Abstract
Background
We investigated the evolutionary relationships, mutations, antigenic epitopes, and structural dynamics of the receptor-binding domain (RBD) of SARS-CoV-2, Omicron and other recently evolved variants.
Methods
The RBD of SARS-CoV-2 and its Omicron, Alpha, Beta, Gamma, Delta, and Mu variants were subjected to pairwise sequence matrix evaluation, antigenic epitope prediction, and phylogenetic relationship and structural dynamics analyses.
Results
The Omicron RBD contained 13–15 amino acid mutations, of which 12 were new and three conserved with other variants. In addition, two mutations found in the Alpha, Beta, Gamma, and Mu variants were not found in the Omicron RBD. The ultrametric clustering unweighted pair group method with arithmetic mean identified Omicron as a novel monophyletic class, but the neighbor-joining method clustered Omicron with Alpha and Delta variants. In the SARS-CoV-2 RBD, five main antigenic epitopes were predicted, and these epitopes were conserved across all SARS-CoV-2 variants tested. Surprisingly, the additional mutations in the Omicron variant increased the size of the expected antigenic sites in two of these antigenic epitopes. Molecular dynamics (MD) simulations revealed higher root-mean-square deviation in the Omicron RBD, greater residue fluctuation at residues 32–42 and 140–160, and increased solvent-accessible surface area.
Conclusions
The Omicron RBD mutations indicate the variant is within a new phylogenetic class of SARS-CoV-2 and significantly impact RBD structure, conformation, and molecular dynamics. However, conserved anticipated antigenic sites may imply partial changes in receptor affinity and response to immune reactions. Omicron RBD binding with the angiotensin-converting enzyme 2 receptor was suggested to be weaker than the original SARS-CoV-2 binding in MD simulations.
Keywords: Omicron variant, SARS-CoV-2, COVID-19, Phylogenetics
1. Introduction
Following the discovery of SARS-CoV-2 in December 2019, several new variants have emerged, including the Alpha variant in the United Kingdom (B.1.1.7) [1], the Beta variant in South Africa (B.1.351) [2], the Gamma variant in Brazil (P.1) [3], California (B.1.429) [4], and New York (B.1.526) [5], the Delta variant in India (B.1.617.2) [6], and most recently, the Omicron variant (B.1.1.529) [7], discovered in Botswana. The creation of novel variants is expected to continue because the virus is subjected to higher immunological pressure as a greater proportion of the host population is vaccinated or acquires spontaneous infection immunity [8].
Reported to the World Health Organization (WHO) by South Africa on November 24, 2021, Omicron was classified as a variant of concern on November 26, 2021 (WHO report). Analyses of the Omicron variant genome have revealed many genomic differences from other variants, particularly in the spike protein, important in virus receptor binding, infection, and the immune response. The Omicron spike protein alone contains at least 32 mutations from the original SARS-CoV-2, compared to only 16 in the highly infectious Delta form. Additional mutations have been detected in proteins essential for viral replication, such as nonstructural proteins 12 and 14 [9]. The Omicron variant has been observed to dramatically outcompete the ubiquitous and highly infectious Delta form. For example, the number of Omicron cases in the UK initially doubled every two days. Furthermore, this variant also has a very high reinfection rate, 5.4 times higher than the Delta variant [10,11]. Researchers have proposed various SARS-CoV-2 evolutionary routes, some of significant concern, based on the robust genomic and evolutionary examination of all variants emerging during the COVID-19 pandemic [10]. The assumptions of these proposed routes include structural constraints that prevent additional spike mutations, as well as insertions, deletions, point mutations and recombination events involving related viruses or even viruses from other genera [10].
A recent mutational scan of SARS-CoV-2 RBDs revealed constraints on angiotensin-converting enzyme 2 receptor (ACE2) folding and binding [12]. Although most of these mutations were detrimental to RBD expression and ACE2 binding, this binding was improved or maintained by multiple alterations, including those in the ACE2 interface residues that differed across coronaviruses related to SARS. However, current SARS-CoV-2 pandemic isolates do not appear to have been selected for ACE2-affinity-enhancing mutations [12].
Many unresolved questions remain about the Omicron variant's origin and its effect on vaccination response and host immunity, relevant clinical aspects, spread potency, and lethality. In this study, we examined the evolution of the Omicron RBD, a crucial component of virus–host interactions. We analyzed the antigenic epitopes of the SARS-CoV-2 variants Omicron, Alpha, Beta, Gamma, Delta, Mu, and SARS-CoV-2 USA to determine epitope differences. The structural stability and variation of RBD residues were also studied.
2. Materials and methods
2.1. Collection of input protein data
The genomes of SARS-CoV-2 variants were retrieved from the Global Initiative on Sharing All Influenza Data (GISAID) (https://www.gisaid.org/) [13]. Only genomes with high coverage were selected and saved as FASTA files. Table 1 provides information about the genomes used. The sequences were analyzed using CLC Genomics Workbench 12.0 (QIAGEN, Aarhus, Denmark) and Geneious Prime (Auckland, New Zealand) [14].
Table 1.
The SARS-CoV-2 variants included in the RBD analyses.
| Variant | Database Accession ID |
|---|---|
| Omicron | EPI_ISL_6640916 |
| Alpha | EPI_ISL_6756515 |
| Beta | EPI_ISL_5416540 |
| Gamma | EPI_ISL_6228367 |
| Delta | EPI_ISL_6832166 |
| hCoV | EPI_ISL_6910522 |
| Mu GH | EPI_ISL_4470504 |
| GH490R | EPI_ISL_6887009 |
2.2. RBD Retrieval and sequence alignment
The RBD protein sequence was determined using the protein sequence tools in the CLC genomics program. The proteins were aligned using the protein alignment wizard with highly accurate alignment settings, including gap cost settings of a ten-gap open cost and a one-gap extension cost. A pairwise comparison matrix was produced to compare the sequences retrieved. After computing the differences, identity percentages, gaps, and mutations, the identity matrix was generated.
2.3. Phylogenetic analyses
The neighbor-joining (NJ) and the unweighted pair group with arithmetic mean (UPGMA) methods were used to construct the phylogenetic tree [15], which was then analyzed for its evolutionary relationships. The CLC genomics software was used by applying the default parameters. The Jones–Taylor–Thornton (JTT) or Whelan and Goldman (WAG) models were used to measure distances. The neighbor-joining approach was subjected to 100 bootstrap resampling replicates.
