Abstract
Break induced replication (BIR) is a pathway specialized in repair of double strand DNA breaks with only one end capable of invading homologous template that can arise following replication collapse, telomere erosion or DNA cutting by site-specific endonucleases. For a long time, yeast remained the only model system to study BIR. Studies in yeast demonstrated that BIR represents an unusual mode of DNA synthesis that is driven by a migrating bubble and leads to conservative inheritance of newly synthesized DNA. This unusual type of DNA synthesis leads to high levels of mutations and chromosome rearrangements. Recently, multiple examples of BIR were uncovered in mammalian cells that allowed the comparison of BIR between organisms. It appeared initially that BIR in mammalian cells is predominantly independent of RAD51, and therefore different from BIR that is predominantly Rad51-dependent in yeast. However, a series of systematic studies utilizing site-specific DNA breaks for BIR initiation in mammalian reporters led to the discovery of highly efficient RAD51-dependent BIR, allowing side-by side comparison with BIR in yeast which is the focus of this review.
Keywords: break induced replication, Double-strand break repair, homologous recombination, alternative lengthening of telomeres, Replication stress
Introduction.
DNA Double strand breaks (DSBs) are a dangerous type of lesion that often happen in a living cell due to its exposure to DNA damaging agents or can be also initiated by replication fork collapse. The repair of a DSB can proceed via either non-homologous end-joining that heals breaks by joining two broken ends, or by homologous recombination (HR), which utilizes a homologous DNA region as a template to copy the DNA region that is necessary for repair [1,2]. When two broken DSB ends are available for coordinated invasion of the homologous template, the repair proceeds either by synthesis-dependent strand-annealing (SDSA) mechanism (when the newly synthesized strand gets unwound and annealed to the second end of the DSB or by double-Holliday junction (dHJ) pathway (when the second end of a DSB anneals to the extended D-loop). DSB repair by SDSA and dHJ pathways leads to gene conversion (GC) involving the copying of a short DNA region to “patch” the break (see Figure 1A, left for the example of GC formed by SDSA), and is traditionally considered “error-free” [3]. However, in some situations only one broken DSB end is available for invasion, and the repair of such “one-ended” breaks often proceeds by break-induced replication (BIR) (Figure 1A, right), initiated by the invasion of one broken DNA end into homologous template followed by long DNA synthesis [reviewed in [4–6]].
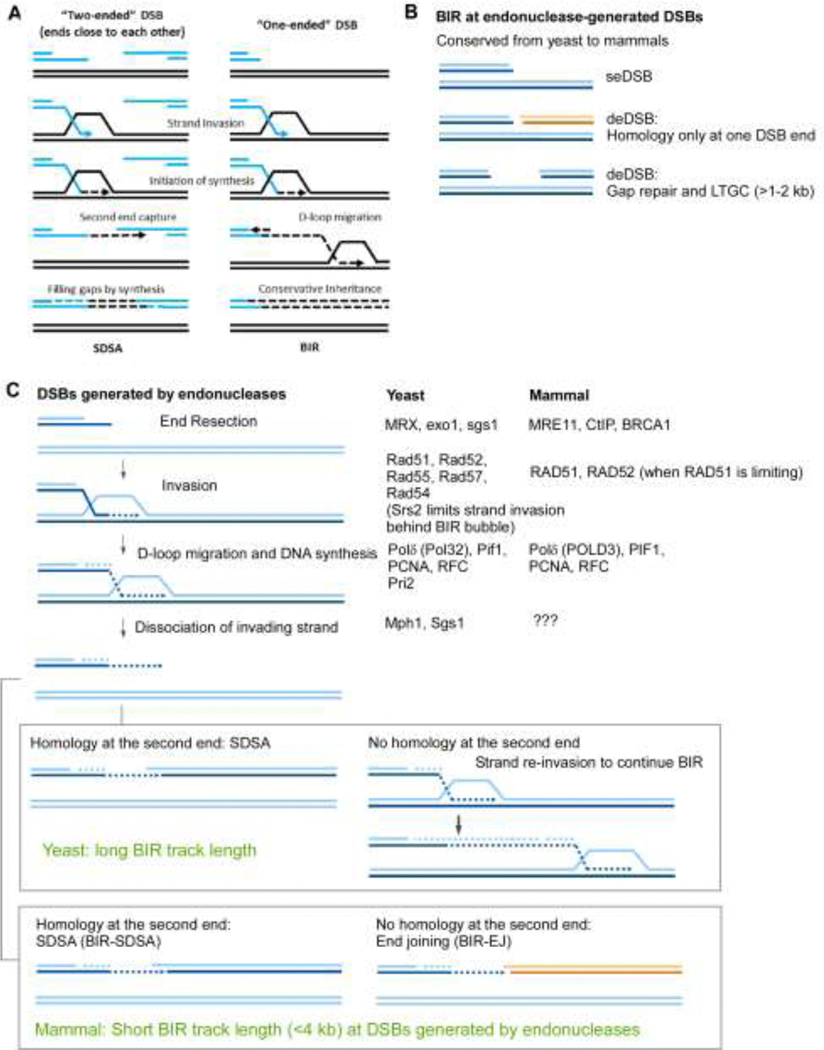
Figure 1. BIR is a conserved mechanism in yeast and mammals.
