Abstract
During an experiment in two laboratory-scale enclosures filled with lake water (130 liters each) we noticed the almost-complete lysis of the cyanobacterial population. Based on electron microscopic observations of viral particles inside cyanobacterial filaments and counts of virus-like particles, we concluded that a viral lysis of the filamentous cyanobacteria had taken place. Denaturing gradient gel electrophoresis (DGGE) of 16S ribosomal DNA fragments qualitatively monitored the removal of the cyanobacterial species from the community and the appearance of newly emerging bacterial species. The majority of these bacteria were related to the Cytophagales and actinomycetes, bacterial divisions known to contain species capable of degrading complex organic molecules. A few days after the cyanobacteria started to lyse, a rotifer species became dominant in the DGGE profile of the eukaryotic community. Since rotifers play an important role in the carbon transfer between the microbial loop and higher trophic levels, these observations confirm the role of viruses in channeling carbon through food webs. Multidimensional scaling analysis of the DGGE profiles showed large changes in the structures of both the bacterial and eukaryotic communities at the time of lysis. These changes were remarkably similar in the two enclosures, indicating that such community structure changes are not random but occur according to a fixed pattern. Our findings strongly support the idea that viruses can structure microbial communities.
Photosynthetically derived organic carbon is one of the major energy sources for heterotrophic bacteria in oceans and lakes (3, 6, 16). This organic carbon is made available to the bacteria through various pathways. Exudation by phototrophs and excretion by their grazers provide rather constant release of carbon. The decline of phytoplankton blooms releases dissolved organic carbon in a short time, which can be rapidly used by heterotrophic bacteria (4, 13, 20, 51). To study the growth of heterotrophic bacteria on exudates released by cyanobacteria, we conducted experiments in laboratory-scale enclosures (LSEs) filled with lake water. However, 15 days after the experiments started nearly all filamentous cyanobacteria lysed. Electron microscopic observations of viruses inside filaments of cyanobacteria and counts of virus-like particles indicated a viral lysis event.
Cell lysis is a major cause of phytoplankton bloom decline (13, 51), and many studies have shown the importance of viruses in phytoplankton mortality (12, 20, 44, 48, 49). These viruses are considered to be important members of the microbial loop (5, 8, 49). High viral abundance and decay rates suggest considerable viral activity (10), which is also indicated by observations of microbial cells containing mature viral particles (42). It has been calculated that 10 to 20% of the marine bacterial community is lysed by viruses on a daily basis (47). Approximately the same mortality was found in a freshwater study (23). Hence, viral lysis is a significant factor in controlling bacterial and primary production (23, 33, 49, 54) and carbon and nutrient flow within the microbial loop (9, 34).
Besides controlling carbon production, viruses are also thought to structure microbial communities (25). Similar to the size-selective grazing of bacterivores (30), viral host specificity could be a very strong structuring force of microbial communities. The lysis and removal of species from the microbial community and the consecutive nutrient release may give other species the opportunity to proliferate. To test the hypothesis that viruses could structure the microbial community, we used denaturing gradient gel electrophoresis (DGGE) (19) to follow the changes in the structure of both the bacterial and eukaryotic communities before and after the lysis event. DGGE analysis of 16S and 18S ribosomal DNA (rDNA) fragments circumvents the problem of underestimating microbial diversity due to noncultivable microorganisms. This molecular technique has been used extensively to profile natural bacterial diversity (18, 36, 50), and statistical analysis of the DGGE patterns can reveal relative changes in the microbial community structure (52, 53).
MATERIALS AND METHODS
Experimental design.
Two LSEs, especially designed to mimic the physical environment of Lake Loosdrecht, The Netherlands (46), were each filled with 130 liters of Lake Loosdrecht water sampled on 26 November 1996. Lake Loosdrecht is a shallow eutrophic lake dominated by filamentous cyanobacteria. The water temperature at the time of sampling was 3.6°C. The temperature was raised to 20°C within 1 day. Both LSEs were supplied with medium at a dilution rate of 0.05 day−1. The incident irradiance was 50 W · m−2 during a 16-h light period. The LSEs were stirred continuously to assure complete mixing. After 1 week of adaptation, both LSEs received elevated light levels of 150 W · m−2 during 4 h around the midpoint of the light period. In one system (LSE 1), stirring was halted during this high-light period. Elevated light levels were used to trigger exudate production.
Chl-a, virus-like particles, and numbers of bacteria.
Chlorophyll a (Chl-a) concentrations were measured after hot-ethanol extraction (35). Virus-like particles were enumerated by YOPRO staining (24). Bacteria were counted by DAPI (4′,6-diamidino-2-phenylindole) staining (41).
DNA extraction, PCR, and DGGE.
