ populations.
A, Comparison of whole transcriptomes of hPSC-derived HE and HPCs (3 biological replicates each). (i) PCA plot demonstrating the similarity between batch-corrected biological replicates of WNTi, RAi, and RAd HE, along with CD34+CD45+ HPCs derived from RAi or RAd HE. (ii) Heatmap and hierarchical clustering showing the Euclidean distance between all biological replicates of WNTd RAi and RAd HE, as in (i). (iii) Selection of significant normalized enrichment scores (NES) from preranked GSEA between RAi HE (green) and RAd HE (blue), as in Extended Data Table 5C. RAi was enriched in embryonic development (NES=−1.80, FDR=0.23), endothelium development (NES=−1.85, FDR=0.23), cellular adhesion (NES=−1.78, FDR=0.24), the epithelial-to-mesenchymal transition (NES=−1.95, FDR 0.14), and response to mechanical stimuli (NES=−1.71, FDR=0.23). In contrast, RAd HE was enriched for RA signaling (NES=1.99, FDR=0.22) and several histone modification pathways (NES=2.12, FDR=0.14). B, (i) Mean expression (log10 FPKM) of hemato-endothelial genes for all biological replicates within hPSC-derived WNTi HE, RAi HE, RAd HE, RAi HPC, and RAd HPC, as in (A), with CD34+CD90+CD43neg cells (“AGM CD34+”), CD34+CD90+CD43+ hematopoietic stem/progenitors (“AGM HSPC”), and CD34+CD90negCD43+ hematopoietic progenitors (“AGM PR”) isolated from the aorta-gonad mesonephros (AGM) region of 5th-week of gestation human embryos6. Grey indicates undetected gene. Scale bar: log10 FPKM. Biological replicates: WNTi HE, RAi HE, RAd HE, RAi HPC, RAd HPC (n=4); AGM CD34+, AGM HSPC, AGM PR (n=1, GEO SuperSeries GSE81102). (ii) Selection of significant normalized enrichment scores (NES) from preranked GSEA comparing RAi HE to AGM 34+ cells (green) and RAd HE to AGM CD34+ cells (blue), as in Extended Data Table 5E,F. RAi and RAd HE was enriched for hematopoietic stem and progenitor differentiation (NES≥1.63, FDR≤0.015), while AGM CD34+ cells were enriched for aorta and vascular development (NES≥1.82, FDR≤0.01), and BMP and VEGF signaling pathways (NES≥1.89, FDR≤0.004).
Extended Data Fig. 8. Establishment of human embryonic dataset for comparative analysis with hPSC-derived hemogenic and arterial endothelial populations.
A, UMAPs visualizing (i) Carnegie Stages and (ii) cell type labels from the complete dataset of Zeng, et al19. Biological replicates: CS10, CS11, CS12, CS14, CS15 (n=1); CS13 (n=2, 1 each for CS13X (10X genomics) and CS13D (Modified STRT-Seq). B, UMAP visualizing the cells categorized as arterial endothelial cells (“AEC”, defined as CDH5+CXCR4+GJA5+DLL4+HEY2+SPNnegPTPRCneg) and hemogenic endothelial cells (“HEC”, defined as CDH5+RUNX1+HOXA+ITGA2BnegSPNnegPTPRCneg), as in Fig. 3B. C, UMAP visualizing the numbered groups for embryonic cells, as in Fig. 3B, within the categorized AEC and HEC. Di, Heatmap visualizing the expression of select broadly and arterial endothelial genes and RUNX1 in human CS10–14 AEC and HEC. Clusters of AEC and HEC are segregated by their relative similarity to hPSC-derived RAi or RAd HE, as in Fig. 3B. Scale bar: gene expression scaled to subset. (ii) Violin plot for scaled expression of select arterial endothelial genes across 6 AEC/HEC clusters, as in (i).
Extended Data Fig. 9. Gating strategy and controls for flow cytometric analyses.
A, Universal gating strategy for all hPSC-derived flow cytometric analyses. B, Single stain controls for markers assessed at the mesodermal stage (day 3 of differentiation). C, Single stain controls for markers assessed at the HE stage (day 8 of differentiation). D, Gating strategy and single stain controls for T cell assay (day 21 of OP9-DL4 co-culture). E, Gating strategy for assessment of xenograft persistence established using human cord blood CD34+ cells.