(A). Schematic drawing of homologous recombination pathways to repair DSBs. (SSA is shown as a representative pathway to repair two-ended DSBs).
(B). The configurations of DSB ends to activate BIR at endonuclease-generated DSBs.
(C). Comparison of BIR at endonuclease-generated DSBs in yeast and mammals. Factors that have been identified important for BIR at endonuclease-generated DSBs are shown.
Break-Induced replication in yeast.
In eukaryotes, BIR has been originally studied in yeast Saccharomyces cerevisiae where several experimental BIR systems were used. The most common way was employing a site-specific endonuclease to induce chromosomal break (Figure 1B) in such a way that only one broken end can invade the homologous region located at an allelic or ectopic position [7–11]. In yeast, two pathways of BIR were described. Both of them require Rad52, the main recombination yeast protein; however one of them also requires Rad51, the main strand invasion protein, while another one is Rad51-independent, but involves Rad59, known to assist Rad52 in single strand annealing [7,9,10,12]. The latter pathway often leads to chromosomal rearrangements [13,14] and therefore is extremely interesting; however, little is known about its molecular details so far, and the mechanistic knowledge that will be presented here predominantly comes from investigation of Rad51-dependent BIR, which is the major pathway in yeast (Figure 1C) [7,10]. From these studies, we know that, different from a replication fork DNA synthesis, BIR is carried by a migrating bubble, with asynchronous synthesis of a leading and lagging strands, and results in conservative inheritance of all newly synthesized DNA [1517].
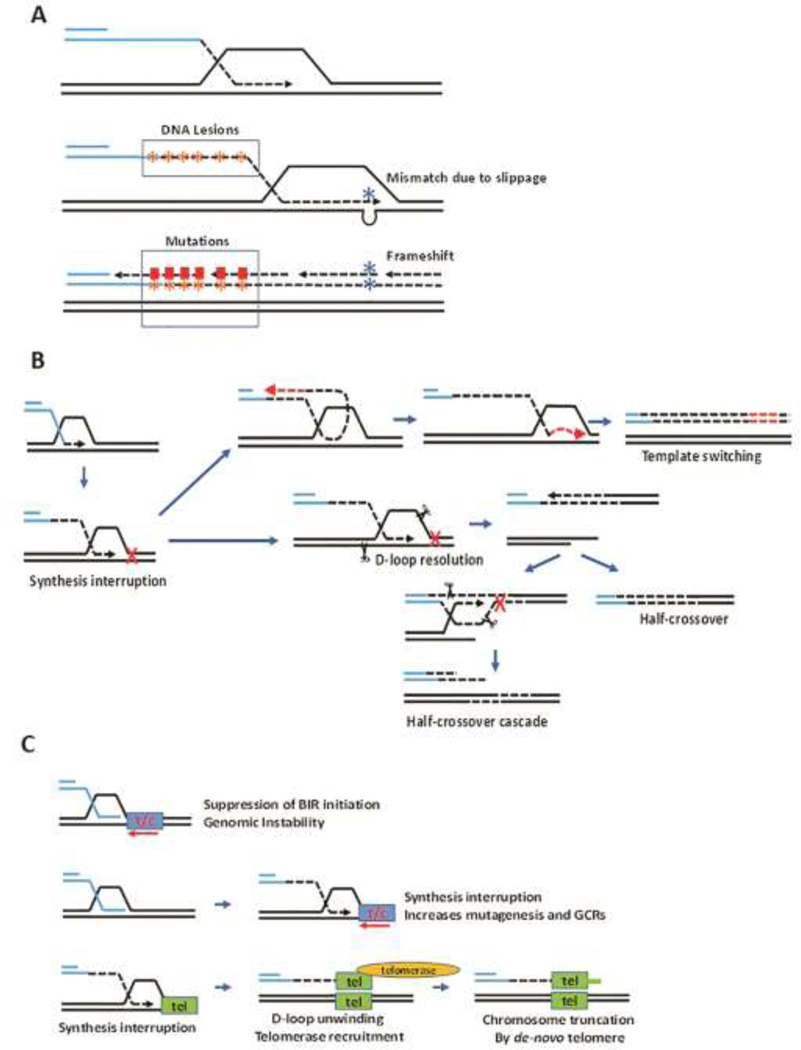
The unusual mode of DNA synthesis is the main cause of genetic instabilities that are frequently associated with BIR. Specifically, the level of mutagenesis is greatly increased resulting from several sources. In particular, base substitutions and frameshift mutations occur hundreds and thousands of times, respectively, more often as compared to S-phase DNA synthesis [15,18]. The main reasons behind increased mutagenesis include unrepaired DNA damage introduced into single-strand (ss) DNA that is accumulated during BIR leading strand synthesis, frequent interruptions of DNA synthesis in the context of a D-loop, and inefficiency of mismatch repair in correction of BIR errors due to rapid dissociation of newly synthesized DNA from its template (Figure 2A) [15,18–20]. When BIR occurs in the presence of ssDNA-specific damaging agents, for example methyl methanesulfonate or APOBEC3A enzyme, it leads to the formation of long mutation clusters [21,22] similar to kataegis described in cancer [23,24]. BIR is also associated with a high level of template switching (Figure 2B) [19,20,25], and frequently leads to chromosome rearrangements explained by a resolution of migrating bubble intermediates leading to translocations as well as to cascades of genome rearrangements (Figure 2B) [22,26–29].
Figure 2. The mechanisms of genetic instabilities associated with BIR in yeast.