2.4. RBD antigenic epitope analysis
The potential antigenic epitopes in the studied variants were searched using the EMBOSS antigenic prediction tool [16], in which the antigenic determination relies on a semi-empirical approach based on the physicochemical properties of amino acid residues and their occurrence frequencies in experimentally determined segmental epitopes. The minimal length of the antigenic region was set to six. The output format was set to EMBOSS motif.
2.5. Simulations of molecular dynamics
The GROMACS (v. 2021) and Desmond software packages were used to execute molecular dynamic (MD) simulations on the proteins. GROMACS simulations incorporated the nucleic AMBER94 force-field and the AMBER99SB-ILDN protein. In a 1.2 nm3 box containing a three-point water model (TIP3P), the complex was ionized using NaCl molecules to simulate neutral pH conditions. Each simulated system was subjected to energy minimization using the steepest descent approach in a maximum of 2000 steps, followed by two equilibration simulation stages with position restraints. At 310 K, the NVT ensemble was used for 100 ps, followed by NPT. The MD simulation was run for 100 ns at 310 K with no position constraints. Periodic boundary conditions were applied in all directions. Bond limitations were modeled using the LINCS algorithm, with a time step of 2 fs. The particle mesh Ewald technique was used to describe short-range nonbonded interactions with a twin-range cutoff of 0.8 nm and long-range electrostatic interactions with a Fourier grid spacing of 0.12 nm. To ensure structural stability, we employed the root-mean-square (RMS) tool (rms). Other protein characteristics were determined using the following tools: secondary structure (do dssp tool), hydrogen bonds (hbond tool), surface accessible area (sasa tool), RMS fluctuation (rmsf tool), and gyration (gyrate tool). The diagrams were created using the Linux application Grace, and the frames were visualized using PyMOL. SARS-CoV-2 and Omicron RBD were compared using MD modeling.
Desmond software (Schrödinger LLC) was used to perform MD for 100 ns. By integrating Newton's classical equation of motion, MD simulations can represent the movements of atoms over time. We used MD simulations to forecast the stability state of the physiological environment. The protein was preprocessed using Maestro's Protein Preparation Wizard, which included substantial optimization and minimization. All of the systems were created using the System Builder tool. The TIP3P orthorhombic box solvent model and the OPLS 2005 force field were employed. The models were made pH neutral using counter ions. To imitate physiological conditions, we incorporated 0.15 M sodium chloride (NaCl). For the entire simulation, the NPT ensemble temperature was 300 K and the pressure 1 atm. The models were relaxed before the simulation. Every 100 ps, the trajectories were examined, and the simulation's stability was demonstrated by comparing the protein and ligand's RMS deviation (RMSD) over time.
3. Results and discussion
3.1. Omicron RBD alignment
The RBD sequence of the first complete Omicron genome isolated in Botswana (accession no. EPI_ISL_6640916) was aligned with the RBD sequences of SARS-CoV-2 and its Alpha, Beta, Gamma, Delta, and Mu variants (Fig. 1 ). The alignment process revealed conserved and new mutations (summarized in Table 2 ). Fifteen Omicron RBD amino acids were mutated, 12 of which were unique to Omicron and three conserved with other variants. The conserved variants were K84N (also in Mu and Beta variants), T145K (SARS-CoV-2 and the Delta variant), and N168Y (Alpha, Beta, Gamma, and Mu variants). The 12 new mutations were G6D, S38L, S40P, S42F, N107K, G113S, S144N, E151A, Q160R, G163S, Q165R, and Y172H. In addition, two mutations identified in the RBDs of SARS-CoV-2 variants were not found in Omicron: R13K (Mu) and K145T (Alpha, Beta, Gamma, and Mu) (Fig. 1).
Fig. 1.
Multiple sequence alignment of RBD from SARS-CoV-2 and its Omicron, Alpha, Beta, Gamma, Delta, and Mu variants. The mutated residues are highlighted in red.
Table 2.
List of Omicron RBD mutations. The mutations are numbered according to the full spike protein length or the RBD residue number. The mutant residue is assigned as new or conserved if found in another SARS-CoV-2 variant.
| Mutation in Omicron (full spike numbering) | Mutation in Omicron (RBD residue no.) | New/conserved mutation | Conserved with other virus |
|---|---|---|---|
| G339D | G6D | New | |
| S371L | S38L | New | |
| S373P | S40P | New | |
| S375F | S42F | New | |
| K417N | K84N | Conserved | Mu variant, Beta variant |
| N440K | N107K | New | |
| G446S | G113S | New | |
| S477N | S144N | New | |
| T578K | T145K | Conserved | SARS-CoV-2, Delta variant |
| E484A | E151A | New | |
| Q493R | Q160R | New | |
| G496S | G163S | New | |
| Q498R | Q165R | New | |
| N501Y | N168Y | Conserved | Alpha variant, Beta variant, Gamma variant, Mu variant |
| Y505H | Y172H | New |
A previous study examined the effect of mutations on RBD affinity and discovered that, like SARS-CoV-1, approximately 46% of single amino acid mutations in SARS-CoV-2 maintained high affinities to ACE2 receptors [12]. Affinity-boosting mutations are particularly common at RBD sites Q493, Q498, and N501. Although these SARS-CoV-2 residues are involved in a dense network of polar contacts with ACE2, mutations at these three sites are tolerated, and even slight changes in polarity will not affect the binding affinity [12]. In a wide-scale analysis of the effects of mutations on RBD–ACE2 affinity, approximately 84.3% of mutations did not affect binding affinity, and only 3.8% of mutations decreased the binding strength [17]. Furthermore, only the N501Y mutation has been linked to lower free energy values, implying stronger affinity [17].
The determined structure of the SARS-CoV-2 RBD–ACE2 interaction [18] revealed a salt bridge between RBD K417 and ACE2 D30. The mutation was expected to also affect a number of hydrogen bonds. In the Omicron RBD, these residues were mutated to K417N, G446S, Q493R, N501Y, and Y505H. These residues are linked to the SARS-CoV-2 RBD and ACE2 through hydrogen bonding.