DNA was released from the cells by mechanical force (bead beating) concomitant with phenol extraction and ethanol precipitation (57). PCR primers against the V2 region were used for the amplification of the 16S rRNA gene. The PCR primers were F357GC (5′-CGCCCG CCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGC AGCAG-3′), which contains a GC-rich clamp and is specific for most Bacteria, and R518 (5′-ATTACCGCGGCTGCTGG-3′), which is specific for most Bacteria, Archaea, and Eucarya (36). PCR amplification was performed in a 50-μl volume containing approximately 100 ng of template DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.01% (wt/vol) gelatin, 1.5 mM MgCl2, 0.5 μM (each) primer, 200 μM (each) deoxynucleotide, 400 ng of bovine serum albumin, and 2.5 U of Taq DNA polymerase (Boehringer Mannheim, Mannheim, Germany). PCR cycling was performed with a Perkin-Elmer 480 thermocycler. The temperature-cycling conditions were as follows. After a preincubation at 94°C for 5 min, a total of 25 cycles were performed at 94°C for 1 min, TA for 1 min, and 72°C for 1 min. In the first 20 cycles, TA decreased by 1°C, stepwise, each two cycles, from 65°C in the first cycle to 56°C in the 20th. In the last five cycles, TA was 55°C. Cycling was followed by 5 min of incubation at 72°C. The primers for the amplification of the 18S rRNA gene were F1427GC (5′-CGCCCGCCGCGCC CCGCGCCCGGCCCGCCGCCCCCGCCCCTCTGTGATGCCCTTAGATGT TCTGGG-3′) and R1616 (5′-GCGGTGTGTACAAAGGGCAGGG-3′). Both primers are specific for eukaryotic aquatic microorganisms (53). The temperature-cycling conditions were as follows: one preincubation step at 94°C for 5 min followed by 25 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min and then a final extension step of 72°C for 5 min. The reaction conditions were as described above.
DGGE was performed as described previously (36, 53). Briefly, similarly sized PCR products were separated on a 1.5-mm-thick vertical gel containing 8% (wt/vol) polyacrylamide (acrylamide-bisacrylamide, 37.5:1) and a linear gradient of the denaturants urea and formamide, increasing from 35% at the top of the gel to 55% at the bottom for the separation of the 16S rDNA fragment and from 30 to 55% for the separation of the 18S rDNA fragment. Here, 100% denaturant is defined as 7 M urea and 40% (vol/vol) formamide. Equal amounts of PCR products were applied to the DGGE gel. The concentrations of PCR products were estimated by separation on 2.0% agarose gels, staining with ethidium bromide (see below), and analysis of digitized images with ImageQuant software (Molecular Dynamics Ltd., Kemsing, England). Fifty microliters of the sample with the largest amount of PCR product was loaded on the DGGE gel. All other samples were loaded in amounts relative to this sample. Electrophoresis was performed at 60°C in a buffer containing 40 mM Tris, 40 mM acetic acid, and 1 mM EDTA at pH 7.6 (0.5× TAE), and 75 V of electricity was applied to the submerged gel for 16 h. Nucleic acids were visualized by staining them for 1 h in 0.5× TAE buffer containing 0.5 mg of ethidium bromide liter−1 followed by destaining for 5 min in demineralized water and photographing the gel with a charge-coupled device camera (The Imager; Appligene, Illkirch, France). Digitized images were inverted with Photostyler software (Aldus Corporation, Seattle, Wash.). The contrast and gray balance of the entire image were adjusted to reduce background.
DGGE marker sequences.
Sequences obtained from a clone library of the 16S rRNA genes from Lake Loosdrecht (56) were used to construct a marker for DGGE analysis. Cyanobacterial sequences in this clone library were aligned against the most similar sequences from the EMBL database (see below).
Sequencing of excised DGGE bands.
Nucleotide sequences of DGGE bands of interest were obtained either by direct sequencing of DNA from excised DGGE bands (18S rDNA) or by sequencing the excised fragment (16S rDNA) ligated in a pGEM-T vector (Promega, Madison, Wis.) and were transfected through heat shock to Epicurian Coli XL1-Blue MRF′ supercompetent Escherichia coli cells (Stratagene, La Jolla, Calif.). Ligation, transformation, and sequencing procedures for the 16S rDNA fragment have been described previously (56). To obtain sequences from the 18S rDNA fragments, a small block of gel from the middle of the target band was excised from the DGGE gel with a surgical knife and placed into a 2-ml screw-cap tube. To extract the DNA from the gel, 0.5 ml of TE (10 mM Tris, pH 7.6, 1 mM EDTA) was added to the tube together with 0.5 g of zirconium beads (0.1-mm diameter). The tubes were then vigorously shaken (5,000 rpm) on a Mini Beadbeater (Biospec Products, Bartlesville, Okla.) for 2 min with intermittent cooling on ice. The released DNA fragment was then amplified in 25 cycles of PCR with eukaryote-specific primers previously described (53). The reverse primer was extended at the 5′ site by the vector-specific M13 forward sequence, yielding 5′-TGTAAAACGACGGCCAGTGCGGTGTGTACAAAGGGCAGGG. Amplification primers and primer-dimers were removed with the Wizard PCR Preps direct purification system (Promega) according to the manufacturer’s instructions. The PCR products were then cycle sequenced with a Texas red-labeled M13 forward primer (5′-TGTAAAACGACGGCCA) and Thermosequenase (Amersham, Little Chalfont, United Kingdom). Fragment separation, detection, and base calling were done with a Vistra DNA sequencer 725 (Amersham).
DGGE pattern analysis.