(A). Mutations associated with BIR: (i) base substitutions and mutation clusters resulting from unrepaired lesions accumulated in the long ssDNA tract behind the BIR bubble; (ii) frameshifts resulting from leading strand slippage within a D-loop.
(B). Chromosome rearrangements initiated by interruption of BIR synthesis: (i) template switching initiated by dissociation of 3’ end from its template followed by annealing at microhomology in the region of ssDNA accumulated behind the BIR bubble, followed by initiation of DNA synthesis that is eventually disrupted causing a second template switch by reannealing to the original track of BIR through microhomology; (ii) Half-crossovers and half-crossover cascades resulting from the resolution of the migrating bubble leading to fusion of the recipient and donor chromosomes.
(C). Obstacles on a way of BIR promoting genome instability. Top: Initiation of BIR synthesis is suppressed in the vicinity of head-on transcription. Middle: Collision of ongoing BIR with head-on transcription promotes genomic instability. Bottom: passage of BIR through interstitial telomere leads to BIR interruption and capping by de-novo telomere. t/c: transcription unit; tel: interstitial telomere.
Initiation of BIR is preceded by a 5’ to 3’ DSB end resection (Figure 1C) [30] followed by strand invasion and priming of DNA synthesis from the invaded 3’-OH end [reviewed in [5,6,31]]. The kinetics of BIR progression was studied for a while [7,17,32,33], but was discerned only recently by combination of DNA purification preserving the nascent ssDNA and employing sensitive approaches to its detection, including proximal ligation and droplet digital PCR [34,35]. BIR synthesis is initiated ~ 1 hour following strand invasion and proceeds with a rate of ~ 0.5 kb/min that is 6-fold slower than S-phase DNA replication [35]. Also, it was demonstrated that without primase, leading strand synthesis is initiated efficiently, but fails to proceed beyond 30 kb, suggesting that primase is needed for stabilization of the nascent leading strand, probably by synthesizing Okazaki fragments. Also, in the absence of Pol32 (Polδ subunit that is essential for BIR, but not for S-phase DNA replication [36]), BIR synthesis can initiate and proceed for up to 15 kb [34,35], while in the absence of Pif1, the main BIR helicase [16], BIR synthesis can start, but is interrupted within the first 5 kb [35]. In addition, it was demonstrated that interstitial telomeric DNA disrupts and terminates BIR progression (Figure 2C) [35,37]. Also, BIR initiation is suppressed by transcription proportionally to the transcription level (Figure 2C), as it was determined by following BIR initiation and progression using droplet digital PCR [35]. In addition, it was observed that collisions between ongoing BIR and transcription lead to mutagenesis and chromosome rearrangements at levels that exceed instabilities induced by transcription during normal replication [35]. Polymerase δ is responsible for synthesizing both leading and lagging BIR strands [38]. Polymerase ε is not required for BIR initiation [36,38], but might participate later on the track of BIR synthesis according to some reports [35,36].