3.2. Pairwise comparison matrix
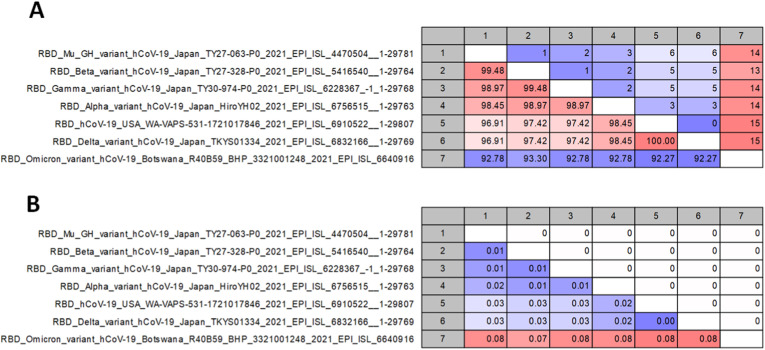
The pairwise comparison matrix revealed that the largest number of RBD amino acid mutations (13–15) were identified in the Omicron variant (Fig. 2 ). The other variants differed from SARS-CoV-2 by only one to a maximum of six mutations. Furthermore, there were no gaps, amino acid insertions, or deletions among the RBD variants.
Fig. 2.
Pairwise comparative matrix of Omicron and SARS-CoV-2 variants. A) The upper diagonal panel shows the differences in amino acid content. The lower diagonal panel depicts percent identity. B) The upper diagonal panel shows the gaps in the sequence alignment. The lower diagonal panel provides the residues' identities.
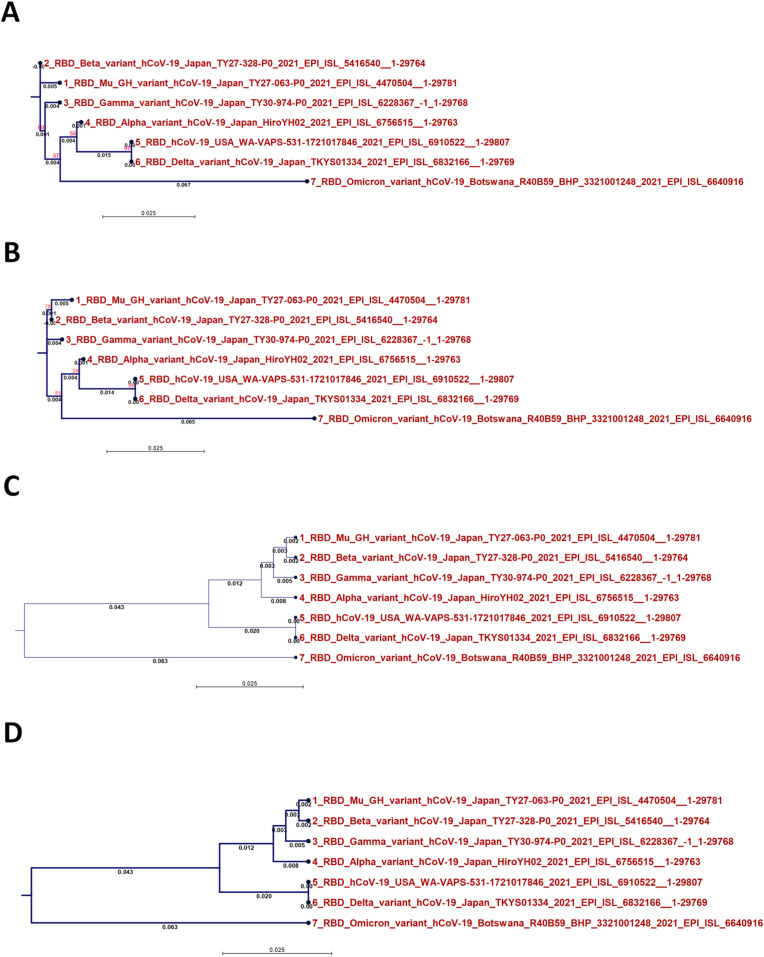
3.3. Phylogenetics of the Omicron variant RBD
The phylogenetic analysis of the Omicron variant RBD is provided in Fig. 3 (NJ/JTT, Fig. 3A; NJ/WAG, Fig. 3B; UPGMA/JTT, Fig. 3C; UPGMA/WAG, Fig. 3D). The UPGMA method positioned Omicron in a new monophyletic class separate from other variants. In contrast, the NJ method classified Omicron RBD with SARS-CoV-2 and the Alpha and Delta variants in a common node.
Fig. 3.
Comparison of different methods of analyzing phylogenetic trees constructed based on RBD analysis results: A) the NJ method and JTT substitution model, B) the NJ method and WAG substitution model, C) the UPGMA method and JTT substitution model, and D) the UPGMA method and WAG substitution model.
Bioinformatics and phylogenetic analyses are gold standards in the study of microbial evolution and the development of novel therapeutics targeting specific biological targets [[19], [20], [21], [22]]; we used a variety of these techniques to gain insights into the evolution of the Omicron variant RBD. Two methods we used to construct a phylogenetic tree are UPGMA and NJ. In the former, the mutation rate is ignored in favor of the assumption that all lineages change at the same rate. Therefore, the distance between two points determines the structure of the tree. In contrast, the NJ technique accounts for the mutation rate. It does not assume that all lineages evolve at the same rate. This allowed the clustering of Omicron RBD with the most closely related variants in the alignment matrix.
The Omicron RBD protein represents a novel monophyletic class, similar to our recent finding regarding the Omicron genome, the outcome of UPGMA analysis [23]. In this context, in terms of the fraction of shared nucleotides, the Omicron variant is the most phylogenetically distant from the SARS-CoV-2 Wuhan-Hu-1 and Gamma variants [24].
Researchers have offered several hypotheses to explain the likely emergence of Omicron. One hypothesis is that the new variant was introduced during the winter wave in some southern African countries and circulated among chronically infected patients. Its introduction was not reported due to reduced genomic sequencing efforts in those countries. Subsequent spike protein mutations may have increased the protein's ability to connect with the ACE2 receptor on host cells. Mutations observed in the Omicron form could also be due to a hidden animal reservoir. Because of Africa's low vaccination rate, the Omicron variant may have more readily circulated [25]. The nature of the Omicron RBD might indicate that the variant represents a new class with complex factors interacting during viral evolution.
