Graphical abstract
Keywords: COVID-19, Cathepsin L, Deep learning, Drug prediction, Daptomycin
Highlights
-
•
Cathepsin L(CTSL) is a promising therapeutic target for COVID-19.
-
•
A new deep learning model was used to predict CTSL inhibitor based on structure.
-
•
5 molecules for inhibiting CTSL and treating COVID-19 at nmol level were identified.
-
•
Daptomycin can distinctly inhibit CTSL and has potential for COVID-19 treatment.
Abstract
Cathepsin L (CTSL), a cysteine protease that can cleave and activate the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein, could be a promising therapeutic target for coronavirus disease 2019 (COVID-19). However, there is still no clinically available CTSL inhibitor that can be used. Here, we applied Chemprop, a newly trained directed-message passing deep neural network approach, to identify small molecules and FDA-approved drugs that can block CTSL activity to expand the discovery of CTSL inhibitors for drug development and repurposing for COVID-19. We found 5 molecules (Mg-132, Z-FA-FMK, leupeptin hemisulfate, Mg-101 and calpeptin) that were able to significantly inhibit the activity of CTSL in the nanomolar range and inhibit the infection of both pseudotype and live SARS-CoV-2. Notably, we discovered that daptomycin, an FDA-approved antibiotic, has a prominent CTSL inhibitory effect and can inhibit SARS-CoV-2 pseudovirus infection. Further, molecular docking calculation showed stable and robust binding of these compounds with CTSL. In conclusion, this study suggested for the first time that Chemprop is ideally suited to predict additional inhibitors of enzymes and revealed the noteworthy strategy for screening novel molecules and drugs for the treatment of COVID-19 and other diseases with unmet needs.
1. Introduction
The emergence of coronavirus disease 2019 (COVID-19), caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has created a massive crisis for global public health. Several effective vaccines have now been put into use[1], [2], [3], but new variants continue to emerge, and in particular, the delta (B.1.617.2) variant of SARS-CoV-2 has spread rapidly across continents, and the omicron variant, a newly emerged SARS-CoV-2 variant, may be more transmissible than all the previous variants and partly resistant to existing vaccines[4], [5], [6], [7]. On the other hand, vaccines are not as convenient and acceptable to people as drugs, and millions of immunocompromised persons are unlikely to respond robustly to vaccination[8]. It remains critical to rapidly develop therapeutic drugs for COVID-19. Remdesivir is the only approved antiviral drug for COVID-19 thus far, which targets of RNA-dependent RNA polymerase (RdRp). This turns out to be not effective as announced by the World Health Organization (WHO) and European Medical Association[9], [10], [11]. Therefore, broadening the spectrum of therapeutic targets is important. Recent studies have attempted to develop antiviral drugs by focusing primarily on the host cell receptor angiotensin-converting enzyme 2 (ACE2) and the surface protease transmembrane protease serine 2 (TMPRSS2), the spike (S) protein, the main protease (Mpro) and the papain-like protease (PLpro), and the results of some of these studies have already been reported, but their efficacy is still under evaluation[8], [11].
Cathepsin L (CTSL), an endosomal cysteine protease, is another host cell protease that has an essential role in coronavirus infection[12]. In our recent study, we revealed that the circulating level of CTSL was elevated after SARS-CoV-2 infection and was positively correlated with disease course and severity[13]. We confirmed that knockdown CTSL by siRNAs led to a significant dose-dependent reduction in SARS-CoV-2 pseudovirus cell entry in Huh7 cells, while overexpression of CTSL using plasmids markedly increased pseudovirus cell entry in a dose-dependent manner[13]. Furthermore, E64D, one of the well-known CTSL inhibitors, significantly inhibited CTSL activity and prevented SARS-CoV-2 pseudovirus infection both in human cells and in humanized mice (hACE2 transgenic mice)[13]. In the current study, we revealed that the lentivirus expressing human CTSL (Lv-CTSL) significantly upregulated CTSL protein level and enhanced SARS-CoV-2 pseudovirus infection in a dose-dependent manner, while the lentivirus expressing shRNA against human CTSL (Lv-shCTSL) significantly reduced CTSL protein level and attenuated pseudovirus infection in a dose-dependent manner both in Huh7 cells and A549 cells (Supplementary Fig. 1A-F). Of note, Nie X et al. found that CTSL expression was significantly upregulated in multiple internal organs of COVID-19 patients and that this upregulation of CTSL expression might contribute to excessive inflammatory activity[14]. We further found that SARS-CoV-2 infection promotes CTSL gene transcription and enzyme activity. Upregulation of CTSL expression, in turn, enhances SARS-CoV-2 infection[13]. Obviously, CTSL is thus a promising therapeutic target for COVID-19. Unfortunately, there is no currently available drug that can specifically inhibit CTSL[15].
Computational drug repurposing screening is an effective approach that can aid in determining new indications for existing drugs[16]. Various computational techniques and software programs are typically used in drug repurposing. Chemprop, a newly trained directed-message passing neural network (MPNN) that was used to discover promising new antibiotics by predicting the likelihood that a molecule would inhibit the growth of E. coli[17], shows strong molecular property prediction capabilities across a range of properties[18]. As a new deep learning approach for drug repurposing, Chemprop can automatically map molecules into continuous vectors to predict their properties, which provides better preservation of molecular information than other methods[18]. In this study, we combined in silico predictions and empirical investigations to discover new CTSL inhibitors. In our efforts to identify small molecules to expand the discovery of CTSL inhibitors for drug development and FDA-approved drugs that can inhibit CTSL for drug repurposing for COVID-19, the Selleck bioactive compound libraries, the ZINC in vitro compound library and the FDA-approved drug library were screened for their ability to inhibit CTSL. We found a number of compounds and several drugs displaying activity inhibition against CTSL, and the most active ones were selected for further investigation of their antiviral activity against SARS-CoV-2.
2. Materials and Methods
2.1. Model training and predictions
We translated every molecule from its simplified molecular input line entry specification (SMILES) string into a molecular graph structure, with nodes representing atoms and edges representing chemical bonds. The feature vectors of nodes and edges were initialized using their corresponding computable features, such as atomic mass and bond type, respectively[18]. We then trained a graph neural network to predict whether a molecule inhibits the activity of CTSL in a supervised fashion[19].
In particular, we trained a bond-level directed-message passing neural network to learn to encode the molecular graph structure into a hidden vector representation[17]. The initial hidden state for each directed bond is the concatenation of the bond vector and the corresponding atom vector encoded by a fully connected layer with nonlinear activation. We aggregated all directed bond information on each step of message passing by summing the messages from neighboring bonds and then concatenating the sum with its message. Another fully connected layer will then encode this hidden vector, learning to understand the local chemistry. This message passing step was repeated a fixed number of times, and all directed bonds' learned hidden states were aggregated and summed to produce a single vector encoding the whole molecule. Finally, a fully connected classifier decoded this learned vector to predict whether the molecule is an inhibitor of CTSL as a binary classification task.
2.2. Deduplication
We applied a data deduplication algorithm to ensure our training set and testing set are mutually exclusive. In detail, SMILES is a specification to represent a chemical structure in strings. There are many possible SMILES strings to describe a chemical structure in most cases. In response to this situation, we utilized the canonicalization algorithm implemented in RDKit[20] to generate the canonical SMILES string for each molecule during the data collection. These canonical SMILES strings are unique for each chemical structure, and therefore, we can remove test molecules that exist in the training set.
2.3. T-distributed stochastic neighbor embedding (t-SNE)
We projected the learned representation of the molecules onto a 2D space using t-SNE for further investigation. T-SNE is a statistical method that first constructs a probability distribution over pairs of high-dimensional data in which similar data are assigned a higher probability and vice versa. Then, it defines another similarity probability distribution in a low-dimensional map and attempts to minimize the Kullback-Leibler divergence between the joint probabilities of the two distributions. Specifically, we used the implementation of scikit-learn with the default values for all parameters to perform the calculation.
2.4. Hyperparameter optimization
We applied the Bayesian hyperparameter optimization scheme to improve our model because hyperparameters are often crucial for the performance of neural network models. Bayesian optimization is an efficient and effective technique based on the Bayes theorem. By building a probabilistic model, all the previous trials will become prior knowledge to effectively identify a better set of hyperparameters for our model. We ran 20 iterations of Bayesian optimization, searching for the number of message-passing steps, neural network hidden size, number of feed-forward classifier layers, and dropout probability. Our search space and the best parameters are shown in Supplementary Table 1.
