Abstract
Background
Post-acute sequelae of SARS-CoV-2 infection, known as long COVID, have severely affected recovery from the COVID-19 pandemic for patients and society alike. Long COVID is characterised by evolving, heterogeneous symptoms, making it challenging to derive an unambiguous definition. Studies of electronic health records are a crucial element of the US National Institutes of Health's RECOVER Initiative, which is addressing the urgent need to understand long COVID, identify treatments, and accurately identify who has it—the latter is the aim of this study.
Methods
Using the National COVID Cohort Collaborative's (N3C) electronic health record repository, we developed XGBoost machine learning models to identify potential patients with long COVID. We defined our base population (n=1 793 604) as any non-deceased adult patient (age ≥18 years) with either an International Classification of Diseases-10-Clinical Modification COVID-19 diagnosis code (U07.1) from an inpatient or emergency visit, or a positive SARS-CoV-2 PCR or antigen test, and for whom at least 90 days have passed since COVID-19 index date. We examined demographics, health-care utilisation, diagnoses, and medications for 97 995 adults with COVID-19. We used data on these features and 597 patients from a long COVID clinic to train three machine learning models to identify potential long COVID among all patients with COVID-19, patients hospitalised with COVID-19, and patients who had COVID-19 but were not hospitalised. Feature importance was determined via Shapley values. We further validated the models on data from a fourth site.
Findings
Our models identified, with high accuracy, patients who potentially have long COVID, achieving areas under the receiver operator characteristic curve of 0·92 (all patients), 0·90 (hospitalised), and 0·85 (non-hospitalised). Important features, as defined by Shapley values, include rate of health-care utilisation, patient age, dyspnoea, and other diagnosis and medication information available within the electronic health record.
Interpretation
Patients identified by our models as potentially having long COVID can be interpreted as patients warranting care at a specialty clinic for long COVID, which is an essential proxy for long COVID diagnosis as its definition continues to evolve. We also achieve the urgent goal of identifying potential long COVID in patients for clinical trials. As more data sources are identified, our models can be retrained and tuned based on the needs of individual studies.
Funding
US National Institutes of Health and National Center for Advancing Translational Sciences through the RECOVER Initiative.
Introduction
Acute COVID-19 affects multiple organ systems, including the lungs, digestive tract, kidneys, heart, and brain.1, 2 The long-term clinical consequences of COVID-19 are still poorly understood and are collectively termed post-acute sequelae of SARS-CoV-2 infection, known as long COVID.3 At this time, this disease is referred to by a number of terms that may or may not represent the same constellation of signs and symptoms; here, we consider post-acute sequelae of SARS-CoV-2 infection synonymous with long COVID. Long COVID can be broadly defined as persistent or new symptoms more than 4 weeks after severe, mild, or asymptomatic SARS-CoV-2 infection.4, 5 Characterising, diagnosing, treating, and caring for patients with long COVID has been challenging due to heterogeneous signs and symptoms that evolve over long trajectories.6 The effect of long COVID on patients' quality of life and ability to work can be profound.
The wide range of symptoms attributed to long COVID was highlighted in an extensive patient-led survey,7 which conducted deep longitudinal characterisation of long COVID symptoms and trajectories in patients with suspected and confirmed COVID-19 who reported illness lasting more than 28 days.8 Evaluation and harmonisation of patient-reported and clinically reported long COVID features using the Human Phenotype Ontology also revealed heterogeneous signs and symptoms, supporting the hypothesis that a complex collection of patient-reported and clinically reported features is necessary to correctly classify and manage patients with long COVID.9 WHO recently published its own case definition of post COVID-19 condition (WHO's term) that includes 12 criteria, which similarly require a wide variety of patient-declared and clinical information.10
Research in context.
Evidence before this study
Initial characterisation of patients with long COVID has contributed to an emerging clinical understanding, but the substantial heterogeneity of disease features makes diagnosing and treating this new disease challenging. This challenge is urgent to address, as many patients report that long COVID symptoms are debilitating and severely affecting their ability to engage in activities of daily life. No formal literature review was done. Few studies have used large-scale databases to understand concordance of clinical patterns and generate data-driven definitions of long COVID. The US National Institutes of Health's RECOVER programme has invested in electronic health record studies to understand the risk factors for, and mechanisms behind, long COVID, accurately identify individuals with long COVID, and prevent and treat long COVID.
Added value of this study
The National COVID Cohort Collaborative (N3C) harmonises patient-level electronic health record data from over 8 million demographically diverse and geographically distributed patients. Here, we describe highly accurate XGBoost machine learning models that use N3C to identify patients with potential long COVID, trained using electronic health record data from patients who attended a long COVID specialty clinic at least once. The most powerful predictors in these models are outpatient clinic utilisation after acute COVID-19, patient age, dyspnoea, and other diagnosis and medication features that are readily available in the electronic health record. The model is transparent and reproducible, and can be widely deployed in individual health-care systems to enable local research recruitment or secondary data analysis.
Implications of all the available evidence
N3C's longitudinal data for patients with COVID-19 provides a comprehensive foundation for the development of machine learning models to identify patients with potential long COVID. Such models enable efficient study recruitment that, in turn, deepen our understanding of long COVID and offer opportunities for hypothesis generation. Moreover, as more patients are diagnosed with long COVID and more data are available, our models can be refined and retrained to evolve the algorithm as more evidence emerges.