3.4. The content of predicted antigenic epitopes
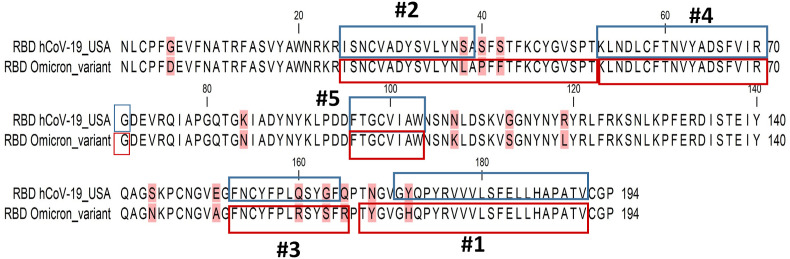
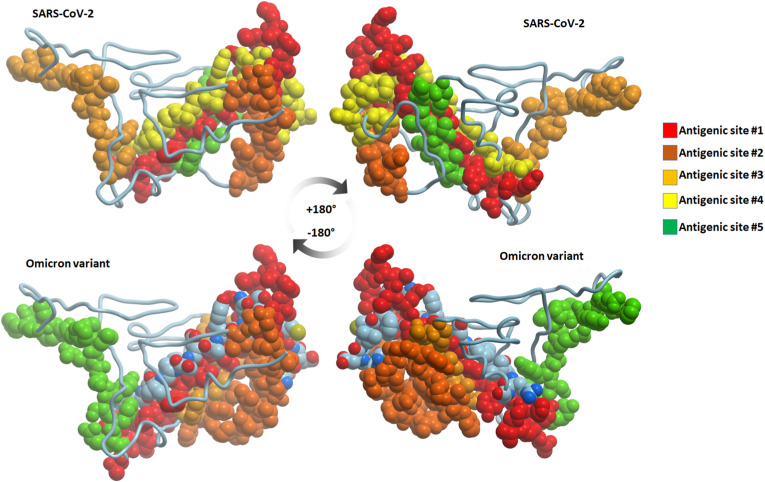
The top five epitopes predicted by the EMBOSS antigenic tool are provided in Table 3 . In the SARS-CoV-2 USA isolate, residues 171–191, 25–39, 152–164, 54–71, and 96–103 (in descending score order) were the predicted antigenic epitopes. The top five epitopes were conserved between the Omicron variant and the SARS-CoV-2 USA isolate (Fig. 4 ). Surprisingly, the additional mutations observed in the Omicron variant enlarged the expected antigenic sites in two of these antigenic epitopes. The N168Y mutation in the Omicron variant was predicted to enlarge the first antigenic epitope by four residues at its N-terminus. Furthermore, due to two additional mutations (S40P and S42F) in the Omicron variant, the second epitope length was predicted to extend from 15 residues in SARS-CoV-2 to 27 residues in Omicron (Fig. 5 ). In SARS-CoV-2 and the Alpha, Beta, Gamma, Delta, and Mu variants, the lengths of the first and second epitopes were consistently conserved. The anticipated length increase, in this case, was unique to the Omicron variant.
Table 3.
The antigenic epitopes predicted by the EMBOSS antigenic detection program in SARS-CoV-2 and Omicron, Alpha, Beta, Gamma, Delta, and Mu variants. The epitopes are in descending order of predicted scores.
| Virus | # | Sequence | length | score | Max_score_pos |
|---|---|---|---|---|---|
| SARS-CoV-2 | 1 | GYQPYRVVVLSFELLHAPATV | 171->191 | 1.214 | 177 |
| 2 | ISNCVADYSVLYNSA | 25->39 | 1.183 | 31 | |
| 3 | GFNCYFPLQSYGF | 152->164 | 1.144 | 158 | |
| 4 | LNDLCFTNVYADSFVIRG | 54->71 | 1.14 | 60 | |
| 5 | FTGCVIAW | 96->103 | 1.126 | 99 | |
| Omicron | 1 | TYGVGHQPYRVVVLSFELLHAPATV | 167->191 | 1.214 | 177 |
| 2 | ISNCVADYSVLYNLAPFFTFKCYGVSPT | 25->52 | 1.183 | 31 | |
| 3 | LNDLCFTNVYADSFVIRG | 54->71 | 1.14 | 60 | |
| 4 | FTGCVIAW | 96->103 | 1.126 | 99 | |
| 5 | GFNCYFPLRSYSFR | 152->165 | 1.123 | 158 | |
| Alpha | 1 | TYGVGHQPYRVVVLSFELLHAPATV | 167->191 | 1.214 | 177 |
| 2 | ISNCVADYSVLYNSA | 25->39 | 1.183 | 31 | |
| 3 | GFNCYFPLQSYGFQ | 152->165 | 1.144 | 158 | |
| 4 | LNDLCFTNVYADSFVIRG | 54->71 | 1.14 | 60 | |
| 5 | FTGCVIAW | 96->103 | 1.126 | 99 | |
| Beta | 1 | GYQPYRVVVLSFELLHAPATV | 171->191 | 1.214 | 177 |
| 2 | ISNCVADYSVLYNSA | 25->39 | 1.183 | 31 | |
| 3 | GFNCYFPLQSYGFQ | 152->165 | 1.144 | 158 | |
| 4 | LNDLCFTNVYADSFVIRG | 54->71 | 1.14 | 60 | |
| 5 | FTGCVIAW | 96->103 | 1.126 | 99 | |
| Gamma | 1 | GYQPYRVVVLSFELLHAPATV | 171->191 | 1.214 | 177 |
| 2 | ISNCVADYSVLYNSA | 25->39 | 1.183 | 31 | |
| 3 | GFNCYFPLQSYGFQ | 152->165 | 1.144 | 158 | |
| 4 | LNDLCFTNVYADSFVIRG | 54->71 | 1.14 | 60 | |
| 5 | FTGCVIAW | 96->103 | 1.126 | 99 | |
| Delta | 1 | GYQPYRVVVLSFELLHAPATV | 171->191 | 1.214 | 177 |
| 2 | ISNCVADYSVLYNSA | 25->39 | 1.183 | 31 | |
| 3 | GFNCYFPLQSYGF | 152->164 | 1.144 | 158 | |
| 4 | LNDLCFTNVYADSFVIRG | 54->71 | 1.14 | 60 | |
| 5 | FTGCVIAW | 96->103 | 1.126 | 99 | |
| Mu | 1 | TYGVGYQPYRVVVLSFELLHAPATV | 167->191 | 1.214 | 177 |
| 2 | ISNCVADYSVLYNSA | 25->39 | 1.183 | 31 | |
| 3 | GFNCYFPLQSYGFQ | 152->165 | 1.144 | 158 | |
| 4 | LNDLCFTNVYADSFVIRG | 54->71 | 1.14 | 60 | |
| 5 | FTGCVIAW | 96->103 | 1.126 | 99 |
Fig. 4.
