Abstract
A novel agar medium, chromogenic Salmonella esterase (CSE) agar, for the differentiation of salmonellae is described. The agar contains peptones and nutrient extracts together with the following (grams per liter unless otherwise specified): 4-[2-(4-octanoyloxy-3,5-dimethoxyphenyl)-vinyl]-quinolinium-1-(propan-3-yl carboxylic acid) bromide (SLPA-octanoate; bromide form), 0.3223; lactose, 14.65; trisodium citrate dihydrate, 0.5; Tween 20, 3.0; ethyl 4-dimethylaminobenzoate, 0.035% (wt/vol), novobiocin, 70 mg liter−1. The key component of the medium is SLPA-octanoate, a newly synthesized ester formed from a C8 fatty acid and a phenolic chromophore. In CSE agar, the ester is hydrolyzed by Salmonella spp. to yield a brightly colored phenol which remains tightly bound within colonies. After 24 h of incubation at 37 or 42°C, colonies of typical Salmonella spp. were burgundy colored on a transparent yellow background, whereas non-Salmonella spp. were white, cream, yellow or transparent. CSE agar was evaluated by using a panel of strains including a high proportion of Salmonella and non-Salmonella strains giving atypical reactions on other differential agars. The sensitivity (93.1%) of CSE agar for non-typhi salmonellae compared favorably with those of Rambach (82.8%), xylose-lysine-deoxycholate (XLD; 91.4%), Hektoen-enteric (89.7%), and SM ID (91.4%) agars. The specificity (93.9%) was also comparable to those of other Salmonella media (SM ID agar, 95.9%; Rambach agar, 91.8%; XLD agar, 91.8%; Hektoen-enteric agar, 87.8%). Strains of Citrobacter freundii and Proteus spp. giving false-positive reactions with other media gave a negative color reaction on CSE agar. CSE agar enabled the detection of >30 Salmonella serotypes, including agona, anatum, enteritidis, hadar, heidelberg, infantis, montevideo, thompson, typhimurium, and virchow, which accounted for 91.8% of the salmonella isolates recorded by the Public Health Laboratory Service (Colindale, London, England) for 1997.
There is a range of selective differential agars which distinguish enteric bacteria and have been applied to the detection of salmonellae. These include xylose-lysine-deoxycholate agar (XLD), brilliant green agar, modified brilliant green agar, Hektoen-enteric agar, mannitol-lysine-crystal violet-brilliant green agar, Salmonella-Shigella agar, deoxycholate-citrate agar, and bismuth sulfite agar (3). The selectivity of these agars is due to the presence of bile salts (or other surfactive compounds) and inhibitors such as brilliant green. Differentiation of most salmonellae from other organisms, particularly members of the family Enterobacteriaceae, relies on the ability to produce hydrogen sulfide and/or the inability to ferment lactose and thus lower the medium pH.
Novobiocin-brilliant green-glucose agar (4) and novobiocin-brilliant green-glycerol-lactose agar (12) also rely on hydrogen sulfide production for detection of salmonellae but include novobiocin (10 mg liter−1) to restrict the growth of Proteus spp. and Citrobacter freundii. In EF-18 agar (5), which contains l-lysine · HCl, sucrose, and bromothymol blue, salmonellae are detected by the ability to ferment sucrose. However, this characteristic is also shared by some strains of Escherichia coli.
Recently, Salmonella agars incorporating chromogenic substrates have become commercially available. Rambach agar (15) incorporates propylene glycol, which is metabolized by Salmonella spp. to give acid products detected by neutral red, together with a chromogenic β-galactosidase substrate (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal]). This is hydrolyzed by many non-Salmonella species to give a blue, insoluble product. SM ID agar (13) also uses a chromogenic β-galactosidase substrate together with a glucuronate which is metabolized by salmonellae.
There has been interest in the use of esterase activity to confirm the presence of salmonellae. Esterases hydrolyze short-chain organic acid esters and are present in all organisms to various extents. Their specificity for organic acid chain length is also variable. Esters of fluorescein derivatives or 4-methylumbelliferone have previously been used to screen for microbial esterase activity; for example, Pancholy and Lynd (11) used 7-butanoyloxymethylumbelliferone to detect esterase-positive soil bacteria and fungi. More recently, Aguirre et al. (2) used a C8-esterase spot test (MUCAP test; Biolife Italiana S.r.l, Milan, Italy) in which a solution of methylumbelliferyl caprylate in ethanol was applied to colonies on Rambach agar. The appearance of blue fluorescence under UV light within 1 min of application was observed for all Salmonella test strains; however, colonies of some Pseudomonas and Acinetobacter spp. also fluoresced (6). Thus, although the sensitivity of the MUCAP test was high (100%), the specificity was low (80%), especially compared to that of a latex slide agglutination test (96%) (9).
The present study examined the use of chromogenic ester substrates (10, 16, 17) in the detection of Salmonella spp. Esters differing in chain length (C4 to C10) were first used to assess the specificity of Salmonella esterase. Subsequently, a novel agar medium for the presumptive identification of salmonellae was developed which incorporated a chromogenic C8 organic acid ester known as 4-[2-(4-octanoyloxy-3,5-dimethoxyphenyl)-vinyl]-quinolinium-1-(propan-3-yl carboxylic acid) bromide (SLPA-octanoate; bromide form). The efficacy of the medium was compared to that of four agars which are dependent on differing phenotypic characteristics for Salmonella detection.