The high level of genetic instabilities resulting from BIR makes it important to channel the repair of two-ended breaks from BIR to gene conversion (GC). Although DNA synthesis associated with GC presents similarities with BIR in its mechanism and the level of mutagenesis [32,39,40], its length is shorter, which makes gene conversion a “safer” choice as compared to BIR. Recent studies [41,42] identified several mechanisms preventing the repair of two-ended DSBs by BIR and channeling them into gene conversion, including (a) second end capture promoted by ssDNA annealing proteins Rad52 and Rad59; (b) D-loop unwinding by Mph1 helicase; (c) synchronous resection of two DSB ends promoted by Mre11-Rad50-Xrs2 complex; and (d) heterochromatic silencing of the template mediated by Sir2. In addition, several factors promote BIR repair, including formation of a hairpin structure by one end or DNA damage checkpoint deficiency [43,7]. Together, BIR is the predominant pathway to repair one-ended DSBs, while its usage for the repair of two-ended breaks is suppressed unless only one end can be engaged.
BIR mechanism is conserved from yeast to mammals.
To model BIR in mammalian cells, EGFP-based reporters have been designed with one available homology end by inserting the recipient and donor cassettes in a reversed orientation [44] or with a long gap (3.8 kb) between two homologous ends [45]. In both systems, endonuclease-induced recombination is dependent on POLD3, the ortholog of yeast Pol32, suggesting that BIR is utilized [44,45]. POLD3-dependent BIR is also used for long-range repeat-mediated deletion when one repeat is present at the DSB end as assayed in another reporter system [46]. In mammalian cells, long track gene conversion (LTGC) refers to gene conversion with track length longer than 1–2 kb [47,48], similar to gap repair in yeast with a large gap size [3]. Consistent with BIR being used for gap repair of the gap sizes greater than 1–2 kb in yeast [32,41], LTGC in mammalian cells is also mediated by BIR [45]. Hence, the mechanism to activate BIR at endonuclease-generated DSBs is conserved, and single DSB ends or two far-apart DSB ends are the signals to be sensed to activate BIR (Figure 1B).
In the absence of PIF1, BIR track length is significantly reduced, suggesting a conserved function of mammalian PIF1 in promoting processive BIR replication [45]. However, while BIR in yeast can proceed for hundreds of kbs to the end of a chromosome [7,10], BIR track length in mammalian cells rarely exceeds 4 kbs at endonuclease-generated DSBs as determined by analyzing BIR events using EGFP-BIR reporters [44,45] (Figure 1C). Another difference is that while being initiated by homologous strand invasion, BIR can be terminated by either SDSA (BIR-SDSA) or end joining (BIR-EJ) in mammalian cells [44,45], but BIR in yeast is either completed by SDSA or proceeds to the end of the chromosome [5]. One possibility is that BIR replisomes are less processive and more easily disassociated from the templates in mammalian cells, and along with more robust end joining activity than in yeast, displaced ends may have a better chance to be ligated to the second break end [44,45,50], rather than repeatedly reinvading the templates as observed in yeast [20] (Figure 1C).
