Abstract
The lipase from Pseudomonas cepacia ATCC 21808 (recently reclassified as Burkholderia cepacia) is widely used by organic chemists for enantioselective synthesis and is manufactured from recombinant P. cepacia harboring on a plasmid the clustered genes for lipase and its chaperone. High levels of expression of inactive lipase (40%) in Escherichia coli were achieved with pCYTEXP1 under the control of the strong, temperature-inducible λPRL promoter. However, no overexpression of the lipase chaperone was achieved in E. coli. Thus, chemical refolding of inactive lipase in the absence of its chaperone yielded only 25 U/mg, compared to 3,470 U of the purified lipase secreted by recombinant P. cepacia per mg. Sequence analysis of the chaperone revealed a high GC content (>90%) in the 5′ region of the gene and the presence of a putative membrane anchor at the N terminus. Hence, the 5′ region of the gene was replaced by a synthetic fragment, and the putative membrane anchor was removed by deletion of the first 34 or 70 N-terminal amino acids. Only truncation of the gene led to overexpression of the chaperone (up to 60%) in E. coli. With this chaperone, it was possible to obtain for the first time in a simple refolding procedure a highly active Pseudomonas lipase (classes I and II) expressed in E. coli with a specific activity of up to 4,850 U/mg and a yield of 314,000 U/g of E. coli wet cells.
Lipases (triacylglycerol acylhydrolases; EC 3.1.1.3), particularly microbial lipases, have been widely used in the hydrolysis and transesterification of triglycerides and in the enantioselective synthesis and hydrolysis of a variety of esters (27, 39).
In recent years, many microbial lipase genes have been isolated, sequenced, modified, and expressed in homologous or heterologous hosts, such as Escherichia coli, filamentous fungi, or yeasts (3, 5, 17, 34, 37). Lipases from Pseudomonas alcaligenes (2, 25) are extensively used as an additive in laundry detergents, and lipases from Pseudomonas species are widely used as catalysts in organic synthesis (12, 36). In addition, these various Pseudomonas lipase preparations are commercially available (e.g., Amano YS, P, and AH; Fluka SAM-II). Among these, P. cepacia lipase preparations are the most predominant and have often been used by organic chemists for enantioselective synthesis (33).
In recent years, the cloning, sequencing, and expression of a variety of Pseudomonas lipase genes have been reported (10, 15, 18, 22, 24, 30, 39). The genes can be divided into three homology groups (assigned as classes I to III), where class III is only distantly related to the other classes. Pseudomonas lipases of classes I and II, including the broadly used lipases of P. cepacia and P. glumae strains (class II) as well as of P. aeruginosa strains (class I), need a chaperone whose gene is located downstream of the lipase gene for efficient secretion and folding of active lipase. The deduced amino acid sequences of the chaperones belong to two homology groups. For a detailed review of the biochemical and molecular properties of Pseudomonas lipases, see references 12, 23, and 36.
The production of Pseudomonas lipases is currently carried out with recombinant Pseudomonas strains harboring both the lipase and its chaperone on a broad-host-range plasmid. With this system, lipase production is increased 40- to 85-fold over that of the wild-type strain (16, 30). For recombinant P. cepacia ATCC 21808 (recently reclassified as Burkholderia cepacia) harboring the large pMMB22-derived plasmid pHES12, only moderate lipase productivity of 200 U/ml is obtained (15, 16). Moreover, lipase production by recombinant strains often decreases over time, possibly because of the large size of the plasmids. In addition, most Pseudomonas strains currently used for lipase production are potential pathogens; thus, special safety directions have to be considered. For these reasons and to further increase lipase productivity, we aimed to develop a heterologous E. coli expression system for the large-scale production of these important lipases.
Unfortunately, the expression of class I and II Pseudomonas lipases in E. coli is hampered by the fact that a lipase chaperone is necessary for effective folding of the lipase. To our knowledge, high-level production of functional class I and II Pseudomonas lipases in E. coli has not yet been achieved.
In this work, we report for the first time the overexpression of both the lipase and its modified and truncated chaperone of P. cepacia ATCC 21808 in E. coli. In a simple and rapid in vitro refolding procedure, functionally active lipase can be obtained in large amounts and with a specific activity comparable to that of the lipase purified from P. cepacia.
MATERIALS AND METHODS
Materials.
Restriction enzymes, DNA-modifying enzymes, T4 DNA ligase, and Taq polymerase were from MBI Fermentas. The Taq Dye Deoxy cycle sequencing kit was from Applied Biosystems. The DNA gel extraction kit, Midi plasmid kit, Prep-spin plasmid kit, and Ni-nitrilotriacetic acid (NTA) matrix were from Qiagen. p-Nitrophenyl-palmitate (pNPP) was from Sigma. Peptone and yeast extract were from Difco. All reagents were of analytical grade unless otherwise stated.
Strains, plasmids, and media.
P. cepacia ATCC 21808 harboring plasmid pHES12 containing the lipase operon was kindly provided by Boehringer Mannheim GmbH (15). E. coli BL321 (hsdS gal [λcIts857 ind1 Dam7 nin5 lacUV5-T7 gene 1]) and E. coli DH5α (supE44 ΔlacU169 [φ80lacZΔM15] hsdR17 recA1 endA1 gyrA96 thi-1 relA1) were used for cloning and gene expression.
Plasmid pCYTEXP1 (4), providing ampicillin resistance, was used for the construction of different expression vectors in E. coli. Plasmid pET20b(+) (Novagen) was used for subcloning.
E. coli was grown at 37°C in Luria-Bertani medium (LB) supplemented with 100 μg of ampicillin per ml for selection of transformants.
Gene expression.
Transformed E. coli BL321 (or E. coli DH5α for expression of truncated and modified chaperone genes) with plasmids derived from pCYTEXP1 containing the strong, temperature-inducible λPRL promoter was cultivated at 30°C until the optical density at 578 nm was 0.8. Next, protein expression was induced by shifting the cultivation temperature to 42°C. After 3 h of induction, cells were harvested by centrifugation (10,000 × g, 10 min, 4°C).
