Abstract
Macrophages are widely distributed immune cells that contribute to tissue homeostasis. Human THP-1 cells have been widely used in various macrophage-associated studies, especially those involving pro-inflammatory M1 and anti-inflammatory M2 phenotypes. However, the molecular characterization of four M2 subtypes (M2a, M2b, M2c, and M2d) derived from THP-1 has not been fully investigated. In this study, we systematically analyzed the protein expression profiles of human THP-1-derived macrophages (M0, M1, M2a, M2b, M2c, and M2d) using quantitative proteomics approaches. The commonly and specially regulated proteins of the four M2 subtypes and their potential biological functions were further investigated. The results showed that M2a and M2b, and M2c and M2d have very similar protein expression profiles. These data could serve as an important resource for studies of macrophages using THP-1 cells, and provide a reference to distinguish different M2 subtypes in macrophage-associated diseases for subsequent clinical research.
Keywords: Macrophage, THP-1 cells, M2 subtype, Proteomics, Biological function
1. Introduction
Macrophages are a group of cells that play key immune functions in a variety of physiological and pathological processes, including inflammation, viral clearance, wound healing, and tissue homeostasis (Wynn et al., 2013). Sources of macrophages include tissue-resident macrophages originating from progenitor cells and monocyte-derived macrophages from bone marrow hematopoietic stem cells (Epelman et al., 2014). Plasticity and heterogeneity are the most critical characteristics of macrophages (Sica and Mantovani, 2012). Three broad pathways (including epigenetic and cell survival pathways that prolong or shorten macrophage development and viability, the tissue microenvironment, and extrinsic factors) are known to control macrophage polarization into different phenotypes with specific biological characteristics and functions (Murray, 2017).
Macrophages are divided into two subsets: classically activated M1 macrophages and alternatively activated M2 macrophages (Shapouri-Moghaddam et al., 2018). M1 macrophages can express high levels of signal transducer and activator of transcription 1 (STAT1) and nitric oxide synthase 2 (NOS2), and are involved mainly in acute pro-inflammatory responses for clearing invading pathogens and bacteria, as well as promoting T helper 1 (Th1) type immunity (Murray, 2017). However, the continued activation of M1 macrophages in the inflammatory area may cause tissue damage and impair wound healing (Liu et al., 2014). In contrast, M2 macrophages secrete considerable amounts of anti-inflammatory cytokines to counteract the inflammatory response maintained by M1 macrophages. M2 macrophages are involved mainly in tissue repair and phagocytic, pro-angiogenic, and pro-fibrotic processes, and promote Th2 type immunity (Wynn and Vannella, 2016).
In response to different stimulating molecules and/or microenvironments, M2 macrophages can be further divided into M2a, M2b, M2c, and M2d subtypes (Ferrante and Leibovich, 2012). Based on previous studies, M2a macrophages secrete large amounts of interleukin-4 (IL-4) and IL-13, and express high levels of mannose receptor C-type 1 (MRC1)/cluster of differentiation 206 (CD206) and fibronectin that contribute to wound healing and the formation of a fibrotic lesion (White and Gomer, 2015); M2b macrophages express high levels of C-C motif chemokine ligand 1 (CCL1) and tumor necrosis factor ligand superfamily member 14 (TNFSF14), and secrete large amounts of anti-inflammatory cytokines to promote Th2 differentiation and humoral immunity (Wang et al., 2019); M2c macrophages exhibit anti‐inflammatory activity by releasing large amounts of IL‐10, and over-express Mer receptor tyrosine kinase (MerTK) to efficiently phagocytize apoptotic cells (Shapouri-Moghaddam et al., 2018); M2d macrophages express high levels of IL-10, transforming growth factor-β (TGF-β), and vascular endothelial growth factor (VEGF) to constitute the main inflammatory component in tumor tissue, and contribute to angiogenesis and cancer metastasis (Wang et al., 2010). The presence of M2 subtypes in macrophage-associated diseases, such as kidney disease, atherosclerosis, and cancers, has been explored in several studies (Lu et al., 2013; Colin et al., 2014; Avila-Ponce de León et al., 2021; Zhao Y et al., 2021). Despite significant advances made in understanding the plasticity and function of macrophages, the molecular characterization of four M2 subtypes (M2a, M2b, M2c, and M2d) has not been fully investigated. Therefore, quantitative proteomic analysis of the four M2 subtypes is of value to predict their potential regulatory effects.
Typically, primary macrophages as an ideal cell model in vitro can maintain many of the important markers and functions seen in vivo. However, they have some disadvantages including their different genetic backgrounds, high individual variations, and limited availabilities. Cell line-derived macrophages have been widely used instead of primary macrophages to circumvent these problems. THP-1 is a human leukemia monocytic cell line, from which derived macrophages resemble primary macrophages in their morphological and functional properties, and differentiation markers. THP-1 has been widely used to study the functions, mechanisms, and signal transduction pathways of macrophages within a homogeneous genetic background with minimal variability (Chanput et al., 2014). In addition, THP-1 macrophages have been used in studies aimed at identifying markers of macrophages, especially in classical M1 and M2 phenotypes (Huang C et al., 2018). Nevertheless, the molecular characterization of the four M2 subtypes derived from THP-1 has not been reported.
In this study, we systematically characterized the proteomes of six different subtypes of macrophages (M0, M1, M2a, M2b, M2c, and M2d) derived from human THP-1 cells. By using bioinformatic and statistical analyses, we identified commonly or specially regulated proteins and their associated biological functions in the four M2 subtypes. In addition, differentially expressed proteins (DEPs) in the global proteomics dataset were randomly selected for analysis by a parallel reaction monitoring (PRM) assay to prove quantification accuracy, and these proteomic data were further used to predict macrophage-associated diseases.
2. Materials and methods
2.1. Materials
Human monocytic cell line THP-1 was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Roswell Park Memorial Institute (RPMI) 1640 medium was purchased from VivaCell (Shanghai, China). Penicillin-streptomycin, bovine serum albumin (BSA), BCA Protein Assay Kit, and phosphate-buffered saline (PBS) were purchased from Solarbio (Beijing, China). Heat-inactivated fetal bovine serum (FBS) was obtained from Biological Industries (Haifa, Israel). Phorbol-12-myristate-13-acetate (PMA), recombinant human interferon-γ (IFN-γ), lipopolysaccharide (LPS), IL-4, and IL-10 were purchased from Beyotime Biotechnology (Shanghai, China). Urea, ammonium bicarbonate (NH4HCO3), dithiothreitol (DTT), iodoacetamide (IAM), ovalbumin (OVA), and rabbit anti-OVA immunoglobulin G (IgG) were purchased from Merck (Darmstadt, Germany). 5'-N-ethylcarboxamidoadenosine (NECA) was purchased from MCE (NJ, USA). Sequencing grade trypsin was purchased from Promega (Madison, WI, USA). C18 column was purchased from Waters (Milford, MA, USA). Acetonitrile (ACN), trifluoroacetic acid (TFA), and formic acid (FA) were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
2.2. Cell culture and stimulation
THP-1 monocytes were cultured in RPMI 1640 medium supplemented with 10% (volume fraction) FBS and 1% (volume fraction) penicillin-streptomycin in an incubator with 5% CO2 at 37 ℃. The cells were treated with PMA (10 ng/mL) for 24 h to differentiate into M0 macrophages. The M0 macrophages were stimulated with IFN-γ (100 ng/mL) and LPS (50 ng/mL) for 24 h to induce the M1 phenotype, and stimulated with IL-4 (20 ng/mL) for 24 h to induce the M2a phenotype. The immune complexes IgG-OVA were prepared by mixing rabbit anti-OVA IgG (150 µg/mL) and OVA (15 µg/mL) for 30 min at room temperature. Then, M0 macrophages were stimulated with LPS (100 ng/mL) and immune complexes (IgG-OVA) for 24 h to obtain the M2b phenotype. The M0 macrophages were stimulated with IL-10 (10 ng/mL) for 24 h to induce the M2c phenotype, while NECA (5 µmol/L) and LPS (100 ng/mL) were added for 24 h to induce the M2d phenotype (Huang X et al., 2018). Images of cells were acquired using an inverted fluorescence microscope (CKX53, Olympus, Japan).