2.5. Analysis of CTSL activity in a cell-free system
The inhibition of CTSL by small molecules or drugs was evaluated in a cell-free system using a commercially available kit (Abcam, Cat. No. ab65306) according to the manufacturer. In detail, 5 μl of CTSL protein (25 mg/L), which was prepared from human liver tissue (Sigma-Aldrich, Cat. No. C6854) was used as the enzyme in this cell-free enzymatic reaction system. For the preliminary screening, all the molecules and drugs were prepared at a concentration of 5 mM. 2 μl of these 5 mM molecules or drugs was added to the set 100 μl system (5 μl of CTSL protein (25 mg/L) + 90 μl of CL buffer + 2 μl of CTSL inhibitor + 1 μl of DTT (1 mM) + 2 μl of CL substrate Ac-FR-AFC (10 mM)) to achieve a working concentration of 100 μM. For further confirmation and determination of the half maximal inhibitory concentration, all the molecules and drugs were prepared with a range of concentrations (5 mM, 1 mM, 0.2 mM, 0.04 mM, 4 μM and 0.4 μM) to achieve a working concentration of 100 μM, 20 μM, 4 μM, 0.8 μM 80 nM and 8 nM, respectively, and tested as described above. The equivalent amount of solvent was used as a control.
2.6. Cell culture and reagents
The human hepatoma cell line Huh7 and the human lung epithelial carcinoma cell line A549 were maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA), 100 units/ml penicillin and 100 mg/ml streptomycin (Thermo Fisher Scientific). The human Calu-3 lung adenocarcinoma cell line was cultured in minimum essential medium (Eagle) with 2 mM L-glutamine and Earle’s BSS adjusted to contain 1.5 g/l sodium bicarbonate, 0.1 mM non-essential amino acids and 1.0 mM sodium pyruvate and 10% FBS. All the cells were maintained at 37 °C in a humidified atmosphere containing 95% air and 5% CO2. Mg-132 (Cat. No. S2619), Z-FA-FMK (Cat. No. S7391), leupeptin hemisulfate (Cat. No. S7380), Mg-101 (Cat. No. S7386), calpeptin (Cat. No. S7396), daptomycin (Cat. No. S1373), beta-lapachone (Cat. No. S7261) and other molecules and drugs were purchased from Selleck (Selleckchem, Houston, TX, USA). The adenovirus expressing human ACE2 (Ad-ACE2, Cat. No. AD-h-ACE2-3flag, Pubmed ID: NM_021804) and the control adenovirus (Ad-Con) were purchased from Vigenebio Ltd (China). The lentivirus expressing human CTSL (Lv-CTSL) and the control lentivirus (Lv-Con), the lentivirus expressing shRNA against human CTSL (Lv-shCTSL) and the control lentivirus (Lv-Scramble) were purchased from XIEBHC BIO (China). The plasmid expressing human TMPRSS2 (pCDH-CMV-MCS-EF1-Puro-TMPRSS2, pCDH-TMPRSS2, NM_005656.3) and the control plasmid (pCDH-CMV-MCS-EF1-Puro-Con, pCDH-Con) were purchased from Vigenebio Ltd (China).
2.7. Pseudovirus
The SARS-CoV-2 pseudovirus used in the current study was purchased from Genomeditech (Shanghai, China). It was generated with the incorporation of SARS-CoV-2 spike protein (SARS-2-S) into a HIV-based pseudovirus system and have been widely used in previous studies[21], [22], [23], [24]. The pseudovirus expresses the SARS-CoV-2 S protein on the surface and contains a defective HIV-1 genome encoding firefly luciferase and a green fluorescent protein as a reporter[21]. Thus, when the pseudovirus infects the host cells, it can express luciferase but cannot replicate or assemble into new viruses. The SARS-CoV-2B.1.351 (Beta) variant pseudovirus containing a luciferase reporter were produced according to our previous study[25]. Therefore, the luciferase activity was used as indicators of pseudovirus infection in the current study.
2.8. Luciferase assay
Huh7 cells were seeded into a 96-well plate at a cell density of 0.5×104 per well and allowed to adhere until the cells were approximately 70% confluent, followed by treatment with different concentrations of drugs or the equivalent amount of solvent for 1 h. In detail, the concentrations of different drugs were as follow: Mg-132 (10 nM, 0.1 μM, 0.5 μM, 1 μM and 5 μM), Z-FA-FMK (8 nM, 80 nM, 4 μM, 20 μM and 100 μM), leupeptin hemisulfate (0.8 μM, 4 μM, 20 μM, 100 μM and 400 μM), Mg-101 (80 nM, 0.8 μM, 4 μM, 20 μM and 100 μM), calpeptin (312.5 nM, 3.125 μM, 12.5 μM, 25 μM and 50 μM), daptomycin (4 μM, 20 μM, 100 μM, 200 μM and 400 μM), and beta-lapachone (8 nM, 80 nM, 0.8 μM and 4 μM). Then, the cells were infected with SARS-CoV-2 pseudovirus (1.3x104 TCID50/ml) in a 5% CO2 environment at 37 °C for 24 h before firefly luciferase activity analysis. The activity of firefly luciferase was measured in cell lysates using luciferase substrate (PerkinElmer, BRITELITE PLUS 100 ml KIT, Cat. No. 6066761) following the manufacturer’s instructions. Briefly, for 96-well plates, the culture supernatant was aspirated gently to leave 100 μl in each well; then, 100 μl of luciferase substrate was added to each well. Two minutes after incubation at 37 °C, 150 μl of lysate was aspirated to a clean 1.5 ml sterile EP tube to rapidly measure the firefly luciferase activity for each well using a luminometer (Turner BioSystems) as described previously[26]. For A549 cells, the cells were treated with 25moi Ad-ACE2 when seeded, then the luciferase assay performed as Huh7 cells as stated above.
2.9. Effect of drug treatment on live SARS-CoV-2 infection
The live SARS-CoV-2 used in this study was described previously[27]. Huh7 cells were seeded into a 96-well plate at a cell density of 2×104 per well and allowed to adhere until the cells were approximately 70% confluent, followed by treatment with different concentrations of drugs or an equivalent amount of solvent for 1 h. Then, the cells were infected with live SARS-CoV-2 at a multiplicity of infection (MOI) of 0.5 at 37 °C for 1 h, followed by changing to fresh medium with the indicated concentrations of drugs. The detection of infected cells was performed 48 h later by using an immunofluorescence assay (IFA) as described previously[27], [28]. In brief, the infection and replication of the virus were determined by detecting the nucleoprotein (N) of SARS-CoV-2 using an N-specific polyclonal antibody (Sino Biological, China), and followed by Alexa Fluor® 488-labelled donkey anti-rabbit secondary antibody (Jackson). All the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma, USA) for nuclear visualization. The average infection ratio was analyzed by a Celigo® Image Cytometer. In brief, DAPI-stained cells were designated total cells, and N-protein-positive stained cells were designated infected cells. The average infection ratio was quantified by N-protein-positive stained cells/DAPI-stained cells, and calculated inhibition rate of different dosages of drugs compared with virus control. All SARS-CoV-2 infection experiments were performed in a biosafety level-3 laboratory.
2.10. Cell viability assay
The effects of Mg-132, Z-FA-FMK, leupeptin hemisulfate, Mg-101, calpeptin, daptomycin and beta-lapachone on cell viability were measured by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Huh7 cells were seeded into a 96-well plate at a cell density of 0.5×104 per well and allowed to adhere until the cells were approximately 70% confluent, followed by treatment with different concentrations of drugs or the equivalent amount of solvent for 24 h. The concentrations of different drugs were as follow: Mg-132 μM (10 nM, 0.1 μM, 0.2 μM, 0.5 μM, 1 μM, 5 μM, 25 μM, 50 μM, 100 μM and 200 μM), Z-FA-FMK (8 nM, 80 nM, 4 μM, 20 μM, 40 μM, 100 μM, 200 μM, 300 μM and 400 μM), leupeptin hemisulfate (0.8 μM, 4 μM, 20 μM, 100 μM, 200 μM, 500 μM, 2.5 mM and 5 mM), Mg-101 (8 nM, 80 nM, 0.8 μM, 4 μM, 20 μM, 100 μM and 200 μM), calpeptin (312.5 nM, 3.125 μM, 6.25 μM 12.5 μM, 25 μM, 50 μM, 100 μM, 200 μM, 400 μM and 800 μM), daptomycin (0.8 μM, 4 μM, 20 μM, 100 μM, 200 μM, 500 μM, 2.5 mM and 5 mM), and beta-lapachone (8 nM, 80 nM, 0.8 μM, 4 μM, 20 μM and 100 μM). Cells without any treatments were used as the blank control. After treatments, MTT was added to the culture medium to a final concentration of 0.5 mg/ml, and then the cells were incubated for 4 h at 37 °C in an incubator. After removing the culture medium, the cells were lysed by gently rotating in 200 μl of DMSO for 10 min in darkness at room temperature. The absorbance at 570 nm was measured using an automatic plate reader. The average absorbance reflected cell viability, with the data normalized to the blank control group. Experiments were performed in quintuplicate and repeated at least three times.