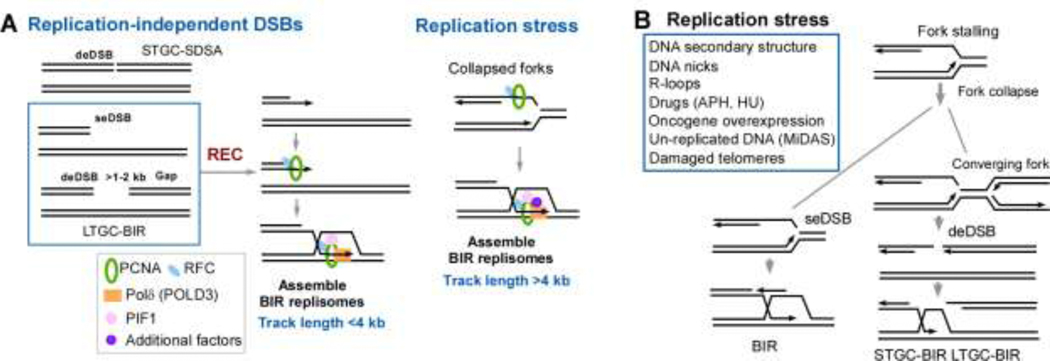
Oncogene overexpression causes replication stress [5,6,51] and induces POLD3-dependent DNA synthesis [44]. Replication restart from collapsed forks in Xenopus egg extracts and in mammalian cells is mediated by BIR [45,52]. Fork breakage induced by Cas9 nickase (Cas9n) or a common fragile site (CFS)-derived structure-prone DNA sequence (Flex1) leads to more frequent use of LTGC than STGC, whereas at DSBs directly generated by Cas9, STGC is used more often [45]. Furthermore, at Cas9-induced DSBs, only LTGC but not STGC is mediated by BIR, but at broken replication forks, both STGC and LTGC utilize BIR. Different from short BIR track length (<4 kb) at endonuclease-generated DSBs in mammalian cells, most BIR events at broken forks are longer than ~4 kb, but the exact track length remains to be determined [45] (Figure 3A).
Figure 3. BIR is involved in repairing DSBs associated with replication stress.
(A). Activation of BIR is differently regulated at DSBs generated by endonucleases (replication-independent) and replication-associated DSBs. REC is only required for BIR activation at replication-independent DSBs.
(B). Replication stress induced by various causes activation of BIR not only at single-ended DSBs (seDSBs) but also at double-ended DSBs (deDSBs) generated after fork converging.
The use of BIR to cope with replication stress is also supported by its involvement in mitotic DNA synthesis (MiDAS), which often occurs at under-replicated CFSs after MUS81 cleavage [53]. MiDAS uses a conservative form of DNA synthesis and depends on POLD3 [53,54]. In yeast, R-loops and structure-prone sequences cause replication fork collapse and activate BIR [43,55,56], and similar R-loops and DNA secondary structures are implicated for inducing CFS breakage in mammalian cells [57]. BIR is also involved in maintaining telomeres by alternative lengthening of telomeres (ALT) [58,59]. However, break-induced telomere synthesis is not unique to ALT cells [58] and can be induced by aphidicolin in non-ALT cells [60]. Fragile telomere formation in BLM- or TRF1-deficient cells due to failure in removing G-quadruplexs (G4) at lagging-strand telomere also involves BIR [61]. Collectively, BIR is a common mechanism for repairing DSBs associated with replication stress (Figure 3B).
A “Recombination execution checkpoint” (REC) was proposed to determine the use of GC or BIR in yeast: if the homologies to the two DSB ends are close to each other, GC is activated, but if homology is only detected at one DSB end, BIR is activated [32]. Based on the recent study, REC is likely operated at a step after strand invasion and the initiation of DNA synthesis [35]. We propose that assembling BIR replisomes, where PIF1 is an important component, may be an important step to activate BIR, but the exact mechanism remains to be determined. In mammalian cells, we speculate that REC is similarly operated to launch STGC or LTGC/BIR at replication-independent DSBs, but at broken forks, BIR appears to be activated immediately without checking the DSB ends (Figure 3A). One model is that PCNA and RFC, which are required for BIR [45,52,58], are already present and modified on forks upon replication stress to activate BIR (Figure 3A, right), but at replication-independent DSBs, PCNA and RFC are not initially present and need to be recruited and/or modified after REC detects a need for BIR (Figure 3A, left). At broken forks, additional accessory factors are likely recruited to make BIR replisomes more processive than that at replication-independent DSBs.