Recombinant DNA techniques.
Standard recombinant DNA methods were carried out as described by Sambrook et al. (32). The fluorescence-based dideoxy DNA cycle sequencing method was used for sequence determination. DNA sequencing was carried out with the Taq Dye Deoxy cycle sequencing kit and with a model 373A DNA sequencing system (Applied Biosystems) in accordance with the manufacturer’s instructions. For PCR, DNA was amplified with the following cycle conditions: first step, 94°C for 4 min, 1 cycle; second step, 94°C for 1.5 min, 64°C for 2 min, and 72°C for 3 min, 25 cycles; and third step, 72°C for 4 min, with 8% dimethyl sulfoxide added to the PCR mixture.
Isolation of lipase inclusion bodies.
E. coli cells from a 500-ml culture (2 g) were suspended in 25 ml of 50 mM Tris buffer (pH 8.0) containing 1 mM EDTA and disrupted by sonification with a Branson Sonifier W-250 (duty cycle, 35%; output control, 3; time, 7 min), and the pellet containing the insoluble inclusion bodies was washed several times with the same buffer. The inclusion bodies were dissolved in 20 ml of buffer B (10 mM Tris-HCl, 8 M urea, 0.1 M sodium phosphate [pH 8.0]) at room temperature for 1 h with mild stirring. Following centrifugation (10,000 × g, 15 min, 4°C), the solubilized and denatured lipase of the supernatant was subjected to refolding.
Purification of the lipase chaperone by Ni-NTA chromatography.
E. coli cells from a 500-ml culture (2 g) were suspended in 20 ml of buffer A (10 mM Tris-HCl, 6 M guanidine-HCl, 0.1 M sodium phosphate [pH 8.0]) and lysed for 1 h at room temperature. Following centrifugation (10,000 × g, 15 min, 4°C), the supernatant was applied (0.2 ml/min) to an Ni-NTA column (1.6 by 5 cm) previously equilibrated with buffer. Unbound protein was eluted by washing the column with 10 volumes of buffer A, 5 volumes of buffer B, and 5 volumes of buffer C (10 mM Tris-HCl, 8 M urea, 0.1 M sodium phosphate [pH 6.3]). Finally, bound chaperone was eluted with buffer D (10 mM Tris-HCl, 8 M urea, 0.1 M sodium phosphate [pH 5.9]). The purity of the eluted chaperone was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
Refolding of lipase in the presence of the chaperone.
One hundred microliters of lipase solution obtained after inclusion body solubilization and containing 1 mg of denatured lipase was refolded in 100 ml of distilled water in the presence of different amounts of denatured (solubilized with 8 M urea) or native (not solubilized with urea) chaperone. After 24 h of refolding at 4°C, the lipase activity in the supernatant was determined with pNPP as the substrate.
In order to investigate the optimal ratio of lipase to chaperone, 0.1, 0.5, 1, and 3 mg (10 to 300 μl) of denatured and purified chaperones Δ70HpHis and ompAΔ70HpHis obtained after Ni-NTA chromatography were used for the refolding of 1 mg of lipase as described above.
For simplified refolding, E. coli cells from a 500-ml culture (2 g of cells) expressing a truncated chaperone (Δ70HpHis, ompAΔ70HpHis, Δ34HpHis, or Δ34HpHis/ompAΔ34HpHis) were lysed in 20 ml of buffer B. After centrifugation (10,000 × g, 15 min, 4°C), the supernatant containing the denatured (solubilized with 8 M urea) chaperone was used for in vitro refolding. The amount of the chaperone in relation to the total protein in the supernatant was estimated by SDS-PAGE analysis. Approximately 1 mg of chaperone was used for the in vitro refolding of 1 mg of lipase as described for the purified chaperone. In a similar way, native (not solubilized with urea) chaperones (Δ70HpHis, ompAΔ70HpHis, and Δ34HpHis) were used for refolding. Instead of cell lysis in the presence of urea, cells from a 500-ml culture were suspended in 25 ml of 50 mM Tris buffer (pH 8.0) containing 1 mM EDTA and disrupted by sonification with a Branson Sonifier W-250 (duty cycle, 35%; output control, 3; time, 7 min). After centrifugation (10,000 × g, 15 min, 4°C), the supernatant containing the soluble chaperone was used for refolding.
Analytical methods.
Preparative gel electrophoresis was carried out with a 12.5% polyacrylamide gel as described by Laemmli (28), and proteins were stained with Coomassie brilliant blue R-250. The expression level was estimated as a percentage of the level of total cellular protein with Imagemaster VDS version 2.0 (Pharmacia).
For amino-terminal sequence analysis, purified proteins were subjected to SDS gel electrophoresis. After blotting, the polyvinylidene difluoride membrane was stained with Coomassie brilliant blue R-250, and the protein bands were cut out and used for amino-terminal sequence analysis. Amino-terminal sequence analysis was performed with a model 470A gas-phase sequencer (Applied Biosystems) in accordance with the manufacturer’s instructions.
Protein concentration was determined with the bicinchoninic acid protein assay kit by the enhanced method in accordance with the manufacturer’s instructions (Pierce instructions 23220/23225) and with bovine serum albumin as the standard.
Enzyme assay.
The enzyme assay was performed with pNPP as the substrate by use of a Biochrom 4060 spectrophotometer (Pharmacia). Cleavage of pNPP was measured at 60°C with 0.1 M Tris buffer (pH 7.5) as described by Schmidt-Dannert et al. (35). One unit was defined as the amount of enzyme which caused the release of 1 μmol of p-nitrophenol per minute under the test conditions.
RESULTS
Construction of plasmids.