2.11. Antibodies
Anti-ACE2 were purchased from Abcam (ab108209), Anti-CTSL were purchased from R&D(AF952-SP), Anti-β-actin were purchased from sigma(A5316), Anti-TMPRSS2 were purchased from Abcam (ab109131).
2.12. Real-time PCR assay
Total RNA was extracted from cultured cells using the RNAprep pure Cell/Bacteria Kit (TIANGEN BIOTECH Corp., Beijing, China), and the reverse transcription was performed with using RevertAid First Strand cDNA Synthesis Kit (Fermentas K1622) according to the manufacturer's instruction. Real-time PCR was performed on a Stratagene Mx3000P real-time quantitative PCR system (Agilent Technologies) using β-actin as the housekeeping gene as described previously[29]. The primer sequences for the quantitative PCR assays are as follow, human ACE2 forward: 5′-CGAAGCCGAAGACCTGTTCTA-3′, human ACE2 reverse: 5′-GGGCAAGTGTGGACTGTTCC-3′. human β-actin forward: 5′-CTACAATGAGCTGCGTGTGG-3′, human β-actin reverse: 5′-CCAGAGGCGTACAGGGATAG-3′. The primer pair 1[30] for TMPRSS2 mRNA whose products showed as TMPRSS21 in supplementary fig. 5A, forward: 5′-TTTGAACTCAGGGTCACCAC-3′, reverse: 5′- CCTCTGAGATGAGTACACCTG-3′. The primer pair 2[31] for TMPRSS2 mRNA whose products showed as TMPRSS22 in supplementary fig. 5A, forward: 5′- CTCTCCCTAACCCCTTGTCC-3′, reverse: 5′- AGAGGTGACAGCTCCATGCT-3′. The primer pair b[32] for TMPRSS2 mRNA whose products showed as TMPRSS2b in supplementary fig. 5A, forward: 5′- CACTGTGCATCACCTTGACC-3′, reverse: 5′- ACACACCGATTCTCGTCCTC-3′.
2.13. Receptor protein selection and preparation
We use human CTSL X-ray structures co-crystallized with a covalent inhibitor from Protein Data Bank (PDB) database (PDB code 5MQY, resolution 1.13 Å) to perform molecule docking[33]. The Schrödinger protein preparation wizard was used to prepare each crystal structure[34]. Hydrogen atoms were added and generated possible metal binding states. Hydrogen bond sampling with adjustment of active site, water molecule orientations was performed using PROPKA at pH 7.4. Delete artifacts from the crystal structure and retain water within 5 Å of the binding site. Finally, the protein-ligand complexes were subjected to geometry refinements using the OPLS3 force field in restrained minimizations[35].
2.14. Ligand preparation
The chemical structures of Z-FA-FMK, Calpeptin, Mg-101(ALLN), Mg-132, Leupeptin Hemisulfate and Daptomycin were obtained from PubChem database. These ligands were subjected to LigPrep (ligand preparation) module of Schrödinger suite. A robust pKa prediction tool, Epik, was used to generate possible ionization and tautomeric states at pH 7.4 and each ligand could generate maximum 32 stereoisomers. The ligands were then minimized by the OPLS3e force field[36], [37].
2.15. Noncovalent docking
Determination of noncovalent interactions of ligands with CTSL was done through extra precision (XP) modules. The 5MQY receptor grid was generated using Receptor Grid Generation module of Schrödinger suite centroid of the co-crystallized inhibitor. The ligands could be flexible and their nitrogen inversions (pyramidal nitrogen atoms) and ring conformations were sampled during the docking process. Only amides were penalized for nonplanar conformations as a default setting. Lastly, Epik state penalties were applied to the docking scores. This is for adopting higher energy states for ligands that were ionized or tautomerized in the preparation stage. The output poses per ligand was set to 1 best pose, while post-docking minimization was also performed.
2.16. Covalent docking
To carry out the covalent docking we used docking modules available in Schrödinger suite: CovDock in Schrödinger 2021–2 version. CovDock considers custom reactions that are present in a list of possible covalent reactions (implemented in the software) using SMARTS pattern, so it is possible to automatically recognize the reactive residue and the portion of the ligand that are involved in the reaction. The prepared ligands were selected from a project table, while a reactive receptor residue (Cys-25) was selected from a 5MQY protein on the workspace. The customized covalent docking algorithm was then selected as the reaction type. No constraints were imposed on the ligand for docking and pose prediction mode was selected. The total energy 2.5 kcal/mol was set as the cutoff to retain poses for further refinement by default, while the maximum number of poses to retain for further refinement, was 200. The output poses per ligand reaction site was set to 1 best pose.
2.17. Statistical analysis
All values are depicted as the mean ± SEM. Statistical analysis was performed using GraphPad Prism software, version 8.0.1. Different treatments were compared with Student's t test and one-way ANOVA for two-group and multiple-group comparisons, respectively. A log(inhibitor) vs. normalized response -- variable slope test was used for IC50, CC50 and EC50 determination. P < 0.05 was considered significant. Representative results from at least three independent experiments are shown unless otherwise stated.
3. Results
3.1. Initial training dataset selection
To identify potential CTSL inhibitors, we first used Chemprop MPNNs to build a robust model that can predict CTSL inhibitors. By searching the publicly available PubChem database with human CTSL (Gene ID: 1514), we obtained 2067 active molecules which can inhibit CTSL and 58,070 negative molecules which cannot inhibit CTSL (https://pubchem.ncbi.nlm.nih.gov/gene/1514#section = Chemicals-and-Bioactivities). Then, we applied a web crawler to collect simplified molecular input line entry specification (SMILES) strings from the PubChem database and preprocessed data by reformatting it and removing compounds with duplicate SMILES strings. Finally, we obtained 58,997 molecules as the initial training dataset, with 1558 compounds (2.64%) showing inhibitory activity against CTSL (Supplementary Table 2).
3.2. Initial model training and predicting potential CTSL inhibitors from bioactive compound libraries
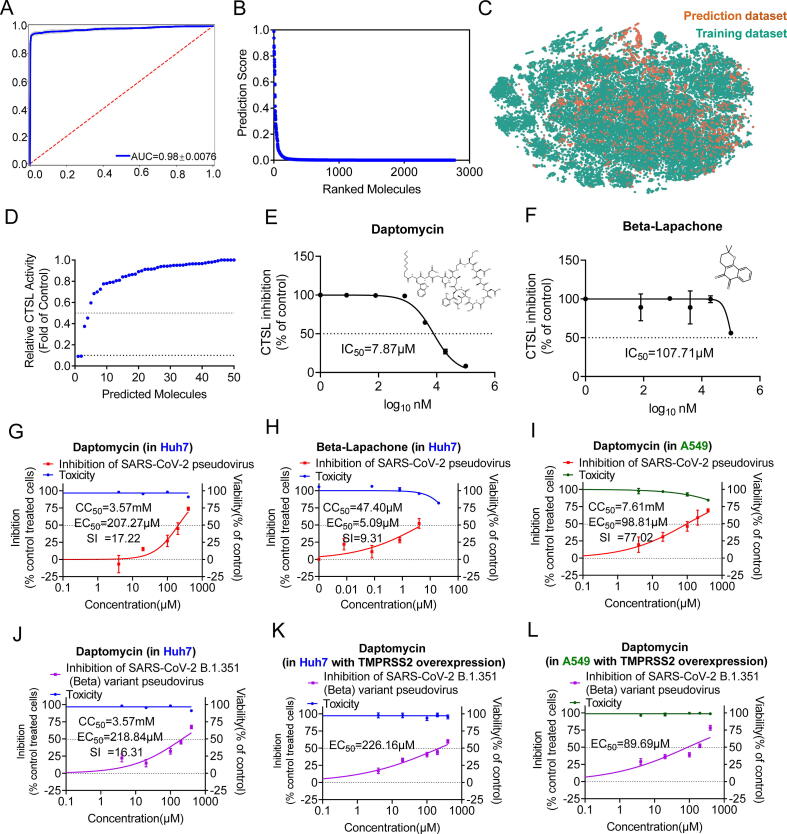
After the establishment of the initial training dataset, we used these data to train a binary classification model that predicts the probability of whether a new compound will inhibit the activity of CTSL. Firstly, we applied the Bayesian hyperparameter optimization scheme to improve our model. We ran 20 iterations of Bayesian optimization to search for the best set of hyperparameters for our model (see Materials and Methods for details). Secondly, with the best set of hyperparameters, we randomly split the dataset into 80% training data, 10% validation data, and 10% test data. We trained our model on the training data for 30 epochs and iterated over the validation data to compute the performance at the end of each epoch. After the training, we selected the model with the best performance on the validation data and evaluated it on the test data. Additionally, we utilized the ensemble modeling technique to further improve our performance. We repeated the procedure mentioned above with 20 different random splits of the data and averaged the prediction results. The resulting model achieved a receiver operating characteristic-area under the curve (ROC-AUC) of 0.98 on the test data (Fig. 1A).