MATERIALS AND METHODS
Cultures.
The test bacteria (see Table 1) were obtained from either the Oxoid stock culture collection (OCC or CMCC; Oxoid Ltd., Basingstoke, United Kingdom), the National Collection of Type Cultures (NCTC; Central Public Health Laboratory, Colindale, London, United Kingdom), or the King’s College London collection (KCL; Division of Life Sciences, Kensington, London, United Kingdom). They were stored in nutrient broth (CM4; Oxoid) plus glycerol (25%) at −70°C.
TABLE 1.
Color reactions of Salmonella and non-Salmonella colonies on streak plates
| Species | Strain | Reactiona on:
|
||||
|---|---|---|---|---|---|---|
| CSE agar | Rambach agar | SMID agar | XLD agar | Hektoen-enteric agar | ||
| Salmonella serotypes | ||||||
| S. agona | OCC06 | √ | √ | √ | √ | √ |
| S. anatum | OCC19 | √ | √ | √ | √ | √ |
| S. anatum | OCC112 | √ | √ | √ | √ | √ |
| S. arizonae | OCC1200 | Weak | × | × | √ | √ |
| S. arizonae | NCTC8297 | Weak | × | × | √ | √ |
| S. choleraesuis | OCC852 | × | × | √ | × | × |
| S. choleraesuis | NCTC5735 | × | √ | √ | × | × |
| S. derby | CMCC63 | √ | √ | √ | √ | √ |
| S. derby | OCC159 | √ | √ | √ | √ | √ |
| S. diarizonae | NCTC10381 | √ | √ | √ | √ | √ |
| S. diarizonae | NCTC12417 | √ | × | × | √ | √ |
| S. dublin | OCC160 | √ | × | √ | √ | √ |
| S. dublin | OCC878 | √ | √ | √ | √ | √ |
| S. dublin | OCC1268 | √ | √ | √ | √ | √ |
| S. dublin | OCC627 | √ | √ | √ | √ | √ |
| S. dublin linton | OCC15 | √ | √ | √ | √ | √ |
| S. eastbourne | OCC05 | √ | √ | √ | √ | √ |
| S. enteritidis | OCC723 | √ | √ | √ | √ | √ |
| S. give | CMCC66 | √ | √ | √ | √ | √ |
| S. hadar | CMCC2414 | √ | √ | √ | √ | √ |
| S. heidelberg | CMCC1785 | √ | √ | √ | √ | √ |
| S. heidelberg | NCTC5717 | √ | √ | √ | √ | √ |
| S. heidelberg | OCC08 | √ | √ | √ | √ | √ |
| S. heidelberg | OCC10 | √ | √ | √ | √ | √ |
| S. indiana | NCTC11304 | × | × | × | × | × |
| S. indiana | OCC597 | × | × | × | × | × |
| S. infantis | OCC03 | √ | √ | √ | √ | √ |
| S. infantis | OCC12 | √ | √ | √ | √ | √ |
| S. infantis | OCC16 | √ | √ | √ | √ | √ |
| S. infantis | OCC18 | √ | √ | √ | √ | √ |
| S. kedougou | OCC13b | √ | √ | √ | √ | √ |
| S. montevideo | CMCC1781 | √ | √ | √ | √ | √ |
| S. napoli | OCC14 | √ | √ | √ | √ | √ |
| S. ovatum | OCC04 | √ | √ | √ | √ | √ |
| S. panama | OCC01 | √ | √ | √ | √ | √ |
| S. panama | OCC20 | √ | √ | √ | √ | √ |
| S. panama | OCC21 | √ | √ | √ | √ | √ |
| S. poona | OCC1162 | √ | √ | √ | √ | √ |
| S. pullorum | NCTC10704 | √ | × | √ | √ | × |
| S. pullorum | OCC09 | √ | × | √ | × | × |
| S. reading | OCC631 | √ | √ | √ | √ | √ |
| S. senftenberg | CMCC114 | √ | √ | √ | √ | √ |
| S. sofia | OCC632 | Weak | √ | √ | Weak | √ |
| S. saint-paul | CMCC1602 | √ | √ | √ | √ | √ |
| S. stanley | CMCC1783 | √ | √ | √ | √ | √ |
| S. teshi | OCC633 | Weak | Weak | √ | Weak | √ |
| S. thompson | OCC02 | √ | √ | √ | √ | √ |
| S. thompson | CMCC1188 | √ | √ | √ | √ | √ |
| S. typhimurium | OCC13311c | √ | √ | √ | √ | √ |
| S. typhimurium | OCC11 | √ | √ | √ | √ | √ |
| S. typhimurium | OCC17 | √ | √ | √ | √ | √ |
| S. typhimurium | OCC22 | √ | √ | √ | √ | √ |
| S. typhimurium | OCC152 | √ | × | √ | √ | √ |
| S. typhimurium | OCC722 | √ | √ | √ | √ | √ |
| S. typhimurium | OCC853 | √ | √ | √ | √ | √ |
| S. uphill | OCC635 | √ | √ | √ | √ | √ |
| S. virchow | OCC703 | √ | √ | √ | √ | √ |
| S. worthington | OCC634 | √ | √ | √ | √ | √ |
| Enterobacteriaceae | ||||||
| Citrobacter freundii | OCC261 | × | × | × | √ | √ |
| C. freundii | OCC370 | × | × | × | × | × |
| Enterobacter cloacae | OCC118 | × | × | × | × | × |
| E. cloacae | OCC720 | × | × | × | × | × |
| E. cloacae | CMCC1887 | √ | × | × | × | × |
| Escherichia coli | OCC122 | × | × | × | × | × |