2.3. Sample preparation
The detailed procedures for sample preparation have been described by Li et al. (2021). Briefly, cells on culturing dishes were washed three times with pre-cooled PBS (pH 7.4) and lysed directly with a lysis buffer with a final concentration of 8 mol/L urea and 1 mol/L NH4HCO3 solution. Lysates were briefly sonicated until the solution was clear, and then centrifuged at 1400g for 15 min to collect the supernatant. The protein concentrations were measured by BCA protein assay reagent. To reduce the biological variance, the samples from three biological replicates were pooled into one sample for further analyses.
The denatured proteins were reduced by 5 mmol/L DTT at 37 ℃ for 1 h, and then alkylated by 15 mmol/L IAM in the dark for 30 min at room temperature. The remaining IAM was quenched by adding 2.5 mmol/L DTT and incubated for 10 min at room temperature. The protein solutions were first diluted two-fold with deionized water and then gently shaken with added sequencing grade trypsin (protein:enzyme=100:1, mass ratio) for 2 h at 37 ℃. The solutions were further diluted four-fold with deionized water, and then digested overnight with sequencing grade trypsin (protein: enzyme=100:1, mass ratio) at 37 ℃. The sample solutions were acidified to pH<2 with TFA, centrifuged at 15 000g for 15 min, and then the digested peptides in the supernatant were purified using a C18 column. The peptide was eluted with 60% (volume fraction) ACN and 0.1% (volume fraction) TFA, and the concentration was measured using an ultra-micro spectrophotometer DS-11 (DeNovix, DE, USA). Finally, peptides were dried by Speed-Vacuum (Marin Christ, Osterode, Germany) and resuspended in 15 µL 0.1% (volume fraction) FA before liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
2.4. LC-MS/MS analysis
The peptide samples were analyzed by LC-MS/MS on an Orbitrap Fusion Lumos Mass Spectrometer (Thermo Fisher Scientific, Bremen, Germany). Peptides were separated on the EASY-nano-LC System (Thermo Fisher Scientific, Bremen, Germany) with an Acclaim PepMap100 pre-column (75 µm×2 cm) and an Acclaim PepMap100 separating column (75 µm×50 cm). The mobile phase consisted of 0.1% FA in water (A) and 0.1% FA in 80% ACN (B), and had a flow rate of 200 nL/min. The spray voltage was set at 2.4 kV. The gradient profile was set as follows: 3%‒7% B for 2 min, 7%‒35% B for 166 min, 35%‒68% B for 40 min, 68%‒99% B for 10 min, and maintained in 99% B for 22 min, for a total of 240 min. The MS1 spectra were collected from 350 to 1800 m/z (mass-to-charge ratio) at a resolution of 60 000, with an automatic gain control (AGC) target of 4×105. The precursor ions were fragmented by higher-energy collisional dissociation (collision energy of 30%), and the MS/MS spectra were collected in data-dependent acquisition (DDA) mode with a resolution of 15 000, charge states range of 2 to 7, isolation window of 1.6 m/z, dynamic exclusion time of 45 s, maximum injection time of 30 ms, and an AGC target of 5×104.
2.5. Parallel reaction monitoring analysis
PRM was used to analyze the expression of specific markers identified in different subtypes of macrophages (Peterson et al., 2012), including previously reported markers of macrophages as well as proteins commonly or specially up-regulated in the four M2 macrophage subtypes. One unique peptide per protein was selected, and a mass list table containing 30 peptides was created for PRM analysis. The gradient profile was set as follows: 3%‒7% B for 2 min, 7%‒35% B for 83 min, 35%‒68% B for 20 min, 68%‒100% B for 5 min, and maintained in 100% B for 20 min, for a total of 130 min. The PRM method was set in time-scheduled acquisition mode with a retention time of ±5 min, higher-energy collisional dissociation (HCD) collision energy of 30%, isolation window of 0.7 m/z, maximum injection time of 100 ms, and an AGC target of 1×105. Targeted MS/MS spectra were collected from 200 to 2000 m/z with a resolution of 30 000. Skyline software was used to search PRM data and perform relative quantification (MacLean et al., 2010).
2.6. Database search for protein identification and quantification
The MS/MS data were searched against the human proteome database from the Uniprot website (https://www.uniprot.org, downloaded in May, 2019) using Thermo Proteome Discoverer 2.3 (Thermo Fisher Scientific, Germany). The database search parameters permitted up to two missed cleavage sites for trypsin digestion. The tolerances for precursor and fragment masses were set at 10 ppm (parts per million) and 0.02 Da, respectively. Carbamidomethylation (C, +57.0215 Da) was set as a fixed modification, while oxidization (M, +15.9949 Da) and N-terminal acetylation (+42.010565 Da) were set as variable modifications. The results were filtered with 1% false discovery rate (FDR) and Score Sequest HT>1.5 at the peptide level, and at least five peptide-spectrum matches (PSMs) were required for a protein identification. Label-free quantification (LFQ) from Proteome Discoverer 2.3 was used for protein quantification. The peptide group ratios between different macrophage subtypes were calculated as the geometric median of all combinations of ratios from all the replicates for the selected study factors. The protein ratio between different macrophage subtypes was subsequently calculated as the geometric median of the above peptide group ratios.
2.7. Bioinformatic analysis
Principal component analysis (PCA) and hierarchical clustering analysis were performed using the "gmodels" and "heatmap" package in R language, respectively. The Database for Annotation, Visualization, and Integrated Discovery (DAVID) V6.8 (https://david.ncifcrf.gov/home.jsp) was used to perform Gene Ontology (GO) with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses by whole genome as background (Dennis et al., 2003). Reactome pathway analysis was performed using ClueGO plug-in and Cluepedia from Cytoscape (Bindea et al., 2009). Protein-protein interaction (PPI) network analysis was performed using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org) (Szklarczyk et al., 2015), and interactions with a combined score of >0.15 were selected to construct the PPI networks using Cytoscape (Snel et al., 2000). The highly interactive modules were constructed with Molecular Complex Detection (MCODE) in Cytoscape (Bader and Hogue, 2003).
2.8. Disease correlation analysis
A network-based prediction method was used to further investigate disease associations in the four M2 macrophage subtypes. Based on the specially regulated proteins in the different M2 macrophage subtypes, disease correlation modules (P<0.05) from the MalaCards database were constructed using GeneAnalytics (http://geneanalytics.genecards.org) (Ben-Ari Fuchs et al., 2016).
3. Results
3.1. Polarization of THP-1 monocytes into different subtypes of macrophages
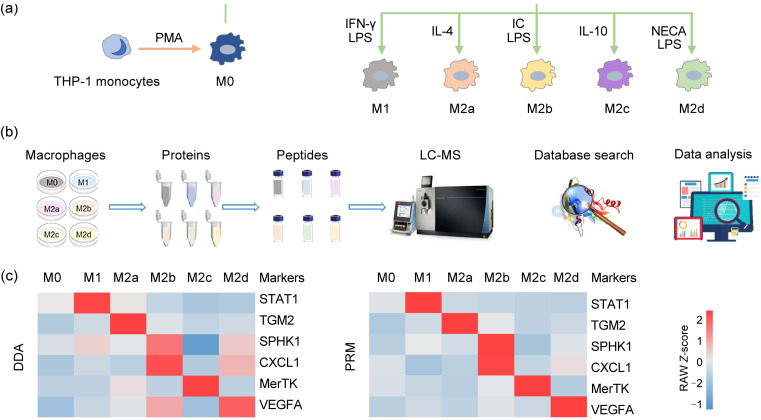
The proteomic characteristics of THP-1 cells might vary when different stimulation protocols are applied. Therefore, the standardization schemes described previously were used for the following processes (Huang X et al., 2018). THP-1 monocytes were first differentiated into a M0 macrophage state using PMA. M0 macrophages were then polarized into M1 by IFN-γ and LPS, into M2a by IL-4, into M2b by LPS and immune complexes, into M2c by IL-10, and into M2d by NECA and LPS (Fig. 1a). Different macrophage phenotypes showed obviously different morphologies. The M0 phenotype showed a round shape, the M1 phenotype had a spindle shape with increased pseudopodia, the M2a phenotype formed numerous filopodia, and the M2b, M2c, and M2d phenotypes all had a spindle or round shape (Fig. S1). After multistep experimental processing, these macrophage subtypes were all characterized by MS-based proteomics (Fig. 1b).