Amino acid sequence alignment of Omicron and SARS-CoV-2 RBD. The top five predicted antigenic epitope sites are highlighted.
Fig. 5.
The sites of antigenic epitopes in SARS-CoV-2 and Omicron RBD. The sites are numbered 1–5 according to their EMBOSS antigenic analysis score.
Multiple epitopes were revealed after applying neutralizing antibodies m396 and 80R [18]. Most of the epitopes were found in SARS-CoV-2 and Omicron RBD conserved areas. The novel Omicron RBD mutations S42F, G113S, Q160R, G163S, Q165R, and Y172H, however, interrupted the resolved epitopes. In a recent study, despite many mutations in Omicron, only one low-prevalence CD8+ T-cell epitope (T95I) from the spike protein contained a single amino acid change in this population [26]. There were no further mutations linked to the previously discovered epitopes. These findings show that almost all anti-SARS-CoV-2 CD8+ T-cell responses should be able to recognize Omicron and that SARS-CoV-2 has not yet generated significant T-cell escape mutations [26]. This result suggests that Omicron RBD may not have a role in immune response modulation.
It was recently found that despite its greater infectivity and transmissibility, the Omicron variant has evolved to have higher antigenicity and decreased pathogenicity in humans. This was determined by the low number of nonself stretches of short constituent sequences (NSC) in both the Omicron and Delta proteomes; however, the number of NSCs was much higher in the RBD of the Omicron spike sequence than in the Delta spike sequence [27]. These NSCs assist in identifying and escaping the immune response. The changes in these amino acids were also part of the altered epitope content in the emerging Omicron variant.
N440K and T478K were two mutations (both to lysine) from polar to positively-charged and larger side-chain residues. These types of modifications may affect the binding affinity between an RBD and an antibody, either by altering protein surface charges or limiting tighter antibody connections [24].
3.5. MD simulations
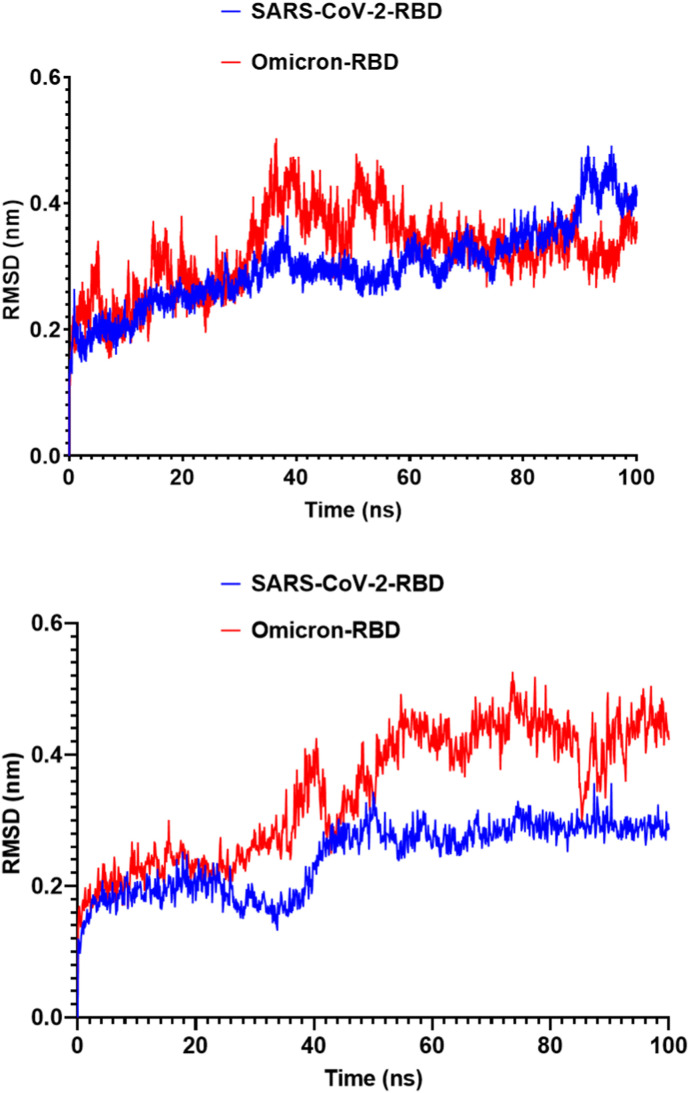
To compare the results, we used two software packages to run MD simulations. The SARS-CoV-2 structure appeared to be more stable than the Omicron RBD after the 100-ns simulation, as evidenced by lower RMSD results using both software packages (Fig. 6 ). In the Omicron RBD, the average RMSD was 0.33 nm, compared to only 0.27 nm in the SARS-CoV-2 RBD.
Fig. 6.
The RMSD of SARS-CoV-2 and Omicron RBD during the 100-ns MD simulation. A) SARS-CoV-2 and Omicron RBD RMSD implemented by GROMACS software analysis. B) SARS-CoV-2 and Omicron RBD RMSD values implemented by Desmond software analysis.