To gain an understanding of the complexities of long COVID, it will be necessary to recruit a large and diverse cohort of research participants. The US National Institutes of Health (NIH)'s RECOVER initiative11 aims to recruit thousands of participants in the USA to answer critical research questions about long COVID, such as understanding pregnancy risk factors, cognitive impairment and mental health, and outcome disparities and comorbidities. Efficient recruitment of cohorts of this size and scope often entails leveraging computable phenotypes12, 13, 14 (ie, electronic cohort definitions) to find sufficient numbers of patients meeting a study's inclusion criteria. Poor cohort definition can result in poor study outcomes.15, 16 For long COVID, as with other novel conditions, the absence of an unambiguous consensus definition and the heterogeneity of the condition's presentation poses a substantial challenge to cohort identification. Machine learning can help to address this challenge by using the rich longitudinal data available in electronic health records to algorithmically identify patients similar to those in a long COVID gold standard.
The National COVID Cohort Collaborative (N3C)17 offers a data-driven solution to quantifying the features of long COVID and an appropriate hypothesis-testing scenario for a machine learning approach.18 N3C is an NIH National Center for Advancing Translational Sciences (NCATS)-sponsored data and analytic environment which compiles and harmonises longitudinal electronic health record data from 65 sites in the USA and over 8 million patients who have tested positive for SARS-CoV-2 infection; have symptoms that are consistent with a COVID-19 diagnosis; or are demographically matched controls who have tested negative for SARS-CoV-2 infection (and have never tested positive) to support comparative studies.19 We aimed to build a foundation for a robust clinical definition of long COVID by linking curated lists of patients who have attended a long COVID clinic from three N3C sites with data in the N3C repository. We used the linked dataset to train and test three machine learning models and applied those models to define a nationwide US cohort of potential patients with long COVID, and to derive a list of prominent clinical features shared among that cohort to help to identify patients for research studies and target features for further investigation.
Methods
Study design and base population
To model long COVID, we used electronic health record data integrated and harmonised in Palantir Foundry inside the secure N3C Data Enclave to identify unique health-care utilisation patterns and clinical features among patients with COVID-19.
We defined our base population (n=1 793 604) as any non-deceased adult patient (age ≥18 years) with either an International Classification of Diseases-10-Clinical Modification COVID-19 diagnosis code (U07.1) from an inpatient or emergency health-care visit, or a positive SARS-CoV-2 PCR or antigen test, and for whom at least 90 days have passed since COVID-19 index date. COVID-19 index date was defined as the earliest date of positive indicator for a patient. For patients with multiple positive tests, we selected the date of the first positive test as the index. Before this analysis, patients from six N3C sites were removed from the cohort due to their sites' use of randomly shifted dates of service, which would have restricted our ability to use temporal logic during analysis.
Because the definition, clinical guidelines, and documentation practices for long COVID are still evolving, there is no gold standard to validate computable phenotypes and to train machine learning models. However, three N3C sites provided lists of locally identified patients who had visited that site's long COVID specialty clinic at least once. These patients represent a silver standard within our base population (n=597, once our base population criteria were applied). Hereafter, we will refer to this group of patients as long COVID clinic patients; patients identified by our trained model as patients with long COVID will be referred to as patients with potential long COVID. This silver standard enabled us to develop a model to identify patients warranting care at a long COVID clinic—a valuable proxy for long COVID until a true gold standard is available.
To train and test the machine learning models, we created a subset of our overall cohort containing only patients originating from the three N3C sites with lists of patients who had visited a long COVID clinic (n=97 995), including the long COVID clinic patients. This subset was stratified further into patients who had been hospitalised with acute COVID-19 (n=19 368) and patients who were not hospitalised (n=78 627). We narrowed the subset further to include only patients who had at least (1) one health-care visit of any type and at least one diagnosis or (2) one medication in their post-COVID-19 window (n=15 621 hospitalised, n=58 351 not hospitalised). The full cohort selection and subsetting process is shown in the appendix (p 2), along with training and test set patient counts (p 6).
Feature selection
Within the three-site subset, we examined demographics, health-care visit details, medical conditions, and prescription drug orders for each patient before and after their period of acute COVID-19. Although its use was considered, laboratory result data proved too sparse among the cohort for use in the models, especially for non-hospitalised patients. Features were selected for inclusion in the model by gathering datapoints in these domains associated with the long COVID clinic patients in the time period of interest (figure 1 ). For each patient, we only counted diagnoses that newly occurred or occurred in greater frequency in the post-COVID-19 period compared with the pre-COVID-19 period, and only counted medications that were newly prescribed in the post-COVID-19 period, with no order records in the pre-COVID-19 period. Detailed feature engineering methods are described in the appendix (pp 3–5).
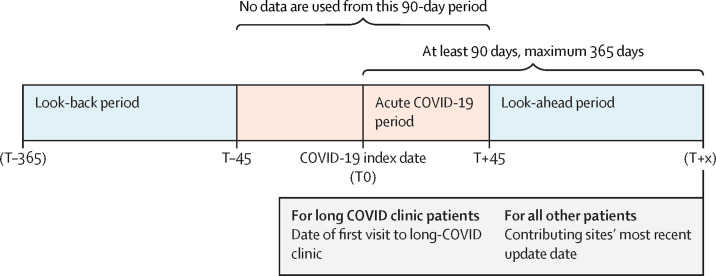
Figure 1.