The DGGE banding patterns were converted to a binary matrix to make the data accessible to statistical analysis (53). The presence or absence of a nucleic acid band at the same height in each lane was marked with a 1 or 0, respectively. Since microorganisms can have multiple copies of their rRNA gene, care should be taken with the interpretation of the number of species present in a DGGE pattern. We therefore refer to a band as a sequence type rather than a species. Gel images were enlarged two times to facilitate band detection. From this binary matrix, a distance matrix was calculated (37). The distance matrix was then analyzed by nonmetric multidimensional scaling (NMDS). This analysis constructs a map showing the relationships among a number of observations given only a table of distances between them. The data is presented in a Euclidean plane such that highly similar measurements are plotted close together. Such a graphical representation is much easier to interpret than the original table of distances. The dimensions (axes) in the map have no special significance and can be rotated or mirrored without influencing the relative distances between the points. As a measure of the goodness of fit of the reproduced distances to the observed distances, the stress value is used. When stress values are <0.1, the NMDS plot is considered to be an acceptable representation of the original data. Interpretation of the NMDS plot can be achieved by explaining single dimensions or by finding structures or patterns in the multidimensional space (7, 31). Applied to DGGE data, the NMDS map shows every banding pattern—the community structure at a particular point in time—as one dated point, and by connecting consecutive points, relative changes in the community structure can be visualized and interpreted. NMDS has been proven to be useful as a tool for analysis of genetic structures (32, 52).
Sequence analysis.
The partial 16S and 18S rRNA gene sequences recovered from the two LSEs were screened against GenBank and EMBL sequences with BLAST (1) (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-blast?Jform=0). The sequences with the highest similarities were then used in a similarity index calculation. Gaps and ambiguities were not included in this calculation. The sequences were aligned by the DCSE program (15).
Nucleotide sequence accession numbers.
All partial sequences from excised bands have been deposited in the EMBL database. Eukaryotic sequences have been deposited under accession no. AJ009546 to AJ009554. Bacterial sequences have been deposited under accession no. AJ009642 to AJ009654.
RESULTS
Cyanobacterial lysis.
During the first 15 days of the experiment the Chl-a concentration increased almost threefold for LSE 1 and fourfold for LSE 2, indicating an actively growing cyanobacterial community (Table 1). Microscopic observations showed that more than 95% of the cyanobacterial population consisted of Oscillatoria c.f. limnetica and Prochlorothrix hollandica. However, 15 days after the experiment started, a decrease in Chl-a occurred in both LSEs: within 4 days, levels dropped to 10% of the maximum levels. Parallel to this decrease in Chl-a, we noticed an increase in virus-like particles and numbers of bacteria (Table 1). Electron microscopic observations showed lambda-like phages in the culture medium, attached to filaments of cyanobacteria and densely packed inside lysed cyanobacterial filaments.
TABLE 1.
Chl-a concentration, virus-like particles, and bacterial abundance for Lake Loosdrecht and both LSEsa
| Parameter | Field | LSE 1
|
LSE 2
|
||
|---|---|---|---|---|---|
| Prelysis | Postlysis | Prelysis | Postlysis | ||
| Chl-a (μg · liter−1) | 113 | 296 | 12 | 401 | 15 |
| Virus-like particles (1010 liter−1) | 6.4 | 9.9 | 20 | 13 | 28 |
| Bacteria (109 liter−1) | 6.5 | 11 | 51 | 19 | 53 |
