Abstract
Background:
Symptoms are a core concept of nursing interest. Large-scale secondary data reuse of notes in electronic health records (EHRs) has the potential to increase the quantity and quality of symptom research. However, the symptom language used in clinical notes is complex. A great need exists for methods designed specifically to identify and study symptom information from EHR notes.
Objectives:
We aim to describe a method that combines standardized vocabularies, clinical expertise, and natural language processing (NLP) to generate comprehensive symptom vocabularies and identify symptom information in EHR notes. We piloted this method with five diverse symptom concepts – constipation, depressed mood, disturbed sleep, fatigue, and palpitations.
Methods:
First, we obtained synonym lists for each pilot symptom concept from the Unified Medical Language System. Then, we used two large bodies of text (n=5,483,777 clinical notes from Columbia University Irving Medical Center and n=94,017 PubMed abstracts containing Medical Subject Headings or key words related to the pilot symptoms) to further expand our initial vocabulary of synonyms for each pilot symptom concept. We used NimbleMiner, an open-source NLP tool, to accomplish these tasks. We evaluated NimbleMiner symptom identification performance by comparison to a manually annotated set of n=449 nurse- and physician-authored common EHR note types.
Results:
Compared to the baseline Unified Medical Language System synonym lists, we identified up to 11 times more additional synonym words or expressions, including abbreviations, misspellings, and unique multi-word combinations, for each symptom concept. NLP system symptom identification performance was excellent (F-measure ranged from 0.80 to 0.96).
Discussion:
Using our comprehensive symptom vocabularies and NimbleMiner to label symptoms in clinical notes produced excellent performance metrics. The ability to extract symptom information from EHR notes in an accurate and scalable manner has the potential to greatly facilitate symptom science research.
Keywords: signs and symptoms, natural language processing, electronic health records
Assessing, monitoring, interpreting, treating, and managing symptoms are central aspects of nursing care. Symptoms are subjective indications of disease and include concepts such as pain, fatigue, disturbed sleep, depressed mood, anxiety, nausea, dry mouth, shortness of breath, and pruritus. Many patients experience one or more symptoms related to a health condition and/or its treatment. Both individual symptoms and symptom clusters, defined as two or more co-occurring, related symptoms (Kim et al., 2005; Miaskowski et al., 2017), can be challenging to manage and influence patient’s mood, psychological status, functional status, quality of life, disease progression, and survival (Armstrong, 2003; Kwekkeboom, 2016).
Consequently, symptom science is a preeminent focus of nursing research (Cashion et al., 2016). Symptom science centers on the patient symptom experience (National Institute of Nursing Research, n.d.). The patient’s symptom experience encompasses multiple dimensions, including occurrence, severity, and distress or bother (Wong et al., 2017). The goal of symptom science is “to be able to precisely identify individuals at risk for symptoms and develop targeted strategies to prevent or mitigate the severity of symptoms” (Dorsey et al., 2019, p. 88). Symptom science considers a wide range of biological, social, societal, and environmental determinants of health (Dorsey et al., 2019).
Secondary data reuse from electronic health records (EHRs), that captures diverse patient symptoms, has the ability to increase the quantity and quality of symptom research. In particular, text-based clinical notes are a rich source of symptom information. Historically, patient symptom information has been manually extracted from notes by clinical experts. This process is labor intensive, time consuming, expensive, and most prominently, lacks the scalability necessary to extract symptom information from large quantities of notes for hundreds to thousands or even millions of patients from data stored in EHRs.
Novel data science approaches, including natural language processing (NLP), can help to overcome scalability challenges related to manual note review. NLP is “any computer-based algorithm that handles, augments, and transforms natural language so that it can be represented for computation” (Yim et al., 2016) and is used to extract information, capture meaning, and detect relationships from language free text through the use of defined language rules and relevant domain knowledge (Doan et al., 2014; Fleuren & Alkema, 2015; Wang, Wang, et al., 2018; Yim et al., 2016).
Members of our team recently synthesized the literature on the use of NLP to process or analyze symptom information from EHR notes (Koleck et al., 2019). This systematic review revealed that NLP systems, methods, and tools are currently being used to extract information from diverse EHR notes (e.g., admission documents, discharge summaries, progress notes, nursing narratives) written by a variety of clinicians (e.g., physicians, nurses) on a wide range of symptoms (e.g., anxiety, chills, constipation, depressed mood, fatigue, nausea, pain, shortness of breath, weakness) across clinical specialties (e.g., cardiology, mental health, oncology). However, the use of NLP to extract symptom information from notes captured in EHRs is still largely in the developmental phase. Moreover, the majority of previous work has focused on the use of symptom information for physician/medicine-focused tasks, predominantly disease prediction, rather than on the investigation of symptoms themselves. Because existing NLP systems, methods, and tools were developed for the purpose of disease prediction, they may be insufficient for symptom-focused tasks. As nurses, it is critical that we develop and use NLP approaches that are designed for the specific purpose of studying core nursing concepts of interest, including symptom documentation in EHR clinical notes.
However, the complexity of the symptom language used in clinical notes makes the application of NLP challenging. The presence of a single symptom concept (e.g., fatigue) can be indicated using many different synonym words and expressions (e.g., feeling tired, drowsiness, energy loss, exhaustion, groggy, sleepy, sluggish, tires quickly, weary, etc.) within notes. The words and expressions used in real world symptom documentation typically go beyond those contained in standardized vocabularies (e.g., wiped out, low energy) and can include common misspellings (e.g., fatugue, faituge, fatiuge) and abbreviations (e.g., tatt - tired all the time). In addition, the presence of a symptom word or expression may not indicate that the patient is experiencing that symptom. For example, a symptom may be negated (e.g., no fatigue, does not complain of fatigue) or occurred in the past (e.g., pmhx fatigue, not currently fatigued).
Advances in NLP of clinical data can help resolve some of these major challenges. Specifically, a new generation of machine learning (ML) models, called language models (Mikolov et al., 2013), can help to discover synonym vocabularies from large bodies of text. For example, a recently developed open-source and free NLP software, NimbleMiner, enables users to mine clinical texts to rapidly discover large vocabularies of synonyms that include abbreviations and misspellings (Topaz, Murga, Bar-Bachar, McDonald, & Bowles, 2019). NimbleMiner was successfully applied to identify a diverse range of clinical concepts in clinical notes, including drug and alcohol abuse (Topaz, Murga, Bar-Bachar, Cato, & Collins, 2019) and patient fall history (Topaz, Murga, Gaddis, et al., 2019), among others. However, new NLP methods (like the one applied by NimbleMiner) have not been used to identify symptom information in clinical notes.
