Abstract
Peptide – polymer complementary pairs can provide useful tools for isolating, organizing and separating biomacromolecules. We describe a procedure for selecting a high affinity complementary peptide-polymer nanoparticle (NP) pair using phage display. A hydrogel copolymer nanoparticle containing a statistical distribution of negatively charged and hydrophobic groups was used to select a peptide sequence from a phage displayed library of >1010 peptides. The NP has low nanomolar affinity for the selected cyclic peptide and exhibited low affinity for a panel of diverse proteins and peptide variants. Affinity arises from the complementary physiochemical properties of both NP and peptide as well as the specific peptide sequence. Comparison of linear and cyclic variants of the peptide established that peptide structure also contributes to affinity. These findings offer a general method for identifying polymer-peptide complementary pairs. Significantly, precise polymer sequences (proteins) are not a requirement, a low information statistical copolymer can be used to select for a specific peptide sequence with affinity and selectivity comparable to that of an antibody. The data also provides evidence for the physiochemical and structural contributions to binding. The results confirm the utility of abiotic, statistical, synthetic copolymers as selective, high affinity peptide affinity reagents.
Keywords: peptide affinity reagents, hydrogel nanoparticles, phage display, molecular recognition, peptide-synthetic polymer complements
Graphical Abstract
1. Introduction.
Aside from their natural role in biology, the structural and functional group diversity of peptides provide opportunities for their use in applications that go far beyond their predicted role. Examples include peptides that bind to both organic and inorganic materials.[1–5] Examples include peptides that bind to specific faces of inorganic crystals [6–9] and semiconductors. [10–11] Peptides have also been used as connectors to metal surfaces[12]. Since inorganic nanoparticles (NPs) are used as delivery vehicles for therapeutics, a substantial effort has focused on the composition and binding kinetics of peptides to these surfaces.[14–17] The interaction of peptides with synthetic organic polymers has also received substantial attention. Selected examples include peptides that discriminate synthetic polymers on the basis of stereochemistry.[18, 19] Their interaction with certain polymers has enabled ordered nanoscale arrays of biomacromolecules on phase separated block copolymers.[20] Contemporary biotechnology routinely brings synthetic organic materials into intimate contact with the biological milieu. Understanding and to the extent possible, controlling factors that contribute to these interactions is of fundamental importance to contemporary biotechnology. [21–24]
Our research has focused on the interaction of synthetic organic copolymers with biomacromolecules. By taking advantage of the variability of synthetic copolymers we select copolymers with high affinity and selectivity for specific peptides and proteins.[25] Noteworthy, the copolymers are not pure substances, they contain statistical distributions of functional monomers. Although statistical copolymers lack the sequence specificity of biomacromolecules, they offer broad chemical group variability. When coupled with their ease of synthesis, robustness and dynamic properties, they provide interesting alternatives to traditional materials with biomacromolecule affinity. There are now a number of examples to support this point. For example Schrader and coworkers designed synthetic copolymers with biomolecule (protein) specificity[27–29] and Rotello and coworkers identified functionality that contribute to the interaction of synthetic copolymers with biological macromolecules.[30, 31] The molecular imprinting community has for many years reported examples of copolymer receptors for small molecules, peptides and proteins.[32, 33] In our approach we identify hydrogel copolymer complements to proteins and peptides.[25] Synthetic copolymer libraries, generated by copolymerization of monomer mixtures containing hydrophobic, charged and hydrogen binding groups, are screened against a target biomacromolecule for lead candidates. Further optimization of the chemical composition by directed chemical evolution results in functional copolymers with high affinity and selectivity for the target. Polymers identified by this process have been developed for protein isolation/purification[25, 34–37] and toxin sequestrants[38–46] that function in the bloodstream of living mice.[40] A statistical copolymer nanoparticle was developed to sequester a vascular growth factor (VEGF A165). The copolymer disrupted angiogenesis and inhibited cancer growth in vivo.[47] More recent efforts have been directed to neutralize the major families of medically relevant protein toxins found in venomous snakes.[38, 39]
The vast compositional diversity of peptides and synthetic copolymers presents both opportunities and challenges to identify complementary affinity pairs. As part of this effort, we are developing tools to accelerate the discovery process. In this report we describe an expedited method for discovery of a peptide sequence that is complementary to a preselected hydrogel copolymer. The availability of peptide - synthetic copolymer complementary pairs affords opportunities for creating biological arrays, peptide-polymer adhesives, supramolecular peptide-polymer assemblies and new tag-affinity ligand combinations for peptide and protein affinity purification.[48] To this end we employed a phage library containing greater than 1010 discrete peptide sequences as the peptide source. The synthetic copolymer selected for this screen is readily synthesized and has a well-defined average chemical composition. The material is best described as a statistical copolymer, with instantaneous sequences determined by the relative reactivity ratios and instantaneous concentration of the monomers.[49] A structure based analysis of affinity finds that copolymer-peptide interactions depend not only on physiochemical properties of peptide and polymer but also on peptide sequence and shape, a finding that strengthens the suggestion that antibody–like affinity and selectivity can be achieved with low-information content, statistical synthetic organic copolymers.
2. Experimental Section
2. 1. Materials.
All chemicals were obtained from commercial sources as follows: N-isopropylacrylamide (NIPAm), N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC), bovine serum albumin (BSA), ammonium persulfate (APS) was obtained from SIGMAALDRICH; acrylic acid (AAc), sodium dodecyl sulfate (SDS) and N-phenylacrylamide (PAA) were from Aldrich Chemical Company; N,N’-methylenebisacrylamide (Bis) and N-hydroxysuccinimide (NHS), Fluka; N-tert-butylacrylamide (TBAm) were obtained from Acros Organics; P14K, PNT and SCP peptides were from Genemed Synthesis. NIPAm was recrystallized from hexane before use. Other chemicals were used as received. Water used in polymerization and characterization was distilled then purified using a Barnstead Nanopure Diamond system.
2. 2. Nanoparticle (NP) synthesis.
NPs 1, 2 and 3 were prepared by precipitation polymerization. Figure 1(a) shows the monomers used for the NP synthesis, and Table S1 summarizes the specific monomer composition of each NP. Solutions of monomers were prepared in 50 mL nanopure water with a total concentration of 65 mM. The solutions were degassed in a sonicatior bath under vacuum for 10 min followed by bubbling nitrogen through the reaction mixture for 30 min. Following the addition of an aqueous solution of ammonium persulfate (APS, 30 mg in 500 μL water), the polymerization was carried out at 60–65 °C for 3 h under a nitrogen atmosphere. The polymerized solutions were purified by dialysis (MWCO from 12 to 14 kD) against an excess of water (changed at least twice a day) for >4 days. The concentrations of NPs were determined by lyophilizing an aliquot of the dialyzed solution and weighing the residue. NPs were monomodal and ranged in size from 40–65 nm.
Figure 1.
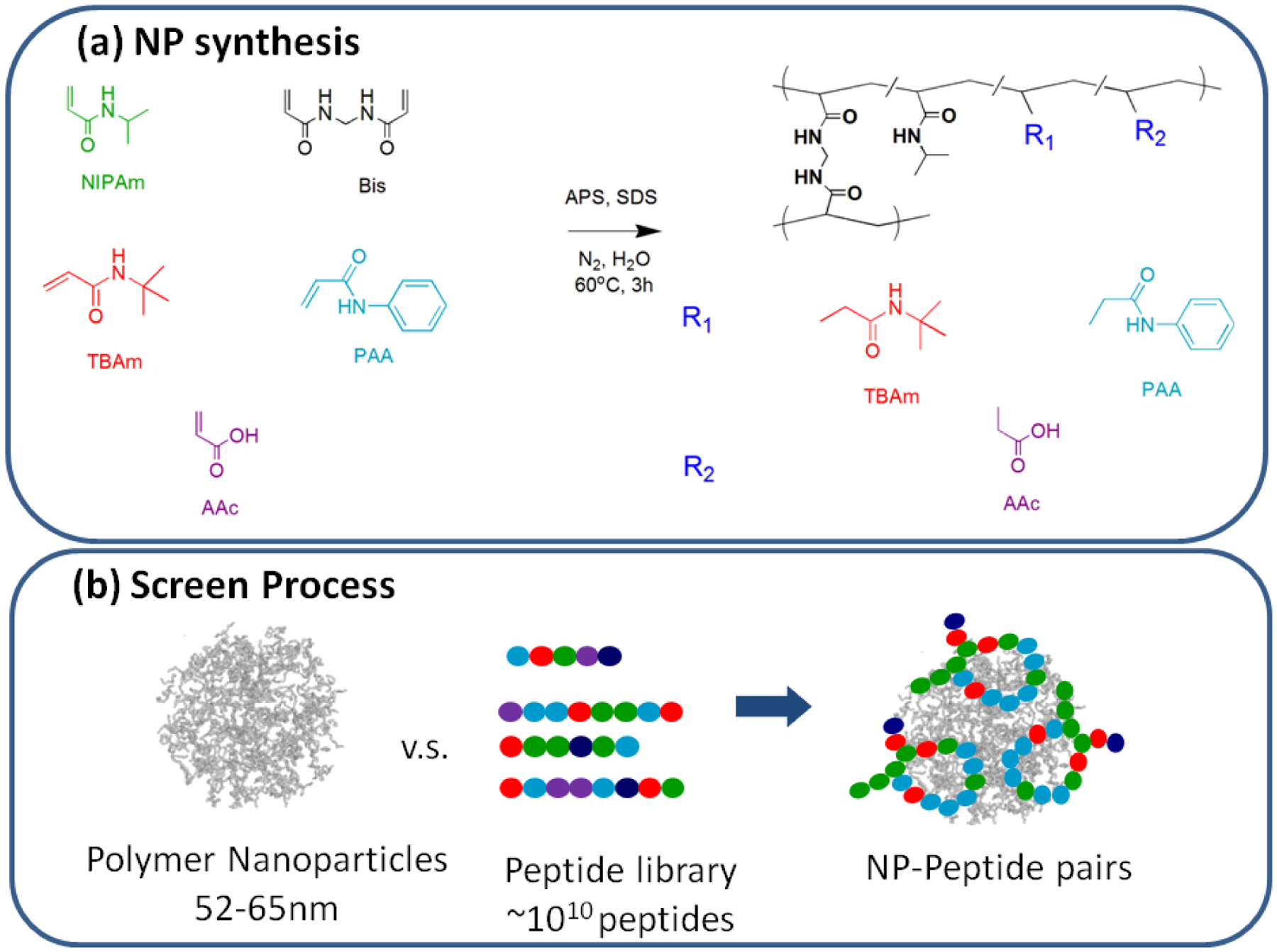
(a) The functional monomers used for nanoparticles synthesis and a schematic showing the general synthesis of polymer NPs and their composition. (b) Schematic of screening process.