Prelysis values were measured on day 15. Postlysis values were measured on day 22.
Bacterial community structure.
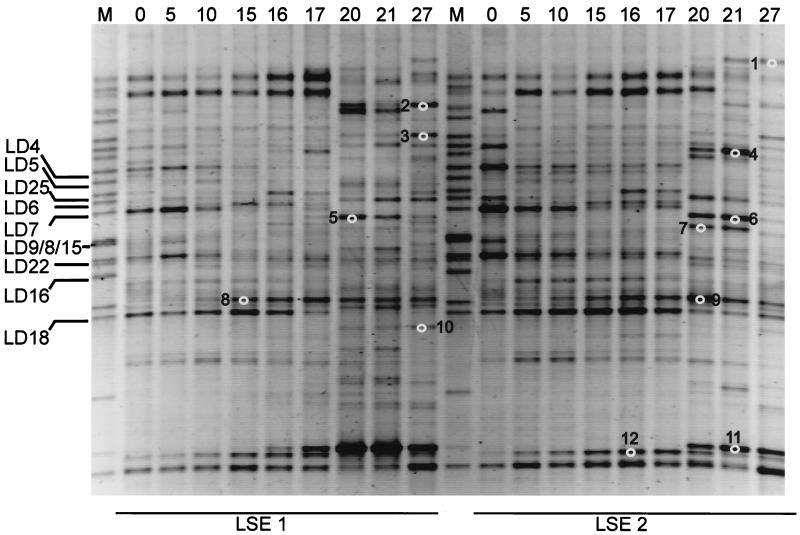
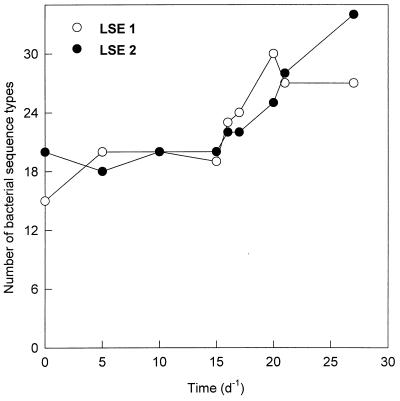
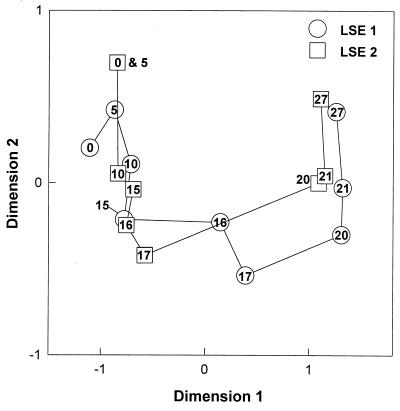
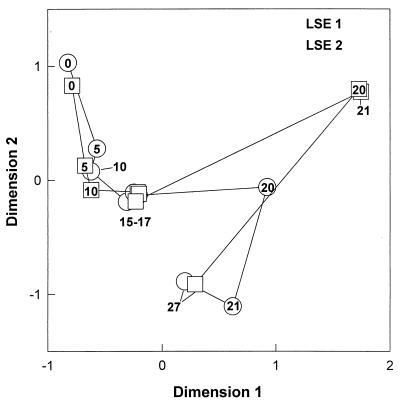
Analysis of the 16S rRNA-defined community revealed that most of the identified bands related to cyanobacteria had disappeared from the DGGE pattern in both LSEs (Fig. 1). Bands related to cyanobacteria were identified by comparison with clone sequences, obtained from Lake Loosdrecht (Table 2), from the marker lanes. At the beginning of the experiment, 35% of the DGGE bands were related to cyanobacteria. Most of these bands decreased in intensity from day 10 on, except for the LD18 band, which increased in intensity until day 15 before it diminished from the pattern. On day 27, the LD18 band was the only band related to cyanobacteria that could be detected. Until day 15, the number of sequence types as detected by DGGE remained almost constant (Fig. 2). After the lysis event (day 15), many new sequence types were appearing in the DGGE patterns of both LSEs. Twelve of these new bands were excised from the gel and sequenced to obtain phylogenetic information. Four of the bands (bands 1, 2, 4, and 7) appeared to be related to the Cytophagales, three (bands 3, 5, and 6) appeared to be related to the β-Proteobacteria, three (bands 10, 11, and 12) appeared to be related to the actinomycetes, and two (bands 8 and 9) appeared to be related to the α-Proteobacteria (Table 3). Analysis of the DGGE patterns by NMDS (Fig. 3) showed a relatively constant bacterial community structure for LSE 1 until day 15. From day 15 to day 20, the community structure showed large changes. Thereafter, changes were relatively small again. The community structure of LSE 2 remained constant until day 17, and then a major change occurred between days 17 and 20. During the last 7 days, changes in the community structure were relatively small again.
FIG. 1.
Negative image of an ethidium bromide-stained DGGE pattern of the bacterial community structures of LSEs 1 and 2. The numbers at the top of the image indicate days from the start of the experiment. The numbered white circles refer to the excised and sequenced bands explained in Table 3. Lane M, marker lane containing the clones from the Lake Loosdrecht clone library. Clones related to cyanobacteria are indicated by “LD” and are elucidated in Table 2.
TABLE 2.
| Clone
|
Closest relative
|
||||
|---|---|---|---|---|---|
| Designation | EMBL accession no. | % Similarity | Species | EMBL accession no. | Taxonomic description |
| LD4 | AJ006279 | 95.8 | O. limnetica | AJ007908 | Filamentous cyanobacterium |
| LD5 | AJ007865 | 91.5 | P. hollandica | AJ007907 | Filamentous cyanobacterium |
| LD6 | AJ006280 | 99.5 | O. limnetica | AJ007908 | Filamentous cyanobacterium |
| LD7 | AJ007864 | 93.0 | P. hollandica | AJ007907 | Filamentous cyanobacterium |
| LD8 | AJ006281 | 94.3 | Nodularia sp. | AJ224447 | Filamentous cyanobacterium |
| LD9 | AJ006282 | 89.2 | Synechococcus sp. strain PCC 6301 | AF001477 | Coccoid cyanobacterium |
| LD15 | AJ006283 | 91.1 | P. hollandica | AJ007907 | Filamentous cyanobacterium |
| LD16 | AJ007866 | 98.9 | P. hollandica | AJ007907 | Filamentous cyanobacterium |
| LD18 | AJ006284 | 99.8 | O. agardhii | X84811 | Filamentous cyanobacterium |
| LD22 | AJ006285 | 99.8 | P. hollandica | AJ007907 | Filamentous cyanobacterium |
| LD25 | AJ006286 | 90.6 | P. hollandica | AJ007907 | Filamentous cyanobacterium |
Sequences were aligned to the closest relative from the EMBL database. The similarity was calculated with gaps and ambiguities not taken into account.
FIG. 2.
Numbers of bacterial sequence types detected by DGGE analysis during the course of the experiment.
