SUMMARY
Enzyme-mediated chemical modifications of nucleic acids are indispensable regulators of gene expression. Our understanding of the biochemistry and biological significance of these modifications has largely been driven by an ever-evolving landscape of technologies that enable accurate detection, mapping, and manipulation of these marks. Here we provide a summary of recent technical advances in the study of nucleic acid modifications with a focus on techniques that allow accurate detection and mapping of these modifications. For each modification discussed (N6-methyladenosine, 5-methylcytidine, inosine, pseudouridine, and N4-acetylcytidine), we begin by introducing the “gold standard” technique for its mapping and detection, followed by a discussion of techniques developed to address any shortcomings of the gold standard. By highlighting the commonalities and differences of these techniques, we hope to provide a perspective on the current state of the field and to lay out a guideline for development of future technologies.
INTRODUCTION
Nucleic acids (RNA and DNA) often carry chemical modifications, with modifications of RNA more numerous and chemically diverse than those of DNA. These modifications can influence the flow of genetic information, which adds a crucial layer of regulation to gene expression (Smith and Meissner, 2013; Roundtree et al., 2017a). Therefore, the ability to map the location of these modifications in the genome/transcriptome is of great interest to this field. Installation and removal of modifications are tightly controlled by several enzymes. The installation process, commonly referred to as modification “writing,” is typically governed by the sequence context (Schibler et al., 1977; Gruenbaum et al., 1982; Dominissini et al., 2012) or the structural context (Bass and Weintraub, 1988) of the base to be modified. Frequently, these modifications take the form of methylations on one of the base pairing surfaces (Watson-Crick-Franklin, Hoogsteen, or sugar), although other modifications can be more elaborate, such as deamination of adenosine to inosine (I) (Figure 1A). Modification produces a chemical signature that is unique compared with the original, unmodified base, often affecting the base pairing (Oakes et al., 2017), reactivity (Frommer et al., 1992), or recognition of the base by protein partners (Wang et al., 2014).
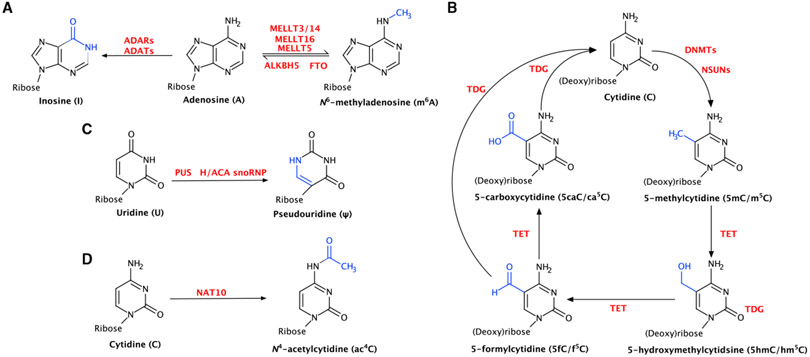
Figure 1. Biogenesis reactions for each modification discussed.
(A) Generation of m6A and inosine from adenosine.
(B) Generation of 5-methylcytidine from cytidine and oxidation reactions of 5-methylcytidine.
(C) Generation of ψ from uridine.
(D) Generation of ac4C from cytidine.
Sequencing technologies capitalize on these unique chemical signatures to differentiate modified from unmodified bases, which has allowed the location of these bases to be mapped. For example, 5-methylcytidine (5mC when in DNA, m5C when in RNA), the most abundant modification in the mammalian genome (Li and Zhang, 2014), does not react with the deaminating reagent sodium bisulfite, whereas unmodified cytidines are changed to uridine upon bisulfite treatment (Frommer et al., 1992). This reactivity difference resulted in creation of bisulfite sequencing (BS-seq), a benchmark technique for mapping the location of DNA methylations. Additionally, some modifications can be detected via enrichment with a modification-specific antibody, such as in the case of methylated RNA immunoprecipitation sequencing (meRIP-seq)/N6-methyladenosine (m6A)-specific immunoprecipitation coupled with high-throughput sequencing (m6A-IP-seq) (Dominissini et al., 2012; Meyer et al., 2012), which is used for transcriptome-wide mapping of m6A, the most abundant internal modification in mammalian poly(A) RNA (Wei et al., 1975b). Techniques such as these have allowed an explosion of research in the fields of epigenetics and epitranscriptomics.
Although these techniques are the gold standard for mapping and identification of their target modification, they are not without shortcomings. BS-seq, although highly useful for detection of cytidines modified at the fifth position, is unable to distinguish 5mC from 5-hydroxymethylcytidine (5hmC), its ten-eleven translocation (TET)-mediated oxidation product (Huang et al., 2010), which has been proposed to have its own epigenetic identity (Kohli and Zhang, 2013). Additionally, methods that rely on antibodies, such as meRIP-seq/m6A-IP-seq, are plagued by problems of antibody cross-reactivity. To address these shortcomings, there is a constant stream of new technologies being developed that can supplement use of these gold standard techniques (Hussain et al., 2013a; Li et al., 2016; Knutson and Heemstra, 2021). Using these techniques, researchers can make novel and important discoveries about the modifications in question, which further spurs technical innovation geared toward not only mapping and detection of these bases but also toward determining the occupancy (stoichiometry) of the modification or toward directly manipulating its presence at a given site.
In this article, we will summarize the current state of technology used for mapping (detection) of several nucleic acid modifications that are abundant in DNA and/or mRNA: m6A, 5mC in DNA, m5C in RNA, and inosine, in addition to two modifications whose roles in mRNA have been investigated more recently: pseudouridine and N4-acetylcytidine. We provide an overview of the underlying principle of each technique and an assessment of its strengths and drawbacks. A summary of all technologies discussed can be found in Table 1. Additionally, we will draw attention to the similarities between techniques developed to study different modifications, revealing the underlying principles on which future technologies can be based. With this overview, we hope to provide a summary of the current state of technology in the fields of epigenetics and epitranscriptomics and to inspire development of future technologies utilizing these principles.
Table 1.
Summary of all technologies discussed
| Technique | Principle | Advantages | Disadvantages | Citation(s) | |
|---|---|---|---|---|---|
| N6-methyladenosine (m6A) | meRIP-seq/m6A-IP-seq | enrichment of m6A-containing fragments via IP with anti-m6A antibody | benchmark technique in this field | antibody cross-reactivity, relatively high input, can’t provide occupancy, low resolution | Meyer et al., 2012; Dominissini et al., 2012 |
| miCLIP-seq/PA-m6A-seq | enrichment of m6A-containing fragments via CLIP with anti-m6A antibody | cross-linking gives m6A a “unique chemical signature” that increases resolution | antibody cross-reactivity, can’t provide occupancy | Linder et al., 2015; Grozhik et al., 2017; Chen et al., 2015 | |
| m6A-LAIC-seq | enrichment of m6A-containing full length RNAs via IP with anti-m6A antibody | isoform aware because of full-length RNA input, spike-ins allow measurement of percent modified RNA | antibody cross-reactivity, doesn’t give information about occupancy of specific sites | Molinie et al., 2016 | |
| MAZTER-seq/m6A-REF-seq | utilizes the differential activity of the MazF nuclease toward m6A/A to distinguish the two | antibody-independent methods, can give a measure of occupancy | MazF only cuts at ACA sites, which are only ~25% of all m6A sites | Garcia-Campos et al., 2019; Zhang et al., 2019 | |
| DART-seq | targets APOBEC1 to m6A sites with a fused YTH domain to leave a C-to-U scar nearby | antibody-independent method, low input | YTH domain of YTHDF2 may have sequence preferences for m6A recognition | Meyer, 2019 | |
| m6A-SEAL-seq | converts m6A to hm6A with FTO, then further modifies with DTT to allow enrichment | antibody-independent method, adaptable to other modifications | low resolution | Wang et al., 2020 | |
| m6A-label-seq | uses allyl-SeAM to produce an m6A derivative that induces RT misincorporations | antibody-independent method | allyl-SeAM may disrupt cellular metabolism, imperfect misincorporation rate | Shu et al., 2020 | |
| SCARLET | measures the ratio of m6A/A at a specific site in RNA using RNase H cleavage and TLC | high specificity of RNaseH cleavage | requires radiation, complex protocol, low throughput | Liu et al., 2013 | |
| SELECT | measures the ratio of m6A/A at a specific site in RNA using differential ligation | easier to implement that SCARLET | low throughput | Xiao et al., 2018 | |
| 5-methylcytosine (5mC/m5C) | BS-seq | utilizes chemical conversion of unmodified Cs detectable by sequencing | benchmark technique in this field | cannot distinguish between 5mC and TET-mediated oxidation products, requires harsh reaction conditions that limit its usefulness for RNA | Frommer et al., 1992 |
| oxBS-seq | chemical oxidation with KRuO4 converts 5hmC to 5fC, which is susceptible to bisulfite deamination | allows differentiation of 5mC and 5hmC | cannot distinguish between 5mC and TET-mediated oxidation products, requires harsh reaction conditions that limit its usefulness for RNA | Booth et al., 2012 | |
| TAB-seq | utilizes the enzymatic activity of TET to convert 5mC to 5caC (which is susceptible to bisulfite) while protecting 5hmC from deamination using βGT | allows differentiation of 5mC and 5hmC | cannot distinguish between 5mC and TET-mediated oxidation products, requires harsh reaction conditions that limit its usefulness for RNA | Yu et al., 2012 | |
| meDIP-seq/m5C meRIP | enrichment of methylated DNA/RNA fragments via IP with anti-5mC/m5C antibody | bisulfite-free method, more amenable to RNA use | antibody cross-reactivity, can’t provide occupancy | Weber et al., 2005 Edelheit et al., 2013 | |
| ACE-seq | utilizes differential deamination by APOBEC to differentiate C and 5mC from 5hmC | can distinguish between 5mC and 5hmC, does not require bisulfite treatment | complex protocol, not currently amenable for RNA | Schutsky et al., 2018 | |
| TAPS | utilizes the enzymatic activity of TET to convert 5mC and 5hmC to 5caC followed by borane reduction, used in combination with βGT protection or KRuO4 oxidation | can distinguish between 5mC and 5hmC, does not require bisulfite treatment | complex protocol, not currently amenable for RNA | Liu et al., 2019c | |
| Aza-IP-seq | 5-azaC analog covalently traps methyltransferases, allowing RNA targets to be identified by pull-down of transferase and sequencing | covalent trapping allows for accurate identification of RNA targets | better suited for some methyltransferases (single cysteine type) than others | Khoddami and Cairns, 2013 | |
| miCLIP | mutant methyltransferases covalently trap RNA targets, allows capture | covalent trapping allows accurate identification of RNA targets | requires overexpression of mutant methyltransferase | Hussain et al., 2013b | |
| Inosine (I) | Inosine mutational profiling (RDD) | Detecting A > G mutations in sequencing (I reads as G in sequencing) | benchmark technique in this field | computationally difficult to differentiate true positives from false positives | Levanon et al., 2004; Li et al., 2011; Bahn et al., 2012; Peng et al., 2012 |
| ICE-seq | modifying inosine with acetylnitrile to induce RT stops | more robust than standard mutational profiling | often unreliable for sites with low (>10%) or high (near 100%) editing frequency | Suzuki et al., 2015 | |
| iSeq | differentiating I from G using glyoxal modification and RNase T1 cleavage | more robust than standard mutational profiling | unsuitable for clusters of inosines because of small fragment size | Cattenoz et al., 2013 | |
| EndoVIPER-seq | using EndoV-MBP fusion protein in the presence of Ca2+ (to prevent catalysis) to selectively bind and IP inosine-containing transcripts | relatively easy to perform, does not require further chemical modification of RNA | cannot give occupancy at specific sites | Knutson et al., 2020 | |
| Pseudouridine (ψ) | Pseudo-seq/CeU-seq/PSI-seq/ψ-seq | chemically modifying ψ with CMCT to cause RT stops | allows high-throughput identification of ψ sites | removal of CMC from other bases may be incomplete | Carlile et al., 2014; Lovejoy et al., 2014; Schwartz et al., 2014; Li et al., 2015 |
| HydraPsiSeq | cleaves RNA at uridine sites (but not ψ) through hydrazine treatment | allows high-throughput identification and occupancy measurement of ψ sites | high read depth required, prone to false positives from highly structured uridines and modified uridines | Marchand et al., 2020 | |
| N4-acetylcytidine (ac4C) | acRIP-seq | enrichment of ac4C-containing fragments via IP | allows high-throughput identification of ac4C sites | antibody cross-reactivity, can’t provide occupancy, low resolution | Arango et al., 2018 |
| ac4C-seq | reduction of ac4C with NaCNB3, resulting in mutational profiling | antibody-independent method | unable to map ac4C in the presence of certain other modifications that also react with NaCNB3 | Sas-Chen et al., 2020 |
N6-METHYLADENOSINE (m6A)
m6A is the most abundant internal modification in polyadenylated (poly(A)) RNA) in mammals, found on approximately 0.2%–0.6% of all adenosines in poly(A) RNA (Dominissini et al., 2012; Meyer et al., 2012). m6A is the result of a methylation at the N6 position on the Watson-Crick-Franklin (WCF) surface of adenosine (Figure 1A). Generally, m6A is installed in poly(A) RNA by the METTL3/14 writer complex (Bokar et al., 1997; Liu et al., 2014), although recent research has found that METTL16 can install m6A on certain mRNAs, long noncoding RNAs (lncRNAs), and the U6 small nuclear RNA (snRNA) (Warda et al., 2017), and work by others has found that METTL5 specifically installs m6A in 18S rRNA (van Tran et al., 2019). When installed, m6A can be removed by one of two eraser enzymes, either AlkB homolog protein 5 (ALKBH5) (Zheng et al., 2013) or fat mass and obesity-associated protein (FTO; also known as ALKBH9) (Jia et al., 2011; Figure 1A). The substrate repertoire of FTO also includes m6Am (N6,2′-O-dimethyladenosine) in poly(A) RNA and snRNA (Mauer et al., 2017, 2019) and N1-methyladenosine (m1A) in tRNA (Wei et al., 2018).