| E. coli | OCC199 | × | × | × | × | × |
| E. coli | OCC402 | × | × | × | × | × |
| E. coli | KCLB35 | × | × | × | × | × |
| E. coli | KCLDH5α | × | √ | √ | × | × |
| E. coli | KCLK12 | × | × | × | × | × |
| Klebsiella oxytoca | OCC323 | × | × | × | × | × |
| Klebsiella pneu-moniae | OCC108 | × | × | × | × | × |
| K. pneumoniae | OCC411 | × | × | × | × | × |
| K. pneumoniae | OCC758 | √ | × | × | × | × |
| K. pneumoniae | KCLK10 | × | × | × | × | × |
| Proteus mirabilis | OCC110 | × | × | × | × | × |
| P. mirabilis | OCC715 | × | √ | × | × | √ |
| P. mirabilis | KCLP3/S0 | × | × | × | × | √ |
| P. mirabilis | KCLP3/S5 | × | × | × | √ | √ |
| P. mirabilis | KCLP0/S1:5 | × | × | × | √ | √ |
| Proteus sp. | OCC311 | × | × | × | √ | √ |
| Providencia sp. | OCC297 | × | × | × | × | × |
| Serratia marcescens | OCC217 | × | × | √ | × | × |
| Shigella sonnei | OCC625 | × | × | × | × | × |
| Pseudomonads | ||||||
| Pseudomonas aeruginosa | OCC201 | × | × | × | × | × |
| P. aeruginosa | OCC467 | × | × | × | × | × |
| P. aeruginosa | OCC484 | × | √ | × | × | × |
| P. aeruginosa | OCC487 | × | × | × | NG | × |
| P. aeruginosa | OCC510 | √ | × | × | × | × |
| P. aeruginosa | KCL10466 | × | × | × | × | × |
| P. fluorescens | NCTC10038 | NG | NG | NG | NG | NG |
| P. fluorescens | OCC292 | NG | NG | NG | NG | NG |
| P. putida | OCC221 | × | √ | × | NG | × |
| Gram-positive bacteria | ||||||
| Enterococcus faecalis | OCC501 | NG | NG | × | NG | × |
| E. faecalis | OCC640 | × | NG | × | NG | × |
| E. faecalis | KCLC01 | × | NG | × | NG | × |
| Staphylococcus aureus | OCC106 | NG | NG | NG | NG | NG |
| S. aureus | OCC198 | × | NG | NG | NG | NG |
| S. epidermidis | OCC691 | × | NG | NG | NG | NG |
| Listeria monocyto-genes | NCTC7973 | NG | NG | NG | NG | NG |
| L. monocytogenes | NCTC7973d | NG | NG | NG | NG | NG |
| L. monocytogenes | NCTC10888 | NG | NG | NG | NG | NG |
| L. monocytogenes | NCTC10890 | NG | NG | NG | NG | NG |
| L. monocytogenes | KCL9493de | NG | NG | NG | NG | NG |
| L. monocytogenes | KCL19118f | NG | NG | NG | NG | NG |
| L. monocytogenes | NCTC10887 | NG | NG | NG | NG | NG |
| L. monocytogenes | KCL9862g | NG | NG | NG | NG | NG |
| L. welshimeri | KCLPHd | NG | NG | NG | NG | NG |
Expected Salmonella color reactions: CSE agar, burgundy; Rambach agar, pink; SM ID agar, pink; XLD agar, black; Hektoen-enteric agar, black. Symbols: √, expected color; ×, other colors; NG, no growth.
Originally obtained from the American Type Culture Collection as ATCC 1360.
Originally obtained as ATCC 13311.
Nonhemolytic Listeria strain; all other strains are hemolytic.
Originally obtained as ATCC 9493.
Originally obtained as ATCC 19118.
Originally obtained as ATCC 9862.
Chromogenic esterase substrates.
The chromogenic substrates used were closely related to the phenols described by Aamlid et al. (1). The butanoate (C4), heptanoate (C7), nonoate (C9), and decanoate (C10) esters of SLPA were synthesized in addition to the octanoate (C8) esters 5-[(4-octanoyloxy)-3,5-dimeth-oxyphenylmethylene]-2-thioxothiazolidin-4-one-3-acetate (SRA)-octanoate, 4-[2-(4-octanoyloxyphenyl)-vinyl]-quinolinium-1-(propan-3-yl carboxylic acid) bromide (PRA)-octanoate, 4-[2-(4-octanoyloxy-3-bromo-5-methoxyphenyl)-vinyl]-quinolium-1-(propan-3-yl carboxylic acid) bromide (BVRAh)-octanoate, 5-[(4-octanoyloxy)-3,5-dimethoxyphenylmethylene]-2--thioxothiozolidin-3-meth-yl-4-one (SRM)-octanoate, and 4-[2-(4-octanoyloxy-3-methoxyphenyl)-vinyl]-quinolinium-1-(propan-3-yl carboxylic acid) bromide (VLPA)-octanoate, and SLPA-octanoate (chloride and bromide forms) (Fig. 1). In water, the substrates generally had low solubility (<2.0 mM). SLPA-octanoate was soluble up to 1.0 mM at room temperature.