TABLE 3.
| Band
|
Closest relative
|
|||
|---|---|---|---|---|
| No. | % Simi-larity | Species | Accession no. | Taxonomic description |
| 1 | 96.5 | Flavobacterium sp. | U63936 | Cytophagales |
| 2 | 86.6 | Flexibacter sancti | M62795 | Cytophagales |
| 3 | 97.4 | Unidentified bacterium LD28 | Z99999 | β-Proteobacteria |
| 4 | 97.4 | Flavobacterium sp. | U63936 | Cytophagales |
| 5 | 98.8 | Blastobacter sp. | U20772 | β-Proteobacteria |
| 5 | 98.8 | Unidentified bacterium | AJ223452 | β-Proteobacteria |
| 6 | 100 | Blastobacter sp. | U20772 | β-Proteobacteria |
| 6 | 100 | Unidentified bacterium | AJ223452 | β-Proteobacteria |
| 7 | 96.8 | Flavobacterium aquatile | M62797 | Cytophagales |
| 8a | 100 | Sphingomonas adhaesiva | X72720 | β-Proteobacteria |
| 8b | 96.4 | Hyphomicrobium vulgare | X53182 | α-Proteobacteria |
| 9 | 96.4 | Hyphomicrobium vulgare | X53182 | α-Proteobacteria |
| 10 | 98.3 | Unidentified bacterium | U85190 | Actinomycetes |
| 11 | 97.1 | Unidentified bacterium | U85190 | Actinomycetes |
| 12 | 99.4 | Mycobacterium sp. | U46146 | Actinomycetes |
Sequences were aligned to the closest relatives from the EMBL database. The similarity was calculated with gaps and ambiguities not taken into account.
FIG. 3.
NMDS map showing the changes in the structures of the bacterial communities during the lysis event. The numbers inside the symbols refer to the days of the experiment.
Eukaryotic community structure.
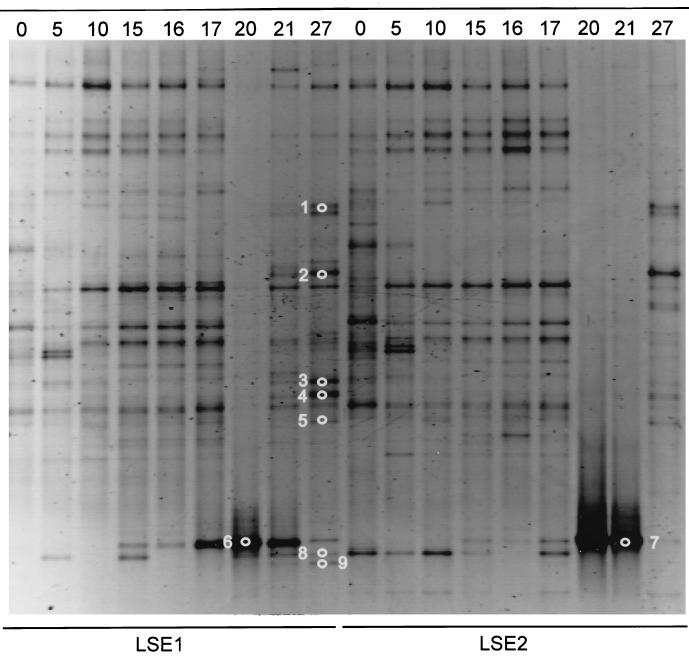
The DGGE analysis of the 18S rDNA fragments of both LSEs showed no notable change in the banding pattern until day 17. The DGGE patterns of both LSEs on day 20, and the DGGE pattern of LSE 2 on day 21, were completely dominated by a single band (Fig. 4). Excision and sequence analysis showed that this band (excised bands 6 and 7) represented a rotifer-like sequence. Rotifer abundance was estimated on days 17, 20, 21, and 27. In LSE 1 the densities were 1 × 104, 3.1 × 104, 1.7 × 104, and 0.1 × 104 individuals · liter−1, respectively. In LSE2 the numbers were 0.1 × 104, 4 × 104, 7 × 104, and 0.8 × 104 individuals · liter−1, respectively. Other dominant bands that appeared after the lysis event were excised from the gel and sequenced. Their similarities to the closest sequences in the EMBL database are shown in Table 4. Three of these bands (1, 3, and 4) showed very low similarity (<90%) to known eukaryotic sequences. Band 2 was related to the Ciliophora. Bands 6, 7, and 9 were very closely related (>99%) to the rotifer Brachionus plicatilis, and band 8 was related to an Arthropoda sequence.
FIG. 4.
Negative image of an ethidium bromide-stained DGGE gel of the eukaryotic community structures of LSEs 1 and 2. The numbers at the top of the image indicate the days from the start of the experiment. The numbered white circles refer to the excised and sequenced bands elucidated in Table 4.
TABLE 4.
| Band
|
Closest relative
|
|||
|---|---|---|---|---|
| No. | % Simi-larity | Species | Accession no. | Taxonomic description |
| 1 | 89.5 | Entrophospora sp. | Z14011 | Fungi |
| 2 | 98.2 | Cyclidium glaucoma | Z22879 | Ciliophora |
| 3 | 84.6 | Gymnodinium beii | U37406 | Dinophyceae |
| 4 | 82.5 | Paulinella chromatophora | X81811 | Euglyphina |
| 5 | 93.1 | Scypha ciliata | L10827 | Porifera |
| 6 | 99.4 | Brachionus plicatilis | U49911 | Rotifera |
| 7 | 99.4 | Brachionus plicatilis | U49911 | Rotifera |
| 8 | 91.4 | Cymindis punctigera | AF002773 | Arthropoda |
| 9 | 97.5 | Brachionus plicatilis | U49911 | Rotifera |
Sequences were aligned to their closest relatives from the EMBL database. The similarity was calculated with gaps and ambiguities not taken into account.
Analysis of the eukaryotic DGGE patterns by NMDS revealed that only small changes occurred in the eukaryotic microbial community during the first 20 days of the experiment (Fig. 5). Five days after the lysis event a major change in the eukaryotic communities of both LSEs was observed. Thereafter, the community structure returned to a state approximately similar to that preceding the lysis event.