The biological effects of m6A are mediated by proteins that bind it, referred to as reader proteins. In mammals, several m6A reader proteins have been identified, most of which are in one of two groups. The first group of proteins contains the canonical m6A-recognizing YT521-B homology (YTH) domain: the YTHDC proteins (YTHDC1 and YTHDC2) and the YTHDF proteins (YTHDF1-3). The other group contains the K homology (KH) domains of the IGF2BP family (Huang et al., 2018). Additionally, the eukaryotic initiation factor eIF3 and the protein FMRP bind m6A through still undiscovered mechanisms (Meyer et al., 2015; Edupuganti et al., 2017).
In humans, YTHDF2 was the first YTH protein characterized as an m6A reader. Upon binding to an m6A-containing mRNA, it recruits the CCR4-NOT (carbon catabolite repression - negative on TATA-less) deadenylase complex to trigger mRNA degradation (Du et al., 2016). In contrast, YTHDF1 and YTHDF3 were initially proposed to bind m6A-containing RNAs and promote their translation (Wang et al., 2015; Shi et al., 2017), although it has been suggested more recently that they can also mediate mRNA decay, similar to YTHDF2 (Lasman et al., 2020). Although the YTHDF proteins are all similar in structure and function, the YTHDC proteins are more diverse. YTHDC1 is the only nuclear YTH protein; it promotes exon inclusion during splicing and promotes mRNA export (Xiao et al., 2016; Roundtree et al., 2017b). YTHDC2 is also an RNA helicase important for proper regulation of spermatogenesis (Bailey et al., 2017; Hsu et al., 2017; Jain et al., 2018). Because of the multiple effects m6A can exert through its readers, much effort has been invested in properly mapping m6A locations to determine which RNAs are subject to these effects.
Antibody-based detection of m6A: MeRIP-seq/m6A-seq
The first and most widely used methods to map transcriptome-wide distribution are m6A-IP-seq and meRIP-seq (Dominissini et al., 2012; Meyer et al., 2012; Figure 3A). This method uses an m6A-specific antibody to enrich for methylated RNA after fragmentation into 100- 200-nt fragments. By comparing the sequencing reads from immunoprecipitated and non-immunoprecipitated libraries, the location of m6A sites can be determined to a resolution of 100–200 nt. Using this technique, m6A has been revealed to be enriched primarily close to the stop codon and within the 3′ untranslated region (3’ UTR) of mRNA. This method is easily integrated into standard RNA sequencing (RNA-seq) pipelines and thus became a foundational technique in the field of m6A research.
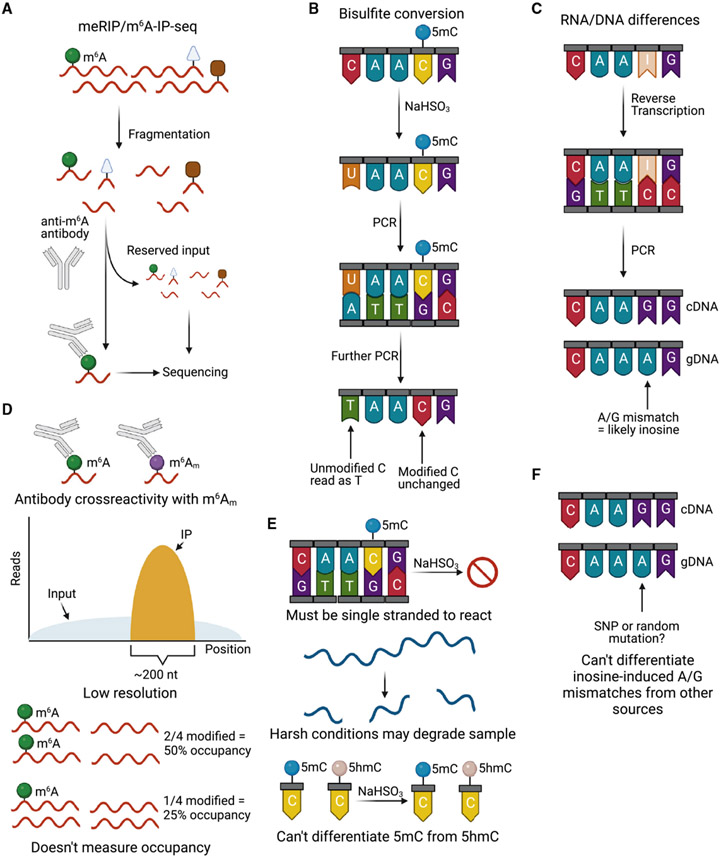
Figure 3. Workflows/principles and drawbacks of the gold standard techniques for each modification.
(A) Workflow of meRIP/m6A-IP-seq.
(B) Principle of bisulfite conversion.
(C) Principle of RNA/DNA differences.
(D) Drawbacks of meRIP/m6A-IP-seq.
(E) Drawbacks of bisulfite conversion.
(F) Drawback of RNA/DNA differences.
Despite its foundational nature, the method is not without drawbacks (McIntyre et al., 2020; Figure 3D). First, as with all antibody-based methods, antibody specificity is a constant concern among researchers. Although the commercial anti-m6A antibodies are great at distinguishing methylated adenosine from unmodified adenosine or other nucleotides, these antibodies often cross-react with m6Am, a related modification that is enriched in mRNA proximal to the m7G cap (Wei et al., 1975a). Recently, efforts have been made to address this concern. By performing an additional meRIP-seq/m6A-IP-seq experiment in cells in which the m6Am writer PCIF1 has been knocked out, bona fide m6A sites can be distinguished from cross-reactive m6Am sites (Akichika et al., 2019; Boulias et al., 2019; Sendinc et al., 2019; Sun et al., 2019). This distinction is especially important for m6A sites in the 5′ UTR because the majority of m6Am sites are in the 5′ UTR. Although this method is very useful, methods that do not require additional sequencing of another cell line are highly desirable. Second, this method requires a substantial amount of input material (~150 ng of poly(A) or 500 ng of total RNA with recent technological advances; Weng et al., 2018; Zeng et al., 2018), which can limit this technique’s usefulness for experiments with scarce material, such as those probing clinical samples. Third, this method is unable to provide a good measure of the occupancy (stoichiometry) of m6A; i.e., how many copies of a transcript have the modification at any given site. Last, although the 100- to 200-nt resolution in combination with the known consensus sequence is sufficient to identify potential sites, it is far from single-nucleotide resolution.
Improved resolution using antibody cross-linking: miCLIP/photo-cross-linking-assisted-m6A-sequencing PA-m6A-seq)
Recent efforts have been made to improve the resolution of meRIP/m6A-IP-seq. For instance, a variation was developed in which antibodies are cross-linked to RNA using UV light (crosslinking IP [CLIP]). This technique, known as methylation-dependent individual-nucleotide-resolution CLIP (miCLIP) (Linder et al., 2015; Ke et al., 2015, 2017; Grozhik et al., 2017), is similar in execution to the previous methods with the exception of the UV light exposure and data analysis. When cross-linked, the antibody is digested with proteinase K, leaving behind a small peptide “scar” on the bound m6A that will cause an reverse transcription (RT) stop or a mutation upon reverse transcription, which is used to increase the resolution of the method. Although an improvement, one downside of this method is that miCLIP can capture m6A and m6Am because of antibody cross-reactivity, although, based on the positions of m6A and m6Am, miCLIP can potentially distinguish the two modifications; a detailed comparison of the identified methylome utilizing the same method actually reveals that the method often mistakenly annotates the two modifications (Linder et al., 2015; Mauer et al., 2017; Boulias et al., 2019). This is perhaps due to the low cross-linking efficiency of miCLIP. Proper annotation of m6A/m6Am using miCLIP and related methods is important because misannotation may result in misattribution of biological functions to some m6A/m6Am sites (Wei et al., 2018).
A more precise method compared to miCLIP is PA-m6A-seq (Chen et al., 2015). This method utilizes 4-thiouridine (4SU), which specifically cross-links to nearby antibodies upon UV exposure in a process known as photoactivatable ribonucleoside-enhanced CLIP (PAR-CLIP) (Hafner et al., 2010). Using this method, single-nucleotide resolution can be achieved effectively because there will likely be only one m6A consensus within the ~30-nt window generated by PAR-CLIP methods. However, this method is slightly limited in its usefulness for m6A without a nearby uridine (Chen et al., 2015), and neither miCLIP nor PA-m6A-seq provide information about occupancy.
Isoform-aware mapping of m6A: m6A-LAIC-seq
m6A-LAIC-seq (m6A level and isoform characterizing sequencing) is an additional variant of the antibody-based gold standard (Molinie et al., 2016) and differs from standard antibody-based sequencing in three major ways: full-length RNA as input (as opposed to fragments), sequencing of the IP eluate and the supernatant (as opposed to sequencing just the eluate along with the reserved input), and use of methylated spike-in RNA. By including full-length RNAs, LAIC-seq can differentiate the methylation status of different isoforms of the same transcript. By sequencing the supernatant and eluate and using methylated spike-ins, some measure of stoichiometry can be taken. Prior to IP, in-vitro-transcribed RNAs that contain various m6A stoichiometries are spiked into the sample. For example, for every 100 molecules of a given spike-in transcript, 50 of them may contain an m6A modification, resulting in 50% stoichiometry. By comparing the known percent occupancy with the measured stoichiometry (calculated from the ratio of enrichment of the spike-in in the eluate to enrichment in the supernatant), a standard curve can be drawn with which the stoichiometry of the endogenous RNAs can be measured. However, this does not truly measure the occupancy of a given modification site; it only measures the percentage of transcript with any m6A modification (i.e., a user would know that isoform 1 of transcript X is more methylated overall than isoform 2 but would not know which specific m6A sites are more or less methylated on that transcript.