Fig. 1. Preparation and validation of different macrophage phenotypes induced from THP-1 monocytes. (a) Processes of inducing THP-1 monocytes into different macrophage phenotypes using different inducers. (b) Workflow of this study, including the cell culture, protein extraction, peptide enrichment, liquid chromatography-mass spectrometry (LC-MS) data generation, database search, and data analysis. (c) Protein expression levels of several recognized markers in M0, M1, M2a, M2b, M2c, and M2d macrophages were analyzed using two MS-based proteomic methods. PMA: phorbol-12-myristate-13-acetate; IFN: interferon; LPS: lipopolysaccharide; IL: interleukin; IC: immune complex; NECA: 5'-N-ethylcarboxamidoadenosine; DDA: data-dependent acquisition; PRM: parallel reaction monitoring; STAT1: signal transducer and activator of transcription 1; TGM2: transglutaminase 2; SPHK1: sphingosine kinase 1; CXCL1: C-X-C motif chemokine ligand 1; MerTK: Mer receptor tyrosine kinase; VEGFA: vascular endothelial growth factor A.
To evaluate the modeling of different phenotypes of macrophages, both discovery and targeted-based MS approaches were used to determine the protein levels of several recognized polarization markers. The M1 marker STAT1 (Ivashkiv, 2018), the M2a marker transglutaminase 2 (TGM2) (Martinez et al., 2013), the M2b markers sphingosine kinase 1 (SPHK1) and C-X-C motif chemokine ligand 1 (CXCL1) (Lee et al., 2014; Wang et al., 2019), the M2c marker MerTK (Zizzo et al., 2012), and the M2d marker VEGFA (Ferrante et al., 2013) were obviously increased in the corresponding phenotypes (Fig. 1c and Table S1). These data confirmed that the THP-1 macrophages were successfully polarized to M1 and the four M2 macrophage subtypes.
3.2. Global proteomic profiling of polarized macrophages
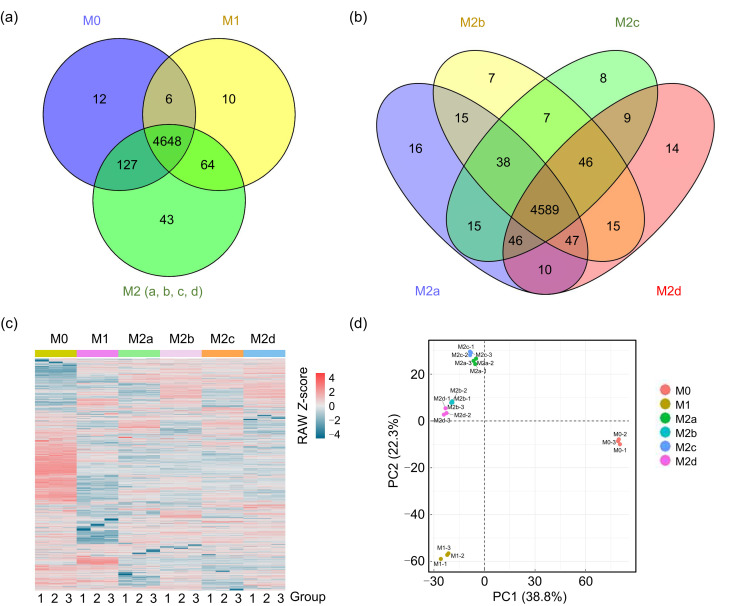
From the six macrophage subtypes, a total of 6616 proteins were identified within a cutoff of 1% FDR. After filtering these proteins with PSM≥5, a total of 4910 proteins were considered eligible for further quantitative analysis (Fig. 2a). By using the same filters, a total of 4882 proteins were used for further quantitative analysis among the four M2 subtypes of macrophages (Fig. 2b). Quantified proteins in the six subtypes are shown in a heatmap (Fig. 2c). In addition, the PCA analysis showed that the four M2 subtypes were clustered in the same quadrant, while M0 and M1 were distributed in different quadrants (Fig. 2d), indicating that the protein expression profiles of four M2 macrophage subtypes had more similarities than those of M0 and M1.
Fig. 2. Quantitative proteomic profiling of M0, M1, M2a, M2b, M2c, and M2d macrophages. (a) Venn diagram of quantifiable proteins among M0, M1, and all four M2 macrophage subtypes. (b) Venn diagram of quantifiable proteins among M2a, M2b, M2c, and M2d macrophages. (c) Heatmap of quantified proteins in macrophages. Groups indicate three technical repeats under each condition. (d) Principal component analysis of quantitative proteomics in six macrophage subtypes. PC: principal component.
3.3. Functional analysis of four M2 macrophage subtypes
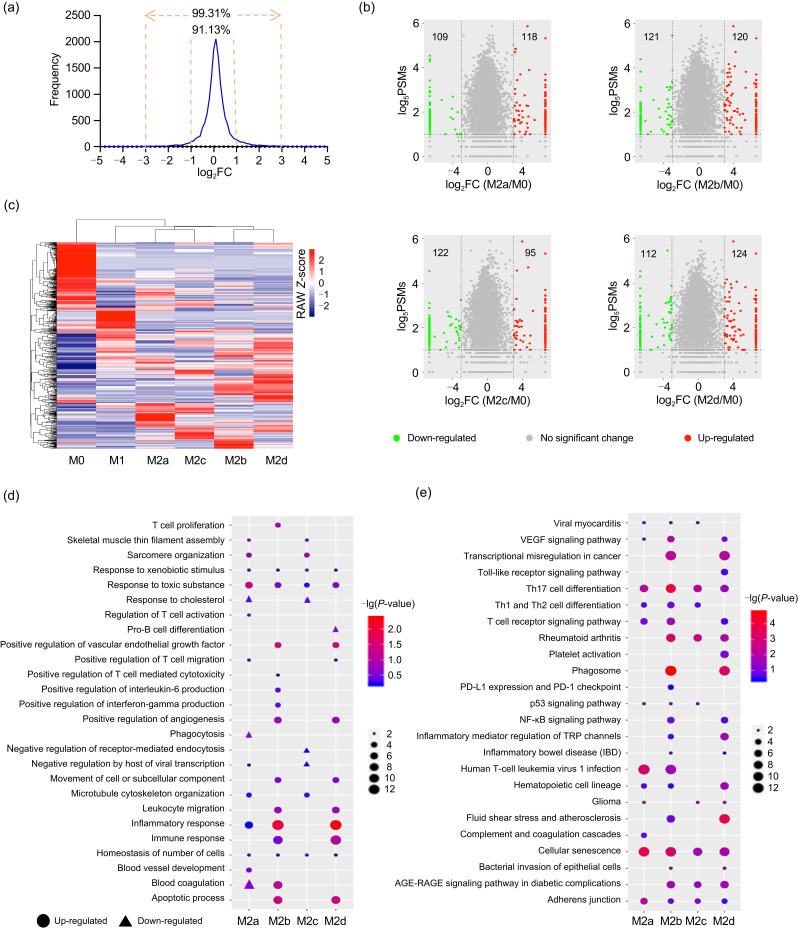
We first determined the criteria for screening DEPs by investigating the technical reproducibility of triplicate MS data from M0 macrophages. The results showed that 90% of proteins were within a two-fold change among triplicate MS data, and more than 99% of proteins were within an eight-fold change (Fig. 3a). By using eight-fold change as a cutoff to determine the DEPs (compared with M0), we identified 118 (109), 120 (121), 95 (122), and 124 (112) up-regulated (down-regulated) proteins in M2a, M2b, M2c, and M2d, respectively (Fig. 3b and Table S2). The hierarchical clustering analysis showed that the four M2 subtypes were tightly clustered. In particular, M2a was clustered with M2c, while M2b was clustered with M2d (Fig. 3c).
Fig. 3. Identification and function analyses of DEPs between M2 subtypes and M0 macrophages. (a) Ratio distributions of quantified proteins among technical replicates of the M0 phenotype. The cutoff was set as two- or eight-fold change. (b) Volcano plot of DEP distributions in the four M2 subtypes. (c) Clustering analysis of respective DEPs in the four subtypes of M2 macrophages. (d) Biological processes analysis of DEPs in the four subtypes of M2 macrophages. (e) KEGG pathway analysis of DEPs in the four subtypes of M2 macrophages. (d, e) The lists of input DEPs were shown in Table S2. DEPs: differentially expressed proteins; PSMs: peptide-spectrum matches; VEGF: vascular endothelial growth factor; Th: T helper; PD-L1: programmed death-ligand 1; NF-κB: nuclear factor-κB; TRP: transient receptor potential; AGE: advanced glycation end-product; RAGE: receptor for AGE.