FIG. 5.
NMDS map showing the changes in the structures of the eukaryotic communities during the lysis event. The numbers inside the symbols refer to the days of the experiment.
DISCUSSION
Quantitative versus qualitative DGGE.
The PCR-based methods we used in this study merely give a qualitative view of the changes in both the bacterial and eukaryotic community structures after the viral lysis event. Many studies have shown that these methods are prone to give a quantitatively incorrect view of the microbial community (17, 22, 27, 39, 45). An example is the apparently increasing dominance of the Oscillatoria agardhii-related band (LD18) during the first 16 days of the experiment. Microscopic estimations revealed that O. agardhii contributed less than 1% to the total cyanobacterial biomass. Another example is the complete dominance of the eukaryotic DGGE patterns by a single rotifer band. Since rotifers are metazoa, every individual contributes many cells, and thus many copies of their rRNA gene, to the PCR. Unless multicellular species can be removed from the sample, eukaryotic DGGE patterns have to be interpreted cautiously. To exclude these errors, we did not use the intensities of the DGGE bands as a measure of abundance. Instead, we reduced the information to simply presence or absence of a sequence type. Thus, only the complete removal or new emergence of sequence types would be detected by NMDS.
Viruses as structuring forces.
We started an experiment to follow the growth of heterotrophic bacteria on exudates of cyanobacteria. However, all filamentous cyanobacteria lysed, and we were able to monitor the appearance of previously undetected bacterial and eukaryotic species on the released carbon. From counts of virus-like particles and observations of numerous free virus particles and viruses attached to filaments of cyanobacteria, we concluded that there had been a massive viral outbreak. The increased numbers of bacteria (Table 1), the disappearance of the bands related to cyanobacteria from the DGGE patterns (Fig. 1), and the increase in bacterial richness (Fig. 2) suggest a change in the bacterial community structure driven by the viral outbreak. This alleged viral control was also shown by the NMDS analysis of the DGGE patterns (Fig. 3), where notable changes in the banding patterns and thus in the community structure were observed during the lysis in both LSEs. This structuring capability of natural viruses was also shown by monitoring the removal of a specific bacterium that was introduced to a microcosm (25). Although the largest changes in the community structure occurred during the lysis of the cyanobacterial populations, small changes in the community structure before and after the lysis of cyanobacteria were apparent. These changes could have been caused by the increase in temperature at the start of the experiment or by the selectivity of the culture medium.
Bacterial community structure.
We not only showed the removal of specific members of the microbial community, but we also detected their replacement by other species. Sequence analysis of the most dominant emerging bands showed that the majority of the newly appearing bacteria belonged to the Cytophagales (bands 1, 2, 4, and 7) and the actinomycetes (bands 10, 11, and 12) (Table 3). Apparently, these bacteria can respond rapidly to the dissolved organic matter released due to the lysis. The Cytophagales are common soil and water bacteria (14) and are well known for their capability to degrade large complex carbohydrates (43). The actinomycetes are also found in freshwater habitats, where they may play an active role in the decomposition of chitin, cellulose, and proteins (26, 28, 29). One sequence (bands 5 and 6, which have identical positions in the gel) was related to the β-Proteobacteria and was 100% identical to that of an unknown bacterium isolated from the surface of a copper pipe used in a potable-water system (11). One sequence (bands 8, 9) was related to the α-Proteobacteria. Both of these α- and β-Proteobacteria are commonly found in freshwater environments (38).
Some bands that appeared at the same position in the DGGE patterns of LSEs 1 and 2 were excised from both patterns and sequenced to validate the identity of the sequences they exhibited. From bands 5 and 6, we did retrieve almost-identical sequences (98.2%). However, from bands 8 and 9 we retrieved different sequences. Additional sequencing of one more clone from each band revealed that of the four sequences obtained from bands 8 and 9, three sequences were nearly identical (99.4 to 100%) and one sequence (band 8a) was only 91.7% similar to the other sequences (bands 8b and 9). Apparently, these two sequence types were not resolved by DGGE.
Eukaryotic community structure.
While the bacterial community showed almost-immediate changes after the lysis event, the eukaryotic community did not react until 5 days later. At that time, the pattern was completely dominated by a single band representing a rotifer-like sequence. After this apparent rotifer dominance, the community returned to a structure similar to that before the lysis. We found high numbers of rotifers in both LSEs, up to 7 × 104 liter−1. While these numbers are commonly found in laboratory cultures, where densities can exceed 106 liter−1 (55), the maximum natural abundance is at least four times lower (21). These rotifers feed on bacteria, phytoplankton, and protozoa (reference 3 and references therein), and the microbial loop after lysis apparently channeled the food for the rapid growth of these rotifer species. Rotifers are considered to be an important link between the microbial loop and higher trophic levels (2, 40). The observed rotifer proliferation suggests that viral lysis can increase the carbon flow to the higher trophic levels, at least in freshwater systems like Lake Loosdrecht, where rotifers can rapidly reach high densities (21). Besides this rotifer-related band, we found one ciliate-related band and five bands that represented sequences with low similarities (<94%) to the limited number of known eukaryotic sequences in the EMBL sequence database. This low similarity may be partly caused by the direct sequencing of excised DGGE bands. We found that this method often yields ambiguous sequences.
Conclusions.