2. 3. Phage display selection.
Evaluation of non- interactions between phage without displayed peptides and NPs. Screening NPs against the phage library first required the immobilization of nanoparticles onto a solid support with sufficient robustness for multiple wash cycles during the selection process. Several methods are commonly used to immobilize nanoparticles onto a solid support; these include covalent immobilization (e.g. NHS/EDC amide coupling chemistry) or physical immobilization between the NP and solid support. The immobilization method was used for all experiments.
Prior to the selection process, non-specific interactions between NPs and phage not having a displayed peptide were evaluated. NPs 1, 3 (100 μL, 80 μg/mL or 160 μg/mL) were incubated in a 96 well Maxisorp plate. After 1 h, the NP solutions were removed, and a BSA blocking solution (300 μL, 0.2% in PBS) was added to each well. After 1 h, the BSA solution was removed and the wells were washed five times with a phosphate-buffered saline (PT) solution (300 μL, PBS with 0.05% Tween 20). Six concentrations of phage without a displayed peptide (100 μL, 12, 4, 1.33, 0.44, 0.15, 0.05 nM) were added to the NP-coated wells. One hour later, the phage solutions were removed, and the wells were washed with PT (300 μL, PBS with 0.05% Tween 20) eight times. a-M13-Horseradish peroxidase (HRP) (1:5000 dilution) in PBT (PBS with 0.1% BSA and 0.1% Tween 20) were added into the wells and incubated for 30 min. The wells were washed by PT solution (PBS with 0.05% Tween 20) five times and PBS solution three times. o-Phenylenediamine (OPD, 0.12 g) solution was prepared in 12 mL citric acid buffer and 100 μL H2O2 (3%). OPD solution (100 μL) was added to the wells and incubated for 10 min. The absorption of the wells was measured at 450 nm.
2. 4. Phage display: Selection of peptides for NPs 1, 3.
Polymer NPs 1, 3 were immobilized on 96 well plates, 4 rounds of selection were performed. For the first selection, the NP solutions (100 μL, 100 μg/mL) were incubated in the wells for 1 h. Then, the NP solutions were removed, and BSA blocking solutions (0.2% BSA in PT) were incubated in the wells for 30 min. The control was treated with identical conditions without the NP immobilization step. The BSA solutions were discarded and PT (PBS with 0.05% Tween 20) solution was used to wash the wells five times. The phage solutions containing the peptide library were incubated in the control wells for 30 min to remove phage with non-specific binding to BSA, and the unbound phages were transferred to the wells containing immobilized NPs. The phage solutions were incubated in the wells for 1 h, and the wells were washed with the PT solution after removing the phage with non-specific binding. Hydrochloric acid (100 μL, 0.1 M) was added to the wells and incubated for 5 min to elute the binding phage from the NPs. The eluted phage were collected and neutralized by Tris-HCl solution (pH 8.0). For the next round of selection, the eluted phage was amplified through propagation in E. coli. The phage-E. coli cultures were titered on LB-carbenillin and LB/no drug plates, and the plates were incubated at 37 °C overnight. Following the previously published procedures,[71] spots with the highest relative intensity for each NP were selected, and PCR was performed for each colony. Following PCR, samples were submitted to DNA sequencing (Genewiz).
2. 5. Peptide synthesis.
P14, P14N and P14S were synthesized by an AAPPTEC APEX 369 peptide synthesizer by the following procedures. Biotin-PEG NovaTag Resin (30 mg, 0.47mmole/g resin) was swelled in DCM for 30 min in each well. Fmoc (0.3 M) protected amino acids (dissolved in 1-methyl-2-pyrrolidinone with 0.3 M HBOt) were prepared and loaded into the amino acid wells. Piperidine (25%) in N-methyl-2-pyrrolidinone was used for deprotection. DIC (25%) in N-methyl-2-pyrrolidinone was used to activate the amino acids. Each coupling involves eight steps: deprotection, wash, deprotection, wash, coupling, wash, coupling and wash. When coupling was finished, the resin was transferred into a syringe reaction vessel and rinsed with DMF and DCM (with saturated sodium bicarbonate) five times each. The resin was then dried in a desiccator under vacuum overnight. The biotinylated-peptide was cleaved from the resin using the cleavage solution (TFA:H2O:EDT:TIS = 94:2.5:2.5:1) for 2 h the peptide was then precipitated into the ether solution and washed with ether three times. The peptide was characterized by HPLC and MALDI TOF-MS.
2. 6. Binding affinity between NP1(40TBAm-20AAc) and P14 by QCM.
An Affinix Q4 quartz crystal microbalance QCM was used (Initium Co. Ltd., Tokyo, http://www.initium2000.com). The instrument has four 500 μL cylindrical cells (10 mm i.d.) each equipped with a 27 MHz QCM plate (8 mm diameter quartz plate and 4.9 mm2 Au electrode) at the bottom of the cell and a horizontal mixer with temperature-control.
Evaluation of interactions between P14 and NP1(40TBAm-20AAc) by QCM.
The gold surface of the QCM was cleaned with piranha solution (H2SO4 : H2O2 = 3:1) (3X) for 10 min. 3,3’-Dithiodipropionic acid in ethanol (200 μL 1 mM) was added to the QCM cells and incubated overnight. After flushing with excess H2O, an aqueous solution of EDC (100 mg/mL, 50 μL) and NHS (100 mg/mL, 50 μL) (1:1) was added into the QCM cells. After 30 min, the cells were washed with water (10X). Water (500 μL) was then added to the QCM cells. Avidin (4 μL, 1mg/mL) was introduced into the cells to saturate the surface. The cells were then washed with water several times and aq. HEPES (500 μL, 10 mM, pH 7) was added into the QCM cells. The peptide was dissolved in two solutions, 20% DMSO in 10 mM aq. HEPES and 50% acetonitrile in water. Based on the MALDI-TOF-MS results (see Figure S5 and S6), the peptides in these two solutions correspond to the cyclic and linear species respectively. The cyclic peptide in 20% DMSO in 10 mM HEPES is labeled “P14C” and the linear peptide in the aqueous solution (containing 50% acetonitrile) is termed “P14L”. Solutions of P14C and P14L (10 μL, 1mg/mL), were injected into QCM cells. Oxidation from linear to cyclic form in the aqueous solution (containing 50% acetonitrile) even with purging air for several h to exclude the possibility of cyclic formation was minimal. The control was treated similarly except for the injection of the peptide. The cells were washed with PBS 5X (35 mM phosphate, 500 μL) and PBS (35mM, 500 μL) was added to each QCM cell. Following baseline stabilization, solutions of NPs (1, 2, 4, 8, 16, or 32 μl) were injected into the cells. Two nanoparticles, NP1(40TBAm-20AAc) and NP2(40TBAm) were used to evaluate peptide affinity.
2. 7. Evaluation of interactions between NP1(40TBAm-20AAc) and other proteins by QCM.
The gold surface was cleaned by incubation in piranha solution (H2SO4 : H2O2 = 3:1) (3X) for 10 min. 3,3’-Dithiodipropionic acid in ethanol (1 mM, 200 μL) was added to the QCM cells and incubated overnight. After flushing with copious amount of H2O, an aqueous solution of EDC (100 mg/mL, 50 μL) and NHS (100 mg/mL, 50 μL) (1:1) was added to the QCM cells. After 30 min, the cells were washed with water (10X). Water (500 μL) was then added to the QCM cells. Solutions of proteins were injected until saturation of QCM sensor was achieved (Fc domain: 0.5 μL 2.3 mg/mL or Avidin: 4 μL 1mg/mL or BSA and Hemoglobin: 1 μL 1mg/mL or TNFa : 3 μL 10 mg/mL). The cells were washed with phosphate buffer (35 mM, 500 μL) five times. Various volumes of the solution of NP1(40TBAm-20AAc) were added to the cells modified with different proteins until saturation was reached.
2. 8. Evaluation of interactions between NP1(40TBAm-20AAc) and P14S, P14M, SCP and PNT by QCM.
Table 1. summarizes the peptides selected for further evaluation of their affinity and specificity to NP1(40TBAm-20AAc). Positive charged, hydrophobic, aromatic and hydrophilic residues are represented in blue, red, cyan and green respectively. A variant of P14 but with scrambled amino acids (SCP) was included in the study. P14, contains two cysteine residues (Cys, C) which form a cyclic peptide upon oxidation. In this study, the interaction between cyclic and linear peptide was also investigated. In addition, P14S, a peptide with two cysteines mutated to serines (Ser, S) to form a linear structure was also synthesized and evaluated. There are four Phe residues in P14. In peptide P14M, three Phe residues were mutated to Asn to investigate the importance of NP-aromatic interactions by QCM. A peptide, PNT, was designed to investigate the specificity of NP1(40TBAm-20AAc); this peptide contains seven hydrophobic, one aromatic, three hydrophilic and five positively charged residues. The net composition of PNT could engender NP1(40TBAm-20AAc) affinity to PNT.
Table 1.
The compositions of synthetic peptides used in this study. Positive charged, hydrophobic, aromatic and hydrophilic residues are represented in blue, red, cyan and green.
| Phages | Peptide Sequence |
|---|---|
| P14a | K P C I S F W Q F W F G F C S S |
| P14Sa | K P S I S F W Q F W F G F S S S |
| P14Na | K P C I S F W Q N W N G N C S S |
| PNTa | G L P A L I S W I K R K R Q Q K |
| SCP | C I S F W K P S S W F G F Q F C |
| P14Kb | A P C I S F W Q F W F G F C S S |
N-terminus coupled to (PEG)2-Biotin
cyclic form
3. Results and Discussion
Prior to selection of the copolymer NP, we were curious as to how many strong binders or “hits” hydrogel copolymers would generate from the library of >1010 peptides. In a preliminary screen we exposed several hydrogel NPs varying in chemical composition to the phage library. The details of the preliminary screen can be found in the SI. An important finding was that relatively few strong binders were picked up across a spectrum of copolymer chemical compositions indicating that these hydrogel copolymers do not engage in broad non-specific binding to the peptide library.