In this paper, we describe a method that utilizes standardized vocabularies, clinical expertise, and NLP tools (i.e., NimbleMiner) to generate comprehensive symptom vocabularies to identify symptom information in EHR clinical notes. We piloted this method using five diverse symptom concepts – constipation, depressed mood, disturbed sleep, fatigue, and palpitations – and report our evaluation of NimbleMiner symptom identification performance using the generated comprehensive symptom vocabularies compared to a manually annotated gold standard note set.
MATERIALS & METHODS
We completed two overarching research activities as part of this study: (1) vocabulary development and (2) evaluation of NimbleMiner symptom identification performance. We outline the steps used to generate the comprehensive symptom vocabularies to identify symptom information in EHR notes and our evaluation of the vocabularies and NimbleMiner symptom identification performance in Figure 1, with additional details in the text. This study was approved by the Columbia University Irving Medical Center Institutional Review Board.
Figure 1.
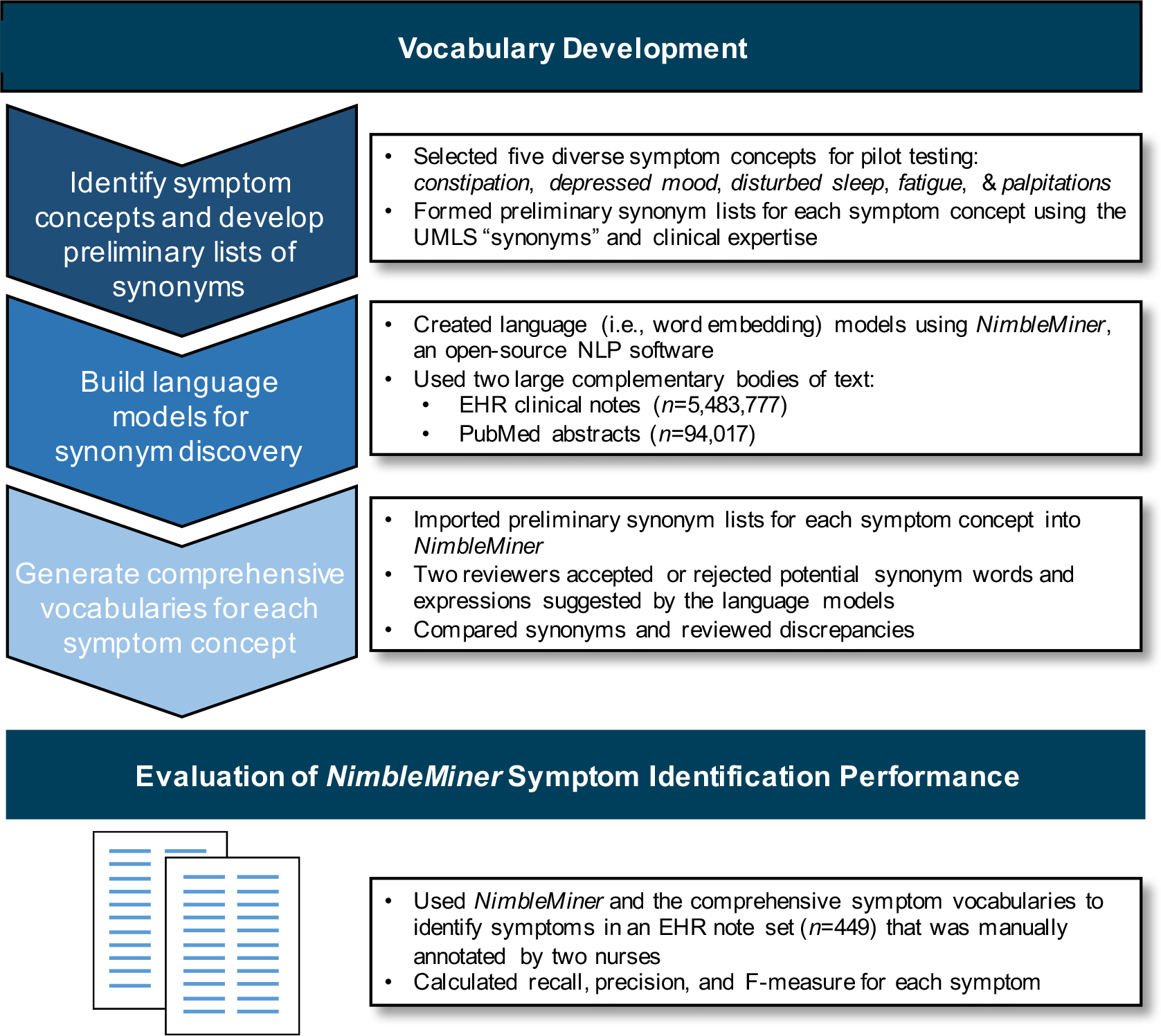
Steps used to generate the comprehensive symptom vocabularies for identifying symptom information in EHR notes and to evaluate the vocabularies and NimbleMiner symptom identification performance
NimbleMiner Natural Language Processing System
NimbleMiner (https://github.com/mtopaz/NimbleMiner) is an open-source and free NLP RStudio Shiny application (https://shiny.rstudio.com/) that enables users to mine clinical texts to rapidly discover large vocabularies of synonyms (Topaz, Murga, Bar-Bachar, McDonald, & Bowles, 2019). Briefly, to build vocabularies within NimbleMiner, the user imports a large body of relevant text and a preliminary list of words and expressions for a concept of interest. The software performs text preprocessing (e.g., removal of punctuation, modification of letter case) and converts frequently co-occurring words to 4-gram (i.e., up to four word) expressions using a phrase2vec algorithm (Topaz, Murga, Bar-Bachar, McDonald, & Bowles, 2019).