Based on the study using reporter systems, BIR with substantial homology (1.3 kb or 0.3 kb) at one DSB end is dependent on RAD51, BRCA1, BRCA2, MRE11 and CtIP, suggesting a highly efficient RAD51-dependent BIR is operated in mammalian cells [45,62] (Figure 1C). However, MiDAS exhibits independence of RAD51 and BRCA2, but requires RAD52 [54]. Different from Rad52 in yeast, RAD52 is not essential for HR in mammalian cells as BRCA2 serves as the RAD51 mediator instead of RAD52 [63]. At the point where cells are undergoing mitosis, end resection and RAD51 filament formation are compromised [64], and conceivably this results in RAD51-independent and RAD52-dependent MiDAS. RAD52-dependent BIR is also a dominant mechanism for ALT and break-induced telomere synthesis, but a RAD52independent pathway also exists [58,65–68]. RAD52 is recruited to ALT telomeres under replication stress at telomeres, and is required for spontaneous ALT telomere synthesis and maintenance, but is dispensable for DSB-induced telomere synthesis [65]. The dominant role of RAD52 in ALT may be attributed to its function to promote telomeric single-strand DNA annealing and D-loop formation at telomere sequences [66].
Conclusions and speculation
While BIR has been more extensively studied in yeast, similar RAD51-dependent and RAD51-independent BIR pathways have been found in mammalian cells, but the detailed mechanisms still require further investigation, especially for RAD51-independent and RAD52-dependent BIR. In yeast, BIR is associated with a hyper mutation rate, APOBEC-induced mutagenesis and complex chromosome rearrangements that are frequently observed in cancer, making it important to evaluate the mutagenic consequence of BIR directly in mammalian cells. Study of copy number variation (CNV) associated with human diseases leads to the proposal of microhomology-mediated break-induced replication (MMBIR) [69]. In yeast, interruption of BIR triggers a switch from BIR to MMBIR [19] and similar microhomology-mediated template switching was also observed in association with gene conversion [70]. It would be extremely interesting to study whether similar microhomology-mediated mechanisms are involved in mammalian cells. While replication stress appears as a key factor to induce BIR at replication forks, under-replicated DNA at CFSs and damaged telomeres in mammalian cells, in yeast, BIR at broken forks seems to be suppressed by converging forks [71]. Side-by-side studies in yeast and mammalian cells will surely uncover new mechanisms governing BIR regulation in response to replication stress. As BIR is stimulated by oncogenic stress to mediate replication restart that is critical for cancer cell survival [45], further exploration of BIR will provide new cancer therapy strategies.
Acknowledgements
AM is funded by NIH grants R35 GM127006, R01 CA232425 and R21 ES030307. XW is funded by R01 CA244912, R01 CA187052 and R35 GM141868.
Footnotes
Declaration of Interests
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
References and recommended reading. Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
- 1.Heyer WD: Regulation of recombination and genomic maintenance. Cold Spring Harb Perspect Biol 2015, 7:a016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jasin M, Rothstein R: Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 2013, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paques F, Haber JE: Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 1999, 63:349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kramara J, Osia B, Malkova A: Break-Induced Replication: The Where, The Why, and The How. Trends Genet 2018, 34:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llorente B, Smith CE, Symington LS: Break-induced replication: what is it and what is it for? Cell Cycle 2008, 7:859–864. [DOI] [PubMed] [Google Scholar]
- 6.Anand RP, Lovett ST, Haber JE: Break-induced DNA replication. Cold Spring Harb Perspect Biol 2013, 5:a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE: RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol 2005, 25:933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosco G, Haber JE: Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 1998, 150:1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malkova A, Ivanov EL, Haber JE: Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A 1996, 93:7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis AP, Symington LS: RAD51-dependent break-induced replication in yeast. Mol Cell Biol 2004, 24:2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrow DM, Connelly C, Hieter P: “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 1997, 147:371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Signon L, Malkova A, Naylor ML, Klein H, Haber JE: Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol 2001, 21:2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A: Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Molecular and Cellular Biology 2007, 27:2601–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downing B, Morgan R, VanHulle K, Deem A, Malkova A: Large inverted repeats in the vicinity of a single double-strand break strongly affect repair in yeast diploids lacking Rad51. Mutat Res 2008, 645:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A: Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 2013, 502:389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. : Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 2013, 502:393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnianni RA, Symington LS: Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci U S A 2013, 110:13475–13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A: Break-Induced Replication Is Highly Inaccurate. Plos Biology 2011, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakofsky CJ, Ayyar S, Deem AK, Chung WH, Ira G, Malkova A: Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Mol Cell 2015, 60:860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith CE, Llorente B, Symington LS: Template switching during break-induced replication. Nature 2007, 447:102–105. [DOI] [PubMed] [Google Scholar]
- 21.Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Malkova A: Break-Induced Replication Is a Source of Mutation Clusters Underlying Kataegis. Cell Reports 2014, 7:1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elango R, Osia B, Harcy V, Malc E, Mieczkowski PA, Roberts SA, Malkova A: Repair of base damage within break-induced replication intermediates promotes kataegis associated with chromosome rearrangements. Nucleic Acids Research 2019, 47:96669684. •This study demonstrates that expression of APOBEC3A during BIR in yeast promotes formation of mutation clusters similar to kataegis.