With assurances that individual hydrogel NPs could offer a high degree of selectivity we chose a lightly cross-linked (2% Bis) carbon backbone hydrogel copolymer of N-isopropylacrylamide (NIPAm) that contained negative charge (acrylic acid, AAc) and aliphatic hydrophobic groups (N-t-Butylacrylamide, TBAm), NP1(40% TBAm, 20% AAc, 2% Bis and 38% NIPAm) for this study. The choice of this specific hydrogel copolymer nanoparticle (NP) to screen for peptides was based on considerations of the potential applications of the peptide-copolymer pair and NP accessibility. The considerations included copolymer biocompatibility, stability, and the ability to control NP size and molecular weight together with ease of synthesis from readily available monomers. NP1 satisfied all of the above criteria.
The hydrogel NP is synthesized in aqueous solution by a modified free radical precipitation polymerization using ammonium persulfate (APS) as initiator. The copolymers were purified by dialysis and are comprised of uniform narrow distributions of stable, nano-sized polymer NPs with average sizes of 52–65 nm and PDIs of 0.07~0.12 (DLS). The NP synthesis is outlined in Figure 1 and their composition is summarized in Table S1.
NP1 was screened against a phage library containing combinations of several sub-libraries of phages with peptides that range in length from 7 to 20 amino acids.[50] All contain two cysteines (C) and under the screening conditions, exist in the cyclic form. The total diversity of the peptide library was 2.5×1010 and over the four rounds of selection, washing and immobilization conditions were progressively more stringent to select for higher affinity phage-displayed binding partners. Details of the library are given in Table S5.
Five phages with high affinity for NP1(40TBAm-20AAc) were selected from ninety single colonies; the peptide sequences are given in Table 2. Positive/negative charged, hydrophobic, aromatic and hydrophilic residues are colored in blue, purple, red, cyan and green. Phages 22, 26 and 80 are composed of over seventy percent hydrophobic residues. These peptides had very low solubility and aggregated under normal working conditions, properties that were deemed to be unsuitable for practical applications so a shorter, hydrophilic sequence was chosen for further study. The sequence selected, peptide P14C, is a 12 AA cyclic peptide that contains a single disulfide linkage (Table 1). A computationally modelled structure of P14C is shown in Figure 2.
Table 2.
The sequences of phage with high affinity for NP1(40TBAM-20AAc).
| Phages | Peptide Sequence |
|---|---|
| 14a | K P C I S F W Q F W F G F C S S |
| 20a | H L G L T Q L C W W M Q T C Y Q S P E M |
| 22a | L S C R W W W M G C G Y |
| 26a | L P L G L C G W V L V S P L C W I T W L |
| 80a | T S C Y Y W W W I V R C L P |
cyclic form
Figure 2.
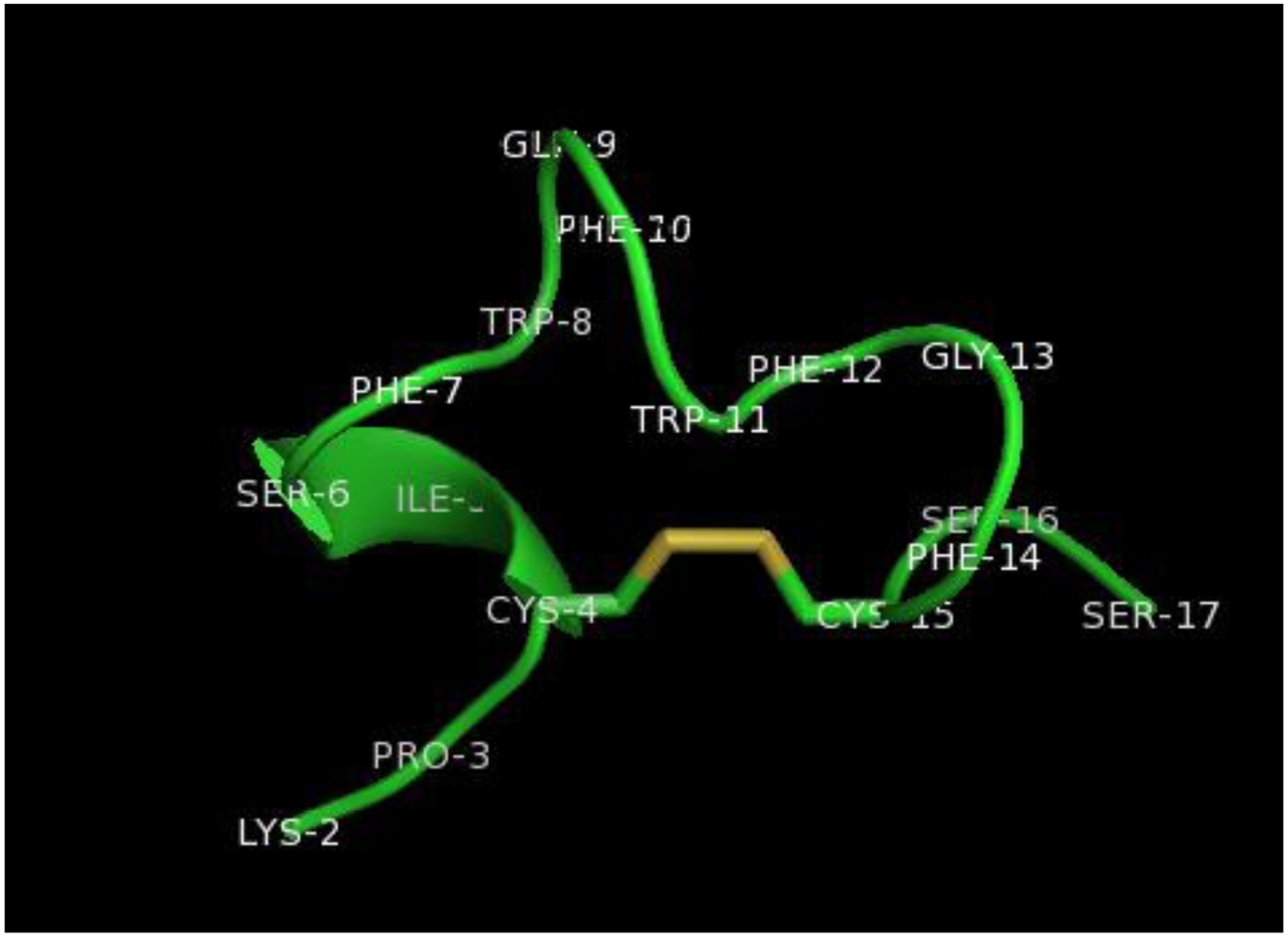
A computed structure of P14C (K P C I S F W Q F W F G F C S S). The structures were calculated by Spartan23.and the surface representation of the complex generated by UCSF Chimera.24 The disulfide linkage is colored gold.
The interaction between NP1(40TBAm-20AAc) and a chemically synthesized biotinylated peptide P14C was evaluated by quartz crystal microbalance (QCM) (Figure 3 and 4). P14C was immobilized on a QCM sensor by an avidin-biotin interaction. QCM cells immobilized only with avidin were used as a control and a NIPAm nanoparticle composed of 40% TBAm without AAc, (NP2(40TBAm)), was used as an additional control to evaluate the importance of electrostatic interactions. There was little interaction between either avidin and NP2(40TBAm) with P14C suggesting the specific chemical composition of NP1(40TBAm-20AAc) is responsible for binding of to P14C. NP1(40TBAm-20AAc) is comprised of statistical distributions of functional groups that can participate in hydrogen bonding, hydrophobic and electrostatic interactions with P14C. The dissociation equilibrium constant of NP-protein complex was calculated from the Hill equation[51, 52] using the NP-bound peptide fraction (θ) and NP concentration:
Figure 3.
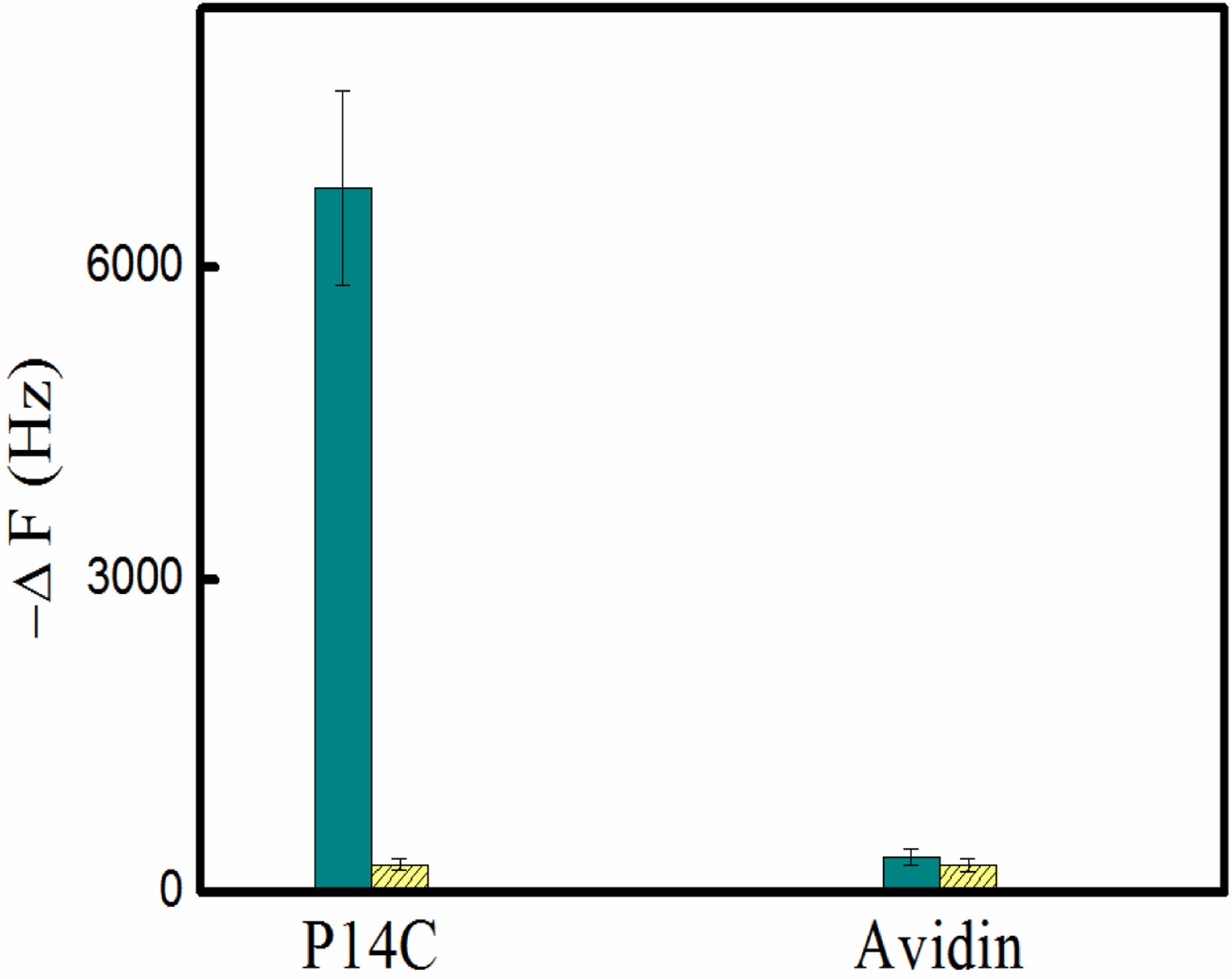
A quartz crystal microbalance (QCM) analysis of the binding between P14C and NP1(40TBAm-20AAc) in phosphate buffer (35mM, pH 7.3). The control is the interaction between NP1(40TBAm-20AAc) and avidin. Cyan column: NP1(40TBAm-20AAc), Yellow column: NP2(40TBAm).