Temporal windows for machine learning model inclusion
We searched for health-care visits, medical conditions, and prescription medication orders before and after each patient's COVID-19 index date, up to a maximum of 365 days post-index. We ignored all data occurring in a buffer period of 45 days before and after the COVID-19 index date to differentiate pre-COVID-19 and post-COVID-19 from acute COVID-19. For patients who attended a long COVID clinic, we ignored all data occurring on or after their first visit to such a clinic to avoid influencing the model with clinical observations occurring as a result of the patient's long COVID assessment.
Modelling and statistical analysis
To reflect that long COVID might look different depending on the severity of the patient's acute COVID-19 symptoms, we built three different machine learning models using the three-site subset: (1) all patients, (2) patients who had been hospitalised with acute COVID-19, and (3) patients who were not hospitalised. Each model aimed to identify the patients who were most likely to have long COVID, using attendance at a long COVID specialty clinic as a proxy for long COVID diagnosis. To train and test each model, patients were randomly sampled to yield similar patient counts in both classes (long COVID clinic patients and patients who did not attend the long COVID clinic). For the all-patients model, data were also sampled to yield similar numbers of hospitalised and non-hospitalised patients. Counts of patients in each group used for training and testing are shown in the appendix (p 6).
The Python package XGBoost was used to construct the models, using 924 features in total. Categorical features were one-hot encoded. Age and health-care visit rates were treated as continuous variables, and medical conditions and prescription drugs were modelled as binary features. Feature engineering details are given in the appendix (pp 3–5). Model hyperparameters were tuned using GridSearchCV (scikit-learn), with ten-fold cross-validation, set to optimise the area under the receiver operating characteristic curve (AUROC). We trained each model using ten-fold cross-validation, repeated five times. To assess performance, we calculated the AUROC, precision, recall, and F-score for each model, with a predictive probability threshold of 0·45. To aid with interpretability, we calculated Shapley values20 for all features to quantify each feature's importance to the classifications made by each model. These features were reviewed by clinical experts to aid in interpretation.
Once trained, to flag patients with potential long COVID in the N3C Data Enclave, we ran the all-patients model over the full base population of patients who had at least one health-care visit and at least one diagnosis or one medication in their post-COVID-19 window (n=846 981). We also validated our models' performance on long COVID clinic patient data submitted by a fourth N3C site (n=32 411) after our initial analysis was complete. Data from this fourth site were not included in our initial modelling work, and thus were used to examine our models' generalisability using new data from different sites.
Role of the funding source
This study was funded by NCATS, which contributed to the design, maintenance, and security of the N3C Data Enclave, and the NIH RECOVER Initiative, which is coordinating the participant recruitment protocol to which this work contributes. The funder of the study had no role in study design, data analysis, data interpretation, or writing of the report. No authors have been paid by a pharmaceutical company or other agency to write this Article.
Results
The combined demographics of patients who attended long COVID clinics at the three N3C sites substantially differ from those of COVID-19 patients at these sites who did not attend a long COVID clinic (table ). In this cohort, non-hospitalised long COVID clinic patients were disproportionately female. Long COVID clinic patients who were hospitalised with acute COVID-19 were disproportionately Black, when compared with all patients hospitalised with acute COVID-19, and were more likely to have a pre-COVID-19 comorbidity (diabetes, kidney disease, congestive heart failure, or pulmonary disease).
Table.
Characteristics of the three-site cohort used for model training and testing
|
Attended a long COVID clinic |
Did not attend a long COVID clinic |
||||
|---|---|---|---|---|---|
| Hospitalised (n=428) | Not hospitalised (n=169) | Hospitalised (n=15 193) | Not hospitalised (n=58 182) | ||
| Sex | |||||
| Female | 237 (55·4%) | 127 (75·1%) | 8465* (55·7%) | 34 771 (59·8%) | |
| Male | 191 (44·6%) | 42 (24·9%) | 6716* (44·2%) | 23 258 (40·0%) | |
| Unknown | 0 | 0 | <20 | 153 (0·3%) | |
| Race | |||||
| Asian | <20 | <20 | 571 (3·8%) | 1361 (2·3%) | |
| Black | 190 (44·4%) | 31 (18·3%) | 3207 (21·1%) | 6370 (10·9%) | |
| Native Hawaiian or Pacific Islander | <20 | <20 | 43 (0·3%) | 138 (0·2%) | |
| Other | <20 | <20 | 85 (0·6%) | 254 (0·4%) | |
| Unknown | 81 (18·9%) | 27 (16·0%) | 2695 (17·7%) | 9842 (16·9%) | |
| White | 142 (33·2%) | 107 (63·3%) | 8592 (56·6%) | 40 217 (69·1%) | |
| Ethnicity | |||||
| Hispanic or Latino | 69* (16·1%) | 26* (15·4%) | 3064 (20·2%) | 11 416 (19·6%) | |
| Not Hispanic or Latino | 354* (82·7%) | 128* (75·7%) | 11 869 (78·1%) | 45 119 (77·5%) | |
| Unknown | <20 | <20 | 260 (1·7%) | 1647 (2·8%) | |
| Age, years | |||||
| 18–25 | <20 | <20 | 790 (5·2%) | 7573 (13·0%) | |
| 26–45 | 86* (20·1%) | 75 (44·4%) | 3824 (25·2%) | 22 732 (39·1%) | |
| 46–65 | 188* (43·9%) | 69 (40·8%) | 5249 (34·5%) | 19 015 (32·7%) | |
| ≥66 | 147* (34·3%) | <20 | 5330 (35·1%) | 8862 (15·2%) | |
| Mean age (SD) | 58·29 (15·03) | 48·14 (14·02) | 56·50 (18·60) | 45·92 (17·32) | |
| Pre-COVID-19 comorbidities | |||||
| Diabetes | 86 (20·1%) | <20 | 2412 (15·9%) | 4842 (8·3%) | |
| Chronic kidney disease | 70 (16·4%) | <20 | 1721 (11·3%) | 2272 (3·9%) | |
| Congestive heart failure | 48 (11·2%) | <20 | 960 (6·3%) | 1133 (1·9%) | |
| Chronic pulmonary disease | 45 (10·5%) | 29 (17·2%) | 1415 (9·3%) | 3698 (6·4%) | |
Data are n (%) unless otherwise stated. All patients shown had acute COVID-19. Diabetes was not separated by type.