Antibody-independent m6A detection via differential enzyme activity: MAZTER-seq/m6A-REF-seq
The E. coli RNA nuclease MazF cleaves preferentially at 5′-ACA-3′ sites as opposed to the methylated form (5′-m6ACA-3′) (Imanishi et al., 2017). Because roughly ~25% of m6A sites contain this sequence, MazF proved to be an attractive option for development of antibody-independent m6A sequencing technology. Two techniques that utilize MazF were first published in 2019 as MAZTER-seq (Garcia-Campos et al., 2019) and m6A-REF-seq (RNA-endoribonuclease faciliated sequencing) (Zhang et al., 2019). In both techniques, RNA is treated with MazF following poly(A) selection. All resulting fragments start with an ACA motif where the initial A was not methylated and end just before the next ACA motif where the initial A is not methylated. Any ACA motifs in the fragment that are not immediately at the 5′ end of the fragment can be inferred to be methylated. For each ACA motif, the methylated:unmethylated ratio can be calculated by comparing the number of times the site is at the 5′ end of a fragment with the number of times the site in the middle of the fragment, either through sequencing analysis (MAZTER-seq) or through qPCR (m6A-REF-seq). However, both techniques are limited in their usefulness to m6A in ACA motifs, and the concentrations of the MazF nuclease used must be controlled carefully because MazF is able to cleave m6A-containing sites at high concentrations (Imanishi et al., 2017).
Repurposed readers and erasers for antibody-independent detection: DART-seq and m6A-SEAL-seq
Additional non-antibody-based m6A detection methods include DART-seq (deamination adjacent to RNA modification targets) and m6A-SEAL-seq (Meyer, 2019; Wang et al., 2020). DART-seq uses a fusion protein between the YTHDF2 reader domain and the cytidine deaminase APOBEC1 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide) to induce C-to-U deamination near m6A sites. This deamination of nearby cytidine bases can be detected by RNA sequencing. Given that all m6A sites are necessarily followed by a cytidine, DART-seq has the potential to detect a high percentage of m6A modifications. Because expression of the YTH-APOBEC1 fusion and the subsequent deamination is not immediately disruptive to cellular processes, the protein can be transiently expressed in living cells, but the effectiveness of the technique is limited by transfection efficiency. Additionally, because APOBEC1 does not digest the RNA, longer RNA pieces than for nuclease-based methods can be subjected to long-read sequencing, cutting the necessary input material down to as little as 10 ng of total RNA. Last, because of the specificity of the YTH domain, off-target effects (false positives) are minimized, and there is little chance of cross-reactivity with m6Am, as is the case with antibody-based methods. However, because this method is reliant on m6A recognition by the YTH domain, any m6A bases not recognized by the YTH domain (because of possible sequence preference) would not be recognized (false negatives). Additionally, control experiments conducted in this study showed that APOBEC alone has a strong preference to deaminate cytidines preceded by an adenosine. Because the method calls for expression of APOBEC alone to estimate the background editing rate, it might set a background rate of modification too high, leading to an increased rate of false negatives.
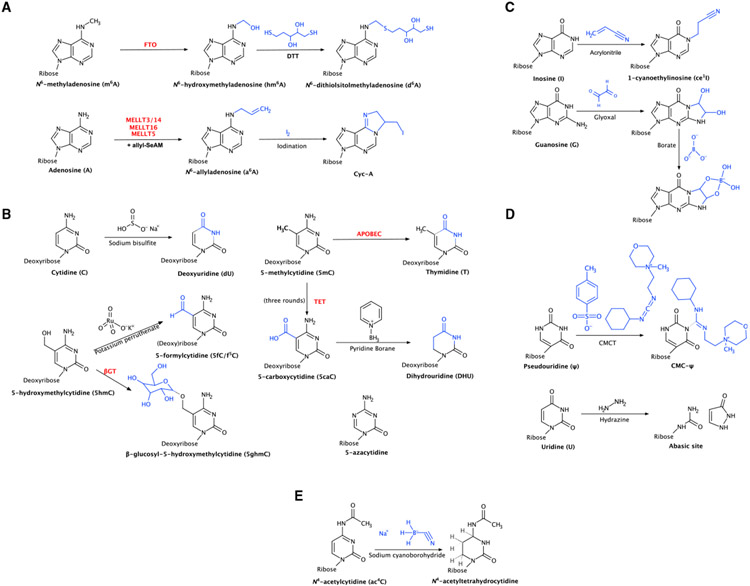
Instead of repurposing reader domains as in DART-seq, Wang et al. (2020) repurposed the m6A eraser FTO to chemically modify m6A in a technique referred to as m6A-SEAL-seq (Figure 2A). Although FTO is an m6A eraser, its exact chemical function is that of a dioxygenase that oxidizes m6A to N6-hydroxymethyladenosine (hm6A). hm6A is highly unstable and will decay to adenosine with a half-life of roughly 3 h (Fu et al., When treated with DTT after FTO oxidation, hm6A is converted to N6-dithiolsitolmethyladenosine (dm6A), which is significantly more stable than hm6A and contains a thiol group for additional labeling. In the case of SEAL, the thiol is labeled with a biotin tag for streptavidin pull-down in a manner analogous to the IP step in antibody-based methods. Because of this similarity, m6A-SEAL-seq has a similar resolution as these antibody-based methods. Additionally, although the method as published is geared toward m6A detection, the authors note that the method can be tuned toward detection of other bases on which FTO is known to act because it has been proposed to act on a wide range of targets beyond m6A.
Figure 2. Chemical and biochemical reactions for each technique discussed.
(A) (Bio)chemical modifications used in the study of m6A: biochemical modification of m6A to hm6A by the enzyme FTO and subsequent chemical modification to d6A by DTT and biochemical modification of A to a6A by writer enzymes and subsequent chemical modification to Cyc-A by iodination.
(B) (Bio)chemical modifications used in the study of 5mC/m5C: chemical deamination of C to dU by bisulfite, chemical oxidation of 5hmC to 5fC by KRuO4 and biochemical modification of 5hmC to 5ghmc by the enzyme β-GT, biochemical deamination of 5mC to T by APOBEC, chemical modification of 5caC to dihydrouridine (DHU) by pyridine borane, and structure of 5-azacytidine.
(C) Chemical modifications used in the study of inosine: chemical modification of inosine to ce1I by acrylonitrile and chemical modification of guanosine with glyoxal and further modification with borate.
(D) Chemical modifications used in the study of ψ: chemical modification of ψ with CMCT and chemical cleavage of uridine with hydrazine.
(E) Chemical modification used in the study of ac4C: chemical reduction of ac4C by NaCNBH3.
m6A detection through antibody-independent mutational profiling: m6A-label-seq
The previous two techniques exploit different m6A-modifying enzymes and reader proteins, but Shu et al. (2020) developed a method that targets a different point in m6A metabolism: the S-adenosyl methionine (SAM)-dependent methylation event. By supplementing cells with a SAM derivative (allyl Se-adenosyl methionine, allyl-SeAM), adenosine bases destined to be methylated are instead converted to N6-allyladenosine (a6A). After enrichment of a6A-containing transcripts via antibody pull-down, a6A is further converted to a cyclic derivative via iodination (Figure 2A). This cyclic derivative (Cyc-A) results in base misincorporation upon reverse transcription. These misincorporations can be detected via sequencing to determine the site of the original Cyc-A with single-nucleotide resolution in a process referred to as mutational profiling. Although this method provides increased resolution over antibody-based methods, use of allyl-SeAM resulted in a moderate stress response in some cells, which may disrupt m6A levels, and the incorporation rate of the allyl moiety onto target adenosine is not 100%. Additionally, Cyc-A does not result in base misincorporations 100% of the time, meaning that some m6A sites may go undetected by this method, although the authors point out that a reverse transcriptase could possibly be evolved to more reliably misincorporate bases when encountering Cyc-A.
m6A occupancy determination at a given site: SCARLET and SELECT
Liu et al. (2013) first reported a method called SCARLET (site-specific cleavage and radioactive labeling followed by ligation-assisted extraction and thin-layer chromatography) that was designed to measure the occupancy of a m6A modification at a known site. This technique capitalized on the cleavage specificity of RNaseH and on the ability of thin-layer chromatography (TLC) to distinguish m6A from A (Liu et al., 2013). Using a chimeric 2′ H and 2′OMe oligo that targets the m6A site of interest, RNaseH is directed to cleave the target RNA immediately 5′ to the modified A. The new 5′ end is then phosphorylated with 32P, and it is ligated to a non-radiolabeled DNA oligo using a DNA splint. This splint ligation allows the RNA of interest to be protected from subsequent digestion with RNase T1/A, which removes contaminating RNA molecules. The DNA/RNA hybrid is then purified from a denaturing gel and digested to mononucleotides with nuclease P1. Following digestion, the radioactive ribonucleotide is separated via TLC. By measuring the difference in density between the pA and pm6A spots, the ratio of A to m6A at the probed site is determined. Using SCARLET, Liu et al. (2013) were able to measure the modification occupancy of several m6A sites in poly(A) RNAs.
More recently, a less complex technique has been developed that allows quantification of m6A stoichiometry: single-base elongation and ligation-based PCR amplification (SELECT) (Xiao et al., 2018). In SELECT, two oligos are designed that sit +1 and −1 from an m6A site (the “up” and “down” oligos, respectively). Following annealing, Bst DNA polymerase is added to attempt to seal the gap between the two oligos at the m6A site. Because m6A sits at the interface between the WCF and Hoogsteen surfaces, it can block base-pairing and will prevent the gap from being filled. After the Bst reaction, SplintR ligase is added to join the two oligos. m6A also hinders the activity of SplintR ligase. Because of these two hindrances, far fewer complete DNA oligos will be formed from m6A-containing template RNAs than from nonmethylated ones, a difference that can be detected via qPCR and used to quantify the percentage of RNAs that contain a m6A at the probed site. This method is also adaptable to detection of other modifications, such as m1A or pseudouridine.
Potential future directions for m6A mapping
The last decade has seen an explosion of interest in m6A, which, in turn, has led to an explosion of techniques for its study. These techniques make an excellent case study for how to exploit multiple proteins and biochemical properties to develop techniques for modification mapping and manipulation. In response to the inherent disadvantages of antibody-based methods, numerous groups quickly developed techniques to supplement meRIP/m6A-IP-seq. Many of the underlying principles utilized by these methods (exploitation of proteins in the modification’s metabolic pathway and of differential enzyme activity, chemical derivatization, and mutational profiling) will appear in our discussion of other modifications. As will always be the case, each of these methods comes with its own drawbacks that can limit each method’s usefulness. For example, the proteins used in MAZTER-seq/m6A-REF-seq and DART-seq have a detection bias that is present in these methods, and introduction of exogenous SAM derivatives in m6A-label-seq results in cellular stress, which could dysregulate m6A levels. Furthermore, very few of the high-throughput methods can measure modification stoichiometry, and neither of the two methods that directly probe stoichiometry are amenable to high-throughput approaches. Ideally, future work will continue to utilize the unique chemistry and protein interactome of m6A to continue to refine our ability to map this modification with high resolution in an unbiased, quantitative way. An alternative future direction is direct sequencing (sequencing without further modification or reverse transcription) of modified nucleic acids via methods like Oxford Nanopore sequencing. Recently, m6A has been detected in synthetic and naturally occurring RNA using Nanopore sequencing (Garalde et al., 2018; Liu et al., 2019a). This detection does not require any modification of the RNA or the sequencing method itself because the modifications were detected via computational analysis of the data.
