Abstract
Transformants of Aspergillus flavus containing the Aequorea victoria gfp gene fused to a viral promoter or the promoter region and 483 bp of the coding region of A. flavus aflR expressed green fluorescence detectable without a microscope or filters. Expression of green fluorescent protein fluorescence was correlated with resistance to aflatoxin accumulation in five corn genotypes inoculated with these transformants.
Aspergillus flavus produces the carcinogen aflatoxin, which can contaminate a number of food crops, including corn, peanuts, cottonseed, and tree nuts (16, 17, 19). To study the infection process and aflatoxin biosynthesis, we developed gene reporter constructs. One construct, a fusion of the promoter of the A. flavus β-tubulin gene to the Escherichia coli uidA reporter gene that encodes β-glucuronidase (GUS) (24), has been used to visualize growth of the fungus in corn tissue (2). A second construct, in which the promoter of the aflatoxin pathway gene ver-1 is fused to uidA, has been used to monitor gene expression and aflatoxin biosynthesis in A. flavus (8, 11). Despite the utility of the GUS reporter gene constructs, they have limitations. Visualization of fungal growth within tissue requires incubation of the sample with the GUS substrate, X-GLUC (5-bromo-4-chloro-3-indolyl-β-d-glucuronide), a procedure that is time consuming and prevents following real-time colonization of tissue.
To overcome some of the limitations of the GUS reporter systems, we examined the feasibility of using the Aequorea victoria green fluorescent protein (GFP) as a tag to follow the growth of A. flavus. GFP has several advantages as an in vivo reporter for monitoring dynamic processes in cells or organisms, as its fluorescence can be measured directly without additional proteins, substrates, or cofactors (4, 7, 10, 12, 20). GFP expression has been widely reported in yeast (1, 14, 18, 22) but in only a few filamentous fungi (6, 13, 21). The objective of this study was to determine if a red-shifted variant, which is 35 times brighter than wild-type GFP (5), could be expressed sufficiently in A. flavus to monitor growth and colonization by the fungus.
A. flavus 86-10 (FGSC A1009) (w arg-7 pyrG afl+), a pyrG mutant of strain 86 (NRRL 60041) (8), was cultured on potato dextrose agar supplemented with 10 mM uracil. This strain was cotransformed as previously described (23) with plasmid B9X2 (15) and either pNuc′Em2 or pGAP33. Cotransformants were selected for uracil prototrophy conferred by plasmid B9X2, and GFP-containing transformants were identified by their green fluorescence under long-wave (365 nm) irradiation.
Plasmid pNuc′Em2 was constructed from pNuc′EM and contains the coding region of the simian virus 40 large T-antigen nuclear localization signal (NLS) and the mammalian-codon-optimized, better folding, red-shifted GFP from plasmid pEGFP-N1 (Clontech Laboratories, Inc., Palo Alto, Calif.). The sequences of the NLS oligonucleotides were 5′-CCG GAC TCA GAT CTG ATC ACC GCC ATG GGC CCC AAG AAG AAG AGA AAG GTG TCG ACG GTA CCG CGG G-3′ and 5′-CCC GCG GTA CCG TCG ACA CCT TTC TCT TCT TCT TGG GGC CCA TGG CGG TGA TCA GAT CTG AGT CCG G-3′. Included in the oligonucleotides was an optimized translational start site for mammalian expression. The oligonucleotides were hybridized, and the duplex NLS oligonucleotide was cloned into the BglII-SalI site of the vector, pEGFP-N1, which is immediately upstream from the open reading frame (ORF) of the gfp gene. This plasmid, which contains the NLS-GFP coding region, was named pNuc′EM, for nuclear expression marker. The BglII-NotI fragment from pNuc′EM, which includes some 5′ untranslated region and the NLS-GFP coding region, was cloned into the BamHI-NotI site of a mammalian expression vector, pcDNA3 (Invitrogen, Carlsbad, Calif.), under the direction of a human cytomegalovirus promoter, cmv, from this plasmid. This new plasmid, which contains the ampicillin resistance gene, was named pNuc′Em2. Plasmid pGAP33 was constructed by subcloning an 864-bp BglII fragment into the BglII-BamHI site of pNuc′Em2, which is immediately upstream from the ORF of gfp gene and does not contain the NLS site. This 864-bp BglII fragment contains the 381-bp untranslated region and 483 bp of the ORF of the aflatoxin biosynthetic regulatory gene, aflR (9).