In accordance with the N3C download policy,21 for demographics where small cell sizes (<20 patients) could be derived from context, we have shifted the counts by a random number between 1 and 5. The accompanying percentages reflect the shifted number.
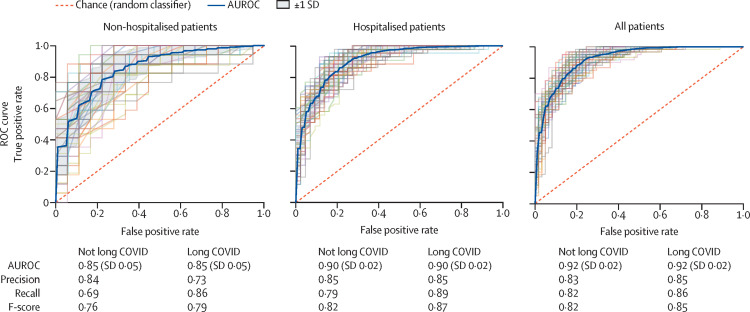
Each model was run against this three-site population, resulting in AUROCs of 0·92 for the all-patients model, 0·90 for the hospitalised model, and 0·85 for the non-hospitalised model (figure 2 ). All three models demonstrate robust performance. For the purpose of calculating these performance metrics, patients who attended a long COVID clinic are considered true positives; patients from the three sites who have not visited the specialty clinic are considered true negatives. Patients labelled by the model as patients with potential long COVID should, therefore, be interpreted as patients warranting care at a specialty clinic for long COVID—a proxy for long COVID diagnosis in the absence of a consensus definition. Our models can be used with a high score threshold for increased precision, or a lower score threshold for increased recall. In figure 2, we selected a score threshold of 0·45 to slightly favour recall, which yielded a precision of 0·85 and recall of 0·86 for the all-patients model. Notably, because long COVID appears to occur in a minority of patients with COVID-19, our model—when applied to large datasets of electronic health records—will always produce a non-trivial number of false positives, especially when tuned for high recall. As more data are available about patients with long COVID over time, we will be better able to characterise false positives and false negatives in future iterations.
Figure 2.
Machine learning model performance in identifying potential long COVID in patients
ROC curves, with 5-fold cross-validation and five repeats, identifying the ability of each of the three models (non-hospitalised, hospitalised, and all patients) to classify patients with long COVID as the discrimination threshold is varied. To emphasise recall of patients with potential long COVID, all models use a predicted probability threshold of 0·45 to generate the precision, recall, and F-score. The threshold can be adjusted to emphasise precision or recall, depending on the use case. AUROC=area under the receiver operating characteristic curve. ROC=receiver operating characteristic.
The three models were validated against an independent dataset from a fourth site. When tested against the patient population of this site qualifying for our base criteria (n=32 411, 125 of whom were long COVID clinic patients, without sampling to address the class imbalance), the AUROCs were 0·82 for the all-patients model, 0·79 for the hospitalised model, and 0·78 for the non-hospitalised model.