5-METHYLCYTIDINE (5mC/m5C)
Another intensively studied nucleic acid modification is 5-methylcytidine (5mC in DNA and m5C in RNA). In DNA, 5mC has long been appreciated as a crucial epigenetic marker. For a thorough review of 5mC in DNA and its dynamics, see Luo et al. (2018). For the purposes of this review, however, demethylation of 5-methylcytidine must be addressed. Demethylation is a multi-step process involving many intermediate modifications. In DNA, demethylation of 5mC first involves oxidation of 5mC to 5hmC, 5-formylcytidine (5fC), and 5-carboxycytidine (5caC) by a set of enzymes in the TET family: TET1, TET2, and TET3 (reviewed in Rasmussen and Helin, 2016; Figure 1B). The final step involves removal of 5fC or 5caC through base excision repair by a thymine-DNA glycosylase (TDG) enzyme to obtain an unmodified cytidine (Kohli and Zhang, 2013).
In addition to its well-appreciated role in DNA, 5-methylcytidine is present in all major species of RNA as well. RNA m5C in humans is installed by members of the NSUN (NOL1/NOP2/Sun domain) methyltransferase family (reviewed in Bohnsack et al., 2019) and DNMT2 (Shanmugam et al., 2015). In tRNA, several m5C sites are known to affect RNA stability and function. For example, m5C in the anticodon loop and stem is involved in ribosomal A-site entry, mRNA translocation, and decoding (Shanmugam et al., 2015). TET2-mediated oxidation of tRNA m5C promotes translation and generation of tRNA fragments (Shen et al., 2021; He et al., 2021). Additionally, ALKBH1 is known to be an RNA-specific dioxygenase that converts m5C into hm5C in mitochondrial tRNAMet (Kawarada et al., 2017) and generate 2′-methoxy forms of hm5C and f5C in cytoplasmic tRNALeu (Takemoto et al., 2009), all at the wobble position, which affects translation. In the yeast 25S rRNA and human 28S rRNA, crucial m5C modifications occur on yeast C2870/human C3761 and yeast C2278/human C4413. Because C2870/C3761 is located near sites of translation, it is predicted to assist with peptidyl transfer during translation (Sharma et al., 2013; Gigova et al., 2014; Schosserer et al., 2015). C2278/C4413 is located at the boundary between the large and small ribosomal subunits and is responsible for proper folding and proper termination at stop codons (Sharma et al., 2013; Bourgeois et al., 2015). In mammalian mRNA, m5C abundance and significance have been somewhat controversial, although there is growing consensus that the modification is present but at low occupancy in mRNA (for a more thorough review of the controversy, see Trixl and Lusser, 2019). In mRNA, m5C is found on approximately 0.03%–0.1% of all cytidines (Huber et al., 2015) and is enriched in the 5′ and 3′ UTRs but depleted in protein-coding regions (Amort et al., 2017). m5C has two proposed reader proteins: ALYREF, which facilitates export of methylated mRNAs from the nucleus (Yang et al., 2017), and YBX1, which stabilizes m5C-modified mRNAs (Chen et al., 2019; Yang et al., 2019).
Mapping 5-methylcytidine with chemical conversion: BS-seq
Initially, mapping of 5mC sites was carried out via restriction enzyme digestion because the presence of an 5mC residue was known to inhibit cleavage (Bird et al., 1979). To develop a method more amenable to high-throughput sequencing, BS-seq was developed to study cytidine methylation in DNA (Hayatsu et al., 1970; Shapiro et al., 1970; Frommer et al., 1992; Figure 2B). The technique works as follows. Bisulfite (HSO3−) reacts with cytidines in single-stranded DNA (ssDNA) under acidic pH to cause deamination and formation of uridine sulfonate, which desulfonates to uridine under basic pH. Because 5-methylcytidine reacts with bisulfite much slower than unmethylated cytidines, bisulfite treatment can help distinguish the two bases through sequencing (5mC will continue to be read as C in sequencing, whereas unmethylated C, which has been converted to U, will read as T). BS-seq incorporates this chemistry upstream of a DNA sequencing pipeline, and this technique has been extended to single-stranded RNA (ssRNA) as RNA BS-seq to identify m5C in RNA. However, the requirement for ssRNA often results in a high false positive rate for RNA in highly structured regions (Figure 3E). Additionally, the conditions of bisulfite treatment (high temperature and basic pH) are not ideal for RNA and even DNA (although to a lesser extent for DNA) because they can cause sample degradation. To address this issue, short reaction times are often used, but this can result in a low conversion rate and a high rate of false positives. Because of this, RNA BS-seq is best suited for study of m5C at high occupancy, such as the m5C sites in tRNA and rRNA (Huang et al., 2010). Another one of the main drawbacks of BS-seq is that it is unable to differentiate 5hmC from 5mC (Huang et al., 2010), although this has been addressed in the techniques listed below.
Improving BS-seq: oxBS-seq and TAB-seq
In 2012, two improvements on BS-seq were published, oxidative BS-seq (oxBS-seq) (Booth et al., 2012) and TET-assisted BS-seq (TAB-seq) (Yu et al., 2012). Both methods allow 5mC and 5hmC to be distinguished from one another in BS-seq; this occurs via an oxidation reaction, either chemical (KRuO4) in oxBS-seq or TET-mediated oxidation in TAB-seq (Figure 2B). In oxBS-seq, chemical oxidation converts 5hmC to 5fC, which is susceptible to deamination by bisulfite. In TAB-seq, 5hmC is first protected by conversion with the enzyme β-glucosyltransferase (βGT), and then 5mC is oxidized to 5caC by TET. 5caC is also susceptible to bisulfite deamination. By comparing sequencing results from each method with standard BS-seq results, the locations of 5mC and 5hmC can be mapped accurately. Although these techniques are improvements over BS-seq alone, they have the same disadvantages toward RNA as standard BS-seq.
Antibody-based detection of 5mC and m5C: MeDIP and m5C meRIP
Like in the antibody-based meRIP/m6A-IP-seq methods for m6A, antibodies have similarly been utilized to enrich for 5-methylcytidine in DNA and RNA. When applied to DNA, this technique is referred to as methylated DNA IP [MeDIP] (Weber et al., 2005), and in RNA it is referred to as m5C-methylated RNA IP (meRIP) (Edelheit et al., 2013). The protocol is effectively identical to the one outlined above for meRIP/m6A-IP-seq and thus has the same advantages and disadvantages. However, the disadvantages of antibody-based methods are most pronounced in the case of m5C in RNA because the anti-m5C antibodies are prone to nonspecific binding and highly prefer ssRNA and, thus, return inconsistent results. Because of this, RNA BS-seq and meRIP datasets often disagree with each other, especially at sites of low m5C occupancy.
Bisulfite-free methods of differentiating 5mC and 5hmC in DNA: ACE-seq and TET-assisted pyridine borane sequencing (TAPS)
APOBEC-coupled epigenetic sequencing (ACE-seq) is a recently developed enzymatic method that uses the AID (activation-induced cytidine deaminase)/APOBEC family of enzymes to selectively identify 5hmC at a high resolution (Schutsky et al., 2018). The AID/APOBEC enzyme can deaminate unmodified cytidine and 5mC to uridine or thymidine, respectively, while leaving 5hmC unmodified (after protection by β-glycosyltransferase in a fashion similar to TAB-seq) (Figure 2B). ACE-seq has proven to be a highly accurate bisulfite-free method for identifying 5hmC with an over 99% conversion rate for cytidine and m5C while leaving 98.5% of 5hmC unconverted. Because of this, ACE-seq provides specific 5hmC profiles BS-seq cannot produce.
Recently, an additional bisulfite-free method was developed that utilizes the oxidative power of TET in combination with chemical modification (Liu et al., 2019c). This method, TAPS (TET-assisted pyridine borane sequencing) involves oxidation of 5mC and 5hmC to 5caC using purified TET, followed by reduction of 5caC with pyridine borane to dihydrouridine, which is read as T during PCR (Figure 2B). The original publication of TAPS also outlines two complimentary methods, TAPSβ and CAPS (chemical-assisted pyridine borane sequencing), in which the enzyme βGT is employed before TET treatment or the oxidizing agent KRuO4 is used instead of TET treatment, respectively. Using these methods (which are essentially fusions of TAPS and TAB/oxBS, respectively) in conjunction with TAPS, m5C and 5hmC can be mapped without the need for bisulfite treatment. Although both of these methods are powerful, neither is amenable to detection of m5C in RNA. Additionally, methods with less complex protocols are desirable.
Mapping the targets of m5C methyltransferases in RNA: Aza-IP and miCLIP:
Unlike the previously discussed methods, which were developed for detection and mapping of 5mC and its oxidation products in DNA, 5-azacytidine-mediated RNA IP (Aza-IP) was developed specifically to map the targets of RNA 5-methylcytidine methyltransferases. The method relies on the covalent bonds formed between the methyltransferase’s active site cysteine and their targets to identify which cytidines are methylated by which methyltransferase. The cytidine derivative 5-azacytidine (Figure 2B) can form the covalent bond, but it will remain bound to the enzyme. This allows the transferase of interest to be enriched by IP and the bound RNA targets to be identified by sequencing (Khoddami and Cairns, 2013). Aza-IP is most well suited for “single-cysteine-type” transferases (those with a single cysteine in their active site, such as DNMT2) as opposed to a double cysteine, like other RNA m5C methyltransferases (King and Redman, 2002). Despite its usefulness, Aza-IP is unable to measure occupancy at a given site, and its protocol may be difficult to perform because it requires a large number of cells and expression of an epitope-tagged transferase for pull-down (Khoddami and Cairns, 2014).
As an alternative approach to Aza-IP, which utilizes a modified nucleotide to trap writer enzymes, a modified writer enzyme can be used to similar effect. This method, referred to as miCLIP (Hussain et al., 2013b), requires mutation of a catalytic cysteine residue to an alanine. When mutated, this cysteine can no longer participate in resolution of the covalently linked intermediate, effectively trapping the enzyme on the RNA (Hussain et al., 2009). Using miCLIP, the RNA targets of NSUN2 (Hussain et al., 2013b), NSUN3 (Van Haute et al., 2016), and NSUN6 (Selmi et al., 2021) have all been mapped. Because this method modifies the methyltransferase directly, it is amenable to use with double cysteine methyltransferase, unlike Aza-IP. However, this method requires overexpression of the mutant methyltransferase, which may result in cellular stress.
Potential future directions of 5mC/m5C detection
In DNA, mapping of 5-methylcytidine can be accomplished reliably by BS-seq. Additionally, as the roles of 5mC’s TET-mediated oxidation products have become more and more appreciated, demand has grown for sequencing methods that can map them in addition to 5mC. This demand has been met by improved versions of BS-seq and by bisulfite-free methods. Although antibody and chemical free methods for detection of 5hmC, such as ACE-seq and TAPS, will no doubt be popular, additional methods will be required that do not require purification of multiple enzymes and that can detect further oxidation products beyond 5hmC. As the roles of TET-mediated oxidation products become appreciated in RNA, techniques should be developed to map and quantify these modifications in RNA as well.