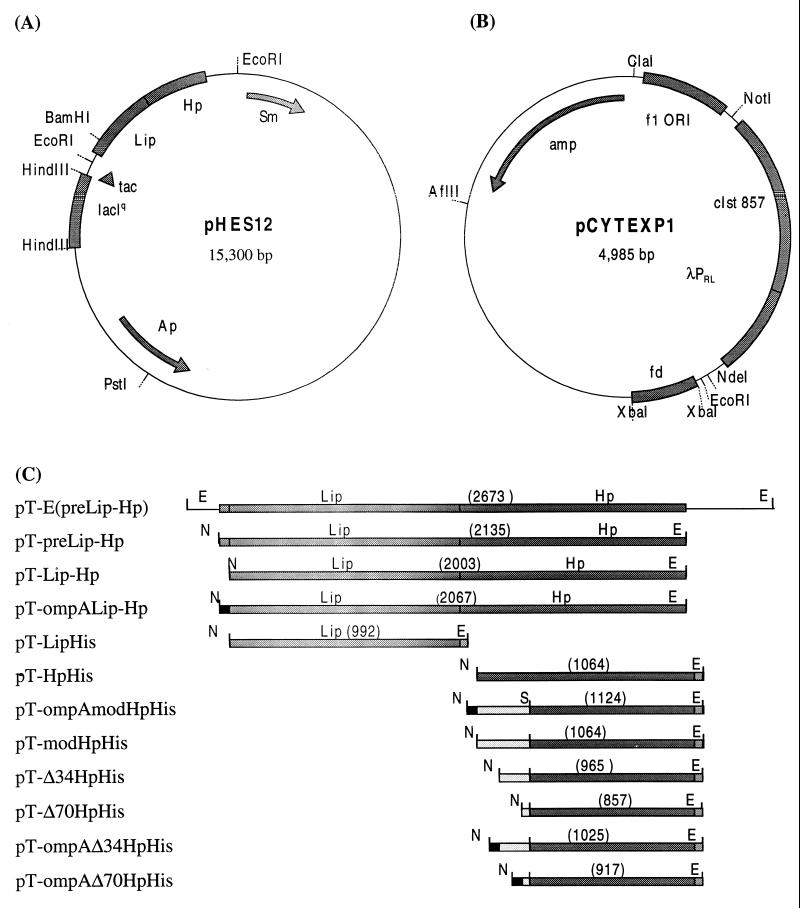
For the initial investigation of protein expression in E. coli, a 2,673-bp fragment containing the genes for the prelipase and the chaperone was cut from pHES12 with EcoRI and ligated into pCYTEXP1 linearized with the same enzyme, yielding pT-E(preLip-Hp) (Fig. 1).
FIG. 1.
Construction of different expression vectors derived from plasmids pHES12 and pCYTEXP1. The inserts and restriction sites used for cloning are given. (A) P. cepacia expression vector (pHES12) containing the complete lipase operon (lipase and chaperone) under the control of the tac promoter. (B) E. coli expression vector (pCYTEXP1) containing the strong, temperature-inducible λPRL promoter. ORI, origin. (C) Inserts of pCYTEXP1-derived expression vectors. N, NdeI; E, EcoRI. Numbers in parentheses are base pairs. See Table 1, footnote a, for explanations of designations.
In order to place the lipase operon (preLip-Hp) at an optimal distance from the λPRL promoter of pCYTEXP1, PCR was performed with pT-E(preLip-Hp) as a template and two oligonucleotides introducing an NdeI site (including the ATG start codon) at the 5′ end of the prelipase gene and an EcoRI site at the 3′ end of the chaperone gene. After digestion of the resulting PCR fragment (containing preLip-Hp) and pCYTEXP1 with NdeI and EcoRI and subsequent ligation of the products, expression vector pT-preLip-Hp was obtained (Fig. 1).
In two additional pCYTEXP1-derived expression plasmids containing complete preLip-Hp, the original signal equence of the lipase gene was removed, resulting in vector pT-Lip-Hp, and replaced by the ompA signal sequence, yielding plasmid pT-ompALip-Hp. pT-ompALip-Hp was obtained by amplifying the ompA signal sequence of expression plasmid pT-ompABTL2 (34) with two oligonucleotides: the first complementary to the 5′ end of the ompA sequence and introducing an NdeI site and the second complementary to the 3′ end of the ompA sequence and to the 5′ end of the mature lipase gene. The PCR fragment thus obtained and an oligonucleotide complementary to an SphI site inside the lipase gene served as primers for a second PCR with pT-preLip-Hp as a template. Digestion of both the obtained PCR fragment and pT-preLip-Hp with NdeI and SphI, followed by ligation, resulted in pT-ompALip-Hp. pT-Lip-Hp was constructed by PCR with pT-preLip-Hp as a template and with one primer complementary to the 5′ end of the mature lipase gene and introducing an NdeI site (including the ATG start codon) and another primer complementary to the 3′ end of the chaperone gene and introducing an EcoRI site. Digestion of both pT-preLip-Hp and the obtained PCR fragment with EcoRI and NdeI, followed by ligation, yielded pT-Lip-Hp (Fig. 1).
To fuse a six-histidine (His6) tag to the mature lipase gene and the chaperone gene, both genes were subcloned in plasmid pET20b(+) harboring a His6 tag (data not shown). The tagged genes were transferred to pCYTEXP1 for expression under the control of the λPRL promoter by amplification with oligonucleotides introducing NdeI and EcoRI sites at the 5′ and 3′ ends, respectively. Cloning of these PCR fragments into pCYTEXP1 also linearized with NdeI and EcoRI resulted in pT-HpHis and pT-LipHis, respectively.