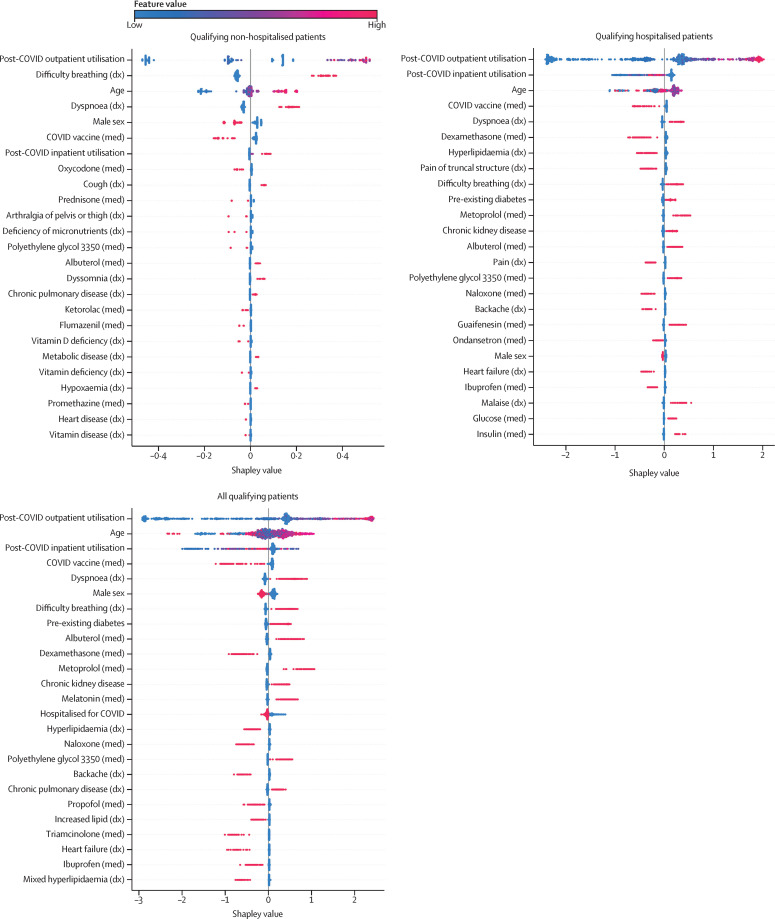
Figure 3 shows the top 20 most important features (as determined using Shapley values) for each model. The top 50 most important features for each model are available in the appendix (pp 7–9). Alhough not every feature can be easily categorised, four themes emerged across the features and models: (1) post-COVID-19 respiratory symptoms and associated treatments, (2) non-respiratory symptoms widely reported as part of long COVID and associated treatments, (3) pre-existing risk factors for greater acute COVID severity, and (4) proxies for hospitalisation.
Figure 3.
Most important model features associated with visits to a long COVID clinic
The top 20 features for each model are shown. Each point on the plot is a Shapley (importance) value for a single patient. The color of each point represents the magnitude and direction of the value of that feature for that patient. The point's position on the horizontal axis represents the importance and direction of that feature for the prediction for that patient. Some features are important predictors in all models (eg, outpatient utilisation, dyspnoea, and COVID-19 vaccine), whereas others are specific to one or two of the models (eg, dyssomnia or dexamethasone). Conditions labelled as chronic were diagnosed in patients before their COVID-19 index. Diabetes was not separated by type. dx=diagnosis. med=medication.
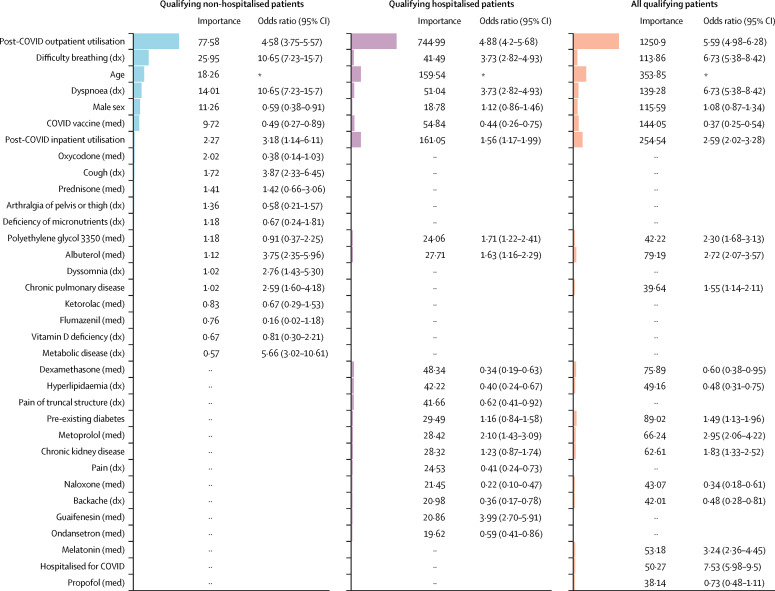
Figure 4 shows the aggregate feature importance and univariate odds ratios for each model. These results illustrate that several of our most important model features are significantly different among patients with potential long COVID and patients without evidence of long COVID.
Figure 4.
Univariate odds ratios for important model features
Shown are the relative feature importance and univariate odds ratios for the top features (union of the 20 most important features) in each model. Regardless of importance, some features are significantly more prominent in the long COVID clinic population, while others are more prominent in the non-long COVID clinic population. ·· denotes that the feature was not in the top 20 features for the model in that column. Conditions labelled chronic were associated with patients before their COVID-19 index. Diabetes was not separated by type. dx=diagnosis. med=medication. *Odds ratios exclude age, which has a non-linear relationship with long COVID.
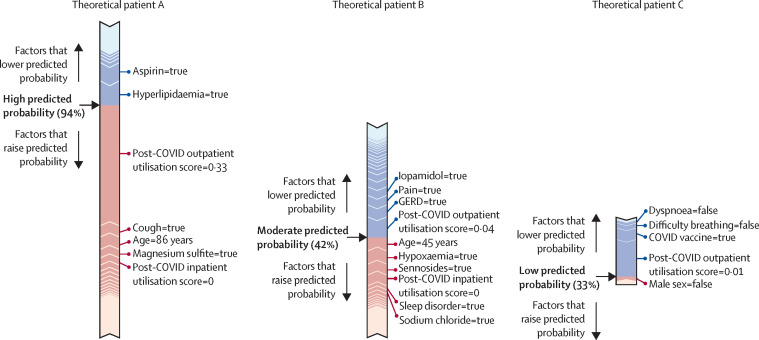
Figure 5 shows the path taken by three hypothetical patients through each of our three models, respectively.
Figure 5.