Because the nature of m5C in RNA remains a matter of current study and debate, there is a great need for techniques that can properly map m5C in RNA. Antibody-based methods are prone to resolution problems (as discussed previously), and the two unique methods discussed (Aza-IP-seq and miCLIP) only capture the methylation targets of the specific methyltransferase being probed. Although these methods are undeniably useful, a method that can detect m5C transcriptome wide in an unbiased manner is necessary. To this end, the two recently discovered reader proteins, ALYREF and YBX1, should be exploited in a similar manner as methods such as DART-seq or endonuclease V inosine precipitation enrichment sequencing (EndoVIPER-seq), which will be discussed in the upcoming section. Alternatively, some of the biochemical techniques applied to DNA (such as ACE-seq and TAPS) may be adaptable for use on RNA as well. Additionally, it is worth noting that the previously mentioned Nanopore sequencing method has been shown to be capable of directly sequencing m5C in synthetic RNA (Garalde et al., 2018).
INOSINE (I)
Beyond monomethylation, nucleic acids can also be modified in more chemically diverse ways. One well-studied example is conversion of adenosine to inosine via deamination (also referred to as A-to-I editing) (Figure 1A). This conversion is catalyzed by an adenosine deaminase enzyme, either ADAT (adenosine deaminase acting on tRNA) when incorporated into tRNA (Torres et al., 2014) or ADAR (adenosine deaminase acting on RNA) when incorporated into other RNA species (Zinshteyn and Nishikura, 2009). In eukaryotic tRNAs, inosine is most commonly incorporated at the wobble position by the heterodimeric ADAT complex, consisting of the catalytic ADAT2 and the target selecting ADAT3 (Gerber and Keller, 1999). I34 has long been known to facilitate tRNA wobble and increase the decoding potential of a given tRNA molecule.
In all other RNA species, A-to-I editing is performed by one of the ADAR proteins, of which three have been found to be functional. The short isoform of ADAR1 (ADAR1p110) and ADAR2 are primarily localized to the nucleus (Zinshteyn and Nishikura, 2009), whereas the long isoform of ADAR1 (ADAR1p150) is localized to the cytoplasm and the nucleus, where it acts as part of the cell’s antiviral defenses (Patterson and Samuel, 1995). All of the functioning ADAR proteins only deaminate adenosines within dsRNA (Ryter and Schultz, 1998), with no strict preference for sequence context. When included in a coding region of an mRNA, inosine is read by the ribosome as a guanosine, which results in genetic recoding. Several of the most well-studied examples of this recoding affecting protein sequence and function are neurotransmitter receptor ion channels in the brain (Seeburg and Hartner, 2003). A-to-I editing also exerts effects on noncoding regions of the transcriptome. For a thorough review of this, see Nishikura (2016).
Detecting inosine sites using mutational profiling: RNA and DNA differences (RDD)
Similar to mutational profiling, reverse-transcribing an inosine-containing RNA will result in incorporation of a cytidine at that position as opposed to a thymidine. When sequenced, the inosine site will be read as a guanosine instead. The sites of A-to-I editing have traditionally been determined using these A-to-G “mutations” (Levanon et al., 2004). When applied to high-throughput sequencing, this approach is referred to as RDD because it relies on comparison of genomics and RNA-seq data to find A-to-G sites (Li et al., 2011; Bahn et al., 2012; Peng et al., 2012; Figure 3C). Studies using RDD have claimed to find a varying number of editing sites in the transcriptome, ranging to upward of 20,000 (Bahn et al., 2012; Li et al., 2011; Peng et al., 2012). As a method, RDD has a high false positive rate because of errors in read mapping and single nucleotide polymorphisms (Kleinman and Majewski, 2012; Lin et al., 2012; Pickrell et al., 2012; Figure 3F), even after a more advanced analysis pipeline was developed to reduce the false positive rate (Piskol et al., 2013). Reanalysis of RDD data generated in glioblastoma cell lines using the I chemical erasing (ICE) method discussed below could only validate just over 50% of the predicted A-to-I edited sites (Sakurai et al., 2014).
Improving detection of inosine using chemical derivatization: ICE
To decrease the false positive rate of the RDD method, Sakurai et al., 2010 capitalized on previously characterized chemistry. When treated with acrylonitrile, a cyanoethyl group will attach to the WCF face of inosine, forming N1-cyanoethylinosine (ce1I) and blocking base pairing across the WCF surface (Yoshida and Ukita, 1968; Figure 1E). Thus, ce1I results in an RT stop (Figure 2C), whereas an unmodified inosine results in the A-to-G substitution. The presence of an inosine could be validated by PCR-amplifying and sequencing the region containing the suspected inosine with and without acrylonitrile treatment. Despite the improvements ICE (inosine chemical erasure) and ICE-seq (Suzuki et al., 2015) made over RDD alone, their dynamic range is somewhat limited. Sites with 100% inosine occupancy may not be detectable via ICE-seq because no unmodified transcript would remain after acrylonitrile treatment, meaning that the base would never be read in the sequencing experiment. Although rare, there are biologically relevant examples of this high occupancy, such as some glutamate receptors (Seeburg et al., 2001). Likewise, the technique has difficulty detecting inosine sites with less than 10% occupancy without the aid of synthetic spike-ins or increased read depth (Suzuki et al., 2015).
Using site specific cleavage to detect inosines: iSeq
In a method that could be described as the reverse complement of ICE-seq, Cattenoz et al. (2013) adapted a method for inosine-specific cleavage developed initially by Morse and Bass (1997) into iSeq (inosine sequencing). With this method, RNA is treated with glyoxal and borate, which will preferentially modify guanosines and not inosines (Figure 2C). The glyoxal-borate modification will prevent cleavage by RNase T1, which normally cleaves 3′ of guanosines (Pace et al., 1991). Inosines, however, are not protected. By biotinylating the 3′ end of the RNA molecules being probed and binding them to streptavidin beads, inosine-containing RNA fragments can be liberated by RNase T1 treatment. The two resulting fractions, free inosine-containing fragments and bead-bound fragments with no inosine, can be interrogated separately by sequencing (Cattenoz et al., 2013). Using this technique to enrich for inosine-containing RNAs, the localization and occupancy of a modification can be determined. However, regions of clustered inosines are hard to detect using iSeq because the fragments generated by digestion will be too small to sequence effectively.
Enriching for inosine using a converted nuclease: EndoVIPER-seq
Knutson et al. (2020) developed a new method that employs a novel biochemical approach to identify I sites. This method, EndoVIPER-seq, makes use of the E. coli homolog of the inosine-specific endonuclease EndoV (eEndoV) in the presence of Ca2+ (which preserves RNA binding activity but ablates cleavage) to bind to and enrich for inosine-containing RNAs. In this way, the eEndoV is effectively rendered a reader protein for inosine, which allows it to be utilized to enrich for inosine-containing RNAs in a manner reminiscent of meRIP-seq/m6A-IP-seq and DART-seq. Taking inspiration from the abovementioned specific cleavage study, the group utilized glyoxal treatment to ensure that the RNA substrates are single stranded, ensuring an optimal target for eEndoV binding. Using EndoVIPER-seq, Knutson et al. (2020) were able to recapitulate previously known inosine sites in the brain transcriptome under developmental and pathological conditions. Although this method is simple to perform and highly specific for inosine detection, it does not provide occupancy information for a given inosine site because it only gives the user information about the location of inosine sites, not the percentage of transcripts that have inosine at any given site.
Potential future directions of inosine detection
The parallel techniques of ICE-seq and iSeq are excellent examples of exploiting the chemistry of a modification to develop techniques that are improvements over the current gold standard in the field. These methods are highly complementary because ICE-seq would have no problem mapping multiple inosines in a row, and iSeq’s dynamic range is not as limited as ICE-seq’s. Future techniques should be geared toward combining these advantages into one cohesive technique. Additionally, the example put forth by EndoVIPER-seq should be studied by those wishing to investigate any modification, not just inosine. Essentially, this technique generates an exploitable reader protein for inosine where there was none simply by swapping metal ions. This creates the possibility of converting other eraser enzymes into being reader proteins through simple manipulations for other modifications besides inosine.
PSEUDOURIDINE (ψ)
First discovered in 1951 (Cohn and Volkin, 1951) and characterized in 1957 (Davis and Allen, 1957) and 1959 (Yu and Allen, 1959), ψ is the most abundant post-transcriptional modification, found in all known RNA species (Boccaletto et al., 2018). ψ is the result of an isomerization reaction (swapping C5 for N1) catalyzed by a ψ synthase (PUS) enzyme, either functioning as a lone enzyme or as a member of a snoRNP (small nucleolar ribonuclear protein) complex (Figure 1E). This isomerization results in an extra hydrogen bond donor on the Hoogsteen face, which is thought to stabilize ψ-A base pairs (Newby and Greenbaum, 2002) and to stabilize helices as a whole through increased stacking (Davis, 1995).
ψ has long been appreciated as having a crucial role in ribosomal biogenesis and function. ψ in ribosomal RNA is installed by the H/ACA box snoRNP, which consists of a H/ACA box small nucleolar RNA (snRNA) which hybridizes to the region of the ribosome to be modified and four protein components: NOP10, NHP2, GAR1, and the catalytic subunit dyskerin (Bachellerie et al., 2002). When installed, ψ affects all aspects of ribosomal function. For example, a ψ near the yeast ribosomal A site is known to be important for amino acid incorporation (King et al., 2003), and another on helix 69 of the large subunit of the yeast ribosome has effects on amino acid incorporation and proper stop codon recognition (Liang et al., 2007).
In addition to rRNA, it has been made clear (using the sequencing methods described below together with mass spectrometry) that ψ is present in mRNA as well (Schwartz et al., 2014; Carlile et al., 2014; Li et al., 2015). These ψs have been found to be installed predominantly by the enzyme PUS1 in an RNA structure-dependent manner (Carlile et al., 2019). In contrast to m6A, ψ lacks any known eraser or reader proteins, suggesting that any effects it exerts on an mRNA transcript are likely due to its intrinsic chemical properties. Indeed, research has found that ψ in the coding region of an mRNA can alter translation by promoting amino acid substitution at codons containing a ψ (Eyler et al., 2019).
Using chemical adducts of ψ to induce RT stops: Pseudo-seq, PSI-seq, ψ-seq, and CeU-seq
An early method for sequencing-based ψ detection involved further chemical modification of the base. First described in 1971 (Ho and Gilham, 1971), the reaction of ψ with N-cyclohexyl-N’-(2-morpholinoethyl)-carbodiimide metho-p-toluene-sulfonate (CMCT) results in addition of a bulky adduct on the WCF face, which, in turn, results in an RT stop (Figure 1D). Although this adduct is also formed on the WCF faces of U and G, they can be removed easily under alkaline conditions, where the ψ adduct is not removable without high heat and highly alkaline conditions (Ho and Gilham, 1971). Initially, this chemical modification, which results in RT stops, was used in combination with low-throughput sequencing to determine sites of pseudouridylation (Bakin and Ofengand, 1993).
More recently, four high-throughput sequencing techniques have been developed based on CMCT modification: Pseudo-seq (Carlile et al., 2014), PSI-seq (pseudouridine site identification sequencing) (Lovejoy et al., 2014), ψ-seq (Schwartz et al., 2014), and CeU-seq (N3-CMC-enriched pseudouridine sequencing) (Li et al., 2015). Although largely similar, each technique has a slight variation that distinguishes it: CeU-seq utilizes an azide-linked CMCT enriched for ψ-containing transcripts via pull-down with a clickable biotin, Pseudo-seq circularizes the cDNA fragments before amplification, ψ-seq fragments RNA after CMCT modification (other techniques do so before), and PSI-seq does not circularize its cDNA, does not further enrich for pseudouridylated RNAs, and fragments before CMCT treatment. These techniques have been used to identify new ψ sites in humans and yeast and in mRNA and noncoding RNA. Although important for the high-throughput study of ψ in mRNA, any incomplete removal of CMC (N-cyclohexyl-N’-(2-morpholinoethyl)carbodiimide) moieties from non-ψ bases would result in false positives.