FIG. 1.
Structures of the chromogenic C8 esters used in this study. In panel A, SLPA-octanoate, the ion X− is either Cl− or Br−. Other panels: B, SRA-octanoate; C, PRA-octanoate; D, BVRA-octanoate; E, SRM-octanoate; F, VLPA-octanoate.
Chromogenic media.
The media consisted of a basal component to which were added the esterase substrate, a UV-absorbing compound (ethyl 4-dimethylaminobenzoate), and novobiocin. The basal medium contained the following (grams per liter): peptone, 4.0; Lab-Lemco powder, 3.0; tryptone, 4.0; lactose, 14.65; l-cystine, 0.128; trisodium citrate dihydrate, 0.5; Tris base, 0.06; Tween 20, 3.0; Roko agar (Industrias Roko S.A., La Coruna, Spain), 12.0. All components were obtained from Oxoid Ltd., Basingstoke, United Kingdom, except trisodium citrate, Tween 20, and ethyl 4-dimethylaminobenzoate, which were obtained from Sigma-Aldrich Company Ltd., Poole, Dorset, United Kingdom. The basal medium was autoclaved (121°C, 15 min) and cooled to about 55°C. The chromogenic substrate (SLPA-octanoate [bromide form] at 0.3223 g liter−1 in the final medium formulation), ethyl 4-dimethylaminobenzoate (0.035%, wt/vol) dissolved in 8 ml of methanol per liter of medium, and novobiocin (70 mg liter−1; Sigma-Aldrich Company Ltd.) were then added. Alternatively, the chromogenic substrate and UV-absorbing compound could be added directly to the basal medium if this was heated only to the boiling point. The complete agar medium was poured into 90-mm-diameter petri dishes. After setting, plates were surface dried for 20 min at 37°C and used immediately or stored in the dark at room temperature for up to 2 weeks. The pH of agar plates was 7.0 ± 0.2, as measured by using a flat pH electrode (Gelplas combination electrode; Merck Ltd., Poole, Dorset, United Kingdom).
During development of the medium, the effects of varying the concentrations of Tris base (0.06 to 0.75 g liter−1), lactose (0.25 to 15.0 g liter−1), ethyl 4-dimethylaminobenzoate (0 to 1.0%, wt/vol), and detergents including Tween 20 (0.5 to 8.0 g liter−1) in the medium were determined. The use of brilliant green (Sigma-Aldrich Company Ltd.) as an inhibitor of Proteus and Shigella species was also investigated, and several substrates were tested as alternatives to lactose. These included d-adonitol, α-amygdalin, d-arabitol, d-arabinose, glucose, maltose, d-mannose, melibiose, sodium pyruvate, sodium malonate, sucrose, l-rhamnose, raffinose, trehalose, and xylose.
Inoculation of media.
Test bacteria were streaked onto chromogenic media after growth for 24 h at 37°C on nutrient agar (CM3; Oxoid) plates, for 4 to 6 h at 37°C in buffered peptone water (nonselective, pre-enrichment broth; CM509; Oxoid), or for 4 to 6 h at 42°C in Rappaport Vassiliadis broth (selective enrichment broth; CM669; Oxoid). To obtain plates showing well-isolated colonies, broth cultures were also serially diluted in Ringer’s solution (BR52; Oxoid) or saline (0.85%), and 100-μl volumes of appropriate dilutions were spread onto plates. Plates were incubated at 37 or 42°C and observed for colony coloration for up to 48 h.
Comparison of CSE agar with commercially available media.
One hundred seven test strains (58 salmonellae and 49 nonsalmonellae) were used. The media evaluated were (i) chromogenic Salmonella esterase (CSE) agar (developed in this study and containing the bromide form of SLPA-octanoate), which was prepared by adding novobiocin to previously boiled medium containing all of the other components (see above); (ii) XLD agar (CM469; Oxoid); (iii) Hektoen-enteric agar (CM419; Oxoid); (iv) Rambach agar (Merck, Darmstadt, Germany); and (v) SM ID agar (BioMerieux S.A., Montalieu-Vercieu, France). Agars ii to iv were prepared in accordance with the manufacturer’s instructions, and SM ID agar was obtained as prepoured plates. Test strains were grown overnight at 37°C on nutrient agar (Oxoid) or brain heart infusion agar (CM375B; Oxoid) (Listeria spp. only). They were then streaked, in duplicate, onto plates of the test media and inoculated for 24 to 48 h at 37 or 42°C (CSE agar only).
RESULTS
Effect of organic acid chain length on esterase activity toward chromogenic substrates.