Example paths taken by the machine learning models to classify patients with potential long COVID
Force plots showing the contribution of individual features to the final predicted probability of long COVID, as generated for individual patients by the all-patients model (A), hospitalised model (B), and non-hospitalised model (C). Features in red increase the predicted probability of long COVID classification by the model, whereas features in blue decrease that probability. The length of the bar for a given feature is proportional to the effect that feature has on the prediction for that patient. The final predicted probability is shown in bold. GERD=gastroesophageal reflux disease.
Discussion
To avoid influencing the model with previous assumptions about the features of long COVID, we took a light-touch approach to feature selection, performing as little manual curation of features as possible before training and testing our models. Because of this approach, the reasons that a given feature might be important to one or more of the models is not always obvious. However, review by clinical experts of the features shown in Figure 3, Figure 4 and in the appendix (pp 7–9) revealed a number of possible themes.
First, post-COVID-19 respiratory symptoms and associated treatments. These features are commonly reported for patients with long COVID.7, 9, 22 A confounding factor that prioritises these features might be that the long COVID clinics at two of the three sites that contributed long COVID clinic patients are based in the pulmonary department. However, given that SARS-CoV-2 is primarily a respiratory virus, it is not surprising that long-term respiratory symptoms were observed. Similar long-term respiratory symptomatology is well described with respiratory viral syndromes, including those from severe acute respiratory syndrome, respiratory syncytial virus, influenza, and COVID-19.23, 24 The high proportion of albuterol use and use of inhaled steroids is consistent with the expected high prevalence of post-viral reactive airways disease. Examples of the most important features include dyspnoea or difficulty breathing, cough, albuterol, guaifenesin, and hypoxaemia.
Second, non-respiratory symptoms widely reported as part of long COVID and associated treatments. Sleep disorders, anxiety, malaise, chest pain, and constipation have all been reported as symptoms of long COVID, and are included in WHO's case definition.10 The example features in this group include symptoms and mitigating treatments. Example features include dyssomnia, chest pain, and malaise, and treatments with lorazepam, melatonin, and polyethylene glycol 3350.
Third, pre-existing risk factors for greater acute COVID severity. Some known risk factors for acute COVID-19 and severity are associated with long COVID—including chronic conditions (such as diabetes, chronic kidney disease, and chronic pulmonary disease), which predispose patients at increased risk for worsened COVID-19 symptoms.25
Fourth, proxies for hospitalisation. Features that are representative of standard hospital admission orders probably contributed to the model as proxies for hospitalisation in general, rather than being individually meaningful. These features were most prominent in patients without long COVID (true negatives), suggesting that the model is correctly differentiating between acute illness requiring hospitalisation and long COVID. Example features include the use of glucose, ketorolac, propofol, and naloxone.
Although there is considerable overlap between the most important features across the three models, there are also distinct differences (Figure 3, Figure 4; appendix pp 7–9). Notable differences include the high importance of dexamethasone in the hospitalised model, which decreased the likelihood of an individual patient being labelled as a potential long COVID patient. Dexamethasone is not present in the top 50 features of the non-hospitalised model. Similarly, cough and dyssomnia, which increased the likelihood of an individual beinglabelled as a potential long COVID patient, are important features in the non-hospitalised model, but do not appear in the hospitalised model. COVID-19 vaccination after acute disease, which is consistently an important feature in all three models, decreased the likelihood of patients being labelled as potentially having long COVID. This result is noteworthy and indicates that not only does vaccination against SARS-CoV-2 protect against hospitalisation and death, but that it might also protect against long COVID.
Rates of outpatient and inpatient utilisation are important features in all three models. This finding can be interpreted in a number of ways—patients who continue to feel unwell long after acute COVID-19 might be more likely to visit their providers repeatedly than those patients who fully recover. Because diagnosing and treating the heterogenous symptoms of long COVID is a challenge, these patients could be referred to one or more specialists, further increasing their utilisation.
Machine learning models do not consider each feature individually; rather, complex relationships between features can greatly influence classification. Each patient has their own path through the model, based on their available data, as shown in figure 5. Information of this type is useful to make the outcomes of the machine learning models interpretable.
Electronic health records were the source of all features used by our model. Although electronic health records contain rich clinical features, these data are also a proxy for health-care utilisation and can be interpreted through that lens. Diagnoses coded in the electronic health record are not representative of the whole patient, but rather are focused on the specific reasons the patient has visited a health-care site on that day. Moreover, the absence of electronic health record data about a patient does not equate to the absence of a disease; it merely represents the absence of a patient seeking care for that disease.
Even as a proxy for health-care utilisation, electronic health record data is well suited to the task of cohort definition by way of computable phenotyping, especially when the end goal is study recruitment. Although there are other methods of identifying potential study participants, a computable phenotype allows us to efficiently narrow the recruitment pool down from everyone available to patients who are likely to qualify— easily eliminating large numbers of patients that do not qualify, and ascertaining patients that might elude human curation.
There are additional advantages to using electronic health record data to identify patients with long COVID. With an evolving definition and no gold standard to compare with, the electronic health record allows us to define proxies for a condition and select on those—in this case, a patient's visit to a long COVID specialty clinic. However, rather than settling for a restrictive criterion of at least one visit to a long COVID specialty clinic, our machine learning models allow us to decouple patients' utilisation patterns from the clinic visit, meaning that we can use the models to identify similar patients who might not have access to a long COVID clinic.