Detection of ψ via uridine-specific chemical cleavage: HydraPsiSeq
Like the CMCT-mediated sequencing techniques discussed above, the ψ detection method developed by Marchand et al. (2020) was also inspired by long-established chemistry. Hydrazine can specifically cleave uridine bases (and, importantly, not ψ or the other three canonical RNA bases) (Levene and Bass, 1926; Peattie, 1979). When hydrazine treated, the affected uridine is reduced to an abasic site and cleaved through β and δ eliminations (Figure 2D), resulting in two RNA strands that begin and end +1 and −1 from the uridine, respectively (Küpfer and Leumann, 2007; Marchand et al. (2020)) capitalized on these chemistries to develop HydraPsiSeq. Sequencing reads will preferentially begin and end +1 and −1 from uridine bases. Any uridines in the middle of reads are thus likely to be ψs. By comparing the number of reads that terminate at a given uridine with the number of reads that read through it, the modification occupancy can be determined, like MAZTER-seq/m6A-REF-seq, as described above. However, this method requires a fairly substantial read depth (~25 million reads) for detection, which may limit the method’s effectiveness on low-abundance transcripts. Additionally, the method is somewhat prone to false positives in two situations: from uridines in highly structured regions that may be protected from cleavage (the method is performed at low temperatures) and from other modified forms of uridine, such as 5-methyluridine.
N4-ACETYLCYTIDINE (ac4C)
Like ψ, ac4C is another modification that has been viewed traditionally as a ribosomal and tRNA modification but has been suggested recently in mRNA. ac4C, which was first discovered in eukaryotic tissue in 1966 (Zachau et al., 1966), is formed by addition of an acetyl group to the N4 position at the intersection of the WCF and Hoogsteen faces (Figure 1D). Despite this placement, ac4C has been shown to strengthen WCF base-pairing of cytidine and guanosine (Kumbhar et al., 2013). In yeast cells, ac4C is installed exclusively by the acetyltransferase Rra1p (Ito et al., 2014a) and in human cells by its homolog NAT10 (Ito et al., 2014b). Currently, there are no known eraser or reader proteins for this modification. In contrast to the other modifications discussed here, which are highly abundant, there are only four bona fide sites in the eukaryotic transcriptome that contain this modification: human 18S rRNA C1337 (Ito et al., 2014b; Sharma et al., 2015) and C1842 (Sharma et al., 2015) and, in the D arm of yeast and human, serine (Kruppa and Zachau, 1972) and leucine (Kowalski et al., 1971)tRNA. NAT10 finds its targets with the help of other factors: snoRNAs are responsible for guiding NAT10 to both of its rRNA sites (Sharma et al., 2015,2017), and the protein THUMPD1 is necessary for NAT10-mediated acetylation of tRNA (Sharma et al., 2015).
Although the presence of ac4C at these sites is well accepted (and crucial for proper ribosome biogenesis in the case of C1842 (Ito et al., 2014a, 2014b)), the abundance of ac4C in eukaryotic mRNA is an ongoing research area in this field. Using the two methods discussed below (acRIP-seq and ac4C-seq), groups came to different conclusions regarding the abundance and effect of mRNA acetylation. Future work (and orthogonal methodologies) will no doubt be required to address these discrepancies. Below, we present a discussion of both methods (and their strengths and weakness) in addition to a summary of the conclusions both groups drew from their implementation. Additional studies (and additional orthogonal methodologies) will no doubt be required to address these discrepancies.
Antibody-based detection of ac4C: acRIP-seq
Arango et al. (2018) utilized an RNA IP-based method to map the location of ac4C in mRNA from Henrietta Lacks’ (HeLa) cells. Using an antibody developed previously by this group (Sinclair et al., 2017), the authors found that NAT10-dependent ac4C was fairly abundant in the CDS of mRNA (after first confirming the presence of the modification using mass spectrometry). Furthermore, the authors found that ac4C modification stabilized mRNA transcripts and enhanced their translation when present at the site complementary to the tRNA wobble position. Technically, acRIP-seq (acetyl RNA IP-seq) is largely similar to meRIP/m6A-IP-seq (Dominissini et al., 2012; Meyer et al., 2012) and m5C-meRIP-seq (Edelheit et al., 2013). Therefore, it is prone to the same strengths (ease of application and data analysis) and drawbacks (problems with antibody cross-reactivity and resolution) as those techniques.
Mapping ac4C via chemically induced mutational profiling: ac4C-seq
Sas-Chen et al. (2020) developed a method using the previously described chemistry of ac4C (Thomas et al., 2018) that could map ac4C at single-base-pair resolution. When treated with sodium cyanoborohydride (NaCNBH3), ac4C is reduced to N4-acetyltetrahydrocytidine (Figure 2E). The presence of this reduced base will result in misincorporation of thymidine in sequencing, which can be used (when compared with an untreated sample or chemically deacetylated sample) to pinpoint the location of ac4C sites. This method, known as ac4C-seq, proved to be exquisitely sensitive, able to detect acetylation at as little as 4% occupancy at a given site. When applied to human RNA, the only sites that could be detected without NAT10 and THUMPD1 overexpression were the four canonical sites discussed previously in rRNA (Ito et al., 2014b; Sharma et al., and tRNA (Kowalski et al., 1971; Kruppa and Zachau, 1972). The group went on, however, to find evidence of the presence of ac4C in archaea. Although this method offers increased resolution and sensitivity compared with acRIP-seq, it is unable to accurately map ac4C in some tRNA contexts. ac4C is sometimes found on the same tRNAs in close proximity to modified-base dihydrouridine. NaCNBH3 also reduces dihydrouridine, meaning that this method will be unable to differentiate these two modifications (Thomas et al., 2019).
Possible future directions for ψ and ac4C
For both modifications, their recent discovery in mRNA (although sometimes controversial) is highly exciting because it add still undiscovered avenues by which mRNAs can be regulated. Although these modifications do not have the same robust library of techniques as the others discussed in this review, researchers aiming to widen their repertoire of tools can look to these other modifications for inspiration. As an example, development of a chemical trapping method for NAT10 (like miCLIP) would be highly advantageous for this modification’s future study because NAT10 is the only enzyme currently known to install this modification. For ψ, it is possible that an Aza-IP-like method could be developed. Several reports on bacterial PUS enzymes (Huang et al., 1998; Gu et al., 1999; Hamilton et al., 2006; Czudnochowski et al., 2014) have shown that the uridine analog 5-flurouridine can covalently trap PUS enzymes. Should this property hold true for human enzymes as well, the Aza-IP workflow could likely be easily adapted toward ψ detection. Additionally, these modifications may benefit from the aforementioned Nanopore-based detection method because ψ has already been shown to be detectable using this technique (Smith et al., 2019).
CONCLUDING REMARKS
For each modification discussed here, there is a need for new technologies that can further our understanding of their localization and occupancy. We would like to draw attention to the latter in particular because accurate occupancy determination is often not a feature of many techniques that map nucleic acid modifications. This is especially important for modifications such as m6A and m5C because the occupancy of a modification at a given site can vary across cellular conditions. The techniques discussed have driven or have the potential to drive their respective subfield forward and to serve as inspiration for development of new techniques that address their shortcomings, as these techniques did for their subfield’s earlier techniques. Ideally, future techniques will be developed with an eye toward reducing detection bias and measuring occupancy and with ease of application in mind.
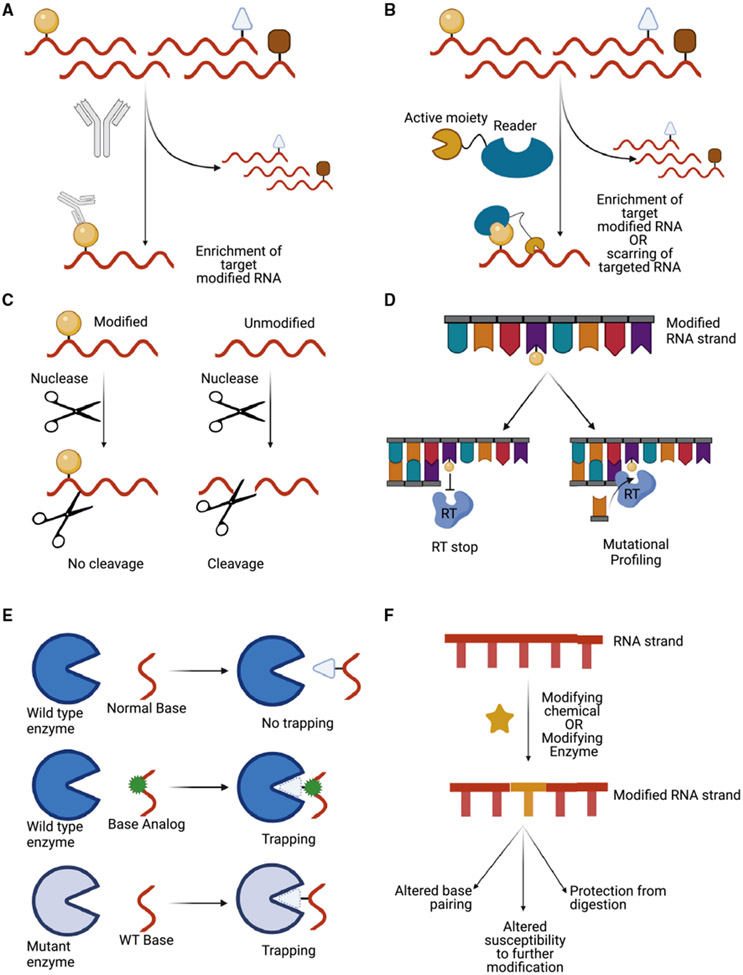
Despite the chemical diversity of their targets, many of the techniques discussed here (and techniques for detection of other modifications) utilize same underlying principles (Figure 4). For example, m6A, 5mC/m5C, and ac4C utilize modification-specific antibodies in their detection (Arango et al., 2018; Dominissini et al., 2012; Edelheit et al., 2013; Meyer et al., 2012; Weber et al., 2005; Figure 4A). m6A and inosine can be detected using proteins that specifically bind them (the YTH domain and EndoV) (Knutson et al., 2020; Meyer, 2019; Figure 4B). Differential cleavage (by enzymes or chemicals) is used for detection of m6A, inosine, and ψ (Garcia-Campos et al., 2019; Zhang et al., 2019; Cattenoz et al., 2013; Marchand et al., 2020; Figure 4C). Techniques for detection of m6A, inosine, ψ, and ac4C utilize reverse transcription inhibition (RT stops) or modification-induced RT error (mutational profiling) (Linder et al., 2015; Ke et al., 2015, 2017; Grozhik et al., 2017; Levanon et al., 2004; Li et al., 2011, 2015; Bahn et al., 2012; Peng et al., 2012; Suzuki et al., 2015; Carlile et al., 2014; Lovejoy et al., 2014; Schwartz et al., 2014; Sas-Chen et al., 2020; Figure 4D), and two separate techniques for detection of m5C in RNA utilize covalent trapping of modifying enzymes (Hussain et al., 2013b; Khoddami and Cairns, 2013) (Figure 4E). Last, all five of these modifications use chemical derivatization to differentiate different modification (Booth et al., 2012; Frommer et al., 1992; Sas-Chen et al., 2020; Suzuki et al., 2015; Wang et al., 2020; Figure 4F). These underlying principles will no doubt continue to be recurring themes as new technologies are developed for study of nucleic acid modifications.
Figure 4. Underlying principles relevant for several techniques.
(A) Enrichment of modified RNA by IP.
(B) Use of reader proteins to target or enrich for modified RNA. Active moieties can be fused to the reader protein to act as an epitope tag for enrichment or to further modify the RNA.
(C) Differential enzyme activity/chemical reactivity as a result of modification.