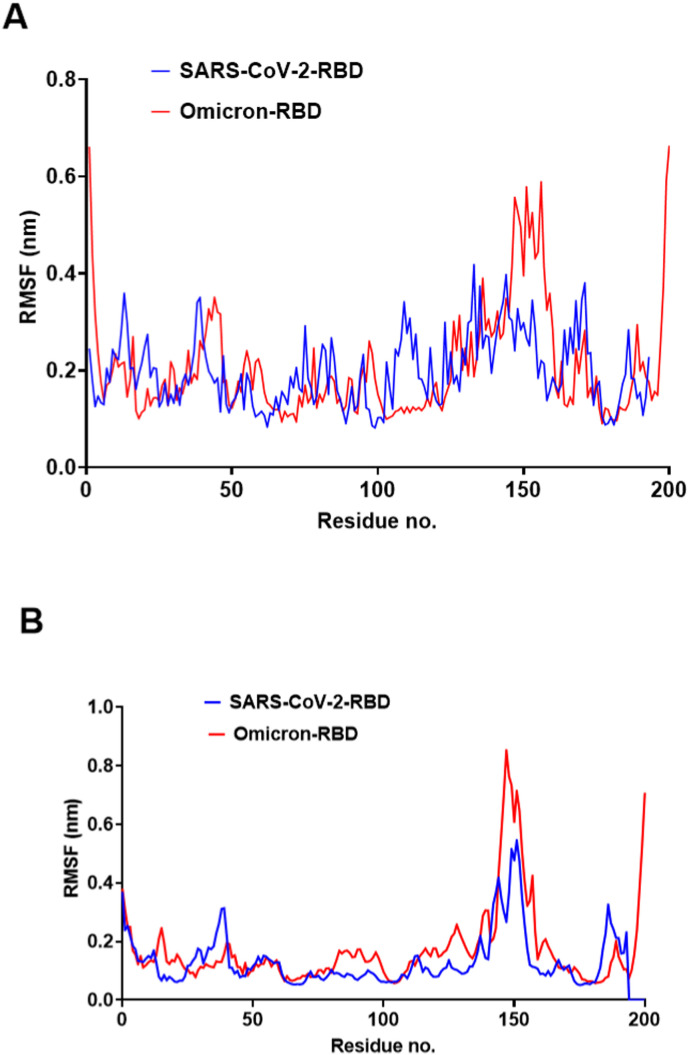
The RMSF profile was nearly identical in SARS-CoV-2 and Omicron RBD, with a notable rise in RMSF values for residues 32–42, which coincides with three novel mutations in the Omicron RBD. These RMSF variations can be overlooked as they occur in a flexible loop that does not interface with the host receptors. The most prominent increase in RMSF was noticed in the residue range 140–160 (Fig. 7 ).
Fig. 7.
The RMSF of SARS-CoV-2 and Omicron RBDs during the 100-ns MD simulation. A) SARS-CoV-2 and Omicron RBD RMSF values implemented by GROMACS software analysis. B) SARS-CoV-2 and Omicron RBD RMSF values implemented by Desmond software analysis.
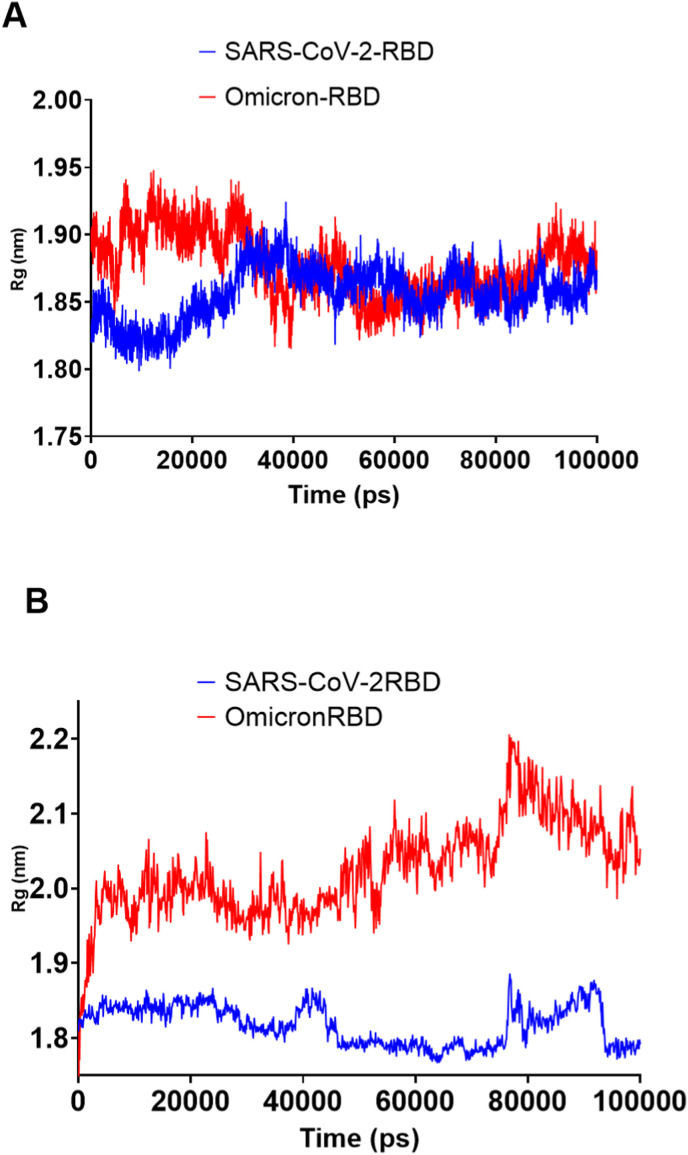
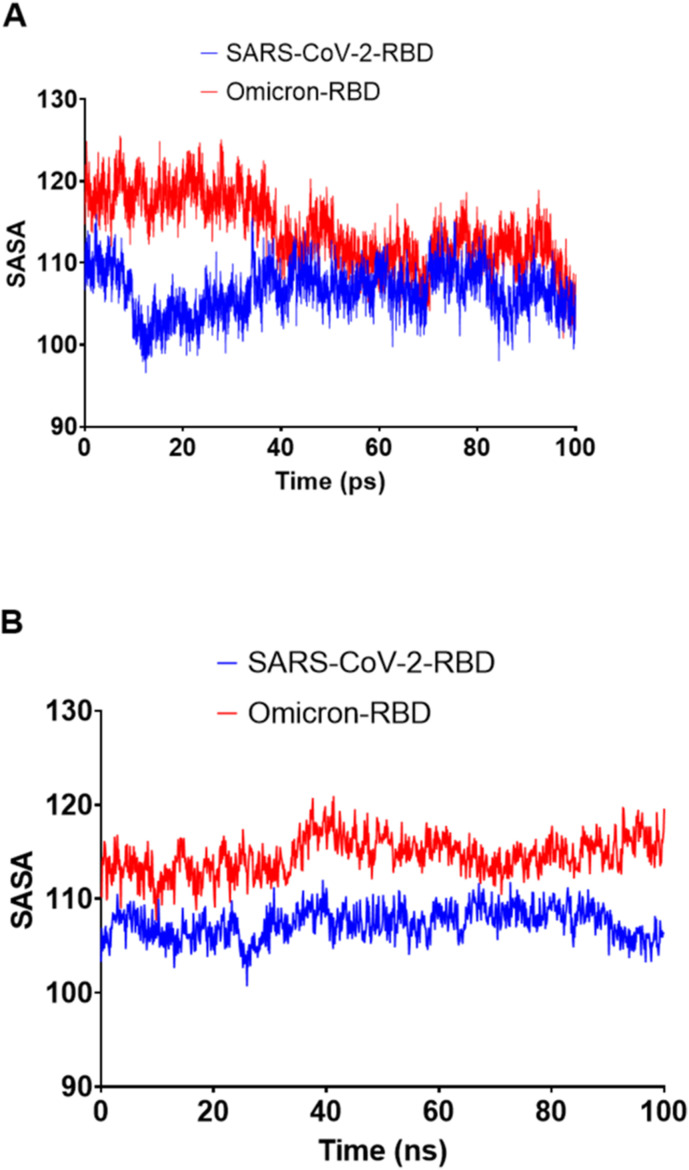
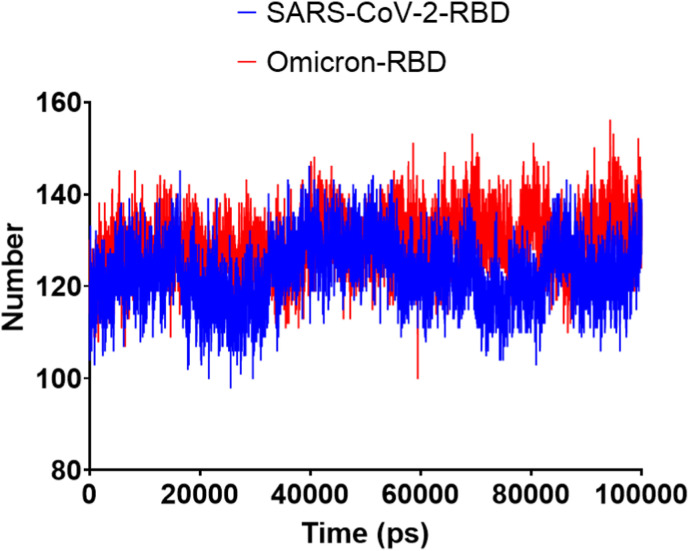
The low radius of gyration (Rg) suggested the general compactness of both SARS-CoV-2 and Omicron RBD (Fig. 8 ). Surprisingly, the solvent-accessible surface area (SASA) was higher for the Omicron RBD than the SARS-CoV-2 RBD (Fig. 9 ). As SASA alterations can be interpreted as resulting from conformational changes caused by protein mutations [28], the mutated residues in the Omicron RBD had resulted in increased SASA. The hydrogen bond analysis (Fig. 10 ) revealed nearly identical profiles, except that the Omicron RBD had an average of 192 hydrogen bonds, six more than the SARS-CoV-2 RBD.
Fig. 8.
The genetic correlation (Rg) of SARS-CoV-2 and Omicron RBD during the 100-ns MD simulation. A) SARS-CoV-2 and Omicron RBD Rg values implemented by GROMACS software analysis. B) SARS-CoV-2 and Omicron RBD Rg values implemented by Desmond software analysis.
Fig. 9.