Fig. 1.
Initial model training and predicting potential CTSL inhibitors from bioactive compound libraries. A, Receiver operating characteristic-area under the curve (ROC-AUC) plot evaluating model performance after training. Blue is the mean of twenty folds (grey). B, Rank-ordered prediction scores of initial prediction dataset that were not present in the training dataset. C, Visualization of all molecules from the initial training dataset (green) and the initial prediction dataset(orange) using t-distributed stochastic neighbor embedding (t-SNE), revealing chemical relationships between these libraries. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
After model development and optimization using the initial training dataset, we applied the best-performing model to identify potential CTSL inhibitors from the training dataset composed of Selleck bioactive compound libraries and the ZINC15 in vitro database[38]. We removed the molecules with the same molecular graphs as the training dataset, leaving 310,283 molecules of diverse structure and function. Then, we determined the prediction scores for each molecule, and molecules were ranked based on their probability of displaying activity inhibition against CTSL (Supplementary Table 3, Fig. 1B). We next employ the t-distributed stochastic neighbor embedding (t-SNE) algorithm to further investigate the low-dimensional node representation learned by our model. The distributions of the training dataset and prediction dataset were similar (Fig. 1C).
Next, among the molecules that we can get currently, we chose 50 molecules that were the most strongly predicted to display CTSL inhibition properties by our model for verification through molecular biology experiments (Supplementary Table 4). Initially, we uniformly chose a single dose of 100 μM to test whether they could inhibit CTSL activity in a cell-free system (see Materials and Methods for details). The results showed that 12 of the 50 predicted molecules displayed over 50% inhibition against CTSL, and the top 5 were Mg-132, Z-FA-FMK, leupeptin hemisulfate, Mg-101 and calpeptin, with inhibition efficiencies greater than 90% (Supplementary Table 4, Fig. 2A). Then, we tested a range of concentrations of Mg-132, Z-FA-FMK, leupeptin hemisulfate, Mg-101 and calpeptin in this cell-free system for further confirmation and determination of the half maximal inhibitory concentration (IC50). Notably, all 5 molecules inhibited CTSL activity in a concentration-dependent manner with IC50 values of 12.28 nM, 54.87 nM, 5.77 nM, 5.77 nM, and 43.98 nM, respectively (Fig. 2B-F). These results verified the feasibility and reliability of the deep learning strategy we used in screening CTSL inhibitors.
Fig. 2.
Prediction dataset validation with a cell-free CTSL activity detection system. A, Among the available molecules, the top 50 molecules from the prediction dataset were chosen for verifying the inhibition effect against CTSL in a cell-free system at a single dose of 100 μM. Twelve of the 50 predicted molecules displayed over 50% inhibition against CTSL, and the top 5 were Mg-132, Z-FA-FMK, leupeptin hemisulfate, Mg-101 and calpeptin, with inhibition efficiencies greater than 90%. The data are expressed as the mean of three individual trials. B-F, Five predicted CTSL inhibitors, Mg-132(B), Z-FA-FMK(C), leupeptin hemisulfate(D), Mg-101(E) and calpeptin(F), with inhibition efficiencies greater than 90% at 100 μM were further tested for determination of the half maximal inhibitory concentration (IC50) in the cell-free system. Corresponding molecular structure was drawn by Chemdraw. Non-linear fit to a variable response curve from one representative experiment with three replicates is shown (black lines). The data are expressed as the mean ± s.e.m.
3.3. The predicted CTSL inhibitors from bioactive compounds prevent SARS-CoV-2 infection in Huh7 cells in vitro
Since Mg-132, Z-FA-FMK, leupeptin hemisulfate, Mg-101 and calpeptin showed very significant inhibitory effects on CTSL activity in cell-free systems, we further explored whether they could inhibit SARS-CoV-2 infection in Huh7 cells in vitro. First, we tested the cytotoxicity of these 5 molecules in Huh7 cells. The results showed that Mg-132, Z-FA-FMK, leupeptin hemisulfate, Mg-101 and calpeptin had 50% cytotoxic concentration (CC50) values of 100.22 μM, 328.10 μM, 6.00 mM, 64.32 μM, and 115.70 μM, respectively (Supplementary Fig. 2A–E).
Since biosafety level 3 (BSL-3) is required for working with live SARS-CoV-2, we first tested the anti-SARS-CoV-2 effect with a pseudovirus system that can be used safely in biosafety level 2 (BSL-2) laboratories (Fig. 3A). As reported in the previous research, the pseudovirus expresses the SARS-CoV-2 spike(S) protein on the surface can faithfully reflect key aspects of SARS-CoV-2 cell entry [39]. We selected a series of concentrations of these 5 molecules that did not cause cytotoxicity for determination. All 5 molecules suppressed SARS-CoV-2 pseudovirus infection with half maximal effective concentration (EC50) values of 212.50 nM, 701.20 nM, 39.29 μM, 3.12 μM and 29.82 μM, respectively. The selectivity index (SI), which was calculated as the ratio of CC50 and EC50, was 471.62, 467.91, 152.71, 20.62, and 3.88, respectively (Fig. 3B–F). To consolidating these findings, we repeat all these in vivo experiments above using the human lung epithelial carcinoma cell line A549. The results showed that Mg-132, Z-FA-FMK, leupeptin hemisulfate, Mg-101 and calpeptin had CC50 values of 477.43 μM, 428.00 μM, 34.84 mM, 355.24 μM, and 595.30 μM, respectively in A549 cells (Supplementary Fig. 3A–E). Considering that A549 cells are far less susceptible to SARS-CoV-2 pseudovirus than Huh7 cells due to the low expression level of ACE2[13], [40], we used adenovirus overexpression human ACE2 (Ad-ACE2) to increase pseudovirus infection rate of A549 cells. Firstly, we demonstrated the overexpression efficiency of 25 moi Ad-ACE2 in Huh7 cells for 24 hours (h) at both the mRNA and the protein level (Supplementary Fig. 4A and B). Then, A549 cells were pretreated with 25moi Ad-ACE2 for 24 h before treated as Huh7 cells stated above. All 5 molecules suppressed SARS-CoV-2 pseudovirus infection in A549 cells with EC50 values of 159.20 nM, 1.13 μM, 4.26 μM, 1.15 μM and 34.81 μM, respectively. The SI was 2998.93, 378.76, 8178.40, 308.90, and 17.10, respectively (Supplementary Fig. 4C–H). In addition, we evaluate the effect of these 5 molecules on B.1.351 (Beta) variant, one of the SARS-CoV-2 variants, in Huh7 cells. The results showed that these 5 molecules had similar or even stronger inhibitory effect against SARS-CoV-2 B.1.351 (Beta) variant pseudovirus infection (Fig. 3G–K). Compared with Calu-3 cells, although CTSL expression is much higher in Huh7 cells and A549 cells, TMPRSS2 expression level was very low in Huh7 cells and even lower in A549 cells (Supplementary Fig. 5A and B). Consider that multiple studies have shown that human airway cells express TMPRSS2 and that this protease is used preferentially to cathepsins[41], [42], [43], we further explored whether these drugs were still effective in the case of TMPRSS2 overexpression both in Huh7 cells and in A549 cells (Supplementary Fig. 6A–C). The results showed that all these 5 molecules suppressed SARS-CoV-2 B.1.351 (Beta) variant pseudovirus infection in Huh7 cells with EC50 values of 901.00 nM, 10.01 μM, 17.08 μM, 0.86 μM and 23.92 μM, respectively (Supplementary Fig. 6D–H). And they suppressed SARS-CoV-2 B.1.351 (Beta) variant pseudovirus infection in A549 cells with EC50 values of 907.60 nM, 5.31 μM, 7.71 μM, 1.89 μM and 36.35 μM, respectively (Supplementary Fig. 6I–M). These data suggest that these drugs remain effective after TMPRSS2 overexpression, although some EC50 are significantly elevated.
Fig. 3.