Figure 4.
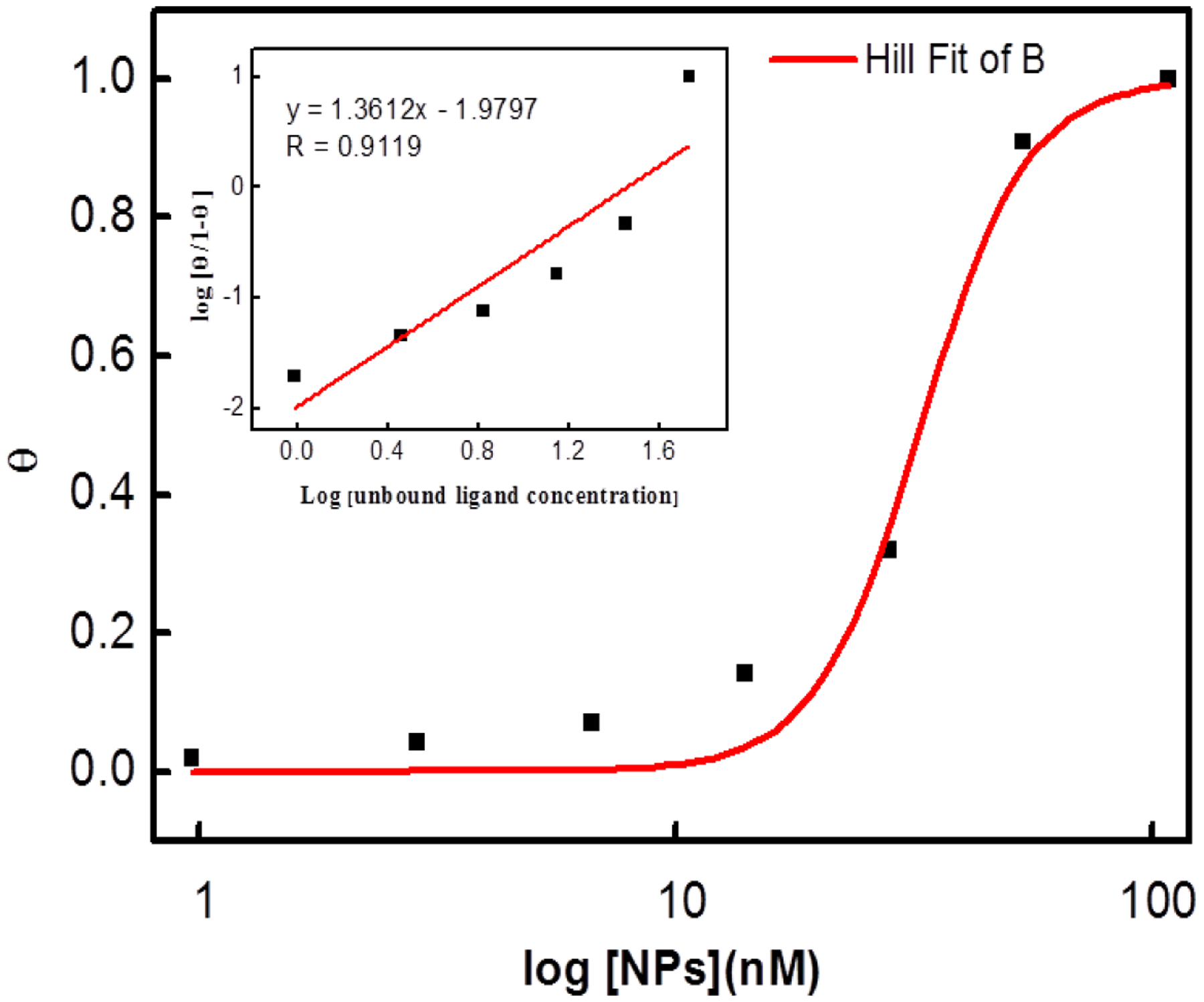
Binding curve of NP-P14C peptide interaction fitted to the Hill equation. The insert graph depicts the Hill plot of the data.
An apparent KD, 95 nM, and Hill coefficient of 1.36 are obtained from the Hill equation fit (Figure 4). Protein and peptide absorption to NP surfaces is a critical factor for their performance in most biomedical applications and has been extensively studied.[53, 54] In some of these studies binding curves were characterized by Hill coefficients n>1. Hill coefficients n>1 are often used as an indication of cooperative binding and can be attributed to stabilizing interactions between adjacent peptide molecules on the nanoparticle surface.[55, 56] Many of these studies involved hard NPs (Au, SiO2, polystyrene, etc). However the NPs in this study present a very different “surface”. NIPAm (2% crosslinked) are ‘soft’ dynamic materials with densities 1/30th to 1/100th that of a protein or hard NPs.[57, 58] They are largely water and because of their average mesh size (~10 nm), biomacromlecule absorption, especially small peptides, can occur throughout the NP. At this time further speculation about the origin of the >1 Hill coefficient is beyond the scope of this study.
In addition to the physiochemical properties of the interacting groups, buffer conditions (pH and ionic strength) can also play a role in the observed NP-biomacromolecule interaction. Figure 5 shows the interaction between P14C and NP1(40TBAm-20AAc) at room temperature and neutral pH at different salt concentrations. The insensitivity of the interaction to added salt suggests that hydrophobic interactions alone do not dominate the binding.[30, 59–61] More likely binding arises from collective weak interactions that characterize typical peptide or protein-protein interactions.[59, 62]
Figure 5.
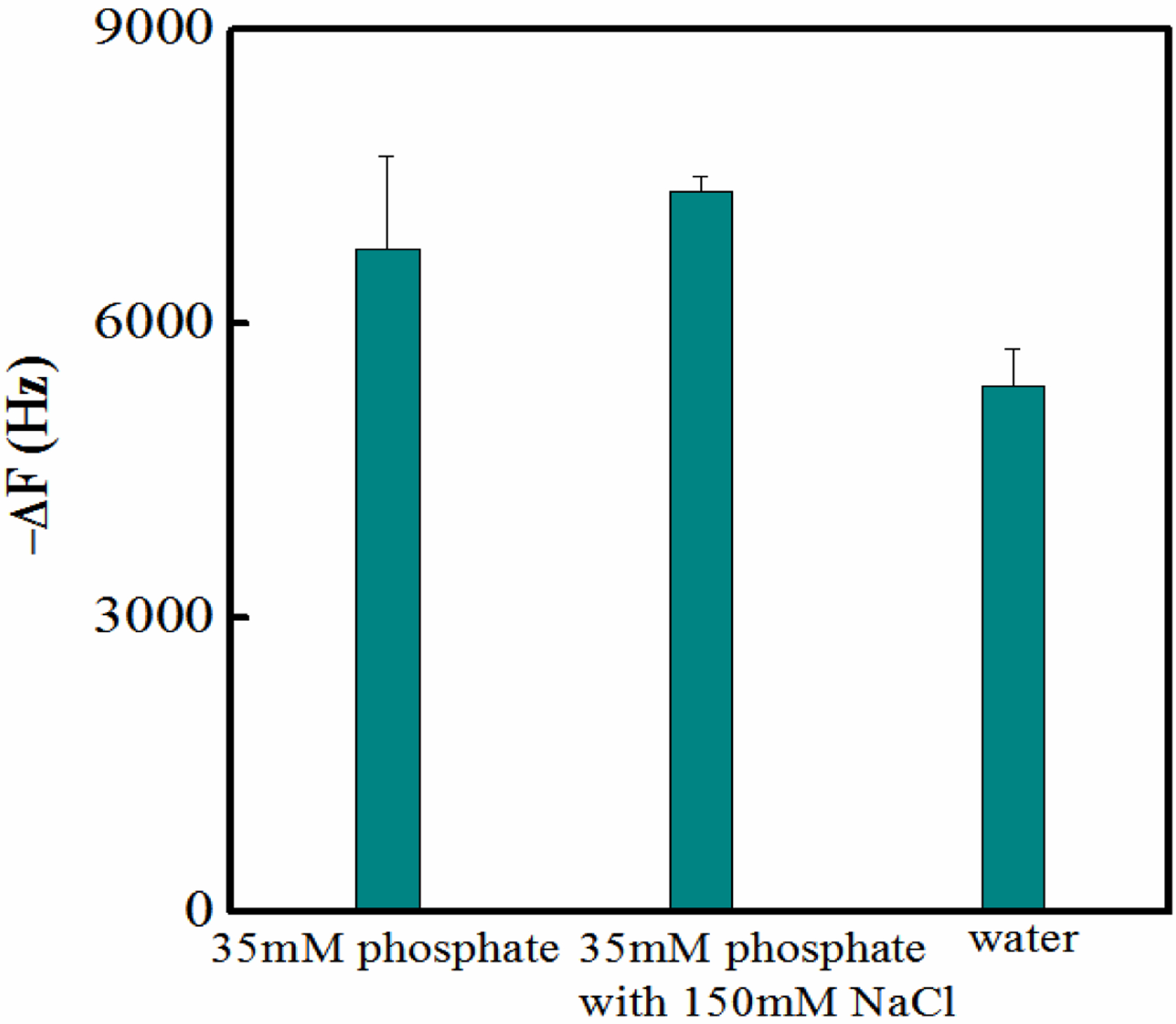
The interaction between P14C and NP1(40TBAm-20AAc) in solutions containing 35 mM phosphate buffer with and without 150 mM NaCl and water.