Then, NimbleMiner builds language models (i.e., statistical representations of a body of text) using a word embedding skip-gram implementation in an R statistical package called word2vec (Mikolov et al., 2013). The word embedding models use neighboring words to identify other potential synonyms (i.e., words or expressions that appear in the same context) for each imported word or expression. The user can iteratively accept (i.e., designate as a synonym) or reject (i.e., designate as an irrelevant term) words or expressions suggested by the system until no new synonyms are identified. Following discovery of synonyms, NimbleMiner is used to identify positive instances of a concept in text using regular expressions (i.e., specially encoded strings of text). NimbleMiner accounts for negated terms as well. For example, the software is able to identify expressions like no palpitations or denies fatigue as negated synonyms. While not a feature employed in this study, NimbleMiner can also use ML algorithms to create predictive models of whether text contains a concept of interest.
Vocabulary Development
Step 1. Identifying symptom concepts and developing preliminary lists of synonyms
First, we reviewed a widely used medical terminology, Systematized Nomenclature of Medicine (SNOMED-CT, clinical findings category), to create a catalog of candidate symptom concepts. Candidate symptom concepts were reviewed by nurse clinician scientists. The nurse clinician scientists who participated in this study have extensive clinical and research expertise in symptoms and chronic conditions that rank among the leading causes of death and disability in the United States, specifically heart disease (SB), cancer (CM), diabetes (AS), and chronic lung disease (MG) (National Center for Chronic Disease Prevention and Health Promotion, n.d.). We identified a list of 57 unique candidate symptom concepts (see Supplemental Table 1 for a full list).
For the purposes of this study, we selected five diverse symptom concepts – constipation, depressed mood, disturbed sleep, fatigue, and palpitations – to pilot-test methods and evaluate NimbleMiner symptom identification performance. These five symptoms were chosen due to their varying degrees of conceptual complexity (i.e., how difficult a symptom concept is to clearly define and distinguish from other symptom concepts) and potential diversity of language used by clinicians to describe these symptoms. Next, we created a preliminary list of words and expressions (further called synonyms) for each of the symptom concepts using the Unified Medical Language System (UMLS). UMLS is a compendium of many health terminologies, including SNOMED, International Classification for Nursing Practice (ICNP), North American Nursing Diagnosis Association (NANDA International), and others (Bodenreider, 2004). Using the UMLS “synonyms” category that includes words and expressions used by different terminologies to describe a concept of interest, we extracted a list of synonyms for each of the five symptom concepts. The nurse clinician scientists had the opportunity to review these lists and make recommendations for changes (e.g., addition of synonyms, removal of synonyms). UMLS/expert-informed synonym lists for the five symptom concepts are displayed in Table 1.
Table 1.
UMLS/expert-informed synonym lists for the five pilot symptom concepts
| Constipation | Depressed Mood | Disturbed Sleep | Fatigue | Palpitations |
|---|---|---|---|---|
| constipate | affect lack | awakening early | daytime somnolence | chest symptom palpitation |
| constipated | affect unhappy | awakening early morning | decrease in energy | heart irregularities |
| constipating | affects lack | bad dream | decreased energy | heart palpitations |
| constipation | anhedonia | bad dreams | drowsiness | heart pounds |
| costiveness | anhedonias | bizarre dreams | drowsy | heart racing |
| defecation difficult | apathetic | bothered by difficulty sleeping | easy fatigability | heart races |
| defecaecation difficult | apathetic behavior | broken sleep | energy decreased | heart throb |
| difficult defaecation | apathetic behaviour | cannot get off to sleep | energy loss | heart throbbing |
| difficult defecation | apathy | chronic insomnia | excessive daytime sleepiness | palpitation |
| difficulty defecating | cannot see a future | complaining of insomnia | excessive daytime somnolence | palpitations |
| difficulty defaecating | decreased interest | complaining of nightmares | excessive sleep | racing heart |
| difficulty in defecating | decreased mood | complaining of vivid dreams | excessive sleepiness | rapid heart beat |
| difficulty in defaecating | demoralisation | delayed onset of sleep | excessive sleepiness during day | |
| difficulty in ability to defaecate | demoralization | difficult sleeping | excessive sleepiness during the day | |
| difficulty in ability to defecate | depressed | difficulty sleeping through the night | excessive sleeping | |
| difficulty to defaecate | depressed mood | difficulties sleeping | exhaustion | |
| difficulty to defecate | depressed state | difficulty falling asleep | exhausted | |
| difficulty opening bowels | depressing | difficulty getting to sleep | extreme exhaustion | |
| difficult passing motion | depression | difficulty in sleep initiation | extreme fatigue | |
| difficulty passing stool | depressions | difficulty in sleep maintenance | fatigability | |
| fecal retention | depression emotional | difficulty sleep | fatigue | |
| have been constipated | depression mental | difficulty sleeping | fatigued | |
| have constipation | depression mental function | difficulty staying asleep | fatigues | |
| depression moods | disturbance in sleep behavior | fatiguing | ||
| depression psychic | disturbance in sleep behaviour | feel drowsy | ||
| depression symptom | disturbed sleep | feel fatigue | ||
| depressive state | disturbances sleep | feel fatigued | ||
| depressive symptom | disturbances of sleep/insomnia | feel tired | ||
| depressive symptoms | dysnystaxis | feeling drowsy | ||
| despair | early awakening | feeling exhausted | ||
| diminished pleasure | early morning awakening | feeling of total lack of energy | ||
| emotional depression | early morning waking | fatigue extreme | ||
| emotional indifference | early waking | feeling tired | ||
| emotionally apathetic | fitful sleep | get tired easily | ||
| emotionally cold | frequent night waking | groggy | ||
| emotionally detached | frightening dreams | had a lack of energy | ||
| emotionally distant | have nightmares | hypersomnia | ||
| emotionally subdued | hyposomnia | I feel fatigued | ||
| feeling blue | initial insomnia | I have a lack of energy | ||
| feeling despair | insomnia | increased fatigue | ||
| feeling depressed | insomnia matutinal | increased need for rest | ||