GO analysis was undertaken to facilitate understanding of the biological significance of the above up- or down-regulated DEPs (the lists of input DEPs are shown in Table S2) in the different M2 macrophage subtypes (Fig. 3d and Table S3). The up-regulated proteins in four subtypes of M2 macrophages were commonly involved in homeostasis of a number of cells, and responded to toxic substances and xenobiotic stimulus; up-regulated proteins in M2a, M2b, and M2d were commonly involved in the response to inflammation; up-regulated proteins in M2a and M2c commonly participated in microtubule cytoskeleton organization, sarcomere organization, and skeletal muscle thin filament assembly; up-regulated proteins in M2b and M2d commonly participated in the apoptotic process, immune response, leukocyte migration, movement of cell or subcellular components, as well as positive regulation of angiogenesis and VEGF production. The down-regulated proteins in M2a and M2c commonly participated in response to cholesterol.
To find out the important pathways of these DEPs in the different M2 macrophage subtypes, all DEPs (the lists of input DEPs are shown in Table S2) of each M2 subtype were analyzed by KEGG pathway analysis (Fig. 3e and Table S3). The results indicated that the DEPs of the different M2 subtypes were commonly involved in Th17 cell differentiation, cellular senescence, and adherens junction; DEPs in M2a, M2b, and M2c commonly participated in signaling pathways of Th1 and Th2 cell differentiation, and viral myocarditis; DEPs in M2a, M2b, and M2d were commonly involved in the VEGF signaling pathway, T cell receptor signaling pathway, and hematopoietic cell lineage; DEPs in M2a, M2c, and M2d participated in glioma; DEPs in M2b, M2c, and M2d commonly participated in rheumatoid arthritis and the advanced glycation end-product (AGE)-receptor for AGE (RAGE) signaling pathway in diabetic complications; DEPs in M2b and M2d were commonly involved in atherosclerosis, inflammatory bowel disease (IBD), inflammatory mediator regulation of transient receptor potential (TRP) channels, nuclear factor-κB (NF-κB) signaling pathway, phagosome, and transcriptional misregulation in cancer.
3.4. Commonly altered proteins in the four subtypes of M2 macrophages
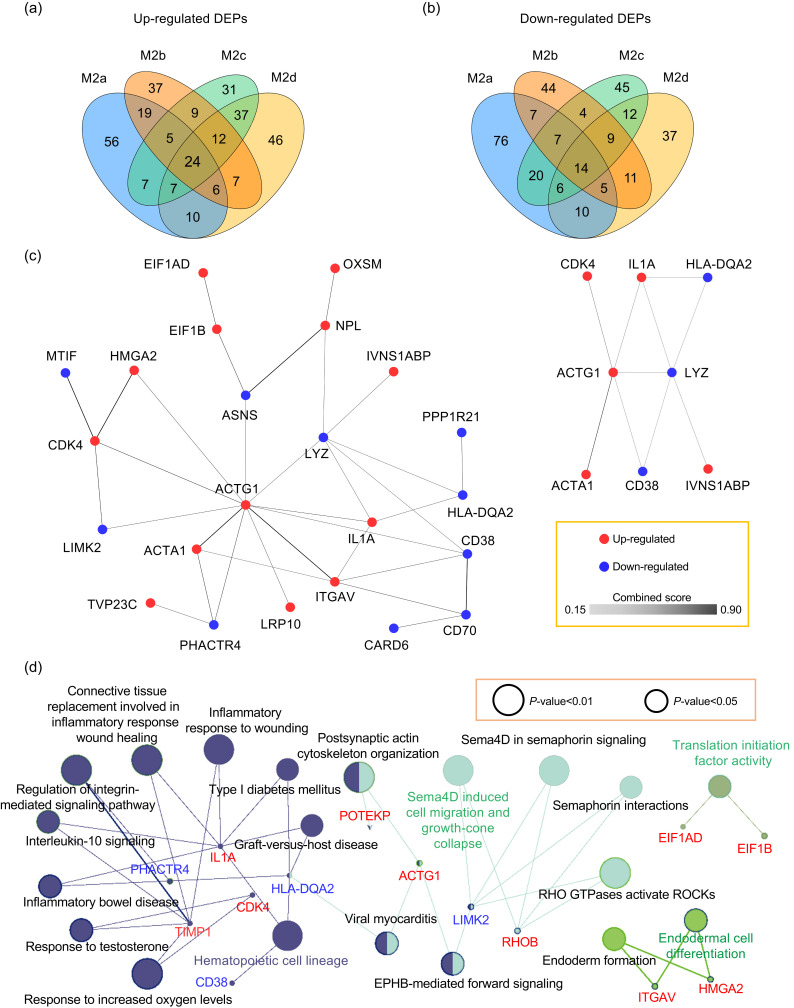
Since some DEPs of the four M2 macrophage subtypes (M0 macrophages as control) may also be differentially expressed in the M1 macrophage, the common DEPs in the four M2 macrophage subtypes were screened using both M0 and M1 macrophages as controls. When using a cutoff of eight-fold, only a few DEPs were common to all M2 subtypes. By adjusting the cutoff to a two-fold change, additional DEPs (including 24 commonly up-regulated proteins and 14 commonly down-regulated proteins) were identified in all M2 subtypes (Figs. 4a and 4b, Table S4). Based on the PPI results, 23 of these had interactions, and the highly interacting proteins included γ-actin 1 (ACTG1), α-actin 1 (ACTA1), cyclin-dependent kinase 4 (CDK4), IL1A, adenosine diphosphate (ADP)-ribosyl cyclase 1 (ADPRC1)/CD38, lysozyme (LYZ), human leukocyte antigen (HLA)-DQA2, and influenza virus non-structural-1A-binding protein (IVNS1ABP) (Fig. 4c).
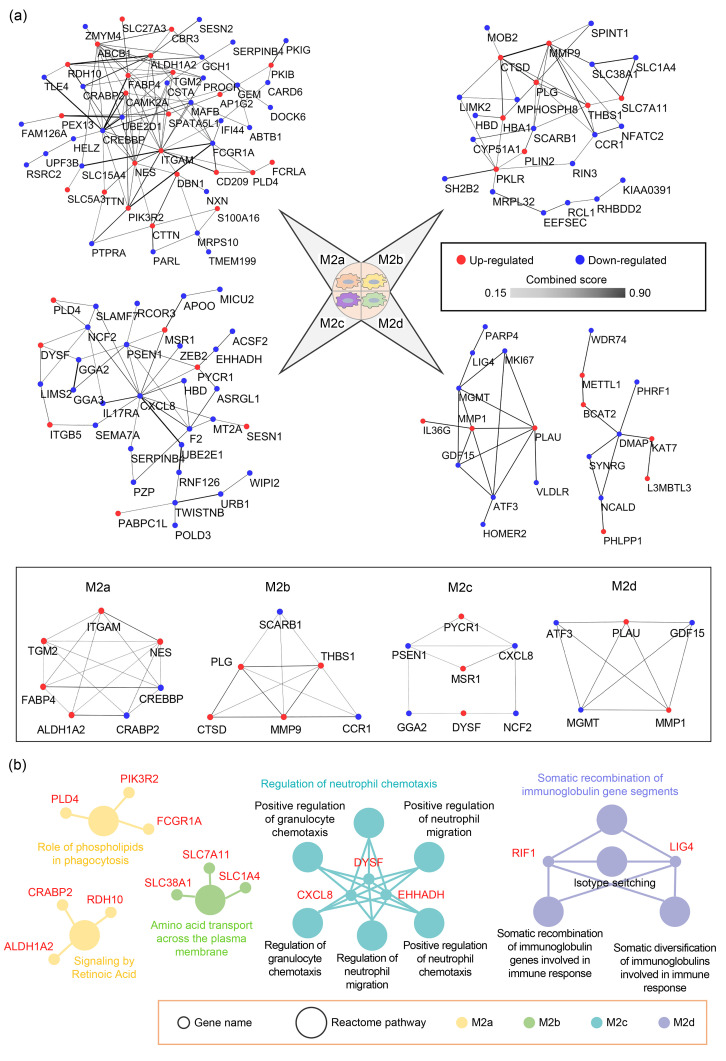
Fig. 4. PPI networks and Reactome pathways involving the common DEPs in the four M2 macrophage subtypes. (a) Venn diagram showing commonly up-regulated proteins of the four M2 subtypes. (b) Venn diagram showing commonly down-regulated proteins of the four M2 subtypes. (c) PPI network of common DEPs (left) and module of highly interacting proteins (right). Edges represent protein‒protein associations, and the colors varying from light to dark represent the combined score of DEPs from 0.15 to 0.90. (d) Reactome pathways of common DEPs in the four M2 subtypes. Common DEPs were screened in the four M2 macrophage subtypes with M0 and M1 macrophages as control groups. (c, d) The list of input DEPs is shown in Table S4. Red indicates commonly up-regulated proteins and blue indicates commonly down-regulated proteins. PPI and Reactome pathways enrichment P-value<0.05. PPI: protein‒protein interactions; DEPs: differentially expressed proteins(Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
To improve quantitation accuracy of the observed common DEPs, five of the commonly up-regulated proteins were randomly selected for further quantification analysis by targeted proteomics based on PRM (Table S5). The results confirmed that all five proteins, including ACTG1, integrin subunit αV (ITGAV), IVNS1ABP, IL1A, and tissue inhibitor of metalloproteinase-1 (TIMP1), were commonly up-regulated in the four M2 macrophage subtypes (relative to M0 and M1) (Table 1). These results indicated that over-expression of the above five proteins is a common characteristic of all four M2 subtype macrophages, and therefore these proteins have potential to serve as additional markers to distinguish any subtypes of M2 from M0 and M1 macrophages.