NP1(40TBAm-20AAc) is a statistical copolymer containing hydrogen-bond donor and acceptor groups as well as hydrophobic (TBAm) and negatively charged (AAc) groups. Although a selection process involving >1010 peptides was used to identify the peptide complement, a clearer understanding of factors that are important to binding is desirable. For example, to what extent is the interaction dependent on peptide sequence and do certain amino acids contribute more than others? Is NP1 capable of discriminating between related peptides with similar physiochemical properties and to what extent does NP1 bind to common abundant proteins? The following experiments were designed to address these questions. Binding to a peptide with the same amino acids as P14 but in a scrambled order (SCP) was evaluated first. Significantly, the interaction of SCP with NP1(40TBAm-20AAc) is ten times weaker than the NP-P14C interaction (Figure 6). To investigate the influence of electrostatic interactions on NP affinity, the single positively charged amino acid lysine (K) in P14C, was mutated to alanine. The mutated peptide P14K, had only a weak interaction with NP1(40TBA-20AAc) implying that lysine plays a significant role in binding to the nanoparticle. NP1(40TBAm-20AAc) affinity to the peptide has strong sequence specificity. This may not be surprising since P14C was selected from a large phage library of very similar cyclic peptides, many comprised of single point mutations of P14 C. The sequence G L P A L I S W I K R K R Q Q K (Peptide for Nanoparticle Test, PNT) contains seven hydrophobic residues, one aromatic residue, three hydrophilic residues and five positively charged residues. Because of its high hydrophobicity and positive charge content, PNT might be expected to engender non-specific binding to NP1(40TBAm-20AAc). However, the result in Figure 6 shows that binding between P14C and NP1(40TBAm-20AAc) is 3.5 times greater than that between NP1(40TBAm-20AAc) and PNT. In another example the three phenylalanines were mutated to asparagine (P14N). Perhaps not surprisingly this change significantly decreases the aromatic hydrophobicity of the peptide. QCM studies indicated that binding of NP1(40TBAm-20AAc) to the mutated peptide P14N in PBS (35 mM, pH 7.3 with 150 mM NaCl) falls to a negligible value (Figure 6). The three phenylalanines are important contributiors to the binding of P14C to NP1(40TBAm-20AAc).
Figure 6.
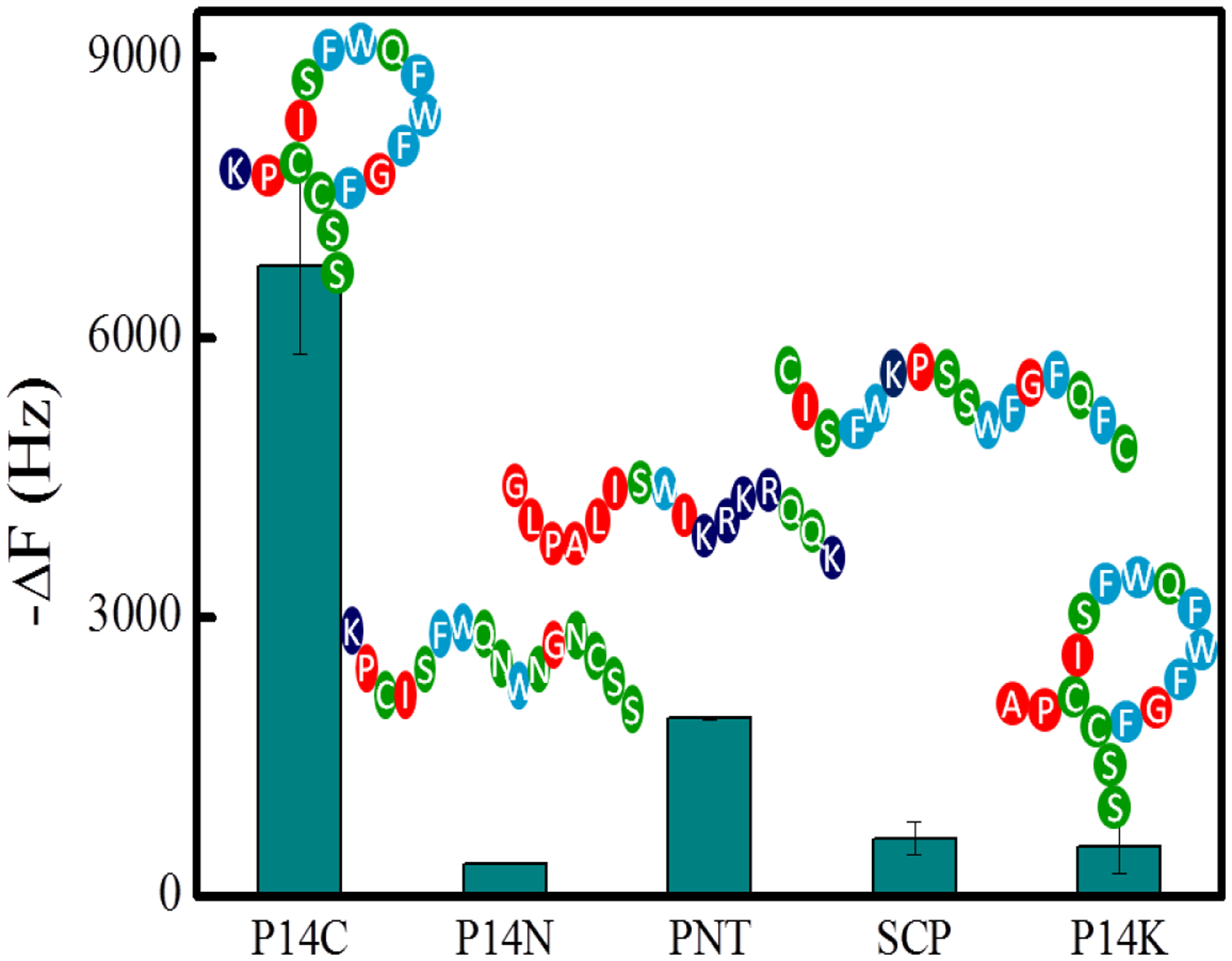
The interaction between NP1(40TBAm-20AAc) and P14C, mutated P14N, PNT, SCP and P14K, respectively. The peptides (P14C, P14N, PNT, SCP or P14K) were immobilized on a QCM surface in PBS (35 mM, pH 7.3 with 150 mM NaCl).
We also note that NP1(40TBAm-20AAc) had relatively low affinity for a panel of proteins that span a range of isoelectric points from 4.7 to 10.5 (Table S2) highlighting the fact that the binding of NP1(40TBAm-20AAc) to P14C is specific (Figure 7) and not easily swamped by common abundant proteins. With the caveat that absolute selectivity is inherently difficult to prove, collectively, these results reinforce the conclusion that the affinity of NP1(40TBAm-20AAc) to P14C is dependent on both physiochemical properties of the peptide and peptide sequence and is not due to non-specific hydrophobic or electrostatic interactions. It is important to note that specificity in this case is achieved by a ternary statistical copolymer (NP1) that discriminates on the basis of a specific peptide sequence. Biomacromolecule affinity is conventionally understood in terms of complementarity derived from the interacting amino acid side chains of the peptide ligand interacting with surface functionality of the receptor. The linear sequences and subsequent structures of the receptor ligands can be traced back to their encoding DNA. This deterministic understanding of the origins of affinity and selectivity suggests one might attempt to describe the high affinity and selectivity of the synthetic polymer NP in this and related studies, to such analysis. For example, one might assume there are “privileged sequences” in the polymer chains that are responsible for binding. Identifying these sequences could define affinity and selectivity within this conventional deterministic framework.
Figure 7.