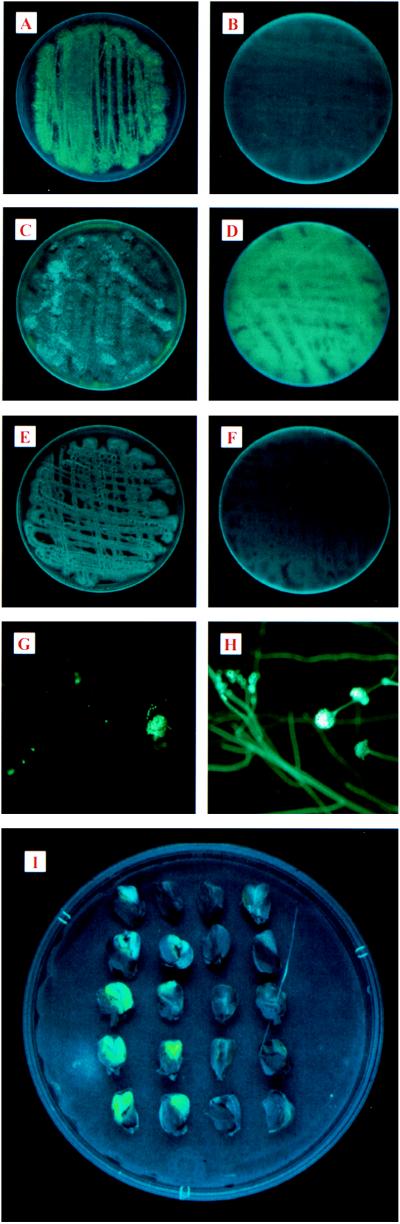
A. flavus was cotransformed with plasmid B9X2 and either pNuc′Em2 or pGAP33. Between 20 and 66% of the resulting transformants fluoresced green under long-wave UV light. Strain 86-10 had negligible autofluorescence at this wavelength (Fig. 1E and F). One transformant containing pNuc′Em2 (GAP32-8) and one transformant containing pGAP33 (GAP33-1) were chosen for further studies. Figures 1A and C show the fluorescence of the upper culture surfaces of GAP32-2 and GAP33-1, respectively, irradiated with UV light. Fluorescence of both sporulating cultures could be detected readily, but the fluorescence of the upper surface of GAP32-8 was more intense than that of GAP33-1. In contrast, fluorescence was observed on the underside of transformant GAP33-1 (Fig. 1D) and not on the underside of GAP32-8 (Fig. 1B). A closer examination of the cultures with a Nikon Optiphot compound microscope using GFP filters providing excitation at 395 nm and emission at 509 nm showed that the bright green fluorescence of the two transformants was most intense in conidia (Fig. 1G and H). Fluorescence could be detected in cultures as young as 3 days old and persisted for more than 12 weeks in cultures maintained at room temperature. Strong expression of GFP in A. flavus transformants harboring pNuc′Em2 is likely due to the modifications made in gfp. It is not clear why fluorescence can be observed on the underside of GAP33-1 cultures. The two constructs differ in that pGAP33 lacks the NLS present in pNuc′EM2, and pGAP33 is a protein fusion with aflR.
FIG. 1.
Expression of green fluorescence by A. flavus transformants GAP32-8 and GAP33-1. (A) Top view of GAP32-8. (B) Underside view of GAP32-8. (C) Top view of GAP33-1. (D) Underside view of GAP33-1. (E) Top view of 86-10 lacking gfp vector. (F) Underside view of 86-10. (G) Bright green fluorescent conidial heads of GAP33-1. (H) Bright green fluorescent conidial heads of GAP32-8. (I) GFP fluorescence in corn kernels of five inbreeds. Columns from left to right show kernels inoculated with GAP33-1, inoculated with GAP32-8, inoculated with strain 86-10, and wounded only. The five corn inbreds from the top to bottom rows are CI2, Tex6, B73, NC232, and 33-16. Kernels were incubated at 28°C for 5 days, sectioned, and observed for GFP fluorescence under a 365-nm UV light.