Table 1.
Targeted proteomics of commonly up-regulated proteins in the four M2 macrophage subtypes
| Gene name | Abundance ratio (relative to M0) | Abundance ratio (relative to M1) | ||||||
|---|---|---|---|---|---|---|---|---|
| M2a | M2b | M2c | M2d | M2a | M2b | M2c | M2d | |
| ACTG1 | 24.29 | 15.12 | 10.44 | 15.59 | 6.87 | 4.28 | 2.95 | 4.41 |
| ITGAV | 7.78 | 6.20 | 9.40 | 7.65 | 4.00 | 3.19 | 4.84 | 3.94 |
| IVNS1ABP | 4.00 | 4.05 | 3.86 | 5.32 | 2.26 | 2.29 | 2.19 | 3.01 |
| IL1A | 10.32 | 8.34 | 14.14 | 7.84 | 13.99 | 11.30 | 19.16 | 10.62 |
| TIMP1 | 4.18 | 3.18 | 2.36 | 2.82 | 3.73 | 2.83 | 2.11 | 2.52 |
ACTG1: γ-actin 1; ITGAV: integrin subunit αV; IVNS1ABP: influenza virus NS1A-binding protein; IL1A: interleukin 1α; TIMP1: tissue inhibitor of metalloproteinase-1.
Reactome pathway analysis showed that the common DEPs in the four M2 subtypes were associated with connective tissue replacement involved in hematopoietic cell lineage, semaphorin 4D (sema4D)-induced cell migration and growth-cone collapse, translation initiation factor activity, and endodermal cell differentiation (Fig. 4d).
3.5. Specially altered proteins in the four subtypes of M2 macrophages
By using the other five macrophage subtypes as controls and a two-fold cutoff, many special DEPs in each of the four M2 macrophage subtypes were identified, including 29 (36), 12 (34), 7 (38), and 11 (20) up-regulated (down-regulated) proteins in M2a, M2b, M2c, and M2d macrophages, respectively (Table S6). The interaction analyses showed that the highly interacting proteins included: integrin subunit αM (ITGAM), TGM2, aldehyde dehydrogenase 1 family member A2 (ALDH1A2), fatty acid-binding protein 4 (FABP4), cyclic adenosine monophosphate (cAMP)-response element-binding protein (CREB)-binding protein (CREBBP), cellular retinoic acid-binding protein 2 (CRABP2), and nestin (NES) in M2a; scavenger receptor class B member 1 (SCARB1), matrix metallopeptidase 9 (MMP9), thrombospondin 1 (THBS1), cathepsin D (CTSD), plasminogen (PLG), and C-C motif chemokine receptor 1 (CCR1) in M2b; macrophage scavenger receptor 1 (MSR1), dysferlin (DYSF), pyrroline-5-carboxylate reductase 1 (PYCR1), CXCL8, neutrophil cytosolic factor 2 (NCF2), Golgi-localized γ-adaptin ear-containing ADP-ribosylation factor-binding protein 2 (GGA2), and presenilin 1 (PSEN1) in M2c; and MMP1, growth differentiation factor 15 (GDF15), activating transcription factor 3 (ATF3), urokinase-type PLG activator (PLAU), and O 6-methylguanine-DNA methyl-transferase (MGMT) in M2d (Fig. 5a). These highly interacting proteins can be considered as the key proteins in each M2 macrophage subtype for further investigation.
Fig. 5. PPI networks and Reactome pathways involving the special DEPs in the four M2 macrophage subtypes. (a) PPI networks (top) and highly interactive proteins (down) of special DEPs in M2a, M2b, M2c, and M2d macrophages. Edges represent protein‒protein associations, and the colors varying from light to dark represent the combined score of DEPs from 0.15 to 0.90. (b) Reactome pathways of special DEPs in the four M2 subtypes. The list of input DEPs is shown in Table S6. PPI and Reactome pathways enrichment P-value<0.05. PPI: protein‒protein interactions; DEPs: differentially expressed proteins(Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
To improve the quantification accuracy of the observed DEPs, 18 DEPs highly expressed in one of the M2 macrophage subtypes were randomly selected for further quantification analysis by targeted proteomics based on PRM (Tables 2 and S7). The PRM results were consistent with DDA-based proteomic data, showing that: FABP4, ITGAM, ALDH1A2, NES, drebrin 1 (DBN1), and adenosine triphosphate (ATP)-binding cassette subfamily B member 1 (ABCB1) were highly expressed in M2a; CTSD, perilipin 2 (PLIN2), MMP9, hemoglobin subunit α1 (HBA1), THBS1, and PLG were highly expressed in M2b; integrin subunit β5 (ITGB5), poly(A)-binding protein cytoplasmic 1 like (PABPC1L), and MSR1 were highly expressed in M2c; and methyltransferase-like protein 1 (METTL1), replication timing regulatory factor 1 (RIF1), and lysine acetyltransferase 7 (KAT7) were highly expressed in M2d. We also observed changes in expression of the above proteins in each polarized subtype (relative to M0 macrophages), with a two-fold change used as a filter (Fig. S2). The results showed that: DBN1 and ABCB1 were solely up-regulated in M2a conditions; CTSD, PLIN2, HBA1, and PLG were solely up-regulated in M2b conditions; and RIF1 and KAT7 were solely up-regulated in M2d conditions. These proteins might be useful as references for distinguishing different subtypes of M2 macrophages. To explore the biological significance of special DEPs in the four M2 subtypes, Reactome pathway analysis was performed (Fig. 5b). The results showed that: M2a participated in roles of phospholipids in phagocytosis and signaling by retinoic acid; M2b participated in amino acid transport across the plasma membrane; M2c participated in regulation of neutrophil chemotaxis; and M2d participated in somatic recombination of immunoglobulin gene segments.
Table 2.