A modified chaperone gene (modHp) was constructed as follows: the first 242 bp (ending at a SacII site) of the gene were replaced by a codon-optimized (for E. coli) and thus less GC-rich nucleotide sequence. To this end, four oligonucleotides of 80 bp each and overlapping by approximately 15 bp were synthesized and assembled by PCR. The terminal oligonucleotides comprised an NdeI site (5′ end) and a SacII site (3′ end inside the chaperone gene) for cloning of the PCR fragment into pT-HpHis cut with the same enzymes, yielding pT-modHpHis (Fig. 1).
5′ truncation of the modified chaperone gene by 34 residues (102 bp) and 70 residues (210 bp) resulted in two plasmids, pT-Δ34HpHis and pT-Δ70HpHis, after PCR with two primers: one introducing an NdeI site (including the ATG start codon) at the new 5′ end of the gene and another introducing an EcoRI site at the 3′ end of the gene. pT-modHpHis was used as a template, and then the EcoRI- and NdeI-digested PCR fragment was cloned into pT-modHpHis cut with the same enzymes (Fig. 1).
Fusion of the ompA signal sequence to the modified and truncated chaperone genes was carried out by PCR in a manner similar to that described for pT-ompALip-Hp. The resulting plasmids were named pT-ompAΔ70HpHis and pT-ompAΔ34HpHis.
Overexpression of lipase in E. coli.
For initial expression studies of recombinant lipase in E. coli, plasmid pT-E(preLip-Hp) containing complete preLip-Hp (EcoRI fragment) of pHES12 under the control of λPRL was constructed. However, after 3 h of induction of transformed cells with this plasmid, no additional bands of expressed lipase or chaperone were detected on Coomassie brilliant blue R-250-stained SDS gels of cell extracts. A small band of 33 kDa representing lipase was visualized after activity staining (data not shown); this band corresponded to the low level of lipase activity (171 U/g of cells) measured after cell breakage (Table 1).
TABLE 1.
Levels of expression of the lipase and the lipase chaperone in E. coli and lipase activity
| Vectora | % Expression ofb:
|
Activity (U/g of cells)c | |
|---|---|---|---|
| Lipase | Chaperone | ||
| pT-E(preLip-Hp) | <1 | <1 | 171 |
| pT-preLip-Hp | <1 | <1 | 293 |
| pT-Lip-Hp | 40 | <1 | 40 |
| pT-ompALip-Hp | 40d | <1 | 547 |
| pT-preLip | <1 | 37 | |
| pT-LipHis | 40 | 0 | |
| pT-HpHis | <1 | ||
| pT-modHpHis | <1 | ||
| pT-Δ34HpHis | 10 | ||
| pT-Δ70HpHis | 10 | ||
| pT-ompAΔ34HpHis | 60d | ||
| pT-ompAΔ70HpHis | 40e | ||
Abbreviations: pre, native lipase signal sequence; ompA, ompA signal sequence; E, EcoRI fragment derived from pHES12; Lip, lipase gene; Hp, chaperone gene; Δ34 or Δ70, chaperone truncated by 34 or 70 amino-terminal amino acids, respectively; His, His6 tag; mod, 5′ end of chaperone gene (bp 1 to 241) replaced by a synthetic, codon-optimized nucleotide sequence.
Estimated by densiometry with Imagemaster VDS version 2.0 and given as a percentage of the level of total cellular protein.
Determined after 3 h of incubation with pNPP as the substrate following cell lysis.
The ompA leader sequence was processed from 50% of expressed ompAΔ34HpHis.
The ompA leader sequence was not processed.
To increase expression levels, preLip-Hp was placed near the strong, temperature-inducible λPRL promoter of pCYTEXP1, yielding pT-preLip-Hp and, after deletion of the chaperone gene, pT-preLip. In addition, the original signal sequence of the lipase gene was deleted in pT-Lip-Hp and replaced by the ompA signal sequence, known to increase expression levels and to transport the protein across the inner membrane, yielding pT-ompALip-Hp.
In contrast to pT-preLip-Hp, both vectors pT-Lip-Hp and pT-ompALip-Hp (data not shown), in which the original presequence is no longer present, led to overexpression of the lipase (expression level, 40%) but not of the chaperone (Table 1). However, the lipase was expressed as inclusion bodies, and most of them were inactive after purification; hence, no processing of the ompA-lipase gene occurred. This conclusion was also confirmed by amino-terminal sequencing. Only slightly increased levels of expressed lipase activity were detected with pT-preLip-Hp (293 U/g of cells) and pT-ompALip-Hp (547 U/g of cells), in which the lipase gene is preceded by a signal sequence and the chaperone gene is present. The purpose of the introduction of the ompA signal sequence in the construct pT-ompALip-Hp was to increase transport of the lipase across the inner membrane and hence to increase lipase activity due to the presence of a small amount of the chaperone in the periplasm. The product was internally retained; however, a small amount of the lipase was exported to the periplasm. This result showed that the lipase activity (547 U/g) of the construct with the ompA signal sequence (pT-ompALip-Hp) was 13 times higher than that of the construct without ompA (pT-Lip-Hp).
Overexpression of lipase chaperone in E. coli.
Although the nucleotide sequence of the lipase gene has been elucidated (15), the nucleotide sequence of the chaperone gene located downstream of the lipase gene has not. Hence, an open reading frame of 1,032 bp, located 3 bp downstream of the lipase gene and encoding the lipase chaperone, was identified (Fig. 2). While the average GC content of the chaperone gene was 73%, a GC content of >90% was calculated for a 250-bp region at the 5′ end. A putative Shine-Dalgarno sequence (GAAG) was found inside the 3′ region of the lipase gene.
FIG. 2.
Complete nucleotide sequence and amino acid sequence of the lipase chaperone of P. cepacia ATCC 21808 and the C-terminal region of the lipase. The putative Shine-Dalgarno sequence is boxed. The modified, codon-optimized 242 bp of the 5′ region is underlined, and exchanged nucleotides are indicated by boldfacing. Arrows indicate positions of truncation.