Bacteria inoculated onto plates prepared with esters with differing chain lengths exhibited dramatic differences in colony coloration. SLPA esters with differing chain lengths were autoclaved added to nutrient agar (plus Tween 20 at 2.0 g liter−1) as supplements in organic solvents. The final concentrations were 0.2 and 0.7 mM. Plates were inoculated with a test panel of the representative salmonellae Salmonella enteritidis OCC723, S. typhimurium OCC722 and OCC626, S. virchow OCC703, S. dublin OCC627, S. worthington OCC634 and the nonsalmonella organisms Klebsiella pneumoniae OCC411, Enterobacter cloacae OCC118 and OCC720, C. freundii OCC370, Shigella sonnei OCC625, E. coli OCC122 and OCC402, Proteus mirabilis OCC715 and OCC110, Staphylococcus aureus OCC198, and Serratia marcescens OCC217 and incubated at 37 and 42°C to test for colony coloration. All of the salmonellae and nonsalmonellae inoculated onto media containing butanoate (C4) produced blue or purple colonies after 24 h of incubation. Similarly, all of the bacteria tested on the heptanoate (C7) media produced red or purple colonies after 24 h and more strongly colored colonies after 48 h incubation. It was only at the C8 level that Salmonella spp. could be differentiated from nonsalmonellae by the production of burgundy-colored colonies. Nonsalmonellae appeared as white or colorless colonies. All of the bacteria inoculated onto media containing the nonoate (C9) and decanoate (C10) esters appeared white or gray after 24 and 48 h of incubation. The results obtained in these experiments suggested that it might be possible to develop a differential medium in which salmonellae were indicated by C8 esterase activity.
Comparison of C8 organic acid esters as substrates for salmonellae.
The C8 esters compared were PRA-octanoate, BVRA-octanoate, SRM-octanoate, VLPA-octanoate, SLPA-octanoate (chloride and bromide forms), and SRA-octanoate (Fig. 1). All of the compounds decomposed upon autoclaving in the base medium and were added as a supplement dissolved in dimethyl formamide or methanol. The final concentrations were 0.2 and 0.7 mM.
All strains in the test panel inoculated onto media containing the BVRA-, PRA-, and SRM-octanoate substrates failed to give a color after 48 h of incubation. When bacteria were inoculated onto agar plates containing VLPA-octanoate, a diffuse blue color appeared in the agar. Colonies of Salmonella spp. on media incorporating SRA-octanoate were a diffuse pink color, and the background agar was a variable pink or yellow. The most effective substrates for the detection of Salmonella spp. were the chloride and bromide forms of SLPA-octanoate in which colonies of Salmonella spp. were burgundy colored on a yellow background. The bromide form of SLPA-octanoate was used in further tests, as it was readily prepared in a good yield and a pure state.
Optimization of SLPA-octanoate medium.
Additional experiments showed that while the SLPA-octanoate acid esters were readily hydrolyzed by the test panel of representative salmonellae, the reaction of the rare serotype S. arizonae was weak. Also, K. pneumoniae OCC411, S. sonnei OCC625, P. mirabilis OCC715 and OCC110, and E. cloacae OCC118 possessed some esterase activity, leading to possible confusion with Salmonella. Therefore, a series of experiments was undertaken to optimize the medium by maximizing Salmonella color production and reducing the color of the false-positive non-Salmonella species.
(i) Substrate concentration.
Increasing the concentration of SLPA-octanoate (bromide form, 0.05 to 1.0 mM) increased the burgundy coloration intensity of Salmonella colonies. Maximal coloration was achieved at ≥0.50 mM, and 0.55 mM was chosen as the most suitable concentration.
(ii) Lactose concentration.
Increasing the lactose concentration (tested at 0.25 to 20 g liter−1) tended to lessen the burgundy color of false-positive organisms and with lactose at 14.65 g liter−1, no color was observed in colonies of K. pneumoniae OCC411 and E. cloacae OCC118. However, colonies of Salmonella spp. (except S. arizonae OCC1200) remained colored. It is likely that lactose-fermenting organisms lower the agar pH at the center of colonies, thus diminishing the intensity of chromophore color. Burgundy coloration in S. sonnei OCC625 and P. mirabilis OCC715 and OCC110 was not reduced by the addition of lactose.
(iii) Addition of citrate.
Citrate was added to the medium, as it is oxidized by salmonellae but not by Shigella spp. Increasing the trisodium citrate concentration (0.25 to 10.0 g liter−1) increased the intensity of the burgundy color of Salmonella spp. and most of the false-positive bacteria. However, citrate (≥0.25 g liter−1) completely inhibited burgundy color formation in P. mirabilis OCC715 and OCC110 and S. sonnei OCC625. Optimal differentiation of salmonellae from other organisms was achieved by using citrate at 0.5 g liter−1 in the presence of lactose at 14.65 g liter−1.
(iv) Addition of detergent.
Detergents are frequently added to Salmonella media to increase selectivity. It was also considered possible that detergents might aid the passage of chromogenic esters across the lipid outer membrane of the cell wall so enhancing color development. In the basal medium plus SLPA-octanoate, Tween 20 (optimum concentration, 3.0 g liter−1) increased the burgundy color of Salmonella colonies and the transparency of the medium. Triton X-100, Niaproof 8, Tyloxopol, and Tween 80 increased colony color to a lesser extent.