Targeted proteomics of specially up-regulated proteins in M2a, M2b, M2c, and M2d macrophages
| Subtype | Gene name | M2a/M0 | M2a/M1 | M2a/M2b | M2a/M2c | M2a/M2d |
|---|---|---|---|---|---|---|
| M2a | FABP4 | 40.63 | 7.86 | 50.67 | 2.64 | 17.88 |
| ITGAM | 50.08 | 16.15 | 14.48 | 2.93 | 10.10 | |
| ALDH1A2 | 120.85 | 35.25 | 49.83 | 20.31 | 44.48 | |
| NES | 29.38 | 29.77 | 9.91 | 2.82 | 33.39 | |
| DBN1 | 2.96 | 14.89 | 10.00 | 2.13 | 11.32 | |
| ABCB1 | 3.33 | 2.32 | 3.25 | 2.40 | 5.82 | |
| M2b | CTSD | 3.81 | 7.86 | 4.77 | 4.39 | 2.80 |
| PLIN2 | 4.69 | 2.80 | 2.76 | 3.50 | 2.41 | |
| MMP9 | 8.32 | 9.18 | 9.98 | 5.70 | 3.17 | |
| HBA1 | 3.00 | 2.36 | 2.32 | 6.80 | 2.97 | |
| THBS1 | 7.41 | 8.05 | 7.26 | 3.82 | 2.49 | |
| PLG | 2.01 | 2.53 | 4.28 | 7.39 | 2.80 | |
| M2c | ITGB5 | 19.54 | 15.75 | 2.35 | 6.37 | 6.19 |
| PABPC1L | 35.33 | 3.33 | 3.32 | 18.67 | 4.81 | |
| MSR1 | 5.36 | 14.31 | 2.16 | 9.36 | 93.21 | |
| M2d | METTL1 | 11.75 | 3.21 | 21.79 | 3.44 | 2.98 |
| RIF1 | 2.54 | 5.30 | 2.77 | 2.22 | 3.43 | |
| KAT7 | 2.92 | 3.66 | 4.52 | 3.26 | 2.97 |
FABP4: fatty acid binding protein 4; ITGAM: integrin subunit αM; ALDH1A2: aldehyde dehydrogenase 1 family member A2; NES: nestin; DBN1: drebrin 1; ABCB1: adenosine triphosphate-binding cassette subfamily B member 1; CTSD: cathepsin D; PLIN2: perilipin 2; MMP9: matrix metallopeptidase 9; HBA1: hemoglobin subunit α1; THBS1: thrombospondin 1; PLG: plasminogen; ITGB5: integrin subunit β5; PABPC1L: poly(A)-binding protein cytoplasmic 1 like; MSR1: macrophage scavenger receptor 1; METTL1: methyltransferase-like protein 1; RIF1: replication timing regulatory factor 1; KAT7: lysine acetyltransferase 7.
3.6. Immune-associated diseases related to the four subtypes of M2 macrophages
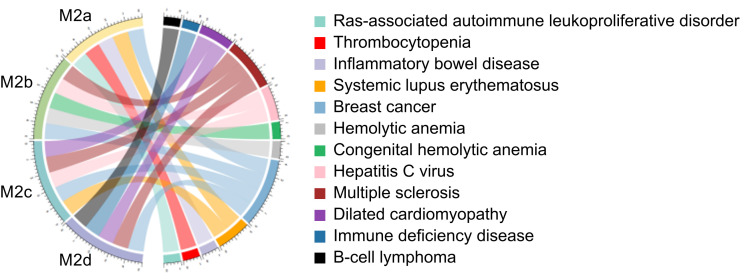
Immune-associated diseases were predicted for the four M2 subtypes based on the above special DEPs (Fig. 6 and Table S8). The results indicated that: M2a macrophages were involved in diseases such as Ras-associated autoimmune leukoproliferative disorder, thrombocytopenia, IBD, systemic lupus erythematosus (SLE), and breast cancer; M2b macrophages participated in anemia diseases, hepatitis C virus (HCV), multiple sclerosis, and breast cancer; M2c macrophages were related to multiple sclerosis, breast cancer, SLE, HCV, and dilated cardiomyopathy; and M2d macrophages were related to immune deficiency disease, B-cell lymphoma, dilated cardiomyopathy, multiple sclerosis, and breast cancer.
Fig. 6. Predicting immune-associated diseases of special DEPs in the four M2 subtypes. Enrichment P-value<0.05. DEPs: differentially expressed proteins.
4. Discussion
Macrophages play key roles in inflammation, immunity, tissue repair, and cancer. Different macrophage phenotypes and associated functions are important characteristics of pathological conditions. Macrophages can be differentiated into classically activated macrophages (M1) and alternative activated macrophages (M2), mainly in relation to their pro-inflammatory and anti-inflammatory functions, respectively (Mosser and Edwards, 2008). In recent years, THP-1-derived macrophages have been widely used in various macrophage-associated studies. However, most of these studies focused mainly on classical M1 and M2 phenotypes. The biological functions of different M2 subtypes (M2a, M2b, M2c, and M2d) have not been systematically delineated.
In this study, we performed quantitative proteome analysis on human THP-1-derived macrophages (M0, M1, M2a, M2b, M2c, and M2d). Our results indicated for the first time that M2a and M2c, and M2b and M2d have very similar expression profiles. In addition, M2a and M2c were shown to be involved in skeletal muscle thin filament assembly and sarcomere organization; M2b and M2d were involved in transcriptional misregulation in cancer.
This study identified a list of common DEPs in the four M2 subtypes (Table S4). Among these DEPs, IL1A, CD38, and HLA-DQA2 were related to hematopoietic cell lineage. di Paolo and Shayakhmetov (2016) showed that IL1A production from macrophages can trigger the recruitment of hematopoietic cells. Prosper et al. (1997) revealed that 10% of long-term culture-initiating cells (LTC-ICs) in mobilized peripheral blood (PB) are CD34+HLA-DR- and CD34+CD38-, and can sustain hematopoiesis for a long time. These results suggest that all four M2 subtypes may play crucial roles in hematopoietic function. In addition, nonmuscle cytoskeletal actin ACTG1, known to be involved in various types of cell motility (Drazic et al., 2018), was commonly up-regulated in the four M2 subtypes, suggesting its role in M2 macrophage migration.
We identified a list of special and highly interactive DEPs in the four M2 subtypes (Table S6), and further explored their links to the underlying biological functions of the M2 subtypes. The special DEPs of M2a were relevant to phagocytosis and retinoic acid. Retinoic acid is reported to increase phagocytosis of myelin by macrophages, and to induce macrophages to develop a noninflammatory phenotype (Wu et al., 2021). In highly interactive DEPs of M2a macrophages, integrin ITGAM is implicated in various adhesive interactions of macrophages, and mediates the uptake of complement-coated particles and pathogens, thereby driving M2 polarization to promote STAT6 activation via IL‐13 and IL‐4 signaling (Song et al., 2019); TGM2 is related to bone development, angiogenesis, and wound healing. The inhibition of TGM2 activity and protein silencing proved that the reduction of TGM2 significantly weakens the efferocytosis of macrophages, reduces the secretion of anti-inflammatory factor IL-10, and triggers a pro-inflammatory phenotype (Eligini et al., 2016); ALDH1A2 is involved in the retinol metabolism pathway and tissue development. In in vitro and mouse models, the inhibition of microRNA-33 induced M2 polarization and increased ALDH1A2 expression, which contributed to the stabilization of plaques in atherosclerosis(Ouimet et al., 2015). In addition, Martinez et al. (2013) confirmed that ITGAM, TGM2, and ALDH1A2 were up-regulated in IL-4 activated human and mouse macrophages, which is consistent with our results. The special DEPs of M2b were involved in amino acid transport across the plasma membrane. Rabinowitz et al. (2021) showed that macrophages can promote antiretroviral activity by regulating amino acid transport. Among the highly interactive DEPs of M2b macrophages, CTSD was related to inflammatory diseases, and inhibiting its expression in intestinal macrophages of inflamed mucosa can ameliorate IBD (Menzel et al., 2006); MMP9 played an essential role in local proteolysis of the extracellular matrix and in leukocyte migration. In the early stages of pulmonary fibrosis, an increase in MMP9 released by macrophages was involved in macrophage-induced fibroblast migration(Li et al., 2019); PLG played an important role in the regression of inflammation. It can combine with its receptor PLG receptor KT (PLG-RKT) to regulate the polarization and endocytosis of macrophages (Vago et al., 2019). The special DEPs of M2c were involved in regulation of neutrophil chemotaxis. There is evidence to indicate that macrophage‐derived cytokines are possible mediators of neutrophil mobilization and chemotaxis during development of sterile peritonitis in the rat (Knudsen et al., 2002). Among the highly interactive DEPs of M2c macrophages, MSR1 is an MSR. Activating MSR1 in M2 macrophages can enhance c-Jun N-terminal kinase (JNK) activation, which promotes the transition of the phenotype from an anti-inflammatory state to a pro-inflammatory state (Guo et al., 2019); PYCR1 was a housekeeping enzyme that catalyzed the last step in proline biosynthesis. Evidence showed that PYCR1 can combine with Lon to induce reactive oxygen species (ROS)-dependent production of inflammatory cytokines in tumors, thereby enhancing epithelial-mesenchymal transition, angiogenesis, and M2 macrophage polarization (Kuo et al., 2020); DYSF is located at monocyte cell adhesion sites and is involved in cell interactions. The absence of DYSF from monocytes and macrophages strongly reduced adhesion and increased the motility of the cells (de Morrée et al., 2013). The special DEPs of M2d were involved in somatic recombination of immunoglobulin gene segments. Immunoglobulin expression in macrophage populations negatively correlates with tumor volume (Busch et al., 2019). Among the highly interactive DEPs of M2d macrophages, MMP1 is involved in various physiological and pathological processes such as wound healing, arthritis, atherosclerosis, and tumor progression. Activation of the MMP1 signaling pathway can promote macrophage infiltration (Liao et al., 2020); Urokinase-type PLG activator (PLAU) is crucial to collagenolysis and neovascularization. Recent evidence showed that IL-6 could promote thrombus resolution by enhancing PLAU expression in macrophages (Nosaka et al., 2020). Overall, the underlying biological functions of the DEPs are principal characteristics of the different THP-1-derived M2 subtypes. Note that the microenvironment may also have an impact on the physiological conditions of different macrophage subtypes. This was not considered in the current study and may need to be explored in future research.