The deduced protein sequence comprises 344 amino acids and, hence, codes for a protein with a molecular mass of 37.4 kDa. Analysis of the protein sequence by the method of Eisenberg et al. (8) identified two hydrophobic stretches of amino acids extending from residues 14 to 34 and residues 50 to 70 and comprising a putative membrane anchor (Fig. 2).
To gain overexpression of the lipase chaperone and, hence, to use the recombinant chaperone for in vitro refolding of the overexpressed lipase, the expression vector pT-HpHis, in which the chaperone gene was placed directly downstream of the λPRL promoter, was constructed. However, no expression of the chaperone was detected on SDS gels.
Both the GC-rich 250-bp 5′ region of the chaperone gene and the amino-terminal hydrophobic stretches of amino acids (residues 14 to 34 and residues 50 to 70), comprising a putative membrane anchor, might adversely affect protein expression in E. coli. Thus, the first 250 bp of the gene was replaced by a synthetic fragment, thereby lowering the GC content from >90 to 60% and resulting in modHp. In addition, the putative membrane anchor was removed by deleting the first 34 (Δ34Hp) and 70 (Δ70Hp) amino-terminal amino acids.
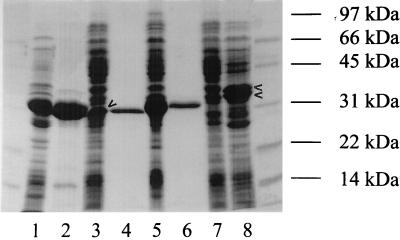
The modified and the truncated chaperone genes were inserted into pCYTEXP1, yielding pT-modHpHis, pT-Δ34HpHis, and pT-Δ70HpHis. Expression levels of 10% were observed for the truncated chaperones Δ34HpHis (34 kDa) and Δ70HpHis (31 kDa) (Fig. 3), whereas with modHp, no additional band was observed after SDS-PAGE analysis of E. coli cells (data not shown). Fusion of the ompA signal sequence to the truncated genes (pT-ompAΔ34HpHis and pT-ompAΔ70HpHis) resulted in further sixfold and fourfold increases in expression, respectively (Table 1 and Fig. 3). Although in both ompA-fused genes the signal cleavage sites were identical (Ala-Ala), cleavage of the ompA leader sequence reached only 50% for ompAΔ34Hp, as seen after SDS-PAGE analysis (Fig. 3, lane 8) and as proved by amino-terminal sequencing. Only ompAΔ34Hp formed insoluble inclusion bodies in E. coli, while all other expressed chaperones were soluble. Both Δ70HpHis and ompAΔ70HpHis were purified by Ni-NTA chromatography to >95% purity (Fig. 3).
FIG. 3.
SDS-PAGE of overexpressed and Ni-NTA-purified lipase and chaperone in E. coli. Proteins were stained with Coomassie brilliant blue R-250. Lanes: 1, E. coli pT-Lip-Hp cell lysate; 2, lipase inclusion bodies isolated from E. coli pT-Lip-Hp cell lysate; 3, E. coli pT-Δ70HpHis cell lysate; 4, Ni-NTA-purified Δ70HpHis; 5, E. coli pT-ompAΔ70HpHis cell lysate; 6, Ni-NTA-purified ompAΔ70HpHis; 7, E. coli pT-Δ34HpHis cell lysate; 8, E. coli pT-ompAΔ34HpHis cell lysate.
In vitro refolding of lipase expressed in E. coli.
In a first attempt, denatured lipase isolated from inclusion bodies was subjected to different refolding procedures without the addition of the chaperone, including those described by Beer et al. (3) for the lipase of Rhizopus oryzae and by Frenken et al. (10) for the lipase of P. glumae (data not shown). However, only a poorly active lipase with a specific activity of 25 U/mg or less was obtained.
Denatured lipase isolated from inclusion bodies, however, was effectively refolded after 24 h of incubation at 4°C in distilled water in the presence of equal amounts (final concentrations, 5 to 10 μg/ml) of truncated chaperone. Refolding of 5 to 10 μg of purified and denatured mature lipase per ml with 5 to 30 μg of purified and denatured chaperone Δ70HpHis or ompAΔ70HpHis per ml yielded a highly active lipase with a specific activity of 3,580 to 4,180 U/mg (Table 2), comparable to or even slightly higher than the activity of the purified lipase secreted from recombinant P. cepacia (3,470 U/mg) and purified as described by Hom (15) and Kordel et al. (26). Increasing the concentration of lipase 10-fold and, hence, also the chaperone concentration in the refolding mixture decreased the refolding efficiency significantly, by a factor of 8 (data not shown). It was found that an excess of chaperone is needed for correct lipase folding, as for other Pseudomonas lipase chaperones (1, 13, 20, 31). In those studies, it was found that at least one lipase chaperone molecule was needed for the correct folding of one lipase molecule, because the chaperone acts toward the lipase noncatalytically.
TABLE 2.
Optimization of the lipase/lipase chaperone molar ratio for in vitro refolding
| Chaperone | Molar ratioa | U/mg ofb:
|
|
|---|---|---|---|
| Lipase | Total protein | ||
| Δ70HpHis | 1:0.1 | 520 | 480 |
| 1:0.5 | 3,680 | 2,622 | |
| 1:1 | 3,750 | 2,520 | |
| 1:3 | 4,180 | 1,450 | |
| ompAΔ70HpHis | 1:0.1 | 410 | 380 |
| 1:0.5 | 2,190 | 1,700 | |
| 1:1 | 3,580 | 2,240 | |
| 1:3 | 3,540 | 1,280 | |
Defined as the ratio of the amount (in moles) of the denatured mature lipase isolated from inclusion bodies to the amount (in moles) of the denatured chaperone (Δ70HpHis or ompAΔ70HpHis) purified by Ni-NTA chromatography.
Specific activity is given as units of lipase per milligram of lipase or milligram of total protein used in the refolding mixture.