This study has several limitations. Electronic health record data is skewed towards patients who make more use of health-care systems, and is further skewed towards high utilisers, patients with more severe symptoms, and hospital inpatients. When researchers train models on N3C's electronic health record data, it is essential to acknowledge whose data is less likely to be represented; for example, uninsured patients, patients with restricted access to or ability to pay for care, or patients seeking care at small practices or community hospitals with scarce data exchange capabilities. Moreover, for patients included in our models, clinic visits and hospitalisations that occur outside of the health-care system (ie, N3C site) for that patient are generally absent from our data. Finally, because our models require an index date for the execution of temporal logic, we cannot make use of cases without a positive indicator (test or diagnosis code) recorded in the electronic health record. This approach excludes the analysis of patients who had COVID-19 early in the pandemic and were not able to be tested.
We did not include race and ethnicity as model features, because we did not believe our three-site sample of long COVID clinic patients to be appropriately representative. As more data on patients with long COVID are available over time, we will be able to balance the cohort based on demographics and, critically, carefully account for race and ethnicity in future iterations of the model.
Because two of the three clinics that provided us with long COVID patient data are based in a pulmonary department, we acknowledge that our lists of important features prominently feature pulmonary conditions and treatments. Feature importance should not necessarily be interpreted as important to the diagnosis and characterisation of long COVID itself, but rather as important as inputs for an accurate electronic health record-based model. As we collect more training data from additional clinics over time, we suspect this set of features might change to provide a fuller picture of the condition. We recommend that readers wishing to utilise the model presented here consult our GitHub repository, where future iterations of the model will be made available.
Beyond identifying cohorts for research studies, the models presented here can be used in various applications and could be enhanced in several ways. Specifically, in future studies, it will be necessary to use a large sample size of patients with long COVID to validate hypotheses relating to social determinants of health and demographics, comorbidities, and treatment implications, and to understand the relationship between acute COVID-19 severity and specific long COVID signs and symptoms and their longitudinal progression. The influence of vaccination in such trajectories will also need to be explored.
It is plausible that long COVID will not have a single definition, and it might be better described as a set of related conditions with their own symptoms, trajectories, and treatments. Thus, as larger cohorts of patients with long COVID are established, future research should identify sub-phenotypes of long COVID by clustering patients with long COVID with similar electronic health record data fingerprints. Such fingerprints might be enhanced by natural language processing of clinical notes, which often include descriptions of signs and symptoms not recorded in structured diagnosis data. Future iterations of our models could discern among these clusters given N3C's large sample size and recurring data feeds.
Data sharing
The N3C data transfer to NCATS is performed under a Johns Hopkins University reliance protocol (IRB00249128) or individual site agreements with the NIH. The N3C Data Enclave is managed under the authority of the NIH; more information can be found at ncats.nih.gov/n3c/resources. Enclave data is protected, and can be accessed for COVID-19-related research with an institutional review board-approved protocol and data use request. A detailed accounting of data protections and access tiers has previously been published.17 Enclave and data access instructions can be found at https://covid.cd2h.org/for-researchers. All code used to produce the analyses in this manuscript is available within the N3C Data Enclave to users with valid login credentials to support reproducibility.
Declaration of interests
ATG is an employee of Palantir Technologies. ERP, JPD, SEJ, RRD, CGC, TDB, JAM, RM, AW, and MAH report research funding from the NIH. ERP and MGK report research funding from PCORI. MAH and JAM are co-founders of Pryzm Health. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
The analyses described in this Article were conducted with data or tools accessed through the NCATS N3C Data Enclave and supported by NCATS (U24 TR002306). This research was also funded in part by the NIH (OT2HL161847–01). The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the NIH. This research was possible because of the patients whose information is included within the data from participating organisations and the organisations and scientists who have contributed to the ongoing development of this community resource.17 We gratefully acknowledge the following contributors: David A Eichmann, Hongfang Liu, Heidi Spratt, Chunlei Wu, Davera Gabriel, Kellie M Walters, Karthik Natarajan, Shyam Visweswaran, Robert Miller, Mary Emmett, Patricia A Francis, Shawn T O'Neil, Christopher P Austin, Leonie Misquitta, Jeremy Richard Harper, Farrukh M Koraishy, and Satyanarayana Vedula.
The N3C Consortium
Carolyn Bramante, David Dorr, Michele Morris, Ann M Parker, Hythem Sidky, Ken Gersing, Stephanie Hong, and Emily Niehaus.
Contributors
ERP, ATG, KK, CGC, and MAH curated the data. ERP, ATG, MGK, KK, and CGC integrated the data. ERP, ATG, MGK, and CGC handled data quality assurance. ERP, KK, and CGC defined the N3C phenotype. ERP, ATG, TDB, MGK, KK, and CGC provided clinical data model expertise. TDB, RRD, and SEJ provided clinical subject matter expertise. ATG, AB, and JPD did the statistical analysis. ATG, AB, JPD, JAM, and MAH were responsible for data visualisation. ERP, ATG, TDB, IMB, RRD, SEJ, MGK, JAM, RM, AW, KK, CGC, and MAH critically revised the manuscript. ERP, ATG, RRD, SEJ, JAM, CGC, and MAH drafted the manuscript. JAM, AW, CGC, and MAH were responsible for governance and regulatory oversight. ERP and ATG accessed and verified all underlying data for these analyses. Authors were not precluded from accessing data in the study, and they accept responsibility to submit for publication.