To further explore the relationship between macrophages and diseases, we predicted macrophage-associated diseases for the four THP-1-derived M2 subtypes. All four M2 subtypes were commonly involved in breast cancer. Previous studies have suggested that macrophages are a major component of various solid tumors (Chen et al., 2019; Zhao et al., 2021), comprising up to about 50% of the tumor mass in breast carcinomas (Lewis et al., 1995). M2b, M2c, and M2d macrophages are involved in multiple sclerosis. Accumulating evidence suggests that M2 macrophages contribute to tissue reparation and can limit inflammation in multiple sclerosis (Batchu, 2020). In addition, M2a is associated with Ras-associated autoimmune leukoproliferative disorder that often triggers thrombocytopenia and SLE (Li et al., 2020; Neven et al., 2021). M2b was associated mainly with hemolytic anemia, which has been reported to be associated with macrophage activation syndrome (Minoia et al., 2021). M2c was associated with dilated cardiomyopathy. It has been suggested that M2 macrophages can reduce cardiac fibrosis in human inflammatory dilated cardiomyopathy (Baumeier et al., 2021). The M2d subtype was related to immune deficiency disease. Evidence showed that macrophages not only are susceptible to human immunodeficiency virus type 1 (HIV-1) infection, but also assist in sustaining viral persistence (Hendricks et al., 2021). These findings imply that the same disease may be accompanied by various subtypes of macrophage, and the composition and interaction of macrophage subtypes can be a key factor in disease progression. However, macrophage subtypes in most diseases have not been fully characterized. The special DEPs generated in this study for each M2 subtype could be candidate markers that may benefit future clinical and pre-clinical research into macrophage-associated disease models.
5. Conclusions
In summary, we revealed for the first time the similarities and differences of biological processes involving four THP-1-derived M2 subtypes based on systematic proteomic analysis. This study confirmed several known markers and generated a list of novel marker candidates which could be used for the identification of M2a, M2b, M2c, and M2d macrophages. A main finding is that M2a and M2c, and M2b and M2d have very similar protein expression profiles. In addition, our study generated a valuable resource for future investigation of the mechanisms of immune regulation in macrophage-related diseases, as well as preclinical studies using THP-1-derived macrophage models.
Availability of data and materials
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD022320.
Supplementary information
Acknowledgments
This work was supported by the National Key Research and Development Program of China (No. 2019YFA0905200), the National Natural Science Foundation of China (Nos. 91853123, 81773180, 81800655, and 21705127), and the China Postdoctoral Science Foundation (Nos. 2019M653715, 2019TQ0260, and 2019M663798).
Author contributions
Pengfei LI and Shisheng SUN designed the experiments. Pengfei LI, Chen MA, Zhifang HAO, and Jing LI performed experiments. Chen MA, Jingyu WU, and Yuan ZHI performed mass spectrometry analysis. Chen MA, Jing LI, Shanshan YOU, and Lin CHEN analyzed data. Pengfei LI, Chen MA, and Shisheng SUN wrote the manuscript. Liuyi DANG and Jun LI revised the manuscript. All authors have read and approved the final manuscript, and therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Pengfei LI, Chen MA, Jing LI, Shanshan YOU, Liuyi DANG, Jingyu WU, Zhifang HAO, Jun LI, Yuan ZHI, Lin CHEN, and Shisheng SUN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Avila-Ponce de León U, Vazquez-Jimenez A, Matadamas-Guzman M, et al. , 2021. Transcriptional and microenvironmental landscape of macrophage transition in cancer: a boolean analysis. Front Immunol, 12: 642842. 10.3389/fimmu.2021.642842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CWV, 2003. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4: 2. 10.1186/1471-2105-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchu S, 2020. Progressive multiple sclerosis transcriptome deconvolution indicates increased M2 macrophages in inactive lesions. Eur Neurol, 83(4): 433-435. 10.1159/000510075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeier C, Escher F, Aleshcheva G, et al. , 2021. Plasminogen activator inhibitor-1 reduces cardiac fibrosis and promotes M2 macrophage polarization in inflammatory cardiomyopathy. Basic Res Cardiol, 116: 1. 10.1007/s00395-020-00840-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Fuchs S, Lieder I, Stelzer G, et al. , 2016. GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS, 20(3): 139-151. 10.1089/omi.2015.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, et al. , 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25(8): 1091-1093. 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S, Talamini M, Brenner S, et al. , 2019. Circulating monocytes and tumor-associated macrophages express recombined immunoglobulins in glioblastoma patients. Clin Transl Med, 8(1): e18. 10.1186/s40169-019-0235-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanput W, Mes JJ, Wichers HJ, 2014. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol, 23(1): 37-45. 10.1016/j.intimp.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Colin S, Chinetti-Gbaguidi G, Staels B, 2014. Macrophage phenotypes in atherosclerosis. Immunol Rev, 262(1): 153-166. 10.1111/imr.12218 [DOI] [PubMed] [Google Scholar]
- Chen YB, Song YC, Du W, et al. , 2019. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci, 26: 78. 10.1186/s12929-019-0568-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Morrée A, Flix B, Bagaric I, et al. , 2013. Dysferlin regulates cell adhesion in human monocytes. J Biol Chem, 288(20): 14147-14157. 10.1074/jbc.M112.448589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, et al. , 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol, 4(5): 3. [PubMed] [Google Scholar]
- di Paolo NC, Shayakhmetov DM, 2016. Interleukin 1α and the inflammatory process. Nat Immunol, 17(8): 906-913. 10.1038/ni.3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Aksnes H, Marie M, et al. , 2018. NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc Natl Acad Sci USA, 115(17): 4399-4404. 10.1073/pnas.1718336115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eligini S, Fiorelli S, Tremoli E, et al. , 2016. Inhibition of transglutaminase 2 reduces efferocytosis in human macrophages: role of CD14 and SR-AI receptors. Nutr Metab Cardiovasc Dis, 26(10): 922-930. 10.1016/j.numecd.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, Randolph GJ, 2014. Origin and functions of tissue macrophages. Immunity, 41(1): 21-35. 10.1016/j.immuni.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante CJ, Leibovich SJ, 2012. Regulation of macrophage polarization and wound healing. Adv Wound Care (New Rochelle), 1(1): 10-16. 10.1089/wound.2011.0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante CJ, Pinhal-Enfield G, Elson G, et al. , 2013. The adenosine-dependent angiogenic switch of macrophages to an M2-like phenotype is independent of interleukin-4 receptor alpha (IL-4Rα) signaling. Inflammation, 36(4): 921-931. 10.1007/s10753-013-9621-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MM, Härtlova A, Gierliński M, et al. , 2019. Triggering MSR1 promotes JNK-mediated inflammation in IL-4-activated macrophages. EMBO J, 38(11): e100299. 10.15252/embj.2018100299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks CM, Cordeiro T, Gomes AP, et al. , 2021. The interplay of HIV-1 and macrophages in viral persistence. Front Microbiol, 12: 646447. 10.3389/fmicb.2021.646447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Lewis C, Borg NA, et al. , 2018. Proteomic identification of interferon-induced proteins with tetratricopeptide repeats as markers of M1 macrophage polarization. J Proteome Res, 17(4): 1485-1499. 10.1021/acs.jproteome.7b00828 [DOI] [PubMed] [Google Scholar]