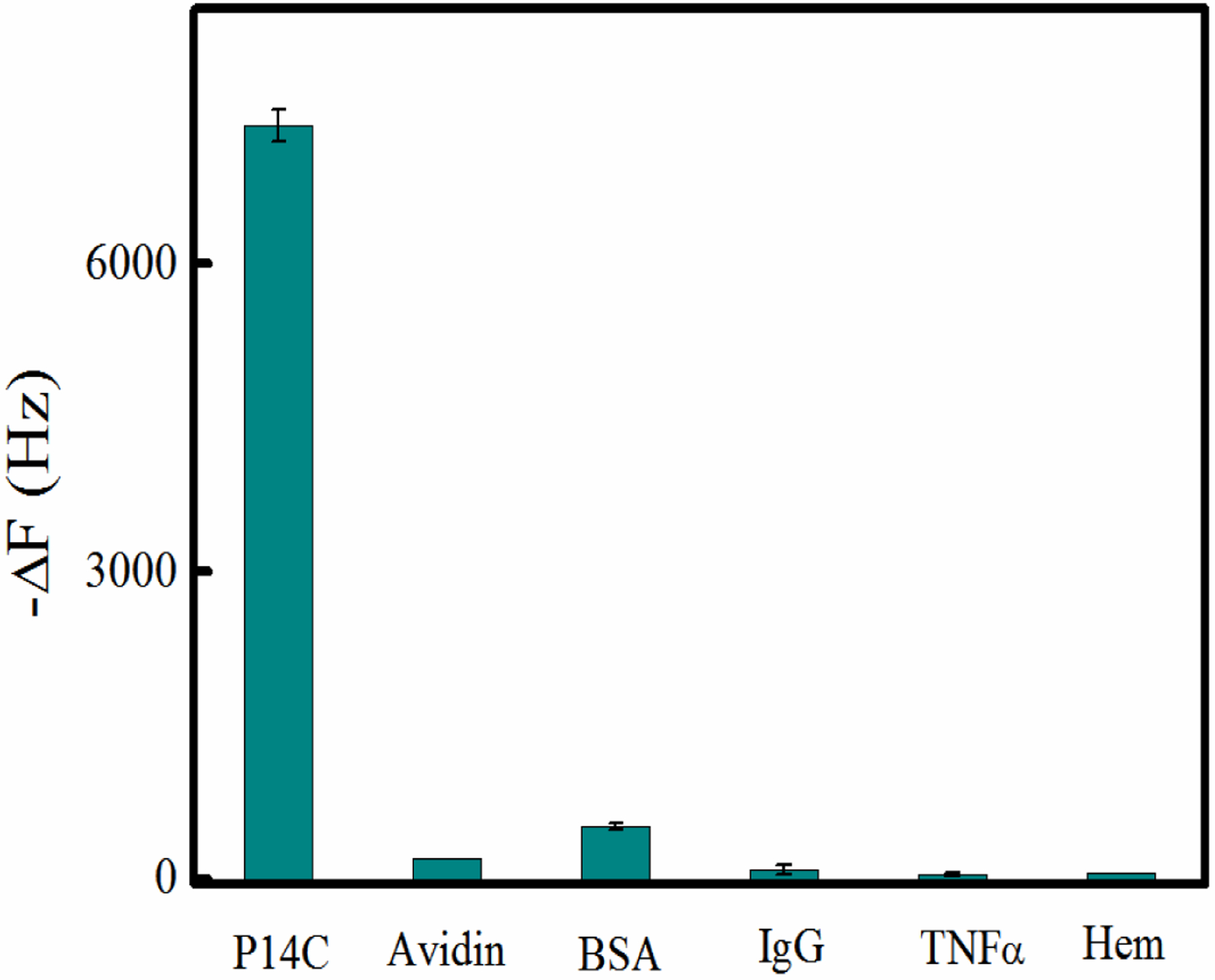
The interaction between NP1(40TBAm-20AAc) and IgG, BSA, TNFα and hemoglobin. In each case the protein was immobilized on a QCM surface in PBS (35 mM, pH 7.3 with 150 mM NaCl) and titrated with a solution of NP1.
However, upon closer analysis, there are a number of practical difficulties that make understanding the molecular-level understanding of NP-peptide binding currently impossible. First, the NP polymers described here are carbon backbone polymers, and unlike biological macromolecules (DNA, RNA, peptides and proteins) they cannot be sequenced by existing methods. Second, the MWs of these copolymers are ~50 mDa. Each polymer NP contains a Bernoulli distribution of ~ 450,000 monomers. In addition, the densities of these hydrogels are ~ 1/30 to 1/50th that of a globular peptide, they are mostly water. They may be described as three-dimensional spider web-like materials. As a result of their low crosslinking, their mesh size allows access to the interior of the NP. The interactions therefore between peptide and polymer chains are not simply along liner sequences on the surface, but also involve interactions throughout the NP between discontinuous segments of polymer chains, further increasing the complexity of interactions.
In view of the complexity of these statistical copolymers and the limited information available to describe the molecular level details of NP-peptide interactions, affinity and selectivity at present is best understood in terms of probabilities. The peptide sequence was selected by screening the copolymer against a library of >1010 peptides, the probability that this and subsequent copolymers prepared in an identical mannor will be selective for that peptide is extremely high. We suggest that at present, a probabilistic approach is an appropriate way to describe binding.
In many examples, peptide structure is an important contributor to binding and function.[63] Cyclization, for example, reduces the conformational freedom of linear peptides. The cyclization of peptides and resultant constraints on their conformational entropy is expected to improve binding affinity, due to the reduced entropic penalty upon binding.[64] It was interesting therefore to examine the effect of peptide structure on binding to NP1.
P14 contains two cysteine residues which under oxidizing conditions forms a disulfide linkage to produce a cyclic structure. When P14 is dissolved in an aqueous solution containing 50% acetonitrile, it exists in the linear form (P14L). In 10 mM HEPES solution containing 20% DMSO, pH 7.3, it exists exclusively in the cyclic form.[65–68] The peptide composition in these two solutions was readily confirmed by MALDI-TOF-MS (Figure S5 and S6). These two solutions were used for the coupling of linear and cyclic forms to the QCM sensor chip. Subsequent binding was evaluated in PBS (35mM phosphate buffer, 150 mM NaCl, pH=7.3). Figure 8 shows that the NP1(40TBAm-20AAc) interaction with cyclic peptide P14C is three times stronger than the linear variant P14L. Since the phage selection process presents the peptide in its cyclic form,[68, 69] the highest affinity is consistent with the structure presented by the phage. This conclusion is supported by binding studies with peptide variants. The variants include a peptide with the cysteines replaced by serine (P14S) to preclude formation of a cyclic structure. The interactions between NP1(40TBAm-20AAc) and P14S/P14L are very similar. The results confirm the stronger interaction with the peptide’s cyclic form (P14C) than with its two linear isoforms (P14L) and P14S. NP1 affinity for a P14C peptide is thus sequence dependent and also reliant on structural constraints resulting from cyclization.
Figure 8.
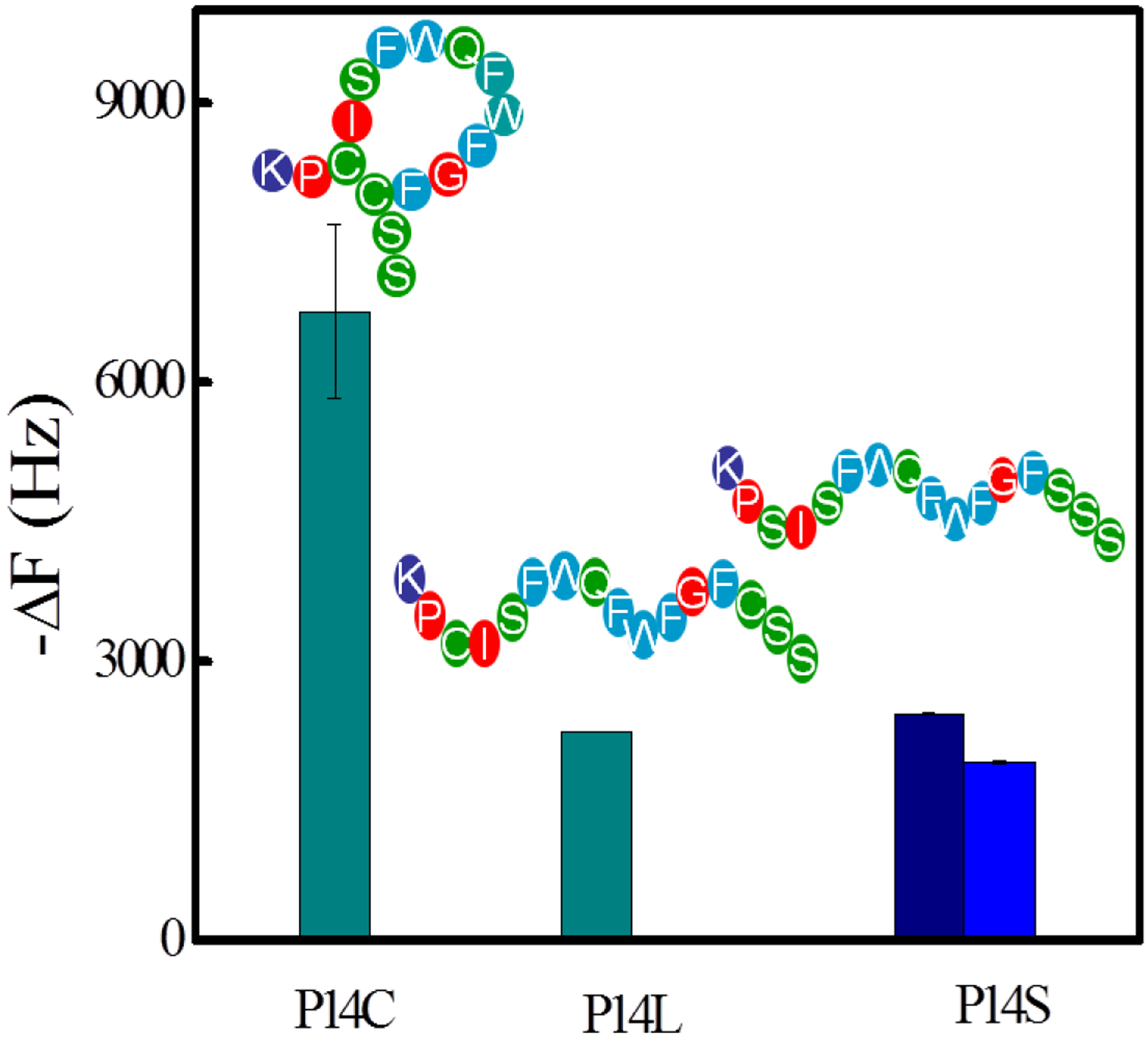
The interactions between NP1(40TBAm-20AAc) and P14C, P14L and P14S. The peptides were immobilized on a QCM surface in PBS (35 mM, pH 7.3 with 150 mM NaCl). Navy column: P14S dissolved in 10 mM HEPES (containing 20% DMSO, pH 7.3). Blue column: P14S dissolved in the aqueous solution (containing 50% acetonitrile)
The selectivity of NP1(40TBAm-20AAc) for the cyclic peptide P14C is an interesting and important finding. Since there are challenges associated with estimating ground state conformation of small peptides computationally,[6, 70, 71] we chose to extract only the solvent accessible area of the peptides used in this study (Table 3). Within the limitations of the method, the data suggests that the four peptides (P14C, P14L, P14S and P14N) have similar solvent accessible surface areas. The result implies that the interaction between P14C and NP1(40TBAm-20AAc) is not due to a difference in solvent accessible area. Although both P14C and P14L have considerable conformational freedom, P14L occupies substantially greater conformational space. By analogy with peptide-protein studies, the structural constraint could contribute to the higher affinity and faster binding kinetics.[9, 30, 64, 62] Since the peptide structure that was presented in phage display is cyclic, these findings are consistent with the experimental results and establish that phage display is an effective strategy for identifying specific peptide-NP affinity pairs.