The predicted CTSL inhibitors from bioactive compounds prevent SARS-CoV-2 pseudovirus infection in Huh7 cells in vitro. A, Schematic of the predicted CTSL inhibitor assay setup. Huh7 cells were pretreated with different drugs 1 hour (h) before infection with SARS-CoV-2 pseudovirus or SARS-CoV-2 B.1.351 (Beta) variant pseudovirus at the same dose (1.3×104 TCID50/ml). Pseudovirus infection and cell viability were evaluated 24 h later by a luciferase activity and MTT assay, respectively. B-F, Inhibition of pseudovirus infection by different doses of Mg-132 (B), Z-FA-FMK (C), Leupeptin Hemisulfate (D), Mg-101 (E), and Calpeptin (F) and viability of Huh7 cells treated with different doses of the drugs as indicated. Non-linear fit to a variable response curve from one representative experiment with four replicates is shown (red lines). Cytotoxic effect on Huh7 cells exposed to increasing concentrations of drugs in the absence of virus is also shown (blue lines). The CC50, EC50, and SI values of this graph are indicated. n = 4. The data are expressed as the mean ± s.e.m. G-K, Inhibition of SARS-CoV-2 B.1.351(Beta) variant pseudovirus infection by different doses of Mg-132 (G), Z-FA-FMK (H), Leupeptin Hemisulfate (I), Mg-101 (J), and Calpeptin (K) and viability of Huh7 cells treated with different doses of the drugs as indicated. Non-linear fit to a variable response curve from one representative experiment with four replicates is shown (purple lines). Cytotoxic effect on Huh7 cells exposed to increasing concentrations of drugs in the absence of virus is also shown (blue lines). The CC50, EC50, and SI values of this graph are indicated. CC50: 50% cytotoxic concentration. EC50: half maximal effective concentration. SI: the selectivity index, which is calculated as the ratio of CC50 and EC50. n = 4. The data are expressed as the mean ± s.e.m. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
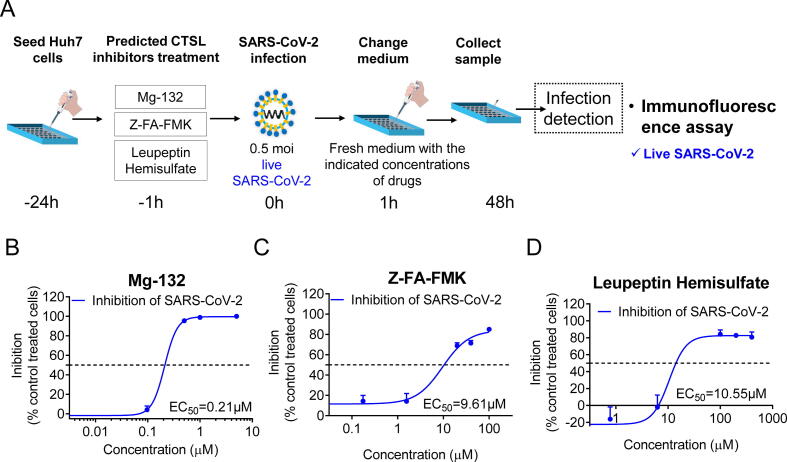
Finally, 3 compounds, Mg-132, Z-FA-FMK and leupeptin hemisulfate, which had a SI greater than 100 in both Huh7 and A549 cells, were further tested for validation using live SARS-CoV-2 in the P3 laboratory (Fig. 4A). Exhilaratingly, all 3 molecules inhibited SARS-CoV-2 infection in a concentration-dependent manner and acted at fairly low concentrations in the micromolar range with EC50 values of 0.21 μM, 9.61 μM and 10.55 μM, respectively (Fig. 4B-D). These results indicated that these 5 small molecules are expected to become therapeutic drugs for COVID-19 by inhibiting CTSL activity.
Fig. 4.
The predicted CTSL inhibitors from bioactive compounds prevent live SARS-CoV-2 infection in Huh7 cells in vitro. A, Schematic of the predicted CTSL inhibitor assay setup. Huh7 cells were pretreated with different drugs 1 h before infection with live SARS-CoV-2 at the same dose (0.5 moi), followed by changing to fresh medium with the indicated concentrations of drugs 1 h later. The detection of infected cells was performed 48 h later by using an immunofluorescence assay. B-D, Inhibition of live SARS-CoV-2 infection by different doses of Mg-132 (B), Z-FA-FMK (C), and Leupeptin Hemisulfate (D). Non-linear fit to a variable response curve from one representative experiment with three replicates is shown (blue lines). The EC50 value of this graph is indicated. n = 3. The data are expressed as the mean ± s.e.m. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Daptomycin predicted from an FDA-approved drug library alleviate SARS-CoV-2 pseudovirus infection in Huh7 cells in vitro
For drug repurposing screening, we applied our model to identify potential CTSL inhibitors from the FDA-approved drug library. First, to increase chemical diversity, we added the experimentally validated molecules from the initial prediction dataset to the training dataset, using 50% CTSL activity inhibition as a hit cutoff. Then, we used these molecular data to train the second binary classification model (Supplementary Table 5). This model also achieved a ROC-AUC of 0.98 on the test data (Fig. 5A). Later, we applied this secondly trained model to the FDA-approved drug library, a unique collection of 3177 drugs that are marketed around the world or have passed at least phase I clinical trials (https://www.selleck.cn/screening/fda-approved-passed-phase-i-drug-library.html), as the prediction dataset. Before prediction, we removed the compounds that had the same molecular graphs as the training dataset, and 2773 drugs with diverse structures and functions remained. We determined the prediction scores for each compound, and they were ranked based on their probability of displaying activity inhibition against CTSL (Supplementary Table 6, Fig. 5B). We also employ t-SNE to further investigate the low-dimensional node representation learned by our secondly trained model. The distribution of the second training dataset and the FDA-approved drug library was similar (Fig. 5C).
Fig. 5.
The second model training and the identification of daptomycin. For drug repurposing screening for COVID-19 from the FDA-approved drug library, we trained the second model by adding the experimentally validated molecules from bioactive compounds aforementioned to the initial training dataset. A, The ROC-AUC plot evaluating the second model performance after training. Blue is the mean of twenty folds (grey). B, Rank-ordered prediction scores of the FDA-approved drug library that were not present in the training dataset. C, Visualization of all molecules from the second training dataset (green) and the second prediction dataset(orange) using t-SNE, revealing chemical relationships between these libraries. D, Among the available drugs, the top 50 drugs from the FDA-approved drug library were chosen for verifying the inhibition effect against CTSL in the cell-free system at a single dose of 100 μM. Four of the 50 predicted drugs displayed over 50% inhibition against CTSL, and the top 2 were daptomycin and beta-lapachone, with inhibition efficiencies greater than 90%. The data are expressed as the mean of three individual trials. E-F, Daptomycin(E) and beta-lapachone(F) were further tested for determination of IC50 in the cell-free system. These 2 drugs were used at a concentration ranging from 8 nM and 80 nM to 100 μM, respectively. The IC50 value of this graph is indicated. n = 3. The data are expressed as the mean ± s.e.m. G-H, Inhibition of pseudovirus infection by different doses of Daptomycin (G), and beta-lapachone (H) and viability of Huh7 cells treated with different doses of the drugs as indicated. Non-linear fit to a variable response curve from one representative experiment with four replicates is shown (red lines). Cytotoxic effect on Huh7 cells exposed to increasing concentrations of drugs in the absence of virus is also shown (blue lines). The CC50, EC50, and SI values of this graph are indicated. n = 4. The data are expressed as the mean ± s.e.m. I, Inhibition of pseudovirus infection by different doses of Daptomycin, and viability of A549 cells treated with different doses of the drugs as indicated. Non-linear fit to a variable response curve from one representative experiment with four replicates is shown (red lines). Cytotoxic effect on A549 cells exposed to increasing concentrations of drugs in the absence of virus is also shown (green lines). The CC50, EC50, and SI values of this graph are indicated. n = 5. The data are expressed as the mean ± s.e.m. J, Inhibition of SARS-CoV-2 B.1.351(Bata) variant pseudovirus infection by different doses of Daptomycin, and viability of Huh7 cells treated with different doses of the drugs as indicated. Non-linear fit to a variable response curve from one representative experiment with four replicates is shown (purple lines). Cytotoxic effect on Huh7 cells exposed to increasing concentrations of drugs in the absence of virus is also shown (blue lines). The CC50, EC50, and SI values of this graph are indicated. n = 5. The data are expressed as the mean ± s.e.m. K, Inhibition of SARS-CoV-2 B.1.351(Bata) variant pseudovirus infection by different doses of Daptomycin, and viability of Huh7 cells with TMPRSS2 overexpression. Non-linear fit to a variable response curve from one representative experiment with four replicates is shown (purple lines). Cytotoxic effect on Huh7 cells exposed to increasing concentrations of drugs as indicated is also shown (blue lines). The EC50 values of this graph are indicated. n = 5. The data are expressed as the mean ± s.e.m. L, Inhibition of SARS-CoV-2 B.1.351(Bata) variant pseudovirus infection by different doses of Daptomycin, and viability of A549 cells with TMPRSS2 overexpression. Non-linear fit to a variable response curve from one representative experiment with four replicates is shown (purple lines). Cytotoxic effect on A549 cells exposed to increasing concentrations of drugs as indicated is also shown (green lines). The EC50 values of this graph are indicated. n = 5. The data are expressed as the mean ± s.e.m. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, among the drugs we obtained, we chose 50 drugs unique to the FDA-approved drug library that were the most strongly predicted to display CTSL activity inhibition properties by our secondly trained model for verification as described above. The results showed that 4 of the 50 predicted drugs displayed over 50% inhibition against CTSL, and the top 2 were daptomycin and beta-lapachone, with inhibition efficiencies over 90% in the cell-free system at a concentration of 100 μM (Supplementary Table 7, Fig. 5D). Therefore, daptomycin and beta-lapachone were chosen as candidate drugs. Intriguingly, daptomycin inhibited CTSL activity in a concentration-dependent manner with an IC50 value of 7.87 μM (Fig. 5E). However, the IC50 value of beta-lapachone was higher than 100 μM (Fig. 5F).