| feeling down | insomnia chronic | increased sleep | ||
| feeling empty | insomnia late | increased sleeping | ||
| feeling helpless | insomnia vesperal | lack of energy | ||
| feeling hopeless | interrupted sleep | lacking energy | ||
| feeling isolated | interrupting sleep | lacking in energy | ||
| feeling lonely | keeps waking up | lacks energy | ||
| feeling of loss of feeling | late insomnia | lethargic | ||
| feeling lost | light sleep | lethargy | ||
| feeling low | light sleeping | loss of energy | ||
| feeling of despair | lights sleeping | quickly exhausted | ||
| feeling of hopelessness | hard to sleep through night | sleep excessive | ||
| feeling of sadness | hard to sleep through the night | sleep too much | ||
| feeling powerless | matutinal insomnia | sleepiness | ||
| feeling sad | middle insomnia | sleeping too much | ||
| feeling sad or blue | my sleep was restless | sleepy | ||
| feeling trapped | nightmares | sluggish | ||
| feeling unhappy | nightmares | sluggishness | ||
| feeling unloved | night wake | somnolence | ||
| feeling unwanted | night wakes | somnolent | ||
| feelings of hopelessness | night waking | time tired | ||
| feelings of worthlessness | not getting enough sleep | tired | ||
| feels there is no future | other insomnia | tired all the time | ||
| helplessness | poor quality sleep | tatt | ||
| hopeless | poor sleep | tired feeling | ||
| hopelessness | poor sleep pattern | tired out | ||
| I feel sad or blue | primary insomnia | tired time | ||
| indifference | problems with sleeping | tiredness | ||
| indifferent mood | prolonged sleep | tires quickly | ||
| lack of interest | restless sleep | tire easily | ||
| lethargic | restless sleeping | too much sleep | ||
| lethargy | short of sleep | weariness | ||
| listless | short sleeping | weary | ||
| listless behavior | sleep difficult | washed out | ||
| listless behaviour | sleep difficulties | worn out | ||
| listless mood | sleep disorder insomnia | |||
| listlessness | sleep disorder insomnia chronic | |||
| loss of capacity for enjoyment | sleep disorders | |||
| loss of hope for the future | sleep disturbance | |||
| loss of interest | sleep disturbances | |||
| loss of pleasure | sleep disturbed | |||
| loss of pleasure from usual activities | sleep dysfunction | |||
| lost feeling | sleep maintenance insomnia | |||
| low mood | sleep is restless | |||
| melancholia | sleep was restless | |||
| melancholic | sleeping difficulty | |||
| melancholy | sleeping difficulties | |||
| mental depression | sleeping disturbances | |||
| miserable | sleep restless | |||
| mood depressed | sleeplessness | |||
| mood depression | terrifying dreams | |||
| mood depressions | terminal insomnia | |||
| morose mood | tosses and turns in sleep | |||
| morosity | tossing and turning during sleep | |||
| negative about the future | trouble falling asleep | |||
| no hope for the future | trouble sleeping | |||
| nothing matters | unpleasant dreams | |||
| powerlessness | vesperal insomnia | |||
| sad | vivid dreams | |||
| sad mood | wakes and cannot sleep again | |||
| sadness | wakes early | |||
| stuporous | waking during night | |||
| symptoms of depression | waking too early | |||
| torpid | waking night | |||
| unhappiness | wakes up at night | |||
| unhappy | wakes up during night | |||
| worthless | ||||
| worthlessness | ||||
Step 2. Building language models for synonym discovery
We used two large bodies of text to generate two corresponding language models in NimbleMiner: (1) EHR clinical notes and (2) PubMed abstracts. These two bodies of text, or corpora, were selected because they would allow us to extract a complementary and diverse range of synonyms. EHR clinical notes include clinical jargon terms while PubMed abstracts have more standardized synonyms used in the scientific literature (Topaz, Murga, Bar-Bachar, McDonald, & Bowles, 2019). For the EHR source, we obtained all available patient clinical notes (n=5,483,777) from the Columbia University Irving Medical Center Clinical Data Warehouse (CDW) authored between January 1, 2016 and December 31, 2016. These notes represent 1,449 unique note types from an array of specialties (e.g., internal medicine, psychiatry, cardiology, nephrology, neurology, surgery, obstetrics), settings (e.g., inpatient, emergency department, ambulatory), and members of the healthcare team, including nurses, physicians, physical therapists, occupational therapists, nutritionists, respiratory therapists, and social workers. Notes were from n=238,026 distinct patient medical record numbers. The number of notes per medical record number ranged from 1 to 2,372 (M=23.04, SD=56.9; median=9). Notes were authored by n=9,863 individuals. The number of notes written by a single clinician ranged from 1 to 9,900 (M=538.1, SD=720.6; median=282).
For the PubMed source, we extracted all available PubMed abstracts (n=94,017) containing Medical Subject Headings (MeSH) or key words related to the pilot symptoms of interest. We used the following query on May 24, 2019, to identify PubMed abstracts:
(“constipation”[MeSH Terms] OR “constipation”[All Fields]) OR (disturbed[All Fields] AND (“sleep”[MeSH Terms] OR “sleep”[All Fields])) OR ((“consciousness disorders”[MeSH Terms] OR (“consciousness”[All Fields] AND “disorders”[All Fields]) OR “consciousness disorders”[All Fields] OR ((“depressed”[All Fields]) AND (“affect”[MeSH Terms] OR “affect”[All Fields] OR “mood”[All Fields])) OR (“fatigue”[MeSH Terms] OR “fatigue”[All Fields]) OR palpitations[All Fields] AND (hasabstract[text] AND “humans”[MeSH Terms] AND English[lang]).
We converted text from each source into a single text (.txt) file and uploaded to NimbleMiner. We used NimbleMiner to perform preprocessing, convert frequently co-occurring words to 4-gram expressions, and build language models.
Step 3. Generating comprehensive vocabularies for each symptom concept
We imported the UMLS/expert-informed preliminary synonyms for each pilot symptom (Table 1) into NimbleMiner. Based on the language models built in Step 2, NimbleMiner suggested 50 similar terms for each imported synonym. Two individuals with expertise in symptoms (TK & MH) independently reviewed and accepted or rejected suggested synonym words or expressions for each symptom concept of interest. This process of NimbleMiner suggesting similar words/expressions and the reviewer accepting/rejecting words/expressions was iteratively repeated for chosen words/expressions until no new relevant synonyms were identified by the reviewers. The two reviewers compared lists of words/expressions and discussed discrepancies. When the two reviewers could not come to an agreement on whether or not a word/expression should be included as a symptom concept synonym, an adjudicator (MT) made the final decision. The output of this step was a comprehensive vocabulary for each symptom concept.