The solvent-accessible surface areas (SASAs) of SARS-CoV-2 and Omicron RBDs during the 100-ns MD simulation. A) SARS-CoV-2 and Omicron RBD SASAs implemented by GROMACS software analysis. B) SARS-CoV-2 and omicron RBD SASAs implemented by Desmond software analysis.
Fig. 10.
The hydrogen bonds of SARS-CoV-2 and Omicron RBDs during the 100-ns MD simulation calculated by GROMACS software.
4. Conclusions
It is critical to explore the Omicron RBD molecular structure changes resulting from RBD mutations. According to the pairwise comparison matrix, the Omicron variant had the highest number of mutations from SARS-CoV-2 and is the first variant in a new monophyletic class. The creation of a new class is supported by 12 novel mutations identified in the Omicron RBD. These mutations extended the length of the conserved antigenic epitopes and affected the protein behavior during MD simulation, especially SASA and RMSF. These findings can assist in determining the link between the identified Omicron RBD mutations and RBD structural alterations, changes in potential antigenic composition, and stability of the variant RBD.
Data availability statement
All data are within the manuscript.
Funding statement
This project is funded by the Deanship of Scientific Research at King Faisal University under Research Groups track (Grant No. 1811016).
Conflict of interest disclosure
None.
Ethics approval statement
Not apply.
Patient consent statement
Not apply.
Permission to reproduce material from other sources
Not apply.
Acknowledgment
The authors acknowledge the financial support from the Deanship of Scientific Research, King Faisal University, under Research Groups track (Grant No. 1811016).