GUS reporter constructs are valuable tools for studying the resistance of corn kernels to colonization by A. flavus (2). To determine if GFP also can be used as a reporter to follow colonization by A. flavus, intact corn kernels of five genotypes were inoculated with GAP32-8 or GAP33-1. Kernels were surface disinfected in 75% ethanol for 5 to 10 min and rinsed three times in sterile water and the kernel endosperm was punctured once to a depth of 0.5 to 1.0 mm with a 20-gauge syringe needle. Two microliters of a conidial suspension (4 × 107 conidia/ml) of strain 86-10, GAP32-8, or GAP33-1 was inoculated into the wound. One kernel was placed in each well of a 24-well tissue culture plate on top of 1 ml of 1% water agar. Kernels were incubated at 28°C for 5 days and cut in half, and one half was examined under UV light in a cabinet designed to view thin-layer chromatography plates (Chromato-Vue Model CC-20, San Gabriel, Calif.). The assay was conducted twice with six replicate plates for each assay. Figure 1I shows the green fluorescence of the cut surface of the corn kernels resulting from the growth of A. flavus. As can be observed in the figure, the area of fluorescence differed among the genotypes, with the more susceptible genotypes (B73, NC232, and 33-16) displaying more fluorescence over the cut surface of the kernels than CI2 and Tex6, the resistant inbreds. Noninoculated kernels and kernels inoculated with 86-10 showed no green fluorescence. Of the five genotypes tested, the genotypes reported to be more susceptible to A. flavus (2, 3) displayed more fluorescence than those reported to be resistant.
Results from these studies demonstrate that the modified GFP encoded by pNuc′Em2 and pGAP33 is highly expressed in A. flavus. The intensity of fluorescence is sufficient to allow the visualization of a GFP-containing strain under a standard laboratory UV light. No special filters or equipment are needed to detect a GFP-expressing strain in culture or to monitor the colonization of corn seeds with the strain. The use of these constructs in strains of A. flavus could facilitate the detection of the fungus in substrates such as soils or foods. GFP expression in the corn kernels shows that these GFP-containing transformants should be useful in screening corn genotypes for resistance to aflatoxin accumulation and making screening faster and more economical.
Acknowledgments
Support for this work was provided by USDA/NRI grant 9601295 and USDA-SCA grant 58-6435-075.
We thank Don White at the University of Illinois and Marty Carson at North Carolina State University for supplying corn seeds. We thank Nina Stromgren Allen and Dana Moxley for the use of their computer-equipped Leica fluorescence stereoscope and Winnel Newman for the use of the fluorescence spectrophotometer. We also thank Dominique Robertson and Steve Nagar for use of their compound fluorescence microscopes and considerable input on analyzing GFP expression in fungal cells and D. Kerr for help in constructing the pNuc′Em2 plasmid.
W.D., Z.H., and J.E.F. contributed equally to this study.
REFERENCES
- 1.Atkins D, Izant J G. Expression and analysis of the green fluorescent protein gene in the fission yeast Schizosaccharomyces pombe. Curr Genet. 1995;28:585–588. doi: 10.1007/BF00518173. [DOI] [PubMed] [Google Scholar]
- 2.Brown R L, Cleveland T E, Payne G A, Woloshuk C P, Campbell K W, White D G. Determination of resistance to aflatoxin production in maize kernels and detection of fungal colonization using an Aspergillus flavus transformant expressing Escherichia coli β-glucuronidase. Phytopathology. 1995;85:983–989. [Google Scholar]
- 3.Campbell K L, White D G. Evaluation of corn genotypes for resistance to Aspergillus ear rot, kernel infection, and aflatoxin production. Plant Dis. 1995;79:1039–1045. [Google Scholar]