Evaluation of NimbleMiner symptom identification performance
Finally, we evaluated NimbleMiner symptom identification performance for the five pilot symptoms. To perform this evaluation, we created a gold standard set of manually annotated clinical notes. In order to increase the probability of positive occurrences of the pilot symptoms in the notes, we first queried the Columbia University Irving Medical Center CDW for patients (n=133) with International Classification of Diseases 10th Revision (ICD10) diagnosis billing codes in 2016 for ≥4 of the pilot symptoms or conditions closely related to a pilot symptom. Included ICD10 codes for each symptom are displayed in Supplemental Table 2. There were n=119 patients with one or more notes (n=27,300). For these patients, we extracted the ten most frequent nurse- and physician-authored note types (n=4,827), including: miscellaneous nursing, medicine follow-up free text, hematology/oncology attending follow-up, ambulatory hematology/oncology nursing assessment, emergency department nursing assessment, medicine resident progress, emergency department disposition, discharge summary, nursing adult admission history, and emergency department adult pre-assessment notes.
Then, we randomly selected n=349 notes, with the counts for each of the ten note types in proportion to their frequency in the overall set of notes. Specific counts for each of the common note types were as follows: miscellaneous nursing – n=87, medicine follow-up free text – n=45, hematology/oncology attending follow-up – n=38, ambulatory hematology/oncology nursing assessment – n=35, emergency department nursing assessment – n=30, medicine resident progress – n=27, emergency department disposition – n=26, discharge summary – n=21, nursing adult admission history – n=20, and emergency department adult pre-assessment notes – n=20. Because documentation of depressed mood and palpitations was limited in these notes, we decided to review and manually label an additional randomly selected n=50 psychiatric consult notes and n=50 cardiology free text notes. Thus, our gold standard note set contained a total of 449 clinical notes. The number of notes per unique patient (n=93) in the gold standard note set ranged from 1 to 30 (M=4.8, SD=5.7; median=3). The number of notes authored by an individual clinician (n=299) ranged from 1 to 10 (M=1.5, SD=1.3; median=1).
Two nurses (TK & SM) manually reviewed each note and annotated the note for the presence or absence of each of the five pilot symptoms. Relative observed agreement (i.e., percent agreement between raters) and inter-rater reliability (i.e., Cohen’s kappa statistic) were calculated for each symptom. Level of agreement for Cohen’s kappa is interpreted as: 0–0.20 – none, 0.21–0.39 – minimal, 0.40–0.59 – weak, 0.60–0.79 – moderate, 0.80–0.90 – strong, and >0.90 – almost perfect (McHugh, 2012). The two nurses plus a third nurse adjudicator (MT) discussed non-agreement to achieve consensus. Overall, the number of notes with positive occurrence of each symptom by manual review was as follows: constipation – n=49, depressed mood – n=62, disturbed sleep – n=77, fatigue – n=84, and palpitations – n=11.
Then, we used NimbleMiner to identify symptoms in the n=449 gold standard note set. We compared the NimbleMiner identification labels to the manual annotation and calculated recall (i.e., NimbleMiner’s ability to identify all notes with positive occurrence of a particular symptom; ), precision (i.e., proportion of notes with a particular symptom endorsed by NimbleMiner that are actually positive occurrences; ) and F-measure (i.e., a measure of test accuracy that considers both precision and recall; ) for each symptom. F measures range from 0 (poor precision and recall) to 1 (perfect precision and recall). We further reviewed all instances of disagreement between NimbleMiner identification labels and gold standard annotations.
RESULTS
Vocabulary Development
We identified additional synonym words and expressions for each symptom concept beyond the inputted UMLS/expert-informed synonym list (Table 2). The increase in synonym vocabulary size ranged from double for disturbed sleep to almost an 11 times increase for constipation. The synonym words and expressions represented abbreviations (e.g., palps), misspellings (e.g., palpations, palipations), and unique multi-word combinations (e.g., feels heart racing, dyspnea palpitations, dizziness palpitations, palpitations holter monitor). For all symptom concepts, a number of synonyms were identified by both users as well as additional unique synonym words or expressions identified by one of the two users. The comprehensive vocabularies for constipation, depressed mood, disturbed sleep, fatigue, and palpitations are available in Supplemental Table 3.
Table 2.
Counts of symptom concept synonym words or expressions
| Symptom concept | Preliminary UMLS/expert- informed synonyms | Additional synonyms identified by both reviewers | Additional unique synonyms identified by Reviewer 1 | Additional unique synonyms identified by Reviewer 2 | Total additional synonyms | Percent increase in synonyms |
|---|---|---|---|---|---|---|
| Constipation | 23 | 22 | 19 | 15 | 56 | 243% |
| Depressed mood | 108 | 494 | 61 | 55 | 610 | 565% |
| Disturbed sleep | 106 | 92 | 35 | 0 | 127 | 120% |
| Fatigue | 75 | 278 | 52 | 35 | 365 | 487% |
| Palpitations | 12 | 111 | 21 | 11 | 143 | 1192% |
Evaluation of NimbleMiner Symptom Identification Performance
Manual annotation of the gold standard clinical note set
Relative observed agreement and inter-rater reliability for manual annotation of the n=449 gold standard note set were as follows for each symptom: constipation – 92.4%, k=0.604; depressed mood – 89.3%, k=0.557; disturbed sleep –90.0%, k=0.630; fatigue – 92.9%, k=0.734; and palpitations – 94.0%, k=0.377. Instances of disagreement were related to extrapolation of medication (e.g., senna or docusate for constipation) or procedure (e.g., cardioversion for palpitations) documentation to the active presence of a symptom. Our team ultimately decided that the symptom itself needed to be documented in the clinical notes to be considered a positive occurrence.
Automated identification of symptoms with NimbleMiner
NimbleMiner symptom identification performance metrics for each pilot symptom are reported in Table 3. Recall ranged from 0.81 to 0.99; precision ranged from 0.75 to 0.96; and F1 ranged from 0.80 to 0.96, all indicating good or excellent system performance.
Table 3.