(v) Photochemical degradation of substrate.
Uninoculated plates were yellow; however, following storage at room temperature for 1 to 7 days in the light, the color became paler, and as a result, poorly colored Salmonella colonies were observed. Protection of the substrate against photochemical degradation in the agar medium was achieved by using the UV-absorbing compound ethyl 4-dimethylaminobenzoate. At the concentration used (0.035%, wt/vol), the viable count of test Salmonella strains in the chromogenic medium was not reduced.
(vi) Novobiocin and brilliant green.
The use of novobiocin to reduce the growth of nonsalmonella strains in the chromogenic medium was investigated by using the test panel of bacteria listed above and additional strains, including Pseudomonas fluorescens NCTC10038 and OCC292; P. aeruginosa OCC487, OCC484, OCC201, OCC467, and OCC510; P. putida OCC221; Staphylococcus epidermidis OCC106; S. sonnei OCC625; and P. mirabilis OCC715. Several pseudomonad strains were tested, as these organisms are frequently confused with salmonellae on commercial Salmonella agars. Complete growth inhibition of P. fluorescens strains and S. epidermidis was observed with novobiocin at 10 mg liter−1. The growth of the remaining pseudomonads, S. sonnei, and P. mirabilis was partially inhibited at 70 mg liter−1. However, higher concentrations could not be used as salmonella growth also became affected. Brilliant green (3 to 9 mg liter−1) partially inhibited the growth of Proteus spp. and S. sonnei OCC625. However, its color reduced the ease with which the chromogenic reaction could be determined, and it was not included in the final medium formulation.
(vii) Incubation time and temperature.
Salmonella species are grown routinely at 37 and 42°C. At the higher temperature, the time required for coloration of most of the test serotypes of salmonellae in the chromogenic medium was reduced from 24 to 18 h, and the intensity of coloration at 24 h increased compared to that observed at 37°C. Additional advantages of the higher incubation temperature were inhibition of the growth of P. fluorescens and of swarming by Proteus spp. Prolonging the incubation time at 42°C to 48 h also increased the coloration of S. arizonae colonies but not that of other salmonellae (S. sofia, S. choleraesuis, and S. indiana), giving poor or no color.
(viii) Replacement sugars for lactose.
A variety of compounds were used as potential replacements for lactose in an attempt to increase the colony coloration of S. arizonae OCC1200 and NCTC8297, S. choleraesuis OCC852 and NCTC5735, and S. indiana OCC597 and NCTC11304. In the absence of lactose, colonies of S. indiana strains became burgundy colored. However, this also caused coloration of E. cloacae OCC118 and K. pneumoniae OCC411 colonies. When salicin and myo-inositol (7.0 to 9.0 g liter−1) were included in the medium, in the absence of lactose, S. indiana colonies remained intensely colored and color development in K. pneumoiae colonies was prevented. However, E. cloacae colonies were still colored. Increasing the concentration of salicin or myo-inositol reduced the burgundy color of all test Salmonella strains. The color of S. arizonae and S. choleraesuis strains was not improved by exclusion of lactose from the medium or addition of replacement compounds (see Materials and Methods).
(ix) Optimized agar.
The optimized chromogenic agar was designated CSE agar and included SLPA-octanoate (bromide form; 0.3223 g liter−1), lactose (14.65 g liter−1), trisodium citrate dihydrate (0.5 g liter−1), Tween 20 (3.0 g liter−1), ethyl 4-dimethylaminobenzoate (0.035%, wt/vol), and novobiocin (70 mg liter−1).
Evaluation of CSE agar.
A total of 107 salmonella and nonsalmonella strains were streak plated onto CSE agar and observed for coloration for up to 48 h at 42°C. Burgundy-colored colonies of Salmonella spp. were typically observed (Table 1; Fig. 2) on the chromogenic Salmonella medium after 18 to 24 h of incubation at 42°C. On densely inoculated streak plates, the surrounding agar tended to change from yellow to green during incubation, but this did not interfere with the coloration of colonies. S. arizonae appeared weakly colored, even after 48 h of incubation. The surrounding medium was, however, very green after incubation. The rarely isolated serotypes S. indiana and S. choleraesuis were noncolored.
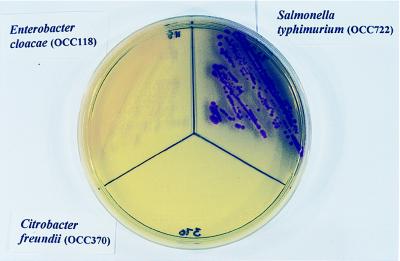
FIG. 2.
CSE agar showing burgundy-colored colonies of S. typhimurium OCC722 and white colonies of K. pneumoniae OCC411 after 24 h of incubation at 42°C. Colonies of C. freundii OCC370 are transparent and difficult to see.