- 4.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–804. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 5.Cormack B P, Valdivia R, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Abalos J M, Fox H, Pitt C, Wells B, Doonan J H. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- 7.Fey P, Compton K, Cox E C. Green fluorescent protein production in the cellular slime molds Polysphondylium pallidum and Dictyostelium discoideum. Gene. 1995;165:127–130. doi: 10.1016/0378-1119(95)00430-e. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty J E, Weaver M A, Payne G A, Woloshuk C P. A beta-glucuronidase reporter gene construct for monitoring aflatoxin biosynthesis in Aspergillus flavus. Appl Environ Microbiol. 1995;61:2482–2486. doi: 10.1128/aem.61.7.2482-2486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaherty J E, Payne G A. Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl Environ Microbiol. 1997;63:3995–4000. doi: 10.1128/aem.63.10.3995-4000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubin A N, Reddy B, Njoroge J M, Miller J L. Long-term, stable expression of green fluorescent protein in mammalian cells. Biochem Biophys Res Commun. 1997;236:347–350. doi: 10.1006/bbrc.1997.6963. [DOI] [PubMed] [Google Scholar]
- 11.Huang Z, White D G, Payne G A. Corn seed proteins inhibitory to Aspergillus flavus and aflatoxin biosynthesis. Phytopathology. 1997;87:622–627. doi: 10.1094/PHYTO.1997.87.6.622. [DOI] [PubMed] [Google Scholar]
- 12.Kain S R, Kitts P. Expression and detection of green fluorescent protein (GFP) Methods Mol Biol. 1997;63:305–324. doi: 10.1385/0-89603-481-X:305. [DOI] [PubMed] [Google Scholar]
- 13.Lehmler C, Steinberg G, Snetselaar K M, Schliwa M, Kahmann R, Bolker M. Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 1997;16:3464–3473. doi: 10.1093/emboj/16.12.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nabeshima K, Saitoh S, Yanagida M. Use of green fluorescent protein for intracellular protein localization in living fission yeast cells. Methods Enzymol. 1997;283:459–471. doi: 10.1016/s0076-6879(97)83037-6. [DOI] [PubMed] [Google Scholar]
- 15.Payne G A, Nystrom G J, Bhatnagar D, Cleveland T E, Woloshuk C P. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl Environ Microbiol. 1993;59:156–162. doi: 10.1128/aem.59.1.156-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payne G A. Process of contamination by aflatoxin-producing fungi and their impact on crops. In: Sinha D B K K, editor. Mycotoxins in agriculture and food safety. New York, N.Y: Marcel Dekker, Inc.; 1998. p. 511. [Google Scholar]
- 17.Payne G A, Brown M P. Genetics and physiology of aflatoxin biosynthesis. Annu Rev Phytopathol. 1998;36:329–362. doi: 10.1146/annurev.phyto.36.1.329. [DOI] [PubMed] [Google Scholar]
- 18.Ram A F, Van den Ende H, Klis F M. Green fluorescent protein-cell wall fusion proteins are covalently incorporated into the cell wall of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1998;162:249–255. doi: 10.1111/j.1574-6968.1998.tb13006.x. [DOI] [PubMed] [Google Scholar]
- 19.Squire R A. Ranking animal carcinogens: a proposed regulatory approach. Science. 1989;214:887–891. doi: 10.1126/science.7302565. [DOI] [PubMed] [Google Scholar]
- 20.Suarez A, Guttler A, Stratz M, Staendner L H, Timmis K N, Guzman C A. Green fluorescent protein-based reporter systems for genetic analysis of bacteria including monocopy applications. Gene. 1997;196:69–74. doi: 10.1016/s0378-1119(97)00197-2. [DOI] [PubMed] [Google Scholar]
- 21.Suelmann R, Sievers N, Fischer R. Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol Microbiol. 1997;25:757–769. doi: 10.1046/j.1365-2958.1997.5131873.x. [DOI] [PubMed] [Google Scholar]
- 22.Walmsley R M, Billinton N, Heyer W D. Green fluorescent protein as a reporter for the DNA damage-induced gene RAD54 in Saccharomyces cerevisiae. Yeast. 1997;13:1535–1545. doi: 10.1002/(SICI)1097-0061(199712)13:16<1535::AID-YEA221>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Woloshuk C P, Seip E R, Payne G A, Adkins C R. Genetic transformation system for the aflatoxin-producing Aspergillus flavus. Appl Environ Microbiol. 1989;55:86–90. doi: 10.1128/aem.55.1.86-90.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woloshuk C P, Foutz K R, Brewer J F, Bhatnagar D, Cleveland T E, Payne G A. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]