The combination of a molecular profiling technique and an ordination method allowed the investigation of the impact of a viral lysis event on the structures of both the bacterial and the eukaryotic communities. Shortly after the cyanobacteria started to lyse, previously undetected bacterial sequence types were emerging in the DGGE profiles. The majority of these sequence types were related to bacterial species that are capable of degrading complex carbons. A few days later, a rotifer species profited from the lysis event and became dominant. Since rotifers are important links between the microbial loop and the classical food chain, this dominance after the viral outbreak could indicate the importance of viruses in channeling carbon through food webs.
The NMDS analysis of DGGE patterns seems to provide us with an interpretable, albeit qualitative, picture of the changes that occurred after the lysis of the cyanobacterial population. The NMDS analysis showed that the changes in the structures of both the bacterial and eukaryotic communities were remarkably similar in both LSEs (Fig. 3 and 5). This supports the notion that changes in community structure following changes in environmental conditions are not random and that these changing conditions act as governing forces of the microbial community structure.
Footnotes
Publication no. 2483 of the Netherlands Institute of Ecology, Centre for Limnology.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arndt H. Rotifers as predators on components of the microbial web (bacteria, heterotrophic flagellates, ciliates)—a review. Hydrobiologia. 1993;255/256:231–246. [Google Scholar]
- 3.Azam F, Fenchel T, Field J G, Gray J S, Meyer-Reil L A, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 4.Baldi F, Minacci A, Saliot A, Mejanelle L, Mozetic P, Turk V, Malej A. Cell lysis and release of particulate polysaccharides in extensive marine mucilage assessed by lipid biomarkers and molecular probes. Mar Ecol Prog Ser. 1997;153:45–57. [Google Scholar]
- 5.Bergh O, Børsheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 6.Bird D F, Kalff J. Empirical relationships between bacterial abundance and chlorophyll concentrations in fresh and marine waters. Can J Fish Aquat Sci. 1984;41:1015–1023. [Google Scholar]
- 7.Borg I, Lingoes J. Multidimensional similarity structure analysis. New York, N.Y: Springer-Verlag; 1987. [Google Scholar]
- 8.Bratbak G, Heldal M, Norland S, Thingstad T F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990;56:1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bratbak G, Heldal M, Thingstad T F, Riemann B, Haslund O H. Incorporation of viruses into the budget of microbial C-transfer. A first approach. Mar Ecol Prog Ser. 1992;83:273–280. [Google Scholar]
- 10.Bratbak G, Thingstad T F, Heldal M. Viruses and the microbial loop. Microb Ecol. 1994;28:209–221. doi: 10.1007/BF00166811. [DOI] [PubMed] [Google Scholar]
- 11.Bremer, P. Unpublished data.
- 12.Brussaard C P D, Kempers R S, Kop A J, Riegman R, Heldal M. Virus-like particles in a summer bloom of Emiliania huxleyi in the North Sea. Aquat Microb Ecol. 1996;10:105–113. [Google Scholar]
- 13.Brussaard C P D, Riegman R, Noordeloos A A M, Cadee G C, Witte H, Kop A J, Nieuwland G, Vanduyl F C, Bak R. Effects of grazing, sedimentation and phytoplankton cell lysis on the structure of a coastal pelagic food web. Mar Ecol Prog Ser. 1995;123:259–271. [Google Scholar]
- 14.Christensen P J. The history, biology and taxonomy of the cytophaga group. Can J Microbiol. 1977;23:1599–1653. doi: 10.1139/m77-236. [DOI] [PubMed] [Google Scholar]
- 15.De Rijk P, De Wachter R. DCSE, an interactive tool for sequence alignment and secondary structure research. Comput Appl Biosci. 1993;9:735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- 16.Egli T. The ecological and physiological significance of the growth of heterotrophic microorganisms with mixtures of substrates. Adv Microb Ecol. 1995;14:305–386. [Google Scholar]
- 17.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1992;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferris M J, Nold S C, Revsbech N P, Ward D M. Population structure and physiological changes within a hot spring microbial community following disturbance. Appl Environ Microbiol. 1997;63:1367–1374. doi: 10.1128/aem.63.4.1367-1374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher S G, Lerman L S. Length-independent separation of DNA restriction fragments in two dimensional gel electrophoresis. Cell. 1979;16:191–200. doi: 10.1016/0092-8674(79)90200-9. [DOI] [PubMed] [Google Scholar]
- 20.Gobler C J, Hutchins D A, Fisher N S, Cosper E M, Sanudowilhelmy S A. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol Oceanogr. 1997;42:1492–1504. [Google Scholar]
- 21.Gulati R D, Ooms-Wilms A L, Van Tongeren O F R, Postema G, Siewertsen K. The dynamics and role of limnetic zooplankton in Loosdrecht Lakes (The Netherlands) Hydrobiologia. 1992;233:69–86. [Google Scholar]
- 22.Hansen M C, Tolker-Nielsen T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 23.Hennes K P, Simon M. Significance of bacteriophages for controlling bacterioplankton growth in a mesotrophic lake. Appl Environ Microbiol. 1995;61:333–340. doi: 10.1128/aem.61.1.333-340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hennes K P, Suttle C A. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol Oceanogr. 1995;40:1050–1055. [Google Scholar]