NimbleMiner symptom identification performance metrics
| Symptom concept | Recall | Precision | F-measure |
|---|---|---|---|
| Constipation | 0.83 | 0.78 | 0.80 |
| Depressed mood | 0.96 | 0.91 | 0.93 |
| Disturbed sleep | 0.81 | 0.96 | 0.87 |
| Fatigue | 0.97 | 0.95 | 0.96 |
| Palpitations | 0.99 | 0.75 | 0.83 |
The most common reason for false positive symptom identification was due to a symptom term being relatively far from a negation term (e.g., no complaint of pain, diarrhea, constipation). Other common causes of false positives included the negation term not being included in the software vocabulary (e.g., pmhx, never exhibited, prior history of, ho, no recent, not a current problem); non-relevant usage of a symptom term (e.g., sluggish referring to pupil response not fatigue, depression referring to a diagnosis and not current mood state); and reference to a potential medication side effect rather than an active problem (e.g., may cause drowsiness). On the other hand, lacking synonym words and expressions for disturbed sleep (e.g., did not sleep well last night, unable to sleep, patient reports change in sleep) and constipation (e.g., no BM, has not had a bm in several days, no significant BM, patient without BM, indication constipation) resulted in the vast majority of false negatives.
DISCUSSION
In this paper, we describe a method that leverages standardized vocabularies, clinical expertise, and NLP tools to create comprehensive vocabularies to identify symptom information documented within EHR notes. The general steps for comprehensive symptom vocabulary development included: 1) identifying symptom concepts and developing preliminary lists of synonyms, 2) building language models for synonym discovery, and 3) generating comprehensive vocabularies for each symptom concept. We piloted this method using five symptom concepts with varying degrees of conceptual complexity and symptom term diversity – constipation, depressed mood, disturbed sleep, fatigue, and palpitations – and evaluated NimbleMiner symptom identification performance for the pilot symptoms against a manually annotated gold standard note set.
Considering that an F-measure reaches its best value at 1 (i.e., perfect precision and recall) and worst at 0, we observed excellent symptom identification performance with the pilot comprehensive symptom vocabularies via the NimbleMiner system. F-measures ranged from 0.80 for constipation to 0.96 for fatigue. It is difficult to compare these results to the literature as extraction/symptom identification performance for individual symptoms are limited and context specific. The systematic review that Koleck et al. (2019) conducted on the use of NLP to process or analyze symptom information from EHR notes identified studies that featured the pilot symptom concepts of constipation (Chase et al., 2017; Hyun et al., 2009; Iqbal et al., 2017; Ling et al., 2015; Nunes et al., 2017; Tang et al., 2017; Wang, Hripcsak, et al., 2009), depressed mood (Chase et al., 2017; Divita et al., 2017; Jackson et al., 2017; Ling et al., 2015; Tang et al., 2017; Wang, Chused, et al., 2008; Weissman et al., 2016; Zhou et al., 2015), fatigue (Chase et al., 2017; Friedman et al., 1999; Hyun et al., 2009; Iqbal et al., 2017; Matheny et al., 2012; Tang et al., 2017; Wang, Hripcsak, et al., 2009), and disturbed sleep (Chase et al., 2017; Divita et al., 2017; Iqbal et al., 2017; Jackson et al., 2017; Tang et al., 2017; Wang, Chused, et al., 2008; Wang, Hripcsak, et al., 2009; Zhou et al., 2015). No NLP investigations specifically named palpitations. Out of these studies, three reported NLP system performance metrics for individual symptoms. Iqbal et al. (2017) created a tool, the Adverse Drug Event annotation Pipeline (ADEPt), to identify adverse drug events from notes; constipation (precision=0.91, recall=0.91, F1=0.91) and insomnia (precision=0.84, recall=0.93, F1=0.88) were included as adverse events. Matheny and colleagues (2012) developed a rule-based NLP algorithm for infectious symptom detection and reported metrics for fatigue (precision=1.00, recall=0.79, F1=0.89). In addition, Jackson et al. (2017) developed a suite of models, comparing a ConText algorithm with or without ML, to identify symptoms of severe mental illness from routine mental health encounters. Symptom model performance against gold standards was reported for the depressed mood synonyms of anhedonia, apathy, and low mood (precision=0–0.96, recall=0–1.00, F1=0–0.96) and the disturbed sleep synonyms of disturbed sleep and insomnia (precision=0.70–0.90, recall=0.84–0.99, F1=0.80–0.94).
While performance for individual symptom concepts was not reported, a study by Divita et al. (2017) had the closest goal to our own – to develop an NLP algorithm that reliably identified mentions of positively asserted symptoms from the free text of clinical notes. Their scalable pipeline features the V3NLP framework, rule-based symptom annotators, and automated ML in Weka and was designed using notes from the Veterans Affairs’ Corporate Data Warehouse. Model performance on a test set of notes was precision=0.80, recall=0.74, and F-measure=0.80. Overall, our method achieved comparable performance to these studies.
While we observed excellent performance, we manually reviewed all discrepancies between our annotated gold standard and NimbleMiner symptom identification. This in-depth exercise revealed additional modifications that could be made to improve performance, including increasing the negation distance, incorporating additional negation terms (e.g., past medical history, no recent), defining irrelevant terms (e.g., sluggish pupil response) and expanding vocabulary terms (e.g., change in sleep, unable to sleep). The irrelevant terms and additional ML features of NimbleMiner may assist with the latter two modifications. Incorporation of irrelevant terms may improve symptom identification performance for symptom term instances of non-relevant usage. For example, we could include sluggish pupil response as an irrelevant expression to correct this instance of sluggish being identified as an occurrence of fatigue. Likewise, we could include major depressive disorder as an irrelevant expression to distinguish between a diagnosis of depression and current depressed mood state. Nevertheless, it may never be possible to create an exhaustive list of synonym or irrelevant words or expressions for a symptom concept. Strategies such as training ML models (e.g., random forest algorithms) that can be used to predict whether a note contains the symptom concept of interest based on characteristics of a note, rather than matching a specific word or expression, may further improve performance (Topaz, Murga, Gaddis, et al., 2019). ML may also help to capture aspects of the symptom experience beyond presence or absence, including frequency, intensity, distress, and meaning (Armstrong, 2003).