References
- 1.Grint D.J., Wing K., Houlihan C., Gibbs H.P., Evans S.J.W., Williamson E., McDonald H.I., Bhaskaran K., Evans D., Walker A.J., Hickman G., Nightingale E., Schultze A., Rentsch C.T., Bates C., Cockburn J., Curtis H.J., Morton C.E., Bacon S., Davy S., Wong A.Y.S., Mehrkar A., Tomlinson L., Douglas I.J., Mathur R., MacKenna B., Ingelsby P., Croker R., Parry J., Hester F., Harper S., DeVito N.J., Hulme W., Tazare J., Smeeth L., Goldacre B., Eggo R.M. Severity of SARS-CoV-2 alpha variant (B.1.1.7) in England. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N. 2020. Emergence and Rapid Spread of a New Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-2) Lineage with Multiple Spike Mutations in South Africa, MedRxiv. [Google Scholar]
- 3.Faria N.R., Claro I.M., Candido D., Franco L.M., Andrade P.S., Coletti T.M., Silva C.A., Sales F.C., Manuli E.R., Aguiar R.S. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2021;372:815–821. [Google Scholar]
- 4.Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in southern California. JAMA. 2021;325:1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West A.P., Jr., Barnes C.O., Yang Z., Bjorkman P.J. BioRxiv; 2021. SARS-CoV-2 Lineage B. 1.526 Emerging in the New York Region Detected by Software Utility Created to Query the Spike Mutational Landscape. [Google Scholar]
- 6.Thangaraj J.W.V., Yadav P., Kumar C.G., Shete A., Nyayanit D.A., Rani D.S., Kumar A., Kumar M.S., Sabarinathan R., Kumar V.S. Predominance of delta variant among the COVID-19 vaccinated and unvaccinated individuals, India, May 2021. J. Infect. 2021;84:94–118. doi: 10.1016/j.jinf.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pulliam J.R., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the omicron variant in South Africa. Science. 2022;376 doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C., Boorla V.S., Banerjee D., Chowdhury R., Cavener V.S., Nissly R.H., Gontu A., Boyle N.R., Vandegrift K., Nair M.S., Kuchipudi S.V., Maranas C.D. Vol. 118. 2021. Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike RBD and human ACE2. (Proceedings of the National Academy of Sciences of the United States of America). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao S.J., Guo H., Luo G. Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert! J. Med. Virol. 2022;94:1255. doi: 10.1002/jmv.27491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amoutzias G.D., Nikolaidis M., Tryfonopoulou E., Chlichlia K., Markoulatos P., Oliver S.G. The remarkable evolutionary plasticity of coronaviruses by mutation and recombination: insights for the COVID-19 pandemic and the future evolutionary paths of SARS-CoV-2. Viruses. 2022:14. doi: 10.3390/v14010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., Zatta F., Kaiser H., Noack J., Farhat N., Czudnochowski N., Havenar-Daughton C., Sprouse K.R., Dillen J.R., Powell A.E., Chen A., Maher C., Yin L., Sun D., Soriaga L., Bassi J., Silacci-Fregni C., Gustafsson C., Franko N.M., Logue J., Iqbal N.T., Mazzitelli I., Geffner J., Grifantini R., Chu H., Gori A., Riva A., Giannini O., Ceschi A., Ferrari P., Cippà P.E., Franzetti-Pellanda A., Garzoni C., Halfmann P.J., Kawaoka Y., Hebner C., Purcell L.A., Piccoli L., Pizzuto M.S., Walls A.C., Diamond M.S., Telenti A., Virgin H.W., Lanzavecchia A., Snell G., Veesler D., Corti D. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starr T.N., Greaney A.J., Hilton S.K., Crawford K.H.D., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data–from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romesburg C. 2004. Cluster Analysis for Researchers, Lulu. Com. [Google Scholar]
- 16.Kolaskar A.S., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 17.Teng S., Sobitan A., Rhoades R., Liu D., Tang Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief Bioinform. 2021;22:1239–1253. doi: 10.1093/bib/bbaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 19.Kandeel M., Albusadah K., Alaydaa S.H., Albokhadaim I., Alhojaily S., Marzok M. Insulin from human, camel and farm animals: comparative bioinformatics and molecular dynamics studies. Pakistan Vet. J. 2022;42:65–73. [Google Scholar]
- 20.Kandeel M., Yamamoto M., Park B.K., Al-Taher A., Watanabe A., Gohda J., Kawaguchi Y., Oh-Hashi K., Kwon H.J., Inoue J.I. Discovery of new potent anti-MERS CoV fusion inhibitors. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.685161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandeel Elsayed Mahmoud, Kwon Hyung-Joo, Al-Nazawi Mohammed, Al-Taher Abdulla. United States Patent Office; 2021. MERS-CoV Inhibitor Peptides. Patent no. US10975126B1. [Google Scholar]
- 22.Kandeel Mahmoud, Park Byoung Kwon, Morsy Mohamed A, Venugopala Katharigatta N, Oh-hashi Kentaro, Al-Nazawi Mohammed, Kwon Hyung-Joo. Virtual screening and inhibition of Middle East respiratory syndrome coronavirus replication by targeting papain-like protease. Dr. Sulaiman Al Habib Med. J. 2021;3(4):179–187. [Google Scholar]
- 23.Kandeel M., Mohamed M.E.M., Abd El-Lateef H.M., Venugopala K.N., El-Beltagi H.S. Omicron variant genome evolution and phylogenetics. J. Med. Virol. 2022;94(4):1627–1632. doi: 10.1002/jmv.27515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford C.T., Jacob Machado D., Janies D.A. Predictions of the SARS-CoV-2 omicron variant (B.1.1.529) spike protein receptor-binding domain structure and neutralizing antibody interactions. Front. Virol. 2022;2 [Google Scholar]
- 25.Kupferschmidt K. Where did 'weird' Omicron come from? Science (New York, N.Y.) 2021;374:1179. doi: 10.1126/science.acx9738. [DOI] [PubMed] [Google Scholar]
- 26.Redd A.D., Nardin A., Kared H., Bloch E.M., Abel B., Pekosz A., Laeyendecker O., Fehlings M., Quinn T.C., Tobian A.A. 2021. Minimal Cross-Over between Mutations Associated with Omicron Variant of SARS-CoV-2 and CD8+ T Cell Epitopes Identified in COVID-19 Convalescent Individuals, bioRxiv : the Preprint Server for Biology. 2012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otaki J.M., Nakasone W., Nakamura M. Nonself mutations in the spike protein suggest an increase in the antigenicity and a decrease in the virulence of the omicron variant of SARS-CoV-2. COVID. 2022;2:407–418. [Google Scholar]
- 28.Marsh J.A., Teichmann S.A. Relative solvent accessible surface area predicts protein conformational changes upon binding. Structure. 2011;19:859–867. doi: 10.1016/j.str.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are within the manuscript.