- 23.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al.: Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 2012, 46:424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. : Mutational processes molding the genomes of 21 breast cancers. Cell 2012, 149:979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anand RP, Tsaponina O, Greenwell PW, Lee CS, Du W, Petes TD, Haber JE: Chromosome rearrangements via template switching between diverged repeated sequences. Genes & Development 2014, 28:2394–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Malkova A: Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep 2014, 7:1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deem A, Barker K, Vanhulle K, Downing B, Vayl A, Malkova A: Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 2008, 179:1845–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CE, Lam AF, Symington LS: Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol 2009, 29:1432–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasan S, Deem A, Ramakrishnan S, Argueso JL, Malkova A: Cascades of genetic instability resulting from compromised break-induced replication. PLoS Genet 2014, 10:e1004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G: Defective Resection at DNA Double-Strand Breaks Leads to De Novo Telomere Formation and Enhances Gene Targeting. Plos Genetics 2010, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakofsky CJ, Malkova A: Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit Rev Biochem Mol Biol 2017, 52:395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE: A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev 2009, 23:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain S, Sugawara N, Mehta A, Ryu T, Haber JE: Sgs1 and Mph1 Helicases Enforce the Recombination Execution Checkpoint During DNA Double-Strand Break Repair in Saccharomyces cerevisiae. Genetics 2016, 203:667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piazza A, Shah SS, Wright WD, Gore SK, Koszul R, Heyer WD: Dynamic Processing of Displacement Loops during Recombinational DNA Repair. Mol Cell 2019, 73:1255–1266 e1254. •This study characterized the dynamics of the initiation of BIR syntehsis in pol32Δ and in POL32 (wild type) yeast cells.
- 35. Liu L, Yan Z, Osia BA, Twarowski J, Sun L, Kramara J, Lee RS, Kumar S, Elango R, Li H, et al. : Tracking break-induced replication shows that it stalls at roadblocks. Nature 2021, 590:655–659 •This study determines the rate and kinetics of BIR progression in yeast, the extent of BIR synthesis in primase-deficient and other BIR-defective mutants, and also characterizes interruption of BIR at interstitial telomere sequences and at highly transcribed regions.
- 36. Lydeard JR, Jain S, Yamaguchi M, Haber JE: Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 2007, 448:820–823. •This study identifies several proteins including Poldelta and its subunit Pol32, Polalphaprimase complex and Polepsilon as required or important for BIR. This study also demonstrates Pol32-dependence of alternative lengthening of telomeres.
- 37. Stivison EA, Young KJ, Symington LS: Interstitial telomere sequences disrupt break-induced replication and drive formation of ectopic telomeres. Nucleic Acids Res 2020, 48:12697–12710. Res 48, 12697–12710 •This study demonstrates interruption of BIR at interstitial telomere sequences.
- 38. Donnianni RA, Zhou ZX, Lujan SA, Al-Zain A, Garcia V, Glancy E, Burkholder AB, Kunkel TA, Symington LS: DNA Polymerase Delta Synthesizes Both Strands during Break-Induced Replication. Molecular Cell 2019, 76:371–381 •This study demonstrates that polymerase delta synthesizes both leading and lagging strands during BIR in yeast.
- 39.Hicks WM, Kim M, Haber JE: Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 2010, 329:82–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ira G, Satory D, Haber JE: Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol Cell Biol 2006, 26:9424–9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mehta A, Beach A, Haber JE: Homology Requirements and Competition between Gene Conversion and Break-Induced Replication during Double-Strand Break Repair. Mol Cell 2017, 65:515–526 e513. •This study demonstrates that D-loop unwinding by Mph1 helicase represents the critical step preventing channeling of repair of two-ended DSBs into BIR.
- 42. Pham N, Yan Z, Yu Y, Faria Afreen M, Malkova A, Haber JE, Ira G: Mechanisms restraining break-induced replication at two-ended DNA double-strand breaks. EMBO J 2021:e104847. •This study identifies second end capture promoted by ssDNA annealing proteins as a critical mechanism preventing the repair of two-endended double-strand breaks by BIR. In addition, it shows that synchronous resection of two DSB ends and heterochromatic silencing of the template contribute to the inhibition of BIR.