This study had a number of strengths, including a text corpus comprising approximately 5.5 million notes from a variety of specialties, settings, and clinicians and over 90,000 relevant PubMed abstracts. Yet, all clinical notes included in this study were obtained from a single medical center. The generalizability of the comprehensive symptom vocabularies will need to be tested, potentially refined, and validated using data from additional medical centers. Another significant strength of this study was the evaluation of NimbleMiner symptom identification performance against a manually annotated gold standard note set from patients with diagnosis billing codes for pilot symptoms or conditions closely related to a pilot symptom. This process helped to ensure that we could test NimbleMiner symptom identification performance on adequate numbers of positive occurrences of pilot symptoms. However, we did limit the gold standard note set and pilot symptom identification evaluation to the 10 most frequent nurse- and physician-authored note types. We do not have strong reason to believe that performance would be drastically different with other note types, especially since all notes were used to generate words and expressions for the comprehensive symptom vocabularies, but this assumption was not evaluated formally. In addition, our evaluation of the NimbleMiner system was limited to five symptom concepts. Pilot symptom concepts were selected by clinical experts based on conceptual complexity and the diversity of language used by clinicians to describe the symptom concept. NimbleMiner symptom identification performance may be different for symptom concepts not included in this pilot study (e.g., pain, anxiety, nausea).
In conclusion, symptoms are a core concept of nursing interest. A great need exists for vocabularies and NLP tools developed specifically for nursing-focused tasks, including studying symptom information documented in the EHR. Therefore, we generated and piloted comprehensive vocabularies for symptoms that can be used to identify symptom information from notes in the EHR. The use of the NLP tool, NimbleMiner, allowed us to enhance standardized vocabularies and clinical expert curation and leverage millions of text documents to develop “real world” EHR vocabularies of relevant words and expressions specific to symptoms. While opportunities exist for refinement, we successfully pilot tested our method and achieved excellent symptom identification performance for five diverse symptoms – constipation, depressed mood, disturbed sleep, fatigue, and palpitations. It is our hope that nurse scientists will be able to take advantage of the comprehensive symptom vocabularies that we are developing, and will continue to refine, for their own work. The ability to extract symptom information from EHR notes in an accurate and scalable manner has the potential to greatly facilitate symptom science research.
Supplementary Material
Acknowledgement:
Research reported in this publication was supported by the National Institute of Nursing Research of the National Institutes of Health under Award Numbers K99NR017651 and P30NR016587. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Kathleen T. Hickey, EdD, FNP, ANP, FAHA, FAAN for her contributions to this project.
Footnotes
The authors have no conflicts of interest to report.
Ethical Conduct of Research: This study was approved by the Columbia University Irving Medical Center Institutional Review Board.
Clinical Trial Registration: N/A
Contributor Information
Theresa A. Koleck, University of Pittsburgh School of Nursing, Pittsburgh, PA.
Nicholas P. Tatonetti, Columbia University Department of Biomedical Informatics, Columbia University Department of Systems Biology, Columbia University Department of Medicine, Columbia University Institute for Genomic Medicine, Columbia University Data Science Institute, New York, NY.
Suzanne Bakken, Columbia University School of Nursing, Columbia University Department of Biomedical Informatics, Columbia University Data Science Institute, New York, NY.
Shazia Mitha, Columbia University School of Nursing, New York, NY.
Morgan M. Henderson, University of Pittsburgh School of Nursing, Pittsburgh, PA.
Maureen George, Columbia University School of Nursing, New York, NY.
Christine Miaskowski, University of California, San Francisco School of Nursing, San Francisco, CA.
Arlene Smaldone, Columbia University School of Nursing, Columbia University College of Dental Medicine, New York, NY.
Maxim Topaz, Columbia University School of Nursing, Columbia University Data Science Institute, New York, NY.
REFERENCES
- Armstrong TS (2003). Symptoms experience: A concept analysis. Oncology Nursing Forum, 30(4), 601–606. 10.1188/03.ONF.601-606 [DOI] [PubMed] [Google Scholar]
- Bodenreider O (2004). The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Research, 32(Database issue), 267–270. 10.1093/nar/gkh061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashion AK, Gill J, Hawes R, Henderson WA, & Saligan L (2016). NIH Symptom Science Model sheds light on patient symptoms. Nursing Outlook, 65(5), 499–506. 10.1016/j.outlook.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase HS, Mitrani LR, Lu GG, & Fulgieri DJ (2017). Early recognition of multiple sclerosis using natural language processing of the electronic health record. BMC Medical Informatics and Decision Making, 17(1), Article 24. 10.1186/s12911-017-0418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divita G, Luo G, Tran L-TT, Workman TE, Gundlapalli AV, & Samore MH (2017). General symptom extraction from VA electronic medical notes. Studies in Health Technology and Informatics, 245, 356–360. [PubMed] [Google Scholar]
- Doan S, Conway M, Phuong TM, & Ohno-Machado L (2014). Natural language processing in biomedicine: A unified system architecture overview. Methods in Molecular Biology (Clifton, N.J.), 1168(Chapter 16), 275–294. 10.1007/978-1-4939-0847-9_16 [DOI] [PubMed] [Google Scholar]
- Dorsey SG, Griffioen MA, Renn CL, Cashion AK, Colloca L, Jackson-Cook CK, Gill J, Henderson W, Kim H, Joseph PV, Saligan L, Starkweather AR, & Lyon D (2019). Working together to advance symptom science in the precision era. Nursing Research, 68(2), 86–90. 10.1097/NNR.0000000000000339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuren WWM, & Alkema W (2015). Application of text mining in the biomedical domain. Methods (San Diego, Calif.), 74, 97–106. 10.1016/j.ymeth.2015.01.015 [DOI] [PubMed] [Google Scholar]