- 25.Hennes K P, Suttle C A, Chan A M. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl Environ Microbiol. 1995;61:3623–3627. doi: 10.1128/aem.61.10.3623-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang C L, Xu H L. Diversity of aquatic actinomycetes in lakes of the middle plateau, Yunnan, China. Appl Environ Microbiol. 1996;62:249–253. doi: 10.1128/aem.62.1.249-253.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson J L. Isolation and purification of nucleic acids. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. West Sussex, England: John Wiley & Sons Ltd.; 1991. pp. 1–19. [Google Scholar]
- 28.Johnston D W, Cross T. The occurrence and distribution of actinomycetes in lakes of the English Lake District. Freshwater Biol. 1976;6:457–463. [Google Scholar]
- 29.Johnston D W, Cross T. Actinomycetes in lake muds: dormant spores or metabolically active mycelium. Freshwater Biol. 1976;6:465–470. [Google Scholar]
- 30.Jürgens K, Güde H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser. 1994;112:169–188. [Google Scholar]
- 31.Kruskal J B, Wish M. Multidimensional scaling. Beverly Hills, Calif: Sage Publications; 1978. [Google Scholar]
- 32.Lessa E P. Multidimensional analysis of geographic genetic structure. Syst Zool. 1990;39:242–252. [Google Scholar]
- 33.Mathias C B, Kirschner A K T, Velimirov B. Seasonal variations of virus abundance and viral control of the bacterial production in a backwater system of the Danube River. Appl Environ Microbiol. 1995;61:3734–3740. doi: 10.1128/aem.61.10.3734-3740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Middelboe M, Jorgensen N O G, Kroer N. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl Environ Microbiol. 1996;62:1991–1997. doi: 10.1128/aem.62.6.1991-1997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moed J R, Hallegraeff G M. Some problems in the estimation of chlorophyll-a and phaeopigments from pre- and post-acidification spectrophotometric measurements. Int Rev Gesamten Hydrobiol. 1978;63:787–800. [Google Scholar]
- 36.Muyzer G, Dewaal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16s rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nei M, Li W H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nold, S. C., and G. Zwart. Patterns and governing forces in aquatic microbial communities. Aquat. Ecol. 32:17–35. (Review.)
- 39.Ogram A V, Sayler G S, Barkay T T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1988;7:57–66. [Google Scholar]
- 40.Porter K G, Paerl H, Hodson R, Pace M, Priscu J, Riemann B, Scavia D, Stockner J. Microbial interactions in lake food webs. In: Carpenter S R, editor. Complex interactions in lake communities. New York, N.Y: Springer-Verlag; 1988. pp. 209–227. [Google Scholar]
- 41.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 42.Proctor L M, Fuhrman J A. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–62. [Google Scholar]
- 43.Reichenbach H. The order Cytophagales. In: Balows A, Truper H G, Dwokin M, Harder W, Schleifer K M, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 3631–3675. [Google Scholar]
- 44.Reisser W. Viruses and virus-like particles of freshwater and marine eukaryotic algae—a review. Arch Protistenkd. 1993;143:257–265. [Google Scholar]
- 45.Reysenbach A, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rijkeboer M, de Bles F, Gons H J. Laboratory scale enclosure: concept, construction and operation. J Plankton Res. 1990;12:231–244. [Google Scholar]
- 47.Suttle C A. The significance of viruses to mortality in aquatic microbial communities. Microb Ecol. 1994;28:237–243. doi: 10.1007/BF00166813. [DOI] [PubMed] [Google Scholar]
- 48.Suttle C A, Chan A M. Dynamics and distribution of cyanophages and their effects on marine Synechococcus spp. Appl Environ Microbiol. 1994;60:3167–3174. doi: 10.1128/aem.60.9.3167-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suttle C A, Chan A M, Cottrell M T. Infection of phytoplankton by viruses and reduction of primary productivity. Nature. 1990;347:467–470. [Google Scholar]
- 50.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Boekel, W. H. M., F. C. Hansen, R. Riegman, and R. P. M. Bak. Lysis-induced decline of a Phaeocystis spring bloom and coupling with the microbial foodweb. Mar. Ecol. Prog. Ser. 81:269–276.
- 52.Van Hannen, E. J., M. Veninga, J. Bloem, H. J. Gons, and H. J. Laanbroek. Genetic changes in the bacterial community structure associated with protistan grazers. Arch. Hydrobiol., in press.
- 53.Van Hannen E J, van Agterveld M P, Gons H J, Laanbroek H J. Revealing genetic diversity of eukaryotic microorganisms in aquatic environments by denaturing gradient gel electrophoresis. J Phycol. 1998;34:206–213. [Google Scholar]
- 54.Weinbauer M G, Hofle M G. Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic lake. Appl Environ Microbiol. 1998;64:431–438. doi: 10.1128/aem.64.2.431-438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yufera M, Navarro N. Population growth dynamics of the rotifer Brachionus plicatilis cultured in non-limiting food conditions. Hydrobiologia. 1995;313:399–405. [Google Scholar]
- 56.Zwart G, Hiorns W D, Methé B A, van Agterveld M P, Huismans R, Nold S C, Zehr J P, Laanbroek H J. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1999;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]
- 57.Zwart G, Huismans R, van Agterveld M P, Van de Peer Y, De Rijk P, Eenhoorn H, Muyzer G, van Hannen E J, Gons H J, Laanbroek H J. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol Ecol. 1998;25:159–169. [Google Scholar]