(D) Induction of RT stops or mutations by modified bases.
(E) Covalent trapping of RNA-modifying enzymes.
(F) Further chemical modification of RNA to alter base-pairing, to alter susceptibility to further modification, or to protect the RNA from digestion.
As the study of epitranscriptomics grows more and more robust, so too grows the demand for methods that can easily and effectively identify modification sites. To this end, techniques have been developed to map multiple modifications at once. For example, Khoddami et al. (2019) have developed a method (modified RNA bisulfite sequencing, RBS-seq) that utilizes bisulfite treatment to detect not only m5C but also m1A and ψ in RNA. Additionally, the Oxford Nanopore sequencing technology is capable of differentiating m6A from A and m5C from C in mRNA and m7G from G and ψ from U in rRNA (Garalde et al., 2018; Smith et al., 2019). Currently, work is ongoing to validate the Nanopore sequencing method for detection of RNA modifications individually and in parallel.
Beyond detection and mapping, direct manipulation of modified bases is a powerful potential tool for study of nucleic acid modifications. Although ψ synthases have been targeted to specific locations using bespoke H/ACA box snoRNAs (Karijolich and Yu, 2011), the advent of CRISPR-Cas-9 technology has the potential to allow direct manipulation of other modified bases. Rauch et al. (2018) have developed a method that couples YTH reader proteins to a catalytically dead Cas13, which allows a reader to be targeted to an RNA regardless of its methylation status, where the reader then exerts its function on the targeted RNA. In contrast, Liu et al. (2019b) coupled catalytically dead Cas9 to m6A writer/eraser enzymes, which allows manipulation of the modification status of the targeted m6A site without disrupting methylation at other sites through knockdown of writers or erasers. Theoretically, any modification for which a writer, reader, or eraser is known can be studied using similar sets of fusion proteins.
Using these and similar techniques, future research will be able to place these modifications more accurately in the epigenome and epitranscriptome and will be able to provide a more complete picture of which modifications are in which RNA molecules or at which genomic loci. When detected and mapped, the effects of these modifications can be investigated in a low-throughput, site-specific manner. By revealing individual targets for further study, these techniques have potential to enhance our mechanistic understanding of how gene expression is regulated by nucleic acid modifications. Furthermore, these techniques may also have implications for human health. Researchers have used liquid biopsy to collect cell-free DNA and analyze its methylation content as a marker for disease (Bronkhorst et al., 2019). Given that the levels and locations of the RNA modifications discussed here can serve as potential biomarkers for diseases such as cancer (Huang et al., 2020), it will be fascinating to see whether these technologies are applied to detection of diagnostic RNA modifications in liquid biopsy samples as well. As new technologies are developed that more accurately and efficiently map these modifications, their diagnostic potential will only increase.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (T32GM132039 to M.C.O. and R35GM133721 to K.F.L.), by the Roy and Diana Vagelos Scholars Program in the Molecular Life Sciences at the University of Pennsylvania (to C.Z.), and by start-up funding from the University of Pennsylvania (to the Liu lab). Marvin was used for drawing, displaying, and characterizing chemical structures, substructures, and reactions in Figures 1 and 2 (Marvin 21.2, 2021, ChemAxon; https://www.chemaxon.com/). Figures 3 and 4 were created with BioRender.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, and Suzuki T (2019). Cap-specific terminal N 6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080. [DOI] [PubMed] [Google Scholar]
- Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, Jia X-Y, Micura R, and Lusser A (2017). Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 18, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, et al. (2018). Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 175, 1872–1886.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie JP, Cavaillé J, and Hüttenhofer A (2002). The expanding snoRNA world. Biochimie 84, 775–790. [DOI] [PubMed] [Google Scholar]
- Bahn JH, Lee J-H, Li G, Greer C, Peng G, and Xiao X (2012). Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 22, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, and Fuller MT (2017). The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6, e26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A, and Ofengand J (1993). Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762. [DOI] [PubMed] [Google Scholar]
- Bass BL, and Weintraub H (1988). An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Bird AP, Taggart MH, and Smith BA (1979). Methylated and unmethylated DNA compartments in the sea urchin genome. Cell 17, 889–901. [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46 (D1), D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack KE, Höbartner C, and Bohnsack MT (2019). Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes (Basel) 10, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, and Rottman FM (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, and Balasubramanian S (2012). Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937. [DOI] [PubMed] [Google Scholar]
- Boulias K, Toczydłowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, Guez T, Vasseur J-J, Debart F, Aravind L, et al. (2019). Identification of the m6Am Methyltransferase PCIF1 Reveals the Location and Functions of m6Am in the Transcriptome. Mol. Cell 75, 631–643.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois G, Ney M, Gaspar I, Aigueperse C, Schaefer M, Kellner S, Helm M, and Motorin Y (2015). Eukaryotic rRNA Modification by Yeast 5-Methylcytosine-Methyltransferases and Human Proliferation-Associated Antigen p120. PLoS ONE 10, e0133321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronkhorst AJ, Ungerer V, and Holdenrieder S (2019). The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect. Quantif 17, 100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, and Gilbert WV (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Martinez NM, Schaening C, Su A, Bell TA, Zinshteyn B, and Gilbert WV (2019). mRNA structure determines modification by pseudouridine synthase 1. Nat. Chem. Biol 15, 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattenoz PB, Taft RJ, Westhof E, and Mattick JS (2013). Transcriptome-wide identification of A > I RNA editing sites by inosine specific cleavage. RNA 19, 257–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Luo G-Z, and He C (2015). High-Resolution Mapping of N6-Methyladenosine in Transcriptome and Genome Using a Photo-Crosslinking-Assisted Strategy. Methods Enzymol. 560, 161–185. [DOI] [PubMed] [Google Scholar]
- Chen X, Li A, Sun B-F, Yang Y, Han Y-N, Yuan X, Chen R-X, Wei W-S, Liu Y, Gao C-C, et al. (2019). 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol 21, 978–990. [DOI] [PubMed] [Google Scholar]
- Cohn WE, and Volkin E (1951). Nucleoside-5′-Phosphates from Ribonucleic Acid. Nature 167, 483–484. [Google Scholar]
- Czudnochowski N, Ashley GW, Santi DV, Alian A, Finer-Moore J, and Stroud RM (2014). The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs. Nucleic Acids Res. 42, 2037–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR (1995). Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 23, 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FF, and Allen FW (1957). Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem 227, 907–915. [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, and Wu L (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun 7, 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, and Sorek R (2013). Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang S-Y, Baltissen MPA, Jansen PWTC, Rossa M, et al. (2017). N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol 24, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler DE, Franco MK, Batool Z, Wu MZ, Dubuke ML, Dobosz-Bartoszek M, Jones JD, Polikanov YS, Roy B, and Koutmou KS (2019). Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. USA 116, 23068–23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, and Paul CL (1992).A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun 4, 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. (2018). Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206. [DOI] [PubMed] [Google Scholar]
- Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis A, et al. (2019). Deciphering the “m6A Code” via Antibody-Independent Quantitative Profiling. Cell 178, 731–747.e16. [DOI] [PubMed] [Google Scholar]
- Gerber AP, and Keller W (1999). An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286, 1146–1149. [DOI] [PubMed] [Google Scholar]
- Gigova A, Duggimpudi S, Pollex T, Schaefer M, and Koš M (2014). A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. RNA 20, 1632–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozhik AV, Linder B, Olarerin-George AO, and Jaffrey SR (2017). Mapping m6A at Individual-Nucleotide Resolution Using Crosslinking and Immunoprecipitation (miCLIP). Methods Mol. Biol 1562, 55–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Cedar H, and Razin A (1982). Substrate and sequence specificity of a eukaryotic DNA methylase. Nature 295, 620–622. [DOI] [PubMed] [Google Scholar]
- Gu X, Liu Y, and Santi DV (1999). The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. USA 96, 14270–14275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr., Jungkamp A-C, Munschauer M, et al. (2010). Transcriptome-wide identification of RNA-binding protein and micro-RNA target sites by PAR-CLIP. Cell 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton CS, Greco TM, Vizthum CA, Ginter JM, Johnston MV, and Mueller EG (2006). Mechanistic investigations of the pseudouridine synthase RluA using RNA containing 5-fluorouridine. Biochemistry 45, 12029–12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu H, Wataya Y, Kai K, and Iida S (1970). Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry 9, 2858–2865. [DOI] [PubMed] [Google Scholar]
- He C, Bozler J, Janssen KA, Wilusz JE, Garcia BA, Schorn AJ, and Bonasio R (2021). TET2 chemically modifies tRNAs and regulates tRNA fragment levels. Nat. Struct. Mol. Biol 28, 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho NW, and Gilham PT (1971). Reaction of pseudouridine and inosine with N-cyclohexyl-N’-beta-(4-methylmorpholinium)ethylcarbodiimide. Biochemistry 10, 3651–3657. [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. (2017). Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Pookanjanatavip M, Gu X, and Santi DV (1998). A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry 37, 344–351. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, and Rao A (2010). The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 5, e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. (2018). Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol 20, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Deng X, and Chen J (2020). RNA modifications in cancer: functions, mechanisms, and therapeutic implications. Annu. Rev. Cancer Biol 4, 221–240. [Google Scholar]
- Huber SM, van Delft P, Mendil L, Bachman M, Smollett K, Werner F, Miska EA, and Balasubramanian S (2015). Formation and abundance of 5-hydroxymethylcytosine in RNA. ChemBioChem 16, 752–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, and Frye M (2009). The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J. Cell Biol 186, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Aleksic J, Blanco S, Dietmann S, and Frye M (2013a). Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 14, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, and Frye M (2013b). NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi M, Tsuji S, Suda A, and Futaki S (2017). Detection of N6-methyl-adenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. (Camb.) 53, 12930–12933. [DOI] [PubMed] [Google Scholar]
- Ito S, Akamatsu Y, Noma A, Kimura S, Miyauchi K, Ikeuchi Y, Suzuki T, and Suzuki T (2014a). A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J. Biol. Chem 289, 26201–26212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, and Suzuki T (2014b). Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J. Biol. Chem 289, 35724–35730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Puno MR, Meydan C, Lailler N, Mason CE, Lima CD, Anderson KV, and Keeney S (2018). ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife 7, e30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang Y-G, and He C (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, and Yu Y-T (2011). Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, and Suzuki T (2017). ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 45, 7401–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr., and Darnell RB, (2017). m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, and Cairns BR (2013). Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol 31, 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, and Cairns BR (2014). Transcriptome-wide target profiling of RNA cytosine methyltransferases using the mechanism-based enrichment procedure Aza-IP. Nat. Protoc 9, 337–361. [DOI] [PubMed] [Google Scholar]
- Khoddami V, Yerra A, Mosbruger TL, Fleming AM, Burrows CJ, and Cairns BR (2019). Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl. Acad. Sci. USA 116, 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MY, and Redman KL (2002). RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry 41, 11218–11225. [DOI] [PubMed] [Google Scholar]
- King TH, Liu B, McCully RR, and Fournier MJ (2003). Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11, 425–435. [DOI] [PubMed] [Google Scholar]
- Kleinman CL, and Majewski J (2012). Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335, 1302, author reply 1302 [DOI] [PubMed] [Google Scholar]
- Knutson SD, and Heemstra JM (2021). Protein-based molecular recognition tools for detecting and profiling RNA modifications. Curr. Opin. Struct. Biol 69, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SD, Arthur RA, Johnston HR, and Heemstra JM (2020). Selective Enrichment of A-to-I Edited Transcripts from Cellular RNA Using Endonuclease V. J. Am. Chem. Soc 142, 5241–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, and Zhang Y (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski S, Yamane T, and Fresco JR (1971). Nucleotide sequence of the “denaturable” leucine transfer RNA from yeast. Science 172, 385–387. [DOI] [PubMed] [Google Scholar]
- Kruppa J, and Zachau HG (1972). Multiplicity of serine-specific transfer RNAs of brewer’s and baker’s yeast. Biochim. Biophys. Acta 277, 499–512. [DOI] [PubMed] [Google Scholar]