- 43.Ramakrishnan S, Kockler Z, Evans R, Downing BD, Malkova A: Single-strand annealing between inverted DNA repeats: Pathway choice, participating proteins, and genome destabilizing consequences. PLoS Genet 2018, 14:e1007543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD: Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 2014, 343:88–91. •This study demonstrates that replication stress caused by cyclin E overexpression induces POLD3-dependent processive DNA synthesis (BIR) that leads to segmental genomic duplications.
- 45. Li S, Wang H, Jehi S, Li J, Liu S, Wang Z, Truong L, Chiba T, Wang Z, Wu X: PIF1 helicase promotes break-induced replication in mammalian cells. EMBO J 2021:e104509. •Authors establish EGFP-based reporters to systematically study BIR in mammalian cells and find that PIF1 has a conserved role in BIR. LTGC is mediated by BIR and BIR is RAD51-dependent in interphase cells. BIR activation is controlled differently at DSBs generated by endonucleases versus those at broken forks induced by structure-prone DNA sequences upon oncogene overexpression.
- 46.Hu Q, Lu H, Wang H, Li S, Truong L, Li J, Liu S, Xiang R, Wu X: Break-induced replication plays a prominent role in long-range repeat-mediated deletion. EMBO J 2019:e101751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson RD, Jasin M: Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 2000, 19:3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puget N, Knowlton M, Scully R: Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst) 2005, 4:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu Q, Lu H, Wang H, Li S, Truong L, Li J, Liu S, Xiang R, Wu X: Break-induced replication plays a prominent role in long-range repeat-mediated deletion. EMBO J 2019, 38:e101751. •This paper shows that SSA is mediated by BIR when the repeats are far apart and when the DSB is close to one repeat.
- 50.Willis NA, Frock RL, Menghi F, Duffey EE, Panday A, Camacho V, Hasty EP, Liu ET, Alt FW, Scully R: Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature 2017, 551:590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malkova A, Ira G: Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev 2013, 23:271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimoto Y, Puddu F, Costanzo V: RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol 2012, 19:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID: Replication stress activates DNA repair synthesis in mitosis. Nature 2015, 528:286–290. [DOI] [PubMed] [Google Scholar]
- 54.Bhowmick R, Minocherhomji S, Hickson ID: RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell 2016, 64:1117–1126. [DOI] [PubMed] [Google Scholar]
- 55.Amon JD, Koshland D: RNase H enables efficient repair of R-loop induced DNA damage. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim JC, Harris ST, Dinter T, Shah KA, Mirkin SM: The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat Struct Mol Biol 2017, 24:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhowmick R, Hickson ID: The “enemies within”: regions of the genome that are inherently difficult to replicate. F1000Res 2017, 6:666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA: Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016, 539:54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roumelioti FM, Sotiriou SK, Katsini V, Chiourea M, Halazonetis TD, Gagos S: Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep 2016, 17:1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ozer O, Bhowmick R, Liu Y, Hickson ID: Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget 2018, 9:15836–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimmermann M, Kibe T, Kabir S, de Lange T: TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev 2014, 28:2477–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da-Re C, Huber F, Padayachy L, Tardy S, Nicati NL, Barriot S, et al. : Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol Cell 2016, 64:1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu J, Heyer WD: Who’s who in human recombination: BRCA2 and RAD52. Proc Natl Acad Sci U S A 2011, 108:441–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hustedt N, Durocher D: The control of DNA repair by the cell cycle. Nat Cell Biol 2016, 19:1–9. [DOI] [PubMed] [Google Scholar]
- 65.Verma P, Dilley RL, Zhang T, Gyparaki MT, Li Y, Greenberg RA: RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev 2019, 33:221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang JM, Yadav T, Ouyang J, Lan L, Zou L: Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep 2019, 26:955–968 e953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Min J, Wright WE, Shay JW: Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes. Mol Cell Biol 2017, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Min J, Wright WE, Shay JW: Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev 2019, 33:814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hastings PJ, Ira G, Lupski JR: A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 2009, 5:e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsaponina O, Haber JE: Frequent Interchromosomal Template Switches during Gene Conversion in S. cerevisiae. Mol Cell 2014, 55:615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR, Ira G: DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science 2015, 349:742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]