In a simplified refolding protocol, E. coli cell extracts containing the different types of truncated chaperones in a denatured or native state were directly used for the in vitro refolding of lipase. With all types of denatured or native chaperones but ompAΔ34HpHis, the lipase could be effectively refolded (Table 3). However, the most effective chaperone for lipase refolding proved to be Δ70HpHis, yielding a highly active lipase (up to 314,000 U of lipase per g of E. coli cells) with the simplified refolding protocol, regardless of whether the chaperone was used in a denatured (4,660 U/mg) or a native (4,850 U/mg) state. In contrast, Δ34HpHis and ompAΔ70HpHis gave rise to a more active lipase when they were used in a denatured state for refolding.
TABLE 3.
Simplified in vitro refolding of denatured mature lipase
| Chaperone | U/mg of lipasea:
|
|
|---|---|---|
| Native chaperone | Denatured | |
| Δ70HpHis | 4,850 | 4,660 |
| Δ34HpHis | 670 | 2,280 |
| ompAΔ70HpHis | 1,040 | 4,170 |
| ompAΔ34HpHis/ompAΔ34HpHisb | 1,730 | 3,580 |
| ompAΔ34HpHisc | ND | 0 |
Specific activity is given as units per milligram of lipase used for refolding. ND, not determined. The inactive lipase was purified from 2 g of wet cells. The amount of the crude chaperone added to the refolding mixture was five to six times higher than that of the crude lipase.
Cell extract from E. coli pT-ompAΔ34HpHis in which 50% of the expressed ompAΔ34HpHis is not processed and is insoluble. Hence, only ompAΔ34HpHis is present in the cell extract after sonification (native chaperone).
Isolated from inclusion bodies produced by E. coli pT-ompAΔ34HpHis.
The refolding efficiency of the unprocessed lipase ompALip, still containing the ompA signal sequence and produced by E. coli pT-ompALip-Hp, was 10 times lower than that of the mature lipase, while lipase LipHis, fused to a His6 tag at the C terminus, could not be refolded (data not shown).
DISCUSSION
In previous work (15), the sequence of the lipase gene of P. cepacia ATCC 21808 was elucidated and was found to be highly homologous (>90%) to those of genes for other class I Pseudomonas lipases, genes including at least five cloned lipases from different P. cepacia strains (22, 24, 30). The sequence of the chaperone gene has not been determined. Sequence analysis revealed a high homology of the protein sequence (>90%) with those of chaperones from P. cepacia DSM 3959 (24) and M-12-33 (30) and Pseudomonas sp. strain KWI-56 (22), as expected.
The role of the chaperone gene located downstream of the lipase gene for the in vivo and in vitro activation of Pseudomonas lipases has been well investigated for lipases of Pseudomonas sp. strain 109 and P. aeruginosa TE3285 (6, 31), belonging to class I, and lipases of P. cepacia DSM 3959 (1, 13, 14, 24), Pseudomonas sp. strain KWI-56 (21, 22), and P. glumae PG1 (9, 11), belonging to class II. Unlike the situation for other industrially relevant microbial lipases, however, no efficient system for the production of large amounts of active Pseudomonas lipase in a biologically safe heterologous host such as E. coli has been described. Since the overexpression of both the lipase and the chaperone needed for in vitro activation is a prerequisite for the economic production of lipase in E. coli, we subcloned the gene cluster for the lipase and the chaperone into E. coli expression vectors pUC19, pET20b(+) (data not shown), and pCYTEXP1. Overexpression of the lipase gene (up to 40%) was achieved only under the control of the strong, temperature-inducible λPRL promoter and when the original signal sequence was either removed or replaced by an ompA signal sequence known to increase protein expression in E. coli (4). Comparable high expression levels for Pseudomonas lipases (classes I and II) in E. coli have been reported for the prelipase of Pseudomonas sp. strain KWI-56 (29) and the mature lipase of P. aeruginosa TE3285 (31). The formation of inactive and insoluble inclusion bodies has also been reported for several Pseudomonas lipases (classes I and II) expressed at higher levels in E. coli (7, 29, 31). Low lipase activities (171 to 547 U/g of cells) were detected in cell lysates of recombinant E. coli cells harboring plasmids containing both the lipase gene and the chaperone gene of P. cepacia; similar observations were made with Pseudomonas lipases expressed in E. coli (9, 11, 20, 22, 24, 31, 39).
Most Pseudomonas lipase chaperones are only poorly expressed in E. coli, and expression had to be observed by a highly sensitive method, such as Western blotting (1, 9, 11, 13, 14, 21). In keeping with these findings, we found no overexpression of unmodified P. cepacia ATCC 21808 lipase chaperone in E. coli, although the gene was placed under the control of the strong λPRL promoter. Sequence analysis identified a GC-rich (>90%) region at the 5′ end of the chaperone gene which might affect transcription in E. coli. However, even when we decreased the GC content to 60% by replacement with a 250-bp synthetic, codon-optimized DNA fragment, we could not increase expression. Frenken et al. (9, 11) showed that the lipase chaperone of P. glumae expressed at low levels in E. coli as well as in wild-type P. glumae is located in the periplasmic space and anchored to the inner membrane by an amino-terminal peptide stretch. It is generally assumed that lipase chaperones of Pseudomonas are involved both in translocation and in folding of lipase during its secretion (6, 9, 11, 20, 22, 24, 41). When we investigated the amino-terminal region of the chaperone, we found two hydrophobic stretches (residues 14 to 34 and residues 50 to 70) which might function as a membrane anchor and in turn hamper protein overexpression by blocking translocation in E. coli. Truncation of the amino terminus by 34 and 70 residues, respectively, led to overexpression of the chaperone in E. coli which could be further increased (from 10% to 60%) by fusion of the truncated genes to an ompA signal sequence.