- Huang X, Li Y, Fu MG, et al. , 2018. Polarizing macrophages in vitro. In: Rousselet G (Ed.), Macrophages: Methods and Protocols. Humana Press, New York, p.119-126. 10.1007/978-1-4939-7837-3_12 [DOI] [Google Scholar]
- Ivashkiv LB, 2018. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol, 18(9): 545-558. 10.1038/s41577-018-0029-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen E, Iversen PO, van Rooijen N, et al. , 2002. Macrophage-dependent regulation of neutrophil mobilization and chemotaxis during development of sterile peritonitis in the rat. Eur J Haematol, 69(5-6): 284-296. 10.1034/j.1600-0609.2002.02657.x [DOI] [PubMed] [Google Scholar]
- Kuo CL, Chou HY, Chiu YC, et al. , 2020. Mitochondrial oxidative stress by Lon-PYCR1 maintains an immunosuppressive tumor microenvironment that promotes cancer progression and metastasis. Cancer Lett, 474: 138-150. 10.1016/j.canlet.2020.01.019 [DOI] [PubMed] [Google Scholar]
- Lee J, French B, Morgan T, et al. , 2014. The liver is populated by a broad spectrum of markers for macrophages. In alcoholic hepatitis the macrophages are M1 and M2. Exp Mol Pathol, 96(1): 118-125. 10.1016/j.yexmp.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CE, Leek R, Harris A, et al. , 1995. Cytokine regulation of angiogenesis in breast cancer: the role of tumor-associated macrophages. J Leukoc Biol, 57(5): 747-751. 10.1002/jlb.57.5.747 [DOI] [PubMed] [Google Scholar]
- Li GM, Li YF, Liu HM, et al. , 2020. Genetic heterogeneity of pediatric systemic lupus erythematosus with lymphoproliferation. Medicine (Baltimore), 99(20): e20232. 10.1097/MD.0000000000020232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GQ, Jin FQ, Du JX, et al. , 2019. Macrophage-secreted TSLP and MMP9 promote bleomycin-induced pulmonary fibrosis. Toxicol Appl Pharmacol, 366: 10-16. 10.1016/j.taap.2019.01.011 [DOI] [PubMed] [Google Scholar]
- Li PF, Hao ZF, Liu HH, et al. , 2021. Quantitative proteomics analysis of berberine-treated colon cancer cells reveals potential therapy targets. Biology, 10(3): 250. 10.3390/biology10030250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CW, Chou CH, Wu XM, et al. , 2020. Interleukin-6 plays a critical role in aldosterone-induced macrophage recruitment and infiltration in the myocardium. Biochim Biophys Acta Mol Basis Dis, 1866(3): 165627. 10.1016/j.bbadis.2019.165627 [DOI] [PubMed] [Google Scholar]
- Liu YC, Zou XB, Chai YF, et al. , 2014. Macrophage polarization in inflammatory diseases. Int J Biol Sci, 10(5): 520-529. 10.7150/ijbs.8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Cao Q, Zheng D, et al. , 2013. Discrete functions of M2a and M2c macrophage subsets determine their relative efficacy in treating chronic kidney disease. Kidney Int, 84(4): 745-755. 10.1038/ki.2013.135 [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, et al. , 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics, 26(7): 966-968. 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Helming L, Milde R, et al. , 2013. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood, 121(9): e57-e69. 10.1182/blood-2012-06-436212 [DOI] [PubMed] [Google Scholar]
- Menzel K, Hausmann M, Obermeier F, et al. , 2006. Cathepsins B, L and D in inflammatory bowel disease macrophages and potential therapeutic effects of cathepsin inhibition in vivo. Clin Exp Immunol, 146(1): 169-180. 10.1111/j.1365-2249.2006.03188.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoia F, Tibaldi J, Muratore V, et al. , 2021. Thrombotic microangiopathy associated with macrophage activation syndrome: a multinational study of 23 patients. J Pediatr, 235: 196-202. 10.1016/j.jpeds.2021.04.004 [DOI] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP, 2008. Exploring the full spectrum of macrophage activation. Nat Rev Immunol, 8(12): 958-969. 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, 2017. Macrophage polarization. Annu Rev Physiol, 79: 541-566. 10.1146/annurev-physiol-022516-034339 [DOI] [PubMed] [Google Scholar]
- Neven Q, Boulanger C, Bruwier A, et al. , 2021. Clinical spectrum of Ras-associated autoimmune leukoproliferative disorder (RALD). J Clin Immunol, 41(1): 51-58. 10.1007/s10875-020-00883-7 [DOI] [PubMed] [Google Scholar]
- Nosaka M, Ishida Y, Kimura A, et al. , 2020. Crucial involvement of IL-6 in thrombus resolution in mice via macrophage recruitment and the induction of proteolytic enzymes. Front Immunol, 10: 3150. 10.3389/fimmu.2019.03150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M, Ediriweera HN, Gundra UM, et al. , 2015. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest, 125(12): 4334-4348. 10.1172/JCI81676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AC, Russell JD, Bailey DJ, et al. , 2012. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics, 11(11): 1475-1488. 10.1074/mcp.O112.020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosper F, Vanoverbeke K, Stroncek D, et al. , 1997. Primitive long-term culture initiating cells (LTC-ICs) in granulocyte colony-stimulating factor mobilized peripheral blood progenitor cells have similar potential for ex vivo expansion as primitive LTC-ICs in steady state bone marrow. Blood, 89(11): 3991-3997. [PubMed] [Google Scholar]
- Rabinowitz J, Sharifi HJ, Martin H, et al. , 2021. xCT/SLC7A11 antiporter function inhibits HIV-1 infection. Virology, 556: 149-160. 10.1016/j.virol.2021.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. , 2018. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol, 233(9): 6425-6440. 10.1002/jcp.26429 [DOI] [PubMed] [Google Scholar]
- Sica A, Mantovani A, 2012. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest, 122(3): 787-795. 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Lehmann G, Bork P, et al. , 2000. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res, 28(18): 3442-3444. 10.1093/nar/28.18.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QQ, Hawkins GA, Wudel L, et al. , 2019. Dissecting intratumoral myeloid cell plasticity by single cell RNA-seq. Cancer Med, 8(6): 3072-3085. 10.1002/cam4.2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, et al. , 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res, 43(D1): D447-D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vago JP, Sugimoto MA, Lima KM, et al. , 2019. Plasminogen and the plasminogen receptor, Plg-RKT, regulate macrophage phenotypic, and functional changes. Front Immunol, 10: 1458. 10.3389/fimmu.2019.01458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LX, Zhang SX, Wu HJ, et al. , 2019. M2b macrophage polarization and its roles in diseases. J Leukoc Biol, 106(2): 345-358. 10.1002/JLB.3RU1018-378RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QS, Ni H, Lan L, et al. , 2010. Fra-1 protooncogene regulates IL-6 expression in macrophages and promotes the generation of M2d macrophages. Cell Res, 20(6): 701-712. 10.1038/cr.2010.52 [DOI] [PubMed] [Google Scholar]
- White MJV, Gomer RH, 2015. Trypsin, tryptase, and thrombin polarize macrophages towards a pro-fibrotic M2a phenotype. PLoS ONE, 10(9): e0138748. 10.1371/journal.pone.0138748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Romero-Ramírez L, Mey J, 2021. Retinoic acid increases phagocytosis of myelin by macrophages. J Cell Physiol, 236(5): 3929-3945. 10.1002/jcp.30137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Vannella KM, 2016. Macrophages in tissue repair, regeneration, and fibrosis. Immunity, 44(3): 450-462. 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW, 2013. Macrophage biology in development, homeostasis and disease. Nature, 496(7446): 445-455. 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang BZ, Zhang QQ, et al. , 2021. Tumor-associated macrophages in osteosarcoma. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 22(11): 885-892. 10.1631/jzus.B2100029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zizzo G, Hilliard BA, Monestier M, et al. , 2012. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J Immunol, 189(7): 3508-3520. 10.4049/jimmunol.1200662 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD022320.