We further explored whether daptomycin and beta-lapachone could inhibit SARS-CoV-2 pseudovirus infection in Huh7 cells in vitro. At the same time, we also tested the cytotoxicity of these 2 drugs. The results showed that daptomycin was much less toxic than beta-lapachone, with CC50 values of 3.57 mM and 47.40 μM, respectively (Supplementary Fig. 7A and B). As expected, daptomycin and beta-lapachone inhibited SARS-CoV-2 pseudovirus infection with EC50 values of 207.27 μM and 5.09 μM, respectively. The SI values were 17.22 and 9.31, respectively (Fig. 5G and H). We further found that daptomycin had a much stronger suppression efficiency in A549 cells (Fig. 5I), and had a similar inhibition efficiency on SARS-CoV-2 B.1.351 (Beta) variant pseudovirus infection in Huh7 cells (Fig. 5J). And we further found that daptomycin remain effective for suppressing SARS-CoV-2 B.1.351 (Beta) variant pseudovirus after TMPRSS2 overexpression both in Huh7 cells and A549 cells (Fig. 5K-L). These results indicated that daptomycin may be a potent therapeutic drug for COVID-19.
3.5. Molecular docking analysis
To further investigate the specific binding pattern of these inhibitors to CTSL, we performed molecular docking calculations using Schrödinger software suite with the human CTSL structures from Protein Data Bank (PDB) database (PDB code 5MQY, resolution 1.13 Å). 5MQY is a high-resolution human CTSL protein structure cocrystallized with a covalent inhibitor, compound 35. Since the Z-FA-FMK, Calpeptin, Mg-101(ALLN), Mg-132 and Leupeptin Hemisulfate are all covalent inhibitors[44], [45], [46], [47], we performed covalent docking available in Schrödinger suite: CovDock in Schrödinger 2021–2 version. To verify the stability of the method, we separated the protein and ligand in 5MQY and performed re-docking calculations with them. The re-docking calculations showed superposition of compound 35 to its crystallographic structure (Supplementary Figure 8) with a Random Mean Square Deviation (RMSD) of 0.141 Å, which is acceptable.
The docking scores and molecular interactions of protein residues and ligands are listed in (Supplementary Table 8). Compound 35 forms a hydrogen bond with Gln19 residue of 5MQY with a docking score of −6.3 kcal/mol (Fig. 6A). Mg-132 forms two hydrogen bonds with Gln19 and Gly68 residues of 5MQY with a docking score of −5.1 kcal/mol (Fig. 6B). Z-FA-FMK forms two hydrogen bonds with Gly68 and Asp162 residues of 5MQY with a docking score of −7.2 kcal/mol (Fig. 6C). Leupeptin Hemisulfate forms three hydrogen bonds with Gln19, Gly68 and Asp162 residues of 5MQY with a docking score of −7.5 kcal/mol (Fig. 6D). Calpeptin forms two hydrogen bonds with Gly68 and Gly164 residues of 5MQY with a docking score of −6.9 kcal/mol (Fig. 6E). Mg-101(ALLN) forms two hydrogen bonds with Gly68 and Asp162 residues of 5MQY with a docking score of −6.5 kcal/mol (Fig. 6F).
Fig. 6.
Molecular docking results of CTSL inhibitors in the crystal structure of human CTSL (5MQY). A-G, 3D structure of the human CTSL (5MQY) showing the main residues involved in the protein-ligand interaction of Compound 35 (A), Mg-132 (B), Z-FA-FMK (C), Leupeptin Hemisulfate (D), Calpeptin (E), Mg-101 (F) and Daptomycin (G). Compound 35 is a covalent inhibitor cocrystallized with human CTSL protein in 5MQY, used here as a positive control. Short intermolecular contacts with distances of <4.0 Å between the ligand fragment (gray) and protein residues (dark green) are shown as dashed yellow lines. Structure visualization was by PyMol. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Finally, we investigate the specific binding pattern of Daptomycin to CTSL. We use Schrödinger extra precision (XP) module to perform molecular docking calculations. Daptomycin forms three hydrogen bonds with Asp160, Met161 and Asp162 residues of 5MQY with a docking score of −4.5 kcal/mol (Fig. 6G).
4. Discussion
Although a number of small molecules and drugs have been reported to resist SARS-CoV-2, no effective drug has yet been developed[8]. CTSL has an essential role in viral infection, inflammatory status, tumor invasion and metastasis, and other chronic diseases, such as atherosclerosis, renal disease, and diabetes[48]. Therefore, CTSL is considered an attractive therapeutic target. To date, many CTSL inhibitors have been synthesized since the first CTSL inhibitor, cystatin, was isolated from Aspergillus in 1981[48]. However, none of these compounds can be clinically used, perhaps mainly due to toxicity and unpredictable side effects[49]. Recently, K777 has completed phase I clinical trials, which reduced SARS-CoV-2 viral infectivity through inhibition of the activity of host CTSL. However, the efficacy and safety still need to be verified[50]. Thus, it is of significance to identify more secure molecules to expand CTSL inhibitor discovery for drug development and identify FDA-approved drugs that can inhibit CTSL for drug repurposing for COVID-19.
To identify more CTSL inhibitors, we applied Chemprop MPNNs to the Selleck bioactive compound libraries and the ZINC15 in vitro database, which contains 310,283 unique molecules that have been reported or inferred to be biologically active. Based on the predicted score ranking and availability, we selected 50 molecules for experimental validation and eventually identified 12 compounds with more than 50% inhibition against CTSL activity at 100 μM in a cell-free system, five of which, Mg-132, Z-FA-FMK, Leupeptin Hemisulfate, Mg-101 and Calpeptin, were able to exert inhibition at nanomolar concentrations. It is reported that proteasome inhibitor, Mg-132, can also cross-inhibit CTSL[51], [52]. Cysteine protease inhibitors, Z-FA-FMK, Leupeptin Hemisulfate, MG-101 and calpain inhibitor, calpeptin were also reported to inhibit CTSL[53], [54], [55], [56], [57], [58], [59], [60], [61]. These data verified the feasibility and reliability of the deep learning strategy we used in screening CTSL inhibitors. Notably, these 5 molecules can significantly inhibit SARS-CoV-2 infection in Huh7 cells and A549 cells in vitro. These results indicated that these 5 small molecules are expected to become therapeutic drugs for COVID-19 by inhibiting CTSL activity. However, the molecules we bought for validation span a ranking of 18 to 2804 (Supplementary Table 4), because many molecules are unavailable due to the impact of the COVID-19 epidemic. According to a study by Stokes JM et al., who trained Chemprop for screening new antibiotics, although the molecule with the best inhibitory effect does not necessarily mean the highest predicted score, higher prediction scores correlated with a greater probability of activity inhibition against CTSL[17]. Thus, there must be more potential CTSL inhibitors in the top-ranked molecules that we did not obtain.