- Friedman C, Knirsch C, Shagina L, & Hripcsak G (1999). Automating a severity score guideline for community-acquired pneumonia employing medical language processing of discharge summaries. Proceedings. AMIA Symposium, 256–260. [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Johnson SB, & Bakken S (2009). Exploring the ability of natural language processing to extract data from nursing narratives. Computers, Informatics, Nursing, 27(4), 215–223. 10.1097/NCN.0b013e3181a91b58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal E, Mallah R, Rhodes D, Wu H, Romero A, Chang N, Dzahini O, Pandey C, Broadbent M, Stewart R, Dobson RJB, & Ibrahim ZM (2017). ADEPt, a semantically-enriched pipeline for extracting adverse drug events from free-text electronic health records. PloS One, 12(11), Article e0187121. 10.1371/journal.pone.0187121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RG, Patel R, Jayatilleke N, Kolliakou A, Ball M, Gorrell G, Roberts A, Dobson RJ, & Stewart R (2017). Natural language processing to extract symptoms of severe mental illness from clinical text: The Clinical Record Interactive Search Comprehensive Data Extraction (CRIS-CODE) project. BMJ Open, 7(1), Article e012012. 10.1136/bmjopen-2016-012012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-J, McGuire DB, Tulman L, & Barsevick AM (2005). Symptom clusters: Concept analysis and clinical implications for cancer nursing. Cancer Nursing, 28(4), 270–282. [DOI] [PubMed] [Google Scholar]
- Koleck TA, Dreisbach C, Bourne PE, & Bakken S (2019). Natural language processing of symptoms documented in free-text narratives of electronic health records: A systematic review. Journal of the American Medical Informatics Association, 26(4), 364–379. 10.1093/jamia/ocy173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwekkeboom KL (2016). Cancer symptom cluster management. Seminars in Oncology Nursing, 32(4), 373–382. 10.1016/j.soncn.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Pan X, Li G, & Hu X (2015). Clinical documents clustering based on medication/symptom names using multi-view nonnegative matrix factorization. IEEE Transactions on Nanobioscience, 14(5), 500–504. 10.1109/TNB.2015.2422612 [DOI] [PubMed] [Google Scholar]
- Matheny ME, Fitzhenry F, Speroff T, Green JK, Griffith ML, Vasilevskis EE, Fielstein EM, Elkin PL, & Brown SH (2012). Detection of infectious symptoms from VA emergency department and primary care clinical documentation. International Journal of Medical Informatics, 81(3), 143–156. 10.1016/j.ijmedinf.2011.11.005 [DOI] [PubMed] [Google Scholar]
- McHugh ML Interrater reliability: The kappa statistic. (2012). Biochemia Medica, 22(3), 276–282. 10.11613/BM.2012.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaskowski C, Barsevick A, Berger A, Casagrande R, Grady PA, Jacobsen P, Kutner J, Patrick D, Zimmerman L, Xiao C, Matocha M, & Marden S (2017). Advancing symptom science through symptom cluster research: Expert panel proceedings and recommendations. Journal of the National Cancer Institute, 109(4), Article djw253. 10.1093/jnci/djw253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolov T, Corrado G, Chen K, & Dean J (2013). Efficient estimation of word representations in vector space (pp. 1–12). Presented at the International Conference on Learning Representations. [Google Scholar]
- National Center for Chronic Disease Prevention and Health Promotion. (n.d.). About chronic diseases. https://www.cdc.gov/chronicdisease/about/index.htm
- National Institute of Nursing Research. (n.d.). Spotlight on symptom science and nursing research. https://www.ninr.nih.gov/researchandfunding/spotlights-on-nursing-research/symptomscience
- Nunes AP, Loughlin AM, Qiao Q, Ezzy SM, Yochum L, Clifford CR, Gately RV, Dore DD, & Seeger JD (2017). Tolerability and effectiveness of exenatide once weekly relative to basal insulin among type 2 diabetes patients of different races in routine care. Diabetes Therapy: Research, Treatment and Education of Diabetes and Related Disorders, 8(6), 1349–1364. 10.1007/s13300-017-0314-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Solti I, Kirkendall E, Zhai H, Lingren T, Meller J, & Ni Y (2017). Leveraging Food and Drug Administration Adverse Event Reports for the automated monitoring of electronic health records in a pediatric hospital. Biomedical Informatics Insights, 9, Article 1178222617713018. 10.1177/1178222617713018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaz M, Murga L, Bar-Bachar O, Cato K, & Collins S (2019). Extracting alcohol and substance abuse status from clinical notes: The added value of nursing data. Studies in Health Technology and Informatics, 264, 1056–1060. 10.3233/SHTI190386 [DOI] [PubMed] [Google Scholar]
- Topaz M, Murga L, Bar-Bachar O, McDonald M, & Bowles K (2019). NimbleMiner: An open-source nursing-sensitive natural language processing system based on word embedding. Computers, Informatics, Nursing, 37(11), 583–590. 10.1097/CIN.0000000000000557 [DOI] [PubMed] [Google Scholar]
- Topaz M, Murga L, Gaddis KM, McDonald MV, Bar-Bachar O, Goldberg Y, & Bowles KH (2019). Mining fall-related information in clinical notes: comparison of rule-based and novel word embedding-based machine learning approaches. Journal of Biomedical Informatics, 90, Article 103103. 10.1016/j.jbi.2019.103103 [DOI] [PubMed] [Google Scholar]
- Wang X, Chused A, Elhadad N, Friedman C, & Markatou M (2008). Automated knowledge acquisition from clinical narrative reports. AMIA Annual Symposium Proceedings. AMIA Symposium, 2008, 783–787. [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hripcsak G, Markatou M, & Friedman C (2009). Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: A feasibility study. Journal of the American Medical Informatics Association, 16(3), 328–337. 10.1197/jamia.M3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, Liu S, Zeng Y, Mehrabi S, Sohn S, & Liu H (2018). Clinical information extraction applications: A literature review. Journal of Biomedical Informatics, 77, 34–49. 10.1016/j.jbi.2017.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman GE, Harhay MO, Lugo RM, Fuchs BD, Halpern SD, & Mikkelsen ME (2016). Natural language processing to assess documentation of features of critical illness in discharge documents of acute respiratory distress syndrome survivors. Annals of the American Thoracic Society, 13(9), 1538–1545. 10.1513/AnnalsATS.201602-131OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ML, Paul SM, Cooper BA, Dunn LB, Hammer MJ, Conley YP, Wright F, Levine JD, Walter LC, Cartwright F, & Miaskowski C (2017). Predictors of the multidimensional symptom experience of lung cancer patients receiving chemotherapy. Supportive Care in Cancer, 25(6), 1931–1939. 10.1007/s00520-017-3593-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim W-W, Yetisgen M, Harris WP, & Kwan SW (2016). Natural language processing in oncology: A review. JAMA Oncology, 2(6), 797–804. 10.1001/jamaoncol.2016.0213 [DOI] [PubMed] [Google Scholar]
- Zhou L, Baughman AW, Lei VJ, Lai KH, Navathe AS, Chang F, Sordo M, Topaz M, Zhong F, Murrali M, Navathe S, & Rocha RA (2015). Identifying patients with depression using free-text clinical documents. Studies in Health Technology and Informatics, 216, 629–633. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