Contributor Information
The N3C Consortium:
Carolyn Bramante, David Dorr, Michele Morris, Ann M Parker, Hythem Sidky, Ken Gersing, Stephanie Hong, and Emily Niehaus
Supplementary Material
References
- 1.Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavriatopoulou M, Korompoki E, Fotiou D, et al. Organ-specific manifestations of COVID-19 infection. Clin Exp Med. 2020;20:493–506. doi: 10.1007/s10238-020-00648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nalbandian A, Sehgal K, Gupta A, et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenhalgh T, Knight M, A'Court C, Buxton M, Husain L. Management of post-acute covid-19 in primary care. BMJ. 2020;370 doi: 10.1136/bmj.m3026. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Pinto MD, Borelli JL, et al. COVID symptoms, symptom clusters, and predictors for becoming a long-hauler: looking for clarity in the haze of the pandemic. medRxiv. 2021 doi: 10.1101/2021.03.03.21252086. published online March 5. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rando HM, Bennett TD, Byrd JB, et al. Challenges in defining long COVID: striking differences across literature, electronic health records, and patient-reported information. medRxiv. 2021 doi: 10.1101/2021.03.20.21253896. published online March 26. (preprint). [DOI] [Google Scholar]
- 7.McCorkell L, Assaf GS, Davis HE, Wei H, Akrami A. Patient-led research collaborative: embedding patients in the long COVID narrative. Pain Rep. 2021;6:e913. doi: 10.1097/PR9.0000000000000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis HE, Assaf GS, McCorkell L, et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. bioRxiv. 2020 doi: 10.1101/2020.12.24.20248802. published online Dec 26. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deer RR, Rock MA, Vasilevsky N, et al. Characterizing long COVID: deep phenotype of a complex condition. bioRxiv. 2021 doi: 10.1101/2021.06.23.21259416. published online June 29. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO A clinical case definition of post COVID-19 condition by a Delphi consensus, 6 October 2021. Oct 6, 2021. https://www.who.int/publications/i/item/WHO-2019-nCoV-Post_COVID-19_condition-Clinical_case_definition-2021.1
- 11.RECOVER Researching COVID to enhance recovery. https://recovercovid.org/research
- 12.Richesson R, Smerek M. Electronic health records-based phenotyping. https://sites.duke.edu/rethinkingclinicaltrials/ehr-phenotyping/ [DOI] [PMC free article] [PubMed]
- 13.Mo H, Thompson WK, Rasmussen LV, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc. 2015;22:1220–1230. doi: 10.1093/jamia/ocv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20:117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richesson RL, Rusincovitch SA, Wixted D, et al. A comparison of phenotype definitions for diabetes mellitus. J Am Med Inform Assoc. 2013;20:e319–e326. doi: 10.1136/amiajnl-2013-001952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beesley LJ, Mukherjee B. Statistical inference for association studies using electronic health records: handling both selection bias and outcome misclassification. Biometrics. 2022;78:214–226. doi: 10.1111/biom.13400. [DOI] [PubMed] [Google Scholar]
- 17.Haendel MA, the N3C Consortium. Chute CG, et al. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021;28:427–443. doi: 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett TD, Moffitt RA, Hajagos JG, et al. Clinical characterization and prediction of clinical severity of SARS-CoV-2 infection among US adults using data from the US National COVID Cohort Collaborative. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Center for Advancing Translational Sciences NIH COVID-19 data warehouse data transfer agreement. Aug 5, 2020. https://ncats.nih.gov/files/NCATS_Data_Transfer_Agreement_05-11-2020_Updated%20508.pdf
- 20.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. arXiv. 2017 doi: 10.48550/arXiv.1705.07874. published online May 22. (preprint). [DOI] [Google Scholar]
- 21.Chute C, Walden A. N3C results download policy. 2021. https://zenodo.org/record/4743236#.YnQ8MOjMJ3g
- 22.Nasserie T, Hittle M, Goodman SN. Assessment of the frequency and variety of persistent symptoms among patients with COVID-19: a systematic review. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngai JC, Ko FW, Ng SS, To K-W, Tong M, Hui DS. The long-term impact of severe acute respiratory syndrome on pulmonary function, exercise capacity and health status. Respirology. 2010;15:543–550. doi: 10.1111/j.1440-1843.2010.01720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fauroux B, Simões EAF, Checchia PA, et al. The burden and long-term respiratory morbidity associated with respiratory syncytial virus infection in early childhood. Infect Dis Ther. 2017;6:173–197. doi: 10.1007/s40121-017-0151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.US Centers for Disease Control and Prevention Science Brief: evidence used to update the list of underlying medical conditions associated with higher risk for severe COVID-19. Feb 15, 2022. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/underlying-evidence-table.html [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The N3C data transfer to NCATS is performed under a Johns Hopkins University reliance protocol (IRB00249128) or individual site agreements with the NIH. The N3C Data Enclave is managed under the authority of the NIH; more information can be found at ncats.nih.gov/n3c/resources. Enclave data is protected, and can be accessed for COVID-19-related research with an institutional review board-approved protocol and data use request. A detailed accounting of data protections and access tiers has previously been published.17 Enclave and data access instructions can be found at https://covid.cd2h.org/for-researchers. All code used to produce the analyses in this manuscript is available within the N3C Data Enclave to users with valid login credentials to support reproducibility.