In vitro refolding of Pseudomonas lipases (classes I and II) expressed so far in E. coli resulted in poorly active lipase preparations, with 5 to 10% of the activity of the native enzyme (13, 19, 21, 33). In contrast, by using overexpressed truncated chaperones either in the native or in the denatured state, we could, for the first time, quantitatively refold a Pseudomonas lipase overexpressed in E. coli (100% specific activity) with high yields (up to 314,000 U/g of E. coli cells) in a simple refolding procedure. We concluded that neither the truncated N-terminal part (70 residues) of the chaperone nor the fused ompA signal sequence (Δ70HpHis) had any influence on the folding activity, while ompAΔ34HpHis (not processed) solubilized (with 8 M urea) from inclusion bodies no longer showed any folding activity. Best refolding results were obtained when the lipase and the chaperone were present in similar amounts in the refolding mixture, as was observed for the in vitro refolding of other lipases from Pseudomonas (1, 33). The fact that denatured chaperones Δ70HpHis, ompAΔ70HpHis, and Δ34HpHis could be applied in the refolding suggested that the active conformation of denatured chaperones is very quickly restored upon dilution in the refolding mixture, thus allowing a simple and economic refolding of denatured lipases by use of crude chaperones overproduced in E. coli and solubilized with urea. With this procedure, as much as 314,000 U of lipase per g of wet cells in a highly pure state and ready to be used in biotechnological applications could be easily obtained. To further simplify the production of active lipase, a coexpression system in which both the lipase and the chaperone are located on one plasmid and each gene is placed under the control of the strong λPRL promoter is being developed.
ACKNOWLEDGMENTS
Dinh Thi Quyen gratefully acknowledges a scholarship from the Konrad Adenauer Foundation, Bonn, Germany. We thank Boehringer Mannheim for providing the recombinant P. cepacia strain.
We thank Volker Nödinger for N-terminal sequencing and H. Atomi for helpful discussions.
REFERENCES
- 1.Aamand J L, Hobson A H, Buckley C M, Jorgensen S T, Diderichsen B, McConnell D J. Chaperone-mediated activation in vivo of a Pseudomonas cepacia lipase. Mol Gen Genet. 1994;245:556–564. doi: 10.1007/BF00282218. [DOI] [PubMed] [Google Scholar]
- 2.Andreoli P M, Cox M M J, Farin F, Wohlfahrt S. European patent 0334462. 1989. [Google Scholar]
- 3.Beer H D, Wohlfahrt G, Schmid R D, McCarthy J E G. Analysis of the catalytic mechanism of a fungal lipase using computer aided design and structural mutants. Biochemistry. 1996;319:351–359. doi: 10.1093/protein/9.6.507. [DOI] [PubMed] [Google Scholar]
- 4.Belev T N, Singh M, McCarthy J E G. A fully modular vector system for the optimization of gene expression in Escherichia coli. Plasmid. 1991;26:147–150. doi: 10.1016/0147-619x(91)90056-3. [DOI] [PubMed] [Google Scholar]
- 5.Catoni E, Schmidt-Dannert C, Schmid R D. Overexpression of lipase A and B of Geotrichum candidum in Pichia pastoris: high-level production and some properties of functional expressed lipase B. Biotechnol Tech. 1997;9:689–695. [Google Scholar]
- 6.Chihara-Siomi M, Yoshikawa K, Oshima-Hirayama N, Yamamoto K, Sogabe Y, Nakatani T, Nishioka T, Oda J. Purification, molecular cloning, and expression of lipase from Pseudomonas aeruginosa. Arch Biochem Biophys. 1992;296:505–513. doi: 10.1016/0003-9861(92)90604-u. [DOI] [PubMed] [Google Scholar]
- 7.Chung G H, Lee P Y, Yoo O J, Rhee J S. Overexpression of a thermostable lipase gene from Pseudomonas fluorescens in Escherichia coli. Appl Microbiol Biotechnol. 1991;35:237–241. [Google Scholar]
- 8.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 9.Frenken L G J, Bos J W, Visser C, Muller W, Tommassen J, Verrips C T. An accessory gene, lipB, required for the production of active Pseudomonas glumae lipase. Mol Microbiol. 1993;9:579–589. doi: 10.1111/j.1365-2958.1993.tb01718.x. [DOI] [PubMed] [Google Scholar]
- 10.Frenken L G J, Egmond M R, Batenburg A M, Bos J W, Visser C, Verrips C T. Cloning of the Pseudomonas glumae lipase gene and determination of the active-site residues. Appl Environ Microbiol. 1992;58:3787–3791. doi: 10.1128/aem.58.12.3787-3791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frenken L G J, de Groot A, Tommassen J, Verrips C T. Role of the lipB gene product in the folding of the secreted lipase of Pseudomonas glumae. Mol Microbiol. 1993;9:591–599. doi: 10.1111/j.1365-2958.1993.tb01719.x. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert E J. Pseudomonas lipases: biochemical properties and molecular cloning. Enzyme Microb Technol. 1993;15:634–645. doi: 10.1016/0141-0229(93)90062-7. [DOI] [PubMed] [Google Scholar]
- 13.Hobson A H, Buckley C M, Aamand J L, Jorgensen S T, Diderichsen B, McConnell D J. Activation of a bacterial lipase by its chaperone. Proc Natl Acad Sci USA. 1993;90:5682–5686. doi: 10.1073/pnas.90.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobson A H, Buckley C M, Jörgensen S T, Diderichsen B, McConnell D J. Interaction of the Pseudomonas cepacia DSM3959 lipase with its chaperone, LimA. J Biochem. 1995;118:575–581. doi: 10.1093/oxfordjournals.jbchem.a124948. [DOI] [PubMed] [Google Scholar]