To find a clinically available inhibitor of CTSL, we applied Chemprop MPNNs to the FDA-approved drug library, a unique collection of 3177 drugs marketed worldwide or that passed a phase I clinical trial. Only 4 of 50 molecules showed over 50% inhibition against CTSL at 100 μM in the cell-free system, and only 2 molecules, daptomycin and beta-lapachone, displayed over 90% inhibition, agreeing with the correspondingly low model prediction scores, which may be due to the relatively small number of drugs in the FDA-approved library. Further investigation demonstrated that daptomycin has an excellent CTSL inhibitory effect (IC50 = 7.87 μM) in a cell-free system. Moreover, daptomycin significantly inhibited SARS-CoV-2 pseudovirus infection with little cytotoxicity (EC50 = 207.27 μM, CC50 = 3.57 mM in Huh7 cells and EC50 = 98.81 μM, CC50 = 7.61 mM in A549 cells). The inhibitory rate of beta-lapachone on pseudovirus infection was 41% at 40 μM, but it showed obvious cytotoxicity. Daptomycin is a semisynthetic compound derived from the fermentation of Streptomyces roseosporus[62]. It is one of a few membrane-active antimicrobial peptides (AMPs) that have been approved by the FDA for clinical use[63]. Hence, we believe that daptomycin may have the potential to become an antiviral drug. However, the EC50 is much higher than the dose of daptomycin for clinical use. We will further modify daptomycin to enhance its antiviral ability and reduce side effects.
To investigate the specific binding pattern of these inhibitors to CTSL, we performed molecular docking calculations using Schrödinger software suite with the human CTSL structures from PDB database (PDB code 5MQY, resolution 1.13 Å). The Leupeptin Hemisulfate, Z-FA-FMK, Calpeptin and Mg-101(ALLN) shows greater binding affinity than the co-crystallized inhibitor compound 35 (Ki = 77 nM[33]), our biological experimental also confirm these results. However, the docking score of Mg-132 is only −5.1 kcal/mol while the IC50 values of CTSL activity is 12.28 nM. This is probably due to the lack of accuracy in Glide docking scoring. We will further conduct molecular dynamics simulations to investigate more precise protein-ligand binding patterns. Finally, we investigate the specific binding pattern of Daptomycin to CTSL. Daptomycin forms three hydrogen bonds with Asp160, Met161 and Asp162 residues of 5MQY while the docking score is just −4.5 kcal/mol. This is consistent with its poor performance in biological experiments. These specific binding sites will help us to modify drugs to achieve better affinity.
In conclusion, we successfully trained a machine learning model that uses Chemprop MPNNs and the publicly available PubChem database to predict CTSL inhibitors. We identified 5 bioactive molecules and one FDA-approved antibiotic, daptomycin, that can significantly inhibit CTSL and alleviate SARS-CoV-2 pseudovirus infection in vitro. Molecule docking results show that the Gln19, Gly68 and Asp162 residues of CTSL are the primary binding site, this provides a reference for the design of specific inhibitors. In the future, experiments using live SARS-CoV-2 viruses in vitro and in vivo using adequate animal models and clinical trials are needed to investigate the role of daptomycin in treating COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (81930019, 8151101058, 81471014), Scientific Project of Beijing Municipal Science & Technology Commission (D171100002817005), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (ZYLX201823) to Jinkui Yang, and National Natural Science Foundation of China (82000825) to Weili Yang.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.05.023.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jackson L.A., Anderson E.J., Rouphael N.G., et al. An mRNA vaccine against SARS-COV-2 — Preliminary report[J] N Engl J Med. 2020;383(20) doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulligan M J, Lyke K E, Kitchin N, et al. Phase 1/2 study of COVID-19 RNA vaccine BNT162b1 in adults[J]. Nature. [DOI] [PubMed]
- 3.Widge A.T., Rouphael N.G., Jackson L.A., et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination[J] N Engl J Med. 2020 doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dos Santos W. Impact of virus genetic variability and host immunity for the success of COVID-19 vaccines[J] Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2021;136 doi: 10.1016/j.biopha.2021.111272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams S V, Vusirikala A, Ladhani S N, et al. An outbreak caused by the SARS-CoV-2 Delta (B.1.617.2) variant in a care home after partial vaccination with a single dose of the COVID-19 vaccine Vaxzevria, London, England, April 2021[J]. Euro Surveill, 2021, 26(27). [DOI] [PMC free article] [PubMed]
- 6.Jiang H.D., Tao Y.Y., Jia S.Y., et al. Coronavirus disease 2019 vaccines: landscape of global studies and potential risks[J] Chin Med J (Engl) 2021 doi: 10.1097/CM9.0000000000001688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torjesen I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear[J] BMJ. 2021 doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 8.Tao K., Tzou P.L., Nouhin J., et al. SARS-CoV-2 Antiviral Therapy[J] Clin Microbiol Rev. 2021:e0010921. doi: 10.1128/CMR.00109-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beigel J., Chung K.K., Colombo R., et al. Remdesivir for the Treatment of Covid-19 — Preliminary Report[J] N Engl J Med. 2020 doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- 10.Pan H, Peto R, Henao-Restrepo A M, et al. Repurposed Antiviral Drugs for Covid-19 – Interim WHO Solidarity Trial Results[J]. 2021. [DOI] [PMC free article] [PubMed]
- 11.Jang W.D., Jeon S., Kim S., et al. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay[J] Proc Natl Acad Sci U S A. 2021;118(30) doi: 10.1073/pnas.2024302118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T., Luo S., Libby P., et al. Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients[J] Pharmacol Ther. 2020;213 doi: 10.1016/j.pharmthera.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao M., Yang W., Yang F., et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development[J] Signal transduction and targeted therapy. 2021;6(1):134. doi: 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie X., Qian L., Sun R., et al. Multi-organ proteomic landscape of COVID-19 autopsies[J] Cell. 2021;184(3):775–791.e714. doi: 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dana P. A Review of Small Molecule Inhibitors and Functional Probes of Human Cathepsin L[J] Molecules. 2020;25(3):698. doi: 10.3390/molecules25030698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J., Zheng S., Chen B., et al. A survey of current trends in computational drug repositioning[J] Briefings Bioinf. 2016;17(1) doi: 10.1093/bib/bbv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stokes J.M., Yang K., Swanson K., et al. A Deep Learning Approach to Antibiotic Discovery[J] Cell. 2020;180(4):688–702.e613. doi: 10.1016/j.cell.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang K., Swanson K., Jin W., et al. Analyzing learned molecular representations for property prediction[J] J Chem Inf Model. 2019;59(8):3370–3388. doi: 10.1021/acs.jcim.9b00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J., Cui G., Hu S., et al. Graph neural networks: A review of methods and applications[J] AI Open. 2020;1:57–81. [Google Scholar]
- 20.Landrum G. Academic Press Cambridge; 2013. RDKit: A software suite for cheminformatics, computational chemistry, and predictive modeling [M] [Google Scholar]
- 21.Wang T., Fang X., Wen T., et al. Synthetic Neutralizing Peptides Inhibit the Host Cell Binding of Spike Protein and Block Infection of SARS-CoV-2[J] J Med Chem. 2021;64(19):14887–14894. doi: 10.1021/acs.jmedchem.1c01440. [DOI] [PubMed] [Google Scholar]
- 22.Ye F., Lin X., Chen Z., et al. S19W, T27W, and N330Y mutations in ACE2 enhance SARS-CoV-2 S-RBD binding toward both wild-type and antibody-resistant viruses and its molecular basis[J] Signal Transduct Target Ther. 2021;6(1):343. doi: 10.1038/s41392-021-00756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He C, Yang J, He X, et al. A bivalent recombinant vaccine targeting the S1 protein induces neutralizing antibodies against both SARS-CoV-2 variants and wild-type of the virus[J]. MedComm (2020), 2021. [DOI] [PMC free article] [PubMed]
- 24.Zheng B., Peng W., Guo M., et al. Inhalable nanovaccine with biomimetic coronavirus structure to trigger mucosal immunity of respiratory tract against COVID-19[J] Chem Eng J. 2021;418 doi: 10.1016/j.cej.2021.129392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao W, Wang Y, Ma D, et al. Circulating SARS-CoV-2 variants B.1.1.7, 501Y.V2, and P.1 have gained ability to utilize rat and mouse Ace2 and altered in vitro sensitivity to neutralizing antibodies and ACE2-Ig[J]. bioRxiv, 2021: 2021.2001.2027.428353.