Table 3.
Solvent accessible areas of the peptides
| Peptide | Solvent accessible surface area (Å2) |
|---|---|
| P14L | 2117 |
| P14S | 2117 |
| P14M | 2059 |
| P14C | 2181 |
4. Conclusion
A general procedure for identifying a synthetic copolymer hydrogel NP and a short peptide complementary pair has been developed. An apparent KD, 95 nM, and Hill coefficient of 1.36 were obtained from the binding isotherm of the Hill equation fit. The high affinity hydrogel NP is a ternary statistical copolymer (NP1) that selects on the basis of physiochemical properties and sequence of the peptide. Furthermore peptide shape is also important. The cyclic peptide selected from the phage display, has greater than three times higher affinity for the nanoparticle then the linear form. Since these carbon backbone polymers each contain a Bernoullian distribution of ~ 450,000 monomers, they cannot be sequenced by existing methods. Furthermore, they can interact with peptides throughout the interior of the NP. The absence of copolymer sequence information and details of the polymer-NP interaction prevents a deterministic explanation of binding. In view of the limited information available at present to describe the molecular level details of polymer peptide interactions, affinity and selectivity is best described in terms of probabilities. Since this specific peptide sequence was selected by a copolymer of specific average chemical composition, from a library of 1010 peptides, the probability that this specific polymer composition will be selective for P14C is extremely high.
Statistical incorporation of a small number of functional groups in a high molecular weight, hydrogel copolymer coupled with a phage-based selection process are sufficient to achieve high affinity and selectivity. The diversity of biological space allows them to interact selectively in a complex biological milieu. High affinity and selectivity of synthetic copolymers for binding to biomacromolecules does not require copolymer sequence homology. Matched affinity pairs of synthetic polymer NPs have important applications at the biological interface and offers possibilities for their use in affinity separations, diagnostics and for therapeutic applications.
Supplementary Material
ACKNOWLEDGMENT
The research was funded in part by the US National Science Foundation (Grant No. DMR-1308363). We gratefully acknowledge the Laser Spectroscopy Laboratories at UC Irvine.
Footnotes
Support information
The supporting information is available free of charge at http://pubs.acs.org
SUPPORTING INFORMATION
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsabm.0c01363.
Composition of NPs, IP of various proteins, phage collection conditions, non-specific interactions between phage and NPs, phage titers after each round of selection, sequences of high affinity phage, library of phages, spot assay-between NP1 and phage, MALD-TOF-MS of P14C under various conditions, 1H NMR of NP1.
REFERENCES
- (1).Sun B; Tao K; Jia Y; Yan X; Zou Q; Gazit E; Li J Photoactive properties of supramolecular assembled short peptides. Chem. Soc. Rev 2019, 48, 4387–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ardoña HAM; Tovar JD Energy transfer within responsive pi-conjugated coassembled peptide-based nanostructures in aqueous environments. Chem. Sci 2015, 6, 1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lakshmanan A; Zhang S; Hauser CAE Short self-assembling peptides as building blocks for modern nanodevices. Trends Biotechnol 2012, 30, 155–165. [DOI] [PubMed] [Google Scholar]
- (4).Tesauro D; Accardo A; Diaferia C; Milano V; Guillon J; Ronga L,; Rossi F Peptide-Based Drug-Delivery Systems in Biotechnological Applications: Recent Advances and Perspectives. Molecules 2019, 24, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lien S; Lowman HB Therapeutic peptides. Trends Biotechnol 2003, 21, 556–562. [DOI] [PubMed] [Google Scholar]
- (6).Brown S Metal-recognition by repeating polypeptides. Nat. Biotechnol 1997, 15, 269–272. [DOI] [PubMed] [Google Scholar]
- (7).Naik RR; Stringer SJ; Agarwal G; Jones SE; Stone MO Biomimetic synthesis and patterning of silver nanoparticles. Nat. Mater 2002, 1, 169–172. [DOI] [PubMed] [Google Scholar]
- (8).Chiu CY; Li YJ; Ruan LY; Ye XC; Murray CB; Huang Y Platinum nanocrystals selectively shaped using facet-specific peptide sequences. Nat.Chem 2011, 3, 393–399. [DOI] [PubMed] [Google Scholar]
- (9).Seker UOS; Wilson B; Dincer S; Kim IW; Oren EE; Evans JS; Tamerler C; Sarikaya M Adsorption Behavior of Linear and Cyclic Genetically Engineered Platinum Binding Peptides. Langmuir 2007, 23, 7895–7900. [DOI] [PubMed] [Google Scholar]
- (10).Whaley SR; English DS; Hu EL; Barbara PF; Belcher AM Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature 2000, 405, 665–668. [DOI] [PubMed] [Google Scholar]
- (11).Lee SW; Mao CB; Flynn CE; Belcher AM Ordering of quantum dots using genetically engineered viruses. Science 2002, 296, 892–895. [DOI] [PubMed] [Google Scholar]
- (12).Park TJ; Lee SY; Lee SJ; Park JP; Yang KS; Lee KB; Ko S; Park JB; Kim T; Kim SK; Shin YB; Chung BH; Ku SJ; Kim DK; Choi IS Protein nanopatterns and biosensors using gold binding polypeptide as a fusion partner. Anal Chem 2006, 78, 7197–7205. [DOI] [PubMed] [Google Scholar]
- (13).Sano K; Sasaki H; Shiba K Utilization of the Pleiotropy of a Peptidic Aptamer To Fabricate Heterogeneous Nanodot-Containing Multilayer Nanostructures. J. Am. Chem. Soc 2006, 128, 1717–1722. [DOI] [PubMed] [Google Scholar]
- (14).Yang ST; Liu Y; Wang YW; Cao A Biosafety and bioapplication of nanomaterials by designing protein-nanoparticle interactions. Small 2013, 9, 1635–1653. [DOI] [PubMed] [Google Scholar]
- (15).Chou LYT; Ming K; Chan WCW Strategies for the intracellular delivery of nanoparticles. Chem. Soc. Rev 2011, 40, 233–245. [DOI] [PubMed] [Google Scholar]
- (16).Shemetov AA; Nabiev I; Sukhanova A Molecular Interaction of Proteins and Peptides with Nanoparticles. ACS nano 2012, 6, 4585–4602. [DOI] [PubMed] [Google Scholar]
- (17).Gorshkov V; Bubis JA; Solovyeva EM; Gorshkov MV; Kjeldsen F Protein corona formed on silver nanoparticles in blood plasma is highly selective and resistant to physicochemical changes of the solution. Environ. Sci. Nano 2019, 6, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Serizawa T; Sawada T; Kitayama T Peptide motifs that recognize differences in polymer-film surfaces. Angew. Chem. Int. Ed 2007, 46, 723–726. [DOI] [PubMed] [Google Scholar]
- (19).Serizawa T; Sawada T; Matsuno H; Matsubara T; Sato T A Peptide Motif Recognizing a Polymer Stereoregularity. J. Am. Chem. Soc 2005, 127, 13780–13781. [DOI] [PubMed] [Google Scholar]
- (20).Zhang X; Gong C; Akakuru OU; Su Z; Wu A; Wei G The design and biomedical applications of self-assembled two-dimensional organic biomaterials. Chem. Soc. Rev 2019, 48, 5564–5595. [DOI] [PubMed] [Google Scholar]
- (21).Ke PC; Lin S; Parak WJ; Davis TP; Caruso F A Decade of the Protein Corona. ACS nano 2017, 11, 11773–11776 [DOI] [PubMed] [Google Scholar]
- (22).Monopoli MP; Åberg C; Salvati A; Dawson KA Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol 2012, 7, 779–786. [DOI] [PubMed] [Google Scholar]
- (23).Casals E; Pfaller T; Duschl A; Oostingh GJ; Puntes V Time Evolution of the Nanoparticle Protein Corona. ACS nano 2010, 4, 3623–3632. [DOI] [PubMed] [Google Scholar]
- (24).Zhang H; Wu T; Yu W; Ruan S; He Q; Gao H Ligand Size and Conformation Affect the Behavior of Nanoparticles Coated with in Vitro and in Vivo Protein Corona. ACS Appl. Mater. Interfaces 2018, 10, 9094–9103. [DOI] [PubMed] [Google Scholar]
- (25).O’Brien J; Shea KJ Tuning the Protein Corona of Hydrogel Nanoparticles: The Synthesis of Abiotic Protein and Peptide Affinity Reagents. Acc. Chem. Res 2016, 49, 1200–1210. [DOI] [PubMed] [Google Scholar]
- (26).Crick F Central Dogma of Molecular Biology. Nature 1970, 227, 561–563. [DOI] [PubMed] [Google Scholar]
- (27).Koch SJ; Renner C; Xie X; Schrader T Tuning Linear Copolymers into Protein‐Specific Hosts. Angew. Chem. Int. Ed 2006, 45, 6352–6355. [DOI] [PubMed] [Google Scholar]
- (28).Latza P; Gilles P; Schaller T; Schrader T Affinity Polymers Tailored for the Protein A Binding Site of Immunoglobulin G Proteins. Chem. Eur. J 2014, 20, 11479–11487. [DOI] [PubMed] [Google Scholar]
- (29).Schrader T; Hamilton A Functional Synthetic Receptors Wiley‐VCH, 2005. [Google Scholar]
- (30).De M; You CC; Srivastava S; Rotello VM Biomimetic Interactions of Proteins with Functionalized Nanoparticles: A Thermodynamic Study. J. Am. Chem. Soc 2007, 129, 10747–10753. [DOI] [PubMed] [Google Scholar]