- Kumbhar BV, Kamble AD, and Sonawane KD (2013). Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochem. Biophys 66, 797–816. [DOI] [PubMed] [Google Scholar]
- Küpfer PA, and Leumann CJ (2007). The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res 35, 58–68. 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasman L, Krupalnik V, Viukov S, Mor N, Aguilera-Castrejon A, Schneir D, Bayerl J, Mizrahi O, Peles S, Tawil S, et al. (2020). Context-dependent functional compensation between Ythdf m6A reader proteins. Genes Dev. 34, 1373–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. (2004). Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol 22, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Li E, and Zhang Y (2014). DNA methylation in mammals. Cold Spring Harb. Perspect. Biol 6, a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene PA, and Bass LW (1926). The action of hydrazine hydrate on uridine. Journal of Biological Chemistry 71, 167–172. [Google Scholar]
- Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, and Cheung VG (2011). Widespread RNA and DNA sequence differences in the human transcriptome. Science 333, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, and Yi C (2015). Chemical pull-down reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol 11, 592–597. [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, and Yi C (2016). Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31. [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu Q, and Fournier MJ (2007). rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 28, 965–977. [DOI] [PubMed] [Google Scholar]
- Lin W, Piskol R, Tan MH, and Li JB (2012). Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335, 1302, author reply 1302. [DOI] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, and Jaffrey SR (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Parisien M, Dai Q, Zheng G, He C, and Pan T (2013). Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol 10, 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, and Novoa EM (2019a). Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun 10, 4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-M, Zhou J, Mao Y, Ji Q, and Qian S-B (2019b). Programmable RNA N6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat. Chem. Biol 15, 865–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Siejka-Zielińska P, Velikova G, Bi Y, Yuan F, Tomkova M, Bai C, Chen L, Schuster-Böckler B, and Song C-X (2019c). Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol 37, 424–429. [DOI] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, and Brown PO (2014). Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 9, e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Hajkova P, and Ecker JR (2018). Dynamic DNA methylation: In the right place at the right time. Science 361, 1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Pichot F, Neybecker P, Ayadi L, Bourguignon-Igel V, Wacheul L, Lafontaine DLJ, Pinzano A, Helm M, and Motorin Y (2020). HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic. Acids Res 48, e110. 10.1093/nar/gkaa769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur J-J, Chen Q, et al. (2017). Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Sindelar M, Despic V, Guez T, Hawley BR, Vasseur J-J, Rentmeister A, Gross SS, Pellizzoni L, Debart F, et al. (2019). FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol 15, 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre ABR, Gokhale NS, Cerchietti L, Jaffrey SR, Horner SM, and Mason CE (2020). Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci. Rep 10, 6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD (2019). DART-seq: an antibody-free method for global m6A detection. Nat. Methods 16, 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, and Jaffrey SR (2015). 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinie B, Wang J, Lim KS, Hillebrand R, Lu Z-X, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, et al. (2016). m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods 13, 692–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, and Bass BL (1997). Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry 36, 8429–8434. [DOI] [PubMed] [Google Scholar]
- Newby MI, and Greenbaum NL (2002). Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. USA 99, 12697–12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K (2016). A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol 17, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes E, Vadlamani P, and Hundley HA (2017). Methods for the Detection of Adenosine-to-Inosine Editing Events in Cellular RNA. Methods Mol. Biol 1648, 103–127. [DOI] [PubMed] [Google Scholar]
- Pace CN, Heinemann U, Hahn U, and Saenger W (1991). Ribonuclease T1: structure, function, and stability. Angew. Chem. Int. Ed. Engl 30, 343–360. [Google Scholar]
- Patterson JB, and Samuel CE (1995). Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol 15, 5376–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie DA (1979). Direct chemical method for sequencing RNA. Proc. Natl. Acad Sci. USA 76, 1760–1764. 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC-M, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. (2012). Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol 30, 253–260. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Gilad Y, and Pritchard JK (2012). Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335, 1302, author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskol R, Peng Z, Wang J, and Li JB (2013). Lack of evidence for existence of noncanonical RNA editing. Nat. Biotechnol 31, 19–20. [DOI] [PubMed] [Google Scholar]
- Rasmussen KD, and Helin K (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch S, He C, and Dickinson BC (2018). Targeted m6a reader proteins to study epitranscriptomic regulation of single rnas. J. Am. Chem. Soc 140, 11974–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, and He C (2017a). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo G-Z, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. (2017b). YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6, e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter JM, and Schultz SC (1998). Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Yano T, Kawabata H, Ueda H, and Suzuki T (2010). Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol 6, 733–740. [DOI] [PubMed] [Google Scholar]
- Sakurai M, Ueda H, Yano T, Okada S, Terajima H, Mitsuyama T, Toyoda A, Fujiyama A, Kawabata H, and Suzuki T (2014). A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 24, 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, Bryson KM, Shachar R, Liman GLS, Burkhart BW, et al. (2020). Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583, 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, and Perry RP (1977). Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol 115, 695–714. [DOI] [PubMed] [Google Scholar]
- Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, et al. (2015). Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun 6, 6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutsky EK, DeNizio JE, Hu P, Liu MY, Nabel CS, Fabyanic EB, Hwang Y, Bushman FD, Wu H, and Kohli RM (2018). Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotechnol 36, 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. (2014).Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg PH, and Hartner J (2003). Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol 13, 279–283. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Single F, Kuner T, Higuchi M, and Sprengel R (2001). Genetic manipulation of key determinants of ion flow in glutamate receptor channels in the mouse. Brain Res. 907, 233–243. [DOI] [PubMed] [Google Scholar]
- Selmi T, Hussain S, Dietmann S, Heiß M, Borland K, Flad S, Carter J-M, Dennison R, Huang Y-L, Kellner S, et al. (2021). Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 49, 1006–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, Navarrete-Perea J, Sheng W, Gygi SP, Adelman K, and Shi Y (2019). PCIF1 Catalyzes m6Am mRNA Methylation to Regulate Gene Expression. Mol. Cell 75, 620–630.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam R, Fierer J, Kaiser S, Helm M, Jurkowski TP, and Jeltsch A (2015). Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 1, 15010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Servis RE, and Welcher M (1970). Reactions of Uracil and Cytosine Derivatives with Sodium Bisulfite. J. Am. Chem. Soc 92, 422–424. [Google Scholar]
- Sharma S, Yang J, Watzinger P, Kötter P, and Entian K-D (2013). Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 41, 9062–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Langhendries J-L, Watzinger P, Kötter P, Entian K-D, and Lafontaine DLJ (2015). Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 43, 2242–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, van Nues R, Watzinger P, Kötter P, Lafontaine DLJ, Granneman S, and Entian K-D (2017). Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 13, e1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Ontiveros RJ, Owens MC, Liu MY, Ghanty U, Kohli RM, and Liu KF (2021). TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J. Biol. Chem 296, 100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, and He C (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Cao J, Cheng M, Xiang S, Gao M, Li T, Ying X, Wang F, Yue Y, Lu Z, et al. (2020). A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat. Chem. Biol 16, 887–895. [DOI] [PubMed] [Google Scholar]
- Sinclair WR, Arango D, Shrimp JH, Zengeya TT, Thomas JM, Montgomery DC, Fox SD, Andresson T, Oberdoerffer S, and Meier JL (2017). Profiling Cytidine Acetylation with Specific Affinity and Reactivity. ACS Chem. Biol 12, 2922–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, and Meissner A (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet 14, 204–220. [DOI] [PubMed] [Google Scholar]
- Smith AM, Jain M, Mulroney L, Garalde DR, and Akeson M (2019). Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS ONE 14, e0216709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhang M, Li K, Bai D, and Yi C (2019). Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 29, 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ueda H, Okada S, and Sakurai M (2015). Transcriptome-wide identification of adenosine-to-inosine editing using the ICE-seq method. Nat. Protoc 10, 715–732. [DOI] [PubMed] [Google Scholar]
- Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T, and Watanabe K (2009). Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 37, 1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JM, Briney CA, Nance KD, Lopez JE, Thorpe AL, Fox SD, Bortolin-Cavaille M-L, Sas-Chen A, Arango D, Oberdoerffer S, et al. (2018). A chemical signature for cytidine acetylation in RNA. J. Am. Chem. Soc 140, 12667–12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JM, Bryson KM, and Meier JL (2019). Nucleotide resolution sequencing of N4-acetylcytidine in RNA. Methods Enzymol. 621, 31–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Piñeyro D, Filonava L, Stracker TH, Batlle E, and Ribas de Pouplana L (2014). A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS Lett. 588, 4279–4286. [DOI] [PubMed] [Google Scholar]
- Trixl L, and Lusser A (2019). The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA 10, e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell A, Rorbach J, Lantaff R, Blanco S, Sauer S, et al. (2016). Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun 7, 12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR, Graille M, and Lafontaine DLJ (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, and He C (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao Y, Dong S, Yu Q, and Jia G (2020). Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat. Chem. Biol 16, 896–903. [DOI] [PubMed] [Google Scholar]
- Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, Sloan KE, and Bohnsack MT, (2017). Human METTL16 is a N6-methyladenosine (m6A) methyitransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, and Schübeler D (2005). Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet 37, 853–862. [DOI] [PubMed] [Google Scholar]
- Wei C, Gershowitz A, and Moss B (1975a). N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, and Moss B (1975b). Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386. [DOI] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. (2018). Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 71, 973–985.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y-L, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, et al. (2018). Epitranscriptomic m6a regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen Y-S, Hao Y-J, Sun B-F, Sun H-Y, Li A, Ping X-L, Lai W-Y, et al. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 61, 507–519. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Wang Y, Tang Q, Wei L, Zhang X, and Jia G (2018). An Elongation- and Ligation-Based qPCR Amplification Method for the Radiolabeling-Free Detection of Locus-Specific N6 -Methyladenosine Modification. Angew. Chem. Int. Ed. Engl 57, 15995–16000. [DOI] [PubMed] [Google Scholar]
- Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y, Li A, Wang X, Bhattarai DP, Xiao W, et al. (2017). 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 27, 606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Han X, Yang W-L, Zhang M, Ma H-L, Sun B-F, Li A, Xia J, Chen J, et al. (2019). RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol. Cell 75, 1188–1202.e11. [DOI] [PubMed] [Google Scholar]
- Yoshida M, and Ukita T (1968). Modification of nucleosides and nucleotides. VII. Selective cyanoethylation of inosine and pseudouridine in yeast transfer ribonucleic acid. Biochim. Biophys. Acta 157, 455–465. [PubMed] [Google Scholar]
- Yu CT, and Allen FW (1959). Studies on an isomer of uridine isolated from ribonucleic acids. Biochim. Biophys. Acta 32, 393–406. [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song C-X, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. (2012). Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachau HG, Dütting D, and Feldmann H (1966). The structures of two serine transfer ribonucleic acids. Hoppe Seylers Z. Physiol. Chem 347, 212–235. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wang S, Gao S, Soares F, Ahmed M, Guo H, Wang M, Hua JT, Guan J, Moran MF, et al. (2018). Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biol. 16, e2006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen L-Q, Zhao Y-L, Yang C-G, Roundtree IA, Zhang Z, Ren J, Xie W, He C, and Luo G-Z (2019). Single-base mapping of m6A by an antibody-independent method. Sci. Adv 5, eaax0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vågbø CB, Shi Y, Wang W-L, Song S-H, et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, and Nishikura K (2009). Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med 1, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]