Supplementary Material
Acknowledgements
S.A.L., C.D., and J.P.C. received support from the NHLBI T32 Training Grant (HL007088-41). S.A.M. is supported by a Vallee Scholar Award and an Allen Distinguished Investigator Award. A.D. is supported by the Telethon Foundation (TIGET grant C4-2016/9 and TIGET grant G3b-2016/9) and San Raffaele Hospital (Seed Grant). C.M.S. is supported by an American Society of Hematology Scholar Award, an American Society of Hematology Bridge Grant, a Washington University Center of Regenerative Medicine Pilot Grant, the Bill & Melinda Gates Foundation INV-002414, and NIH R01HL145290 and R01HL151777. This publication was made possible, in part, by Grant Number UL1 RR024992 from the NIH-National Center for Research Resources (NCRR). R.S. conducted this study as partial fulfillment of an international Ph.D. in Molecular Medicine, Vita-Salute San Raffaele University.
Footnotes
Competing Interests
The methodology described in this publication is subject to patent PCT/US2020/014626 (Inventors: A.D. and C.M.S.).
References
- 1.Chanda B, Ditadi A, Iscove NN & Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell 155, 215–227 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Sturgeon CM, Ditadi A, Awong G, Kennedy M. & Keller G. Wnt Signaling Controls the Specification of Definitive and Primitive Hematopoiesis From Human Pluripotent Stem Cells. Nat Biotechnol 32, 554–561, doi: 10.1038/nbt.2915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy M. et al. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep 2, 1722–1735, doi: 10.1016/j.celrep.2012.11.003. (2012). [DOI] [PubMed] [Google Scholar]
- 4.Dege C. et al. Potently Cytotoxic Natural Killer Cells Initially Emerge from Erythro-Myeloid Progenitors during Mammalian Development. Dev Cell 53, 229–239 e227, doi: 10.1016/j.devcel.2020.02.016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditadi A. et al. Human Definitive Haemogenic Endothelium and Arterial Vascular Endothelium Represent Distinct Lineages. Nat Cell Biol 17, 580–591, doi: 10.1038/ncb3161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng ES et al. Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat Biotechnol 34, 1168–1179, doi: 10.1038/nbt.3702 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Gao L. et al. RUNX1 and the endothelial origin of blood. Exp Hematol 68, 2–9, doi: 10.1016/j.exphem.2018.10.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldie LC, Lucitti JL, Dickinson ME & Hirschi KK Cell signaling directing the formation and function of hemogenic endothelium during murine embryogenesis. Blood 112, 3194–3204, doi: 10.1182/blood-2008-02-139055 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcelo KL, Goldie LC & Hirschi KK Regulation of endothelial cell differentiation and specification. Circ Res 112, 1272–1287, doi: 10.1161/CIRCRESAHA.113.300506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou DR et al. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nat Cell Biol 18, 595–606, doi: 10.1038/ncb3354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu C. et al. Retinoic acid enhances the generation of hematopoietic progenitors from human embryonic stem cell-derived hemato-vascular precursors. Blood 116, 4786–4794 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Ronn RE et al. Retinoic acid regulates hematopoietic development from human pluripotent stem cells. Stem Cell Reports 4, 269–281, doi: 10.1016/j.stemcr.2015.01.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creamer JP et al. Human definitive hematopoietic specification from pluripotent stem cells is regulated by mesodermal expression of CDX4. Blood 129, 2988–2992, doi: 10.1182/blood-2016-11-749382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Sandell LL, Trainor PA, Koentgen F. & Duester G. Alcohol and aldehyde dehydrogenases: retinoid metabolic effects in mouse knockout models. Biochim Biophys Acta 1821, 198–205, doi: 10.1016/j.bbalip.2011.04.004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Jong JL et al. Interaction of retinoic acid and scl controls primitive blood development. Blood 116, 201–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyser RCV et al. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature, doi: 10.1038/s41586-021-04158-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugimura R. et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438, doi: 10.1038/nature22370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanovs A, Rybtsov S, Anderson RA, Turner ML & Medvinsky A. Identification of the niche and phenotype of the first human hematopoietic stem cells. Stem Cell Reports 2, 449–456, doi: 10.1016/j.stemcr.2014.02.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng Y. et al. Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res 29, 881–894, doi: 10.1038/s41422-019-0228-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Q. et al. Developmental trajectory of pre-hematopoietic stem cell formation from endothelium. Blood, doi: 10.1182/blood.2020004801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.North TE et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16, 661–672 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Hou S. et al. Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res 30, 376–392, doi: 10.1038/s41422-020-0300-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez RE, Putzke AP, Myers JP, Margaretha L. & Moens CB Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development 134, 177–187, doi: 10.1242/dev.02706 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JH, Protze SI, Laksman Z, Backx PH & Keller GM Human Pluripotent Stem Cell-Derived Atrial and Ventricular Cardiomyocytes Develop from Distinct Mesoderm Populations. Cell Stem Cell 21, 179–194 e174, doi: 10.1016/j.stem.2017.07.003 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Tanaka Y. et al. Early ontogenic origin of the hematopoietic stem cell lineage. Proc Natl Acad Sci U S A 109, 4515–4520, doi: 10.1073/pnas.1115828109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka Y. et al. Circulation-independent differentiation pathway from extraembryonic mesoderm toward hematopoietic stem cells via hemogenic angioblasts. Cell Rep 8, 31–39, doi: 10.1016/j.celrep.2014.05.055 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Dzierzak E. & Bigas A. Blood Development: Hematopoietic Stem Cell Dependence and Independence. Cell Stem Cell 22, 639–651, doi: 10.1016/j.stem.2018.04.015 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Chen MJ et al. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 9, 541–552, doi: 10.1016/j.stem.2011.10.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dignum T. et al. Multipotent progenitors and hematopoietic stem cells arise independently from hemogenic endothelium in the mouse embryo. Cell Rep 36, 109675, doi: 10.1016/j.celrep.2021.109675 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson JA et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Park IH et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Kennedy M, D’Souza SL, Lynch-Kattman M, Schwantz S. & Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood 109, 2679–2687 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dege C. & Sturgeon CM Directed Differentiation of Primitive and Definitive Hematopoietic Progenitors from Human Pluripotent Stem Cells. J Vis Exp, doi: 10.3791/55196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ditadi A. & Sturgeon CM Directed differentiation of definitive hemogenic endothelium and hematopoietic progenitors from human pluripotent stem cells. Methods 101, 65–72, doi: 10.1016/j.ymeth.2015.10.001 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Sturgeon CM et al. Primitive erythropoiesis is regulated by miR-126 via nonhematopoietic Vcam-1+ cells. Dev Cell 23, 45–57, doi: 10.1016/j.devcel.2012.05.021 (2012). [DOI] [PubMed] [Google Scholar]
- 36.La Motte-Mohs RN, Herer E. & Zuniga-Pflucker JC Induction of T-cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood 105, 1431–1439 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Schmitt TM et al. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol 5, 410–417 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Kinsella RJ et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011, bar030, doi: 10.1093/database/bar030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaspi A. & Ziemann M. mitch: multi-contrast pathway enrichment for multi-omics and single-cell profiling data. BMC Genomics 21, 447, doi: 10.1186/s12864-020-06856-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aran D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20, 163–172, doi: 10.1038/s41590-018-0276-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alles J. et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol 15, 44, doi: 10.1186/s12915-017-0383-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All gene expression analysis datasets are available in the Gene Expression Omnibus (GEO) under the accession numbers GSE139853 (scRNA-seq in Fig. 1, Fig. 3, and Extended Data Fig. 2; CXCR4+/− mesoderm RNA-seq in Fig. 1 and Extended Data Fig. 3; RAi/RAd HE RNA-seq in Fig. 4, Extended Data Fig. 1, Extended Data Fig. 3, and Extended Data Fig. 7; RAi/RAd HPC RNA-seq in Fig. 4, Extended Data Fig. 7), or BioProjects #PRJNA352442 (WNTi mesoderm RNA-seq in Fig. 1, Extended Data Fig. 1, and Extended Data Fig. 3; WNTd mesoderm RNA-seq in Extended Data Fig. 1) and #PRJNA525404 (WNTi HE RNA-seq in Fig. 4, Extended Data Fig. 1, Extended Data Fig. 3, and Extended Data Fig. 8). Publicly-available datasets used include: GEO SuperSeries GSE81102, “AGM CD34+” (GSM2142333), “AGM PR” (GSM2142334), “AGM HSPC” (GSM2142332) for Human 5 week AGM (Fig. 4 and Extended Data Fig. 7); GEO GSE135202 for Human CS10-CS15 embryo (Fig. 4 and Extended Data Fig. 8); ArrayExpress E-MTAB-9388 for Human CS7 embryo (Fig. 3). Genome alignments were performed with GENCODE GRCh38.p3 version 23. All source data has been deposited within the San Raffaele Open Research Data Repository and are available at doi: 10.17632/pbbn55mhy9.1