- 15.Hom S S M. European patent application Ep 0 443 063 A1. 1991. [Google Scholar]
- 16.Hom S S M, Scott E M, Atchison R E, Picataggio S, Mielenz J R. Characterization and over-expression of a cloned Pseudomonas lipase gene. GBF Monogr. 1991;16:267–270. [Google Scholar]
- 17.Huge-Jensen B, Andreasen F, Christensen T, Christensen M, Thim L, Boel E. Rhizomucor miehei triglyceride lipase is processed and secreted from transformed Aspergillus oryzae. Lipids. 1989;24:781–785. doi: 10.1007/BF02544584. [DOI] [PubMed] [Google Scholar]
- 18.Ihara F, Kageyama Y, Hirata M, Nihira T, Yamada Y. Purification, characterization, and molecular cloning of lactonizing lipase from Pseudomonas species. J Biol Chem. 1991;266:18135–18140. [PubMed] [Google Scholar]
- 19.Ihara F, Okamoto I, Akao K, Nihira T, Yamada Y. Lipase modulator protein (LimL) of Pseudomonas sp. strain 109. J Bacteriol. 1995;177:1245–1258. doi: 10.1128/jb.177.5.1254-1258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihara F, Okamoto I, Nihira T, Yamada Y. Requirement in trans of the downstream limL gene for activation of lactonizing lipase from Pseudomonas sp. 109. J Ferment Bioeng. 1992;73:337–342. [Google Scholar]
- 21.Iizumi T, Fukase T. Role of the gene encoding lipase activator from Pseudomonas sp. strain KWI-56 in in vitro activation of lipase. Biosci Biotechnol Biochem. 1994;58:1023–1027. doi: 10.1271/bbb.58.1023. [DOI] [PubMed] [Google Scholar]
- 22.Iizumi T, Nakamura K, Shimada Y, Sugihara A, Tominaga Y, Fukase T. Cloning, nucleotide sequencing, and expression in Escherichia coli of a lipase and its activator gene from Pseudomonas sp. KWI-56. Agric Biol Chem. 1991;55:2349–2357. [PubMed] [Google Scholar]
- 23.Jaeger K-E, Ransac S, Dijkstra B W, Colson C, van Heuvel M, Misset O. Bacterial lipases. FEMS Microbiol Rev. 1994;15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 24.Jörgensen S, Skov K W, Diderichsen B. Cloning, sequence, and expression of a lipase gene from Pseudomonas cepacia: lipase production in a heterologous host requires two Pseudomonas genes. J Bacteriol. 1991;173:559–567. doi: 10.1128/jb.173.2.559-567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloster F. European patent 0 574 050 A1. 1993. [Google Scholar]
- 26.Kordel M, Hofman B, Schomburg D, Schmid R D. Extracellular lipase of Pseudomonas sp. strain ATCC 21808: purification, characterization, crystallization, and preliminary X-ray diffraction data. J Bacteriol. 1991;173:4836–4841. doi: 10.1128/jb.173.15.4836-4841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kötting J, Eibl H. Lipases and phospholipases in organic synthesis. In: Woolley P, Petersen S B, editors. Lipases: their structure, biochemistry and application. Cambridge, England: Cambridge University Press; 1994. pp. 289–314. [Google Scholar]
- 28.Laemmli U K. Cleavage of structure proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura K, Iizumi T, Fukase T. Hyperproduction of thermostable lipase by genetically engineered Pseudomonas species. Ann N Y Acad Sci. 1992;672:100–102. [Google Scholar]
- 30.Nakanishi Y, Watanabe H, Washizu K, Narahashi Y, Kurono Y. Cloning, sequencing and regulation of the lipase gene from Pseudomonas sp. M-12-33. Gesellschaft Biotechnol Forsch mbH Monogr. 1991;16:263–266. [Google Scholar]
- 31.Oshima-Hirayama N, Yoshikawa K, Takaaki N, Oda J. Lipase from Pseudomonas aeruginosa. Production in Escherichia coli and activation in vitro with a protein from the downstream gene. Eur J Biochem. 1993;215:239–246. doi: 10.1111/j.1432-1033.1993.tb18028.x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Santaniello E, Ferraboschi P, Grisenti P, Manzocchi A. The biocatalytic approach to the preparation of enantiomerically pure chiral building blocks. Chem Rev. 1992;92:1071–1140. [Google Scholar]
- 34.Schmidt-Dannert C, Rua M L, Atomi H, Schmid R D. Thermoalkalophilic lipase of Bacillus thermocatenulatus. I. Molecular cloning nucleotide sequence, purification and some properties. Biochim Biophys Acta. 1996;1301:105–114. doi: 10.1016/0005-2760(96)00027-6. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt-Dannert C, Sztajer H, Stöcklein W, Menge U, Schmid R D. Screening, purification and properties of a thermophilic lipase from Bacillus thermocatenus. Biochim Biophys Acta. 1994;1214:43–53. doi: 10.1016/0005-2760(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 36.Soberon-Chavez G I, Palmeros B. Pseudomonas lipases: molecular genetics and potential industrial applications. Crit Rev Microbiol. 1994;20:95–105. doi: 10.3109/10408419409113549. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiya A, Nakazawa H, Toida J, Ohnishi K. Cloning and nucleotide sequence of the mono- and diacylglycerol lipase gene (mdlB) of Aspergillus oryzae. FEMS Microbiol Lett. 1996;143:63–67. doi: 10.1111/j.1574-6968.1996.tb08462.x. [DOI] [PubMed] [Google Scholar]
- 38.Vulfson E N. Industrial applications of lipases. In: Woolley P, Petersen S B, editors. Lipases: their structure, biochemistry and application. Cambridge, England: Cambridge University Press; 1994. pp. 271–288. [Google Scholar]
- 39.Wohlfahrt S, Hoessche C, Strunk C, Winkler U. Molecular genetics of the extracellular lipase of Pseudomonas aeruginosa PAO1. J Gen Microbiol. 1992;138:1325–1335. doi: 10.1099/00221287-138-7-1325. [DOI] [PubMed] [Google Scholar]