- 26.Yang W., Wang J., Chen Z., et al. NFE2 Induces miR-423-5p to Promote Gluconeogenesis and Hyperglycemia by Repressing the Hepatic FAM3A-ATP-Akt Pathway[J] Diabetes. 2017;66(7):1819–1832. doi: 10.2337/db16-1172. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H.L., Li Y.M., Sun J., et al. Evaluating angiotensin-converting enzyme 2-mediated SARS-CoV-2 entry across species[J] J Biol Chem. 2021;296 doi: 10.1016/j.jbc.2021.100435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Zeng E., Zhang L., et al. Potent prophylactic and therapeutic efficacy of recombinant human ACE2-Fc against SARS-CoV-2 infection in vivo[J] Cell Discov. 2021;7(1):65. doi: 10.1038/s41421-021-00302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Yang W., Chen Z., et al. Long Noncoding RNA lncSHGL Recruits hnRNPA1 to Suppress Hepatic Gluconeogenesis and Lipogenesis[J] Diabetes. 2018;67(4):581–593. doi: 10.2337/db17-0799. [DOI] [PubMed] [Google Scholar]
- 30.Polson E.S., Lewis J.L., Celik H., et al. Monoallelic expression of TMPRSS2/ERG in prostate cancer stem cells[J] Nat Commun. 2013;4:1623. doi: 10.1038/ncomms2627. [DOI] [PubMed] [Google Scholar]
- 31.Herman-Edelstein M., Guetta T., Barnea A., et al. Expression of the SARS-CoV-2 receptorACE2 in human heart is associated with uncontrolled diabetes, obesity, and activation of the renin angiotensin system[J] Cardiovasc Diabetol. 2021;20(1):90. doi: 10.1186/s12933-021-01275-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esumi M., Ishibashi M., Yamaguchi H., et al. Transmembrane serine protease TMPRSS2 activates hepatitis C virus infection[J] Hepatology. 2015;61(2):437–446. doi: 10.1002/hep.27426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn B., Tichý M., Wang L., et al. Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors[J] J Med Chem. 2017;60(6):2485–2497. doi: 10.1021/acs.jmedchem.6b01881. [DOI] [PubMed] [Google Scholar]
- 34.Sastry G.M., Adzhigirey M., Day T., et al. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments[J] J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 35.Harder E., Damm W., Maple J., et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins[J] J Chem Theory Comput. 2016;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 36.Shelley J.C., Cholleti A., Frye L.L., et al. Epik: a software program for pK a prediction and protonation state generation for drug-like molecules[J] J Comput Aided Mol Des. 2007;21(12):681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen W.L., Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin[J] J Am Chem Soc. 1988;110(6):1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 38.Sterling T., Irwin J.J. ZINC 15–Ligand Discovery for Everyone[J] J Chem Inf Model. 2015;55(11):2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nie J., Li Q., Wu J., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2[J] Emerg Microbes Infect. 2020;9(1):680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Fan J, Chen Z, et al. Nonmuscle myosin heavy chain IIA facilitates SARS-CoV-2 infection in human pulmonary cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(50). [DOI] [PMC free article] [PubMed]
- 41.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor[J] Cell. 2020;181(2):271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ou T., Mou H., Zhang L., et al. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2[J] PLoS Pathog. 2021;17(1):e1009212. doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laporte M., Raeymaekers V., Van Berwaer R., et al. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways[J] PLoS Pathog. 2021;17(4):e1009500. doi: 10.1371/journal.ppat.1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S., Shi Y., Jin H., et al. Covalent complexes of proteasome model with peptide aldehyde inhibitors MG132 and MG101: docking and molecular dynamics study[J] J Mol Model. 2009;15(12):1481–1490. doi: 10.1007/s00894-009-0515-0. [DOI] [PubMed] [Google Scholar]
- 45.Lawrence C P, Kadioglu A, Yang A-L, et al. The cathepsin B inhibitor, z-FA-FMK, inhibits human T cell proliferation in vitro and modulates host response to pneumococcal infection in vivo[J]. Journal of immunology (Baltimore, Md : 1950), 2006, 177(6): 3827-3836. [DOI] [PubMed]
- 46.Ray S.K., Wilford G.G., Matzelle D.C., et al. Calpeptin and methylprednisolone inhibit apoptosis in rat spinal cord injury[J] Ann N Y Acad Sci. 1999;890:261–269. doi: 10.1111/j.1749-6632.1999.tb08001.x. [DOI] [PubMed] [Google Scholar]
- 47.Fu L., Shao S., Feng Y., et al. Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs[J] mBio. 2021;12(5):e0222021. doi: 10.1128/mBio.02220-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomes C.P., Fernandes D.E., Casimiro F., et al. Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics[J] Front Cell Infect Microbiol. 2020;10 doi: 10.3389/fcimb.2020.589505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y.W., Xie Y., Tang L.S., et al. Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies[J] Signal Transduct Target Ther. 2021;6(1):317. doi: 10.1038/s41392-021-00733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellott D.M., Tseng C.T., Drelich A., et al. A Clinical-Stage Cysteine Protease Inhibitor blocks SARS-CoV-2 Infection of Human and Monkey Cells[J] ACS Chem Biol. 2021;16(4):642–650. doi: 10.1021/acschembio.0c00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garrison P., Bangs J.D. p97 Inhibitor CB-5083 Blocks ERAD in Trypanosoma brucei[J] Mol Biochem Parasitol. 2020;239 doi: 10.1016/j.molbiopara.2020.111313. [DOI] [PubMed] [Google Scholar]
- 52.Costanzi E., Kuzikov M., Esposito F., et al. Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L[J] Int J Mol Sci. 2021;22(21) doi: 10.3390/ijms222111779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roscow O., Ganassin R., Garver K., et al. Z-FA-FMK demonstrates differential inhibition of aquatic orthoreovirus (PRV), aquareovirus (CSRV), and rhabdovirus (IHNV) replication[J] Virus Res. 2018;244:194–198. doi: 10.1016/j.virusres.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 54.Shen J., Cai Q., Yan L., et al. Cathepsin L is an immune-related protein in Pacific abalone (Haliotis discus hannai)–Purification and characterization[J] Fish Shellfish Immunol. 2015;47(2):986–995. doi: 10.1016/j.fsi.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Millest A., Breen S., Loveday B., et al. Effects of an inhibitor of cathepsin L on bone resorption in thyroparathyroidectomized and ovariectomized rats[J] Bone. 1997;20(5):465–471. doi: 10.1016/s8756-3282(97)00014-8. [DOI] [PubMed] [Google Scholar]
- 56.Ebisui C., Tsujinaka T., Kido Y., et al. Role of intracellular proteases in differentiation of L6 myoblast cells[J] Biochem Mol Biol Int. 1994;32(3):515–521. [PubMed] [Google Scholar]
- 57.Guo M., Mathieu P., Linebaugh B., et al. Phorbol ester activation of a proteolytic cascade capable of activating latent transforming growth factor-betaL a process initiated by the exocytosis of cathepsin B[J] The Journal of biological chemistry. 2002;277(17):14829–14837. doi: 10.1074/jbc.M108180200. [DOI] [PubMed] [Google Scholar]
- 58.Haspel J., Shaik R.S., Ifedigbo E., et al. Characterization of macroautophagic flux in vivo using a leupeptin-based assay[J] Autophagy. 2011;7(6):629–642. doi: 10.4161/auto.7.6.15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li S.Z., Zhang H.H., Zhang J.N., et al. ALLN hinders HCT116 tumor growth through Bax-dependent apoptosis[J] Biochem Biophys Res Commun. 2013;437(2):325–330. doi: 10.1016/j.bbrc.2013.06.088. [DOI] [PubMed] [Google Scholar]
- 60.Sasaki T., Kishi M., Saito M., et al. Inhibitory effect of di- and tripeptidyl aldehydes on calpains and cathepsins[J] J Enzyme Inhib. 1990;3(3):195–201. doi: 10.3109/14756369009035837. [DOI] [PubMed] [Google Scholar]
- 61.Lopez-Hernandez F.J., Ortiz M.A., Bayon Y., et al. Z-FA-fmk inhibits effector caspases but not initiator caspases 8 and 10, and demonstrates that novel anticancer retinoid-related molecules induce apoptosis via the intrinsic pathway[J] Mol Cancer Ther. 2003;2(3):255–263. [PubMed] [Google Scholar]
- 62.Debono M., Abbott B.J., Molloy R.M., et al. Enzymatic and chemical modifications of lipopeptide antibiotic A21978C: the synthesis and evaluation of daptomycin (LY146032)[J] J Antibiot. 1988;41(8):1093–1105. doi: 10.7164/antibiotics.41.1093. [DOI] [PubMed] [Google Scholar]
- 63.Chen C H, Lu T K. Development and Challenges of Antimicrobial Peptides for Therapeutic Applications[J]. Antibiotics (Basel, Switzerland). 2020. 9(1). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.