Colonies of non-Salmonella spp. typically appeared cream, transparent, white, or yellow with a yellow background. There were two false-positive strains, E. cloacae OCC1887 and K. pneumoniae OCC758, which both gave a moderate burgundy color. Growth of Proteus spp., S. sonnei, and the pseudomonad strains was generally poor, and colonies appeared colorless or white. The growth of two of the nine Pseudomonas strains (OCC292 and NCTC10038) was completely inhibited. Notably, colonies of strains of C. freundii, a common false-positive organism on H2S-detecting Salmonella agars, were not colored on CSE agar.
Comparison of CSE agar with four commercially available media.
The sensitivity and specificity of CSE agar were compared with those of Rambach, XLD, Hektoen-enteric, and SM ID agars. Reactions of 58 salmonellae and 49 nonsalmonellae on the test agars are given in Table 1, and the data are summarized in Table 2. The sensitivity of the CSE agar was 93.1% and compared favorably with those of Rambach (82.8%), SM ID (91.4%), XLD (91.4%), and Hektoen-enteric (89.7%) agars. S. diarizonae NCTC12417 was detectable on CSE agar but not on the two commercial chromogenic agars SM ID agar and Rambach agar (which gave blue colonies, indicating the presence of β-galactosidase). In addition, both strains of S. pullorum were readily detectable on the CSE medium but produced false-negative reactions on Rambach, XLD (strain OCC09 only), and Hektoen-enteric agars. None of the media tested detected the two strains of S. indiana. SM ID agar was the only one to detect S. choleraesuis OCC852 within 24 h. One strain (OCC152) of the common serotype S. typhimurium was not detectable on Rambach agar.
TABLE 2.
Comparison of the sensitivities and selectivities of CSE, Rambach, SM ID, XLD, and Hektoen-enteric agars for Salmonella detection
| Parameter | Resulta obtained with:
|
||||
|---|---|---|---|---|---|
| CSE agar | Rambach agar | SM ID agar | XLD agar | Hektoen-enteric agar | |
| No. of Salmonella strains showing positive color reactions (including weak reactions) | 54/58 | 48/58 | 53/58 | 53/58 | 52/58 |
| Sensitivity (%) | 93.1 | 82.8 | 91.4 | 91.4 | 89.7 |
| No. of non-Salmonella strains showing negative color reactions | 46/49 | 45/49 | 47/49 | 45/49 | 43/49 |
| Specificity (%) | 93.9 | 91.8 | 95.9 | 91.8 | 87.8 |
The data were derived from Table 1. Sensitivity is the number of Salmonella strains giving the expected color reaction divided by the total number of Salmonella strains tested. Specificity is the number of non-Salmonella strains giving a negative color reaction or failing to grow divided by the total number of non-Salmonella strains tested.
The specificity of CSE agar (93.9% [Table 2]) was higher than those of Rambach (91.8%), XLD (91.8%), and Hektoen-enteric (87.8%) agars and only slightly lower than that of SM ID agar (95.9%). E. cloacae OCC1887 and K. pneumoniae OCC758 were the false-positive strains on CSE agar. P. aeruginosa OCC510 produced a weak brown color in the center of colonies and could possibly be mistaken for salmonella by the untrained eye; thus, it was also counted as a false positive. The strains giving false-positive color reactions on the other test media included H2S-producing Proteus sp. strains OCC311, OCC715, KCLP3/S0, KCLP3/S5, and KCLP0/S1:5 and C. freundii OCC261. On Rambach and SM ID agars, false-positive pink colonies were observed with Pseudomonas sp. strains OCC484 and OCC221, E. coli KCLDHα5, and S. marcescens OCC217.
DISCUSSION
In standard protocols, two diagnostic media are usually specified for the presumptive identification of salmonellae from food and clinical specimens (3). CSE agar is based on the detection of C8-esterase activity in salmonellae and appears to be a suitable complementary medium for those based on other biochemical activities. The medium has a high degree of sensitivity (low number of false negatives) and specificity (low number of false positives) compared with other Salmonella agars. CSE medium also appears to have higher specificity (93.9%) than the MUCAP spot test (80%; 7). The medium is particularly effective for the detection of the more common Salmonella serotypes (S. typhimurium and S. enteriditis, which together account for 85.03% of the serotypes reported to the Public Health Laboratory Service [PHLS] in 1997 [14]).
During the development of the medium, it became apparent that burgundy coloration of colonies was dependent not only upon esterase activity but also on the presence of sugars and other energy substrates in the medium. In organisms possessing C8-esterase activity, color intensity will be modified by pH. The chromophore of SLPA-octanoate becomes protonated at lower pH values, and the protonated form is only weakly colored (16). Organisms producing acid products from sugar fermentation lower the medium pH, particularly at the center of colonies. Thus, including lactose in the medium prevented color formation by some lactose-fermenting Klebsiella and Enterobacter strains. Unfortunately, it also blocked color formation by S. indiana (lactose fermenting), which was shown to be esterase positive in CSE agar without lactose. Citrate promoted color formation by salmonellae, presumably acting as an energy source which, by being oxidized rather that fermented, would not tend to reduce the pH of the medium.