- (31).Zhang X; Huang R; Gopalakrishnan S; Cao-Milán R; Rotello VM Bioorthogonal Nanozymes: Progress towards Therapeutic Applications. Trends Chem 2019, 1, 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Cieplak M; Kutner W Artificial Biosensors: How Can Molecular Imprinting Mimic Biorecognition? Trends Biotechnol 2016, 34, 922–941. [DOI] [PubMed] [Google Scholar]
- (33).Yan M; Ramström O Molecularly Imprinted Materials: Science and Technology, CRC Press: Boca Raton, FL, 2004. [Google Scholar]
- (34).Yoshimatsu K; Lesel BK; Yonamine Y; Beierle JM; Hoshino Y; Shea KJ Temperature‐Responsive “Catch and Release” of Proteins by using Multifunctional Polymer‐Based Nanoparticle. Angew. Chem. Int. Ed 2012, 51, 2405–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yoshimatsu K Yamazaki T; Hoshino Y; Rose PE; Epstein LF; Miranda LP; Tagari P; Beierle JM; Yonamine Y; Shea KJ Epitope Discovery for a Synthetic Polymer Nanoparticle: A New Strategy for Developing a Peptide Tag. J. Am. Chem. Soc 2014, 136, 1194–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Yonamine Y; Yoshimatsu K; Lee SH; Hoshino Y; Okahata Y; Shea KJ Polymer Nanoparticle–Protein Interface. Evaluation of the Contribution of Positively Charged Functional Groups to Protein Affinity. ACS Appl. Mater. Interfaces 2013, 5, 374–379. [DOI] [PubMed] [Google Scholar]
- (37).Lee SH; Hoshino Y; Randall A; Zeng ZY; Baldi P; Doong RA; Shea KJ Engineered Synthetic Polymer Nanoparticles as IgG Affinity Ligands. J. Am. Chem. Soc 2012, 134, 15765–15772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).O’Brien J; Lee SH; Onogi S; Shea KJ Engineering the Protein Corona of a Synthetic Polymer Nanoparticle for Broad-Spectrum Sequestration and Neutralization of Venomous Biomacromolecules. J. Am. Chem. Soc 2016, 138, 16604–16607. [DOI] [PubMed] [Google Scholar]
- (39).O’Brien J; Lee SH; Gutiérrez JM; Shea KJ Engineered nanoparticles bind elapid snake venom toxins and inhibit venom-induced dermonecrosis. PLOS Neglected Tropical Diseases, 2018, 12, e0006736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hoshino Y; Koide Y; Furuya K; Haberaecker WW; Lee SH; Kodama T; Kanazawa H; Oku X; Shea KJ The rational design of a synthetic polymer nanoparticle that neutralizes a toxic peptide in vivo. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wada Y; Lee H; Hoshino Y; Kotani S; Shea KJ; Miura Y Design of multi-functional linear polymers that capture and neutralize a toxic peptide: a comparison with cross-linked nanoparticles. J. Mater. Chem. B, 2015, 3, 1706–1711. [DOI] [PubMed] [Google Scholar]
- (42).Yoshimatsu K; Koide H; Hoshino Y; Shea KJ Preparation of abiotic polymer nanoparticles for sequestration and neutralization of a target peptide toxin. Nature Protocols, 2015, 10, 595–604. [DOI] [PubMed] [Google Scholar]
- (43).Weisman A; Chen YA; Hoshino Y; Zhang HT; Shea KJ Engineering Nanoparticle Antitoxins Utilizing Aromatic Interactions. ACS Biomacromolecules 2014, 15, 3290–3295. [DOI] [PubMed] [Google Scholar]
- (44).Weisman A; Chou B; O’Brien J; Shea KJ Polymer antidotes for toxin sequestration. Adv. Drug Delivery Rev 2015, 90, 81–100. [DOI] [PubMed] [Google Scholar]
- (45).Hoshino Y; Kodama T; Okahata Y; Shea KJ Peptide Imprinted Polymer Nanoparticles: A Plastic Antibody. J. Am. Chem. Soc 2008, 130, 15242–15243. [DOI] [PubMed] [Google Scholar]
- (46).Hoshino Y; Urakami T; Kodama T; Koide H; Oku N; Okahata Y; Shea KJ Design of synthetic polymer nanoparticles that capture and neutralize a toxic peptide. Small, 2009, 5, 1562–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Koide H; Yoshimatsu K; Hoshino Y; Lee SH; Okajima A; Ariizumi S; Narita Y; Yonamine Y; Weisman AC; Nishimura Y; Oku N; Miura Y; Shea KJ A polymer nanoparticle with engineered affinity for a vascular endothelial growth factor (VEGF165). Nat. Chem 2017, 9, 715–722. [DOI] [PubMed] [Google Scholar]
- (48).Gómez-Arribas LN; Urraca JL; Benito-Peña E; Moreno-Bondi MC Tag-Specific Affinity Purification of Recombinant Proteins by Using Molecularly Imprinted Polymers. Anal. Chem 2019, 91, 4100–4106. [DOI] [PubMed] [Google Scholar]
- (49).Odian G Principles of Polymerization, 4th Edition, John Wiley & Sons, Hoboken, New Jersey, 2004, pp. 464–543. [Google Scholar]
- (50).Ogata A; Edgar JM; Majumdar S; Briggs JS; Patterson SV; Tan MX; Kudlacek ST; Schneider CA; Weiss GA; Penner PM Virus-Enabled Biosensor for Human Serum Albumin. Anal. Chem 2017, 89, 1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Stefan MI; Novere NL Cooperative Binding. PLoS Comput Biol 2013, 9, e1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Goutelle S; Maurin M; Rougier F; Barbaut X; Bourguignon L; Ducher M; Maire P The Hill equation: a review of its capabilities in pharmacological modeling. Fundam. Clin. Pharmacol 2008, 22, 633–648. [DOI] [PubMed] [Google Scholar]
- (53).Dominguez-Medina S; McDonough S; Swanglap P; Landes CF; Link S In Situ Measurement of Bovine Serum Albumin Interaction with Gold Nanospheres. Langmuir 2012, 28, 9131–9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Deng ZJ; Liang MT; Toth I; Monteiro MJ; Minchin RF Molecular Interaction of Poly(acrylic acid) Gold Nanoparticles with Human Fibrinogen. ACS Nano 2012, 6, 8962–8969. [DOI] [PubMed] [Google Scholar]
- (55).Jiang X; Weise S; Hafner M; Röcker C; Zhang F; Parak WJ; Nienhaus GU Quantitative analysis of the protein corona on FePt nanoparticles formed by transferrin binding. J. R. Soc. Interface 2010, 7, S5–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Gebauer JS; Malissek M; Simon S; Knauer SK; Maskos M; Stauber RH; Peukert W; Treuel L Impact of the Nanoparticle–Protein Corona on Colloidal Stability and Protein Structure. Langmuir 2012, 28, 9673–9679. [DOI] [PubMed] [Google Scholar]
- (57).Smith MH; Herman ES; Lyon LA Network Deconstruction Reveals Network Structure in Responsive Microgels. J. Phys. Chem. B 2011, 115, 3761–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Chou B; Mirau P; Jiang T; Wang SW; Shea KJ Tuning Hydrophobicity in Abiotic Affinity Reagents: Polymer Hydrogel Affinity Reagents for Molecules with Lipid-like Domains. Biomacromolecules 2016, 17, 1860–1868. [DOI] [PubMed] [Google Scholar]
- (59).Zeng Z; Patel J; Lee SH; McCallum M; Tygi A; Yan M; Shea KJ Synthetic Polymer Nanoparticle–Polysaccharide Interactions: A Systematic Study. J. Am. Chem. Soc 2012, 134, 2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Lin FY; Chen CS; Chen WY; Yamamoto S Microcalorimetric studies of the interaction mechanisms between proteins and Q-sepharose at pH near the isoelectric point (pI) effects of NaCl concentration, pH value, and temperature. J. Chromatogr. A 2001, 912, 281–289. [DOI] [PubMed] [Google Scholar]
- (61).Zhang SP; Sun Y Further studies on the contribution of electrostatic and hydrophobic interactions to protein adsorption on dye–ligand adsorbents. Biotechnol. Bioeng 2001, 75, 710–717. [DOI] [PubMed] [Google Scholar]
- (62).Li G; Li J; Wang W; Yang M; Zhang Y; Sun P; Yuan Z; He B; Yu Y Adsorption mechanism at the molecular level between polymers and uremic octapeptide by the 2D 1H NMR technique. Biomacromolecules 2006, 7, 1811–1818. [DOI] [PubMed] [Google Scholar]
- (63).Murase K; Morrison KL; Tam PY; Stafford RL; Jurnak F; Weiss GA EF-Tu Binding Peptides Identified, Dissected, and Affinity Optimized by Phage Display. Chem. Biol 2003, 10, 161–168. [DOI] [PubMed] [Google Scholar]
- (64).Hill TA; Shepherd NE; Diness F; Fairlie DP Constraining cyclic peptides to mimic protein structure motifs. Angew. Chem. Int. Ed 2014, 53, 13020–13041. [DOI] [PubMed] [Google Scholar]
- (65).Chen L; Annis I; Barany G Introduction to Peptide Synthesis. Curr. Protoc. Protein Sci 2001, 18.6.1. [DOI] [PubMed] [Google Scholar]
- (66).Andreu1 D; Albericio F; Solé NA; Munson MC; Ferrer M; Barany G Peptide Synthesis Protocols 1994, 35, 91. [DOI] [PubMed] [Google Scholar]
- (67).Tam JP; Wu CR; Liu W; Zhang JW Disulfide bond formation in peptides by dimethyl sulfoxide. Scope and applications. J. Am. Chem. Soc 1991, 113, 6657–6662. [Google Scholar]
- (68).Lima AR; Juliano L; Juliano MA Cyclic, Linear, Cycloretro-Isomer, and Cycloretro-Inverso Peptides Derived from the C-Terminal Sequence of Bradykinin as Substrates or Inhibitors of Serine and Cysteine Proteases. Protein J 2004, 23, 287–294. [DOI] [PubMed] [Google Scholar]
- (69).Thorstholm L; Craik DJ Discovery and applications of naturally occurring cyclic peptides. Drug Discov. Today 2012, 9, e13–e21. [DOI] [PubMed] [Google Scholar]
- (70).Colombo G; Curnis F; De Mori GMS; Gasparri A; Longoni C; Sacchi A; Longhi R; Corti A Structure-Activity Relationships of Linear and Cyclic Peptides Containing the NGR Tumor-homing Motif. J. Biol. Chem 2002, 277, 47891–47897. [DOI] [PubMed] [Google Scholar]
- (71).Thévenet P; Shen Y; Maupetit J; Guyon F; Derreumaux P; Tufféry P PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res 2012, 40, W288–W293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