In addition to S. indiana, S. choleraesuis was not detected on CSE agar, and S. arizonae OCC1200 and OCC8297 were weakly colored. The detection of these strains also presents difficulties when other Salmonella agars are used. S. indiana and S. arizonae are lactose-fermenting organisms and on chromogenic media such as Rambach (8) and SM ID agars, they give a false color reaction (violet or blue). XLD and Hektoen-enteric agars can usually identify S. arizonae but are unable to detect other lactose-fermenting serotypes, including some strains of S. montevideo and S. virchow (18). However, it should be noted that these atypical strains represent only a small proportion of those isolated. For example, in 1997, only 0.11% of the Salmonella isolates recorded by the PHLS were S. arizonae. The corresponding proportions for S. choleraesuis and S. indiana were 0.003 and 0.15%, respectively. S. sofia, which gave only a weak positive reaction on CSE and XLD agars, is also a rarely isolated serotype (0.006%, of the isolates recorded by the PHLS in 1997).
ACKNOWLEDGMENTS
This work was partly supported by Oxoid Ltd., Basingstoke, United Kingdom, and a SMART development project award from the Department of Trade and Industry, United Kingdom.
We thank the late Clive Bird for advice on UV-absorbing compounds and Oxoid Ltd. for the kind gift of bacterial strains and agar media.
REFERENCES
- 1.Aamlid K H, Lee G, Price R G, Richardson A C, Smith B V, Taylor S A. Development of improved chromogenic substrates for the detection and assay of hydrolytic enzymes. Chem. Ind. (London) 1989. pp. 106–108. [Google Scholar]
- 2.Aguirre P M, Cacho J B, Folgueira L, Lopez M, Garcia J, Velasco A C. Rapid fluorescence method for screening Salmonella spp. from enteric differential agars. J Clin Microbiol. 1990;28:148–149. doi: 10.1128/jcm.28.1.148-149.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. The Oxoid manual. Basingstoke, Hampshire, United Kingdom: Oxoid Ltd.; 1990. [Google Scholar]
- 4.Devenish J A, Ciebin B W, Brodsky M H. Novobiocin-brilliant green-glucose agar: new medium for isolation of salmonellae. Appl Environ Microbiol. 1986;52:539–545. doi: 10.1128/aem.52.3.539-545.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Entis P. Improved hydrophobic grid membrane filter method, using EF-18 agar, for detection of Salmonella in foods: collaborative study. J Assoc Off Anal Chem. 1990;73:734–742. [PubMed] [Google Scholar]
- 6.Freydiere A M, Gille Y. Detection of salmonellae by using Rambach agar and by a C8 esterase spot test. J Clin Microbiol. 1991;29:2357–2359. doi: 10.1128/jcm.29.10.2357-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruenwald R, Henderson R W, Yappow S. Use of Rambach propylene glycol agar for identification of Salmonella spp. J Clin Microbiol. 1991;29:2354–2356. doi: 10.1128/jcm.29.10.2354-2356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn H, Barbel W, Rabsch W, Reissbrodt R. Evaluation of Rambach agar for the detection of Salmonella subspecies I to VI. Appl Environ Microbiol. 1994;60:749–751. doi: 10.1128/aem.60.2.749-751.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manafi M, Sommer R. Comparison of three rapid screening methods for Salmonella spp.: MUCAP Test, MicroScreen Latex and Rambach Agar. Lett Appl Microbiol. 1992;14:163–166. [Google Scholar]
- 10.Miles R J, Siu E L T, Carrington C, Richardson A C, Smith B V, Price R G. The detection of lipase activity in bacteria using novel enzyme substrates. FEMS Microbiol Lett. 1992;90:283–288. doi: 10.1016/0378-1097(92)90661-7. [DOI] [PubMed] [Google Scholar]
- 11.Pancholy S K, Lynd J Q. Microbial esterase detection with ultraviolet fluorescence. Appl Microbiol. 1971;22:939–941. doi: 10.1128/am.22.5.939-941.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poisson D M. Novobiocin, brilliant green, glycerol, lactose agar: a new medium for the isolation of Salmonella strains. Res Microbiol. 1992;143:211–216. doi: 10.1016/0923-2508(92)90010-l. [DOI] [PubMed] [Google Scholar]
- 13.Poupart M C, Mounier M, Denis F, Sirot J, Couturier C, Villeval F. Abstracts of the Fifth European Congress of Clinical Microbiology and Infectious Diseases. 1991. A new chromogenic ready-to-use medium for Salmonella detection, abstr. 1254. [Google Scholar]
- 14.Public Health Laboratory Service. Salmonella serotypes recorded in the PHLS Salmonella data set: January to December. 1997. p. 103. , 216, 338, and 444. Public Health Laboratory Service, London, England. [Google Scholar]
- 15.Rambach A. New plate medium for facilitated differentiation of Salmonella spp. from Proteus spp. and other enteric bacteria. Appl Environ Microbiol. 1990;56:301–303. doi: 10.1128/aem.56.1.301-303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson A C, Cooke V M. United Kingdom patent application 9802817.8. 1998. [Google Scholar]
- 17.Richardson A C, Smith B V, Price R G, Praill P F G. U.S. patent, 5,221,606. 1993. [Google Scholar]
- 18.Ruiz J, Lunez M, Diaz J, Lorente I, Perez J, Gomez J. Comparison of five plating media for isolation of Salmonella species from human stools. J Clin Microbiol. 1996;34:686–688. doi: 10.1128/jcm.34.3.686-688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]