Abstract
A phylogenetic analysis of the sequences of 60 clones of archaeal small-subunit rRNA genes amplified from the termite Reticulitermes speratus revealed that most of them (56 clones) clustered in the genus Methanobrevibacter. Three clones were classified in the order Thermoplasmales. The Methanobrevibacter-related symbionts were detected by in situ hybridization analysis.
Termites, both lower and higher ones, harbor methanogenic Archaea in their hindguts (7, 13). The H2 consumption of methanogens is expected to promote anaerobic cellulose decomposition in the termite hindgut. Methanogenic symbionts of termites are present in two different niches: endosymbiotic methanogens are present in flagellates, and nonendosymbiotic ones adhere to hindgut surfaces (6, 7). Recently, Leadbetter and Breznak (6) reported on the isolation and characterization of two novel species of methanogenic bacteria from Reticulitermes flavipes which belong to the genus Methanobrevibacter. These species were related to the cells colonizing the hindgut epithelium but not to the cells in the symbiotic flagellates, as determined by morphological observation. In the present investigation we surveyed (by PCR) the phylogenetic diversity of methanogens in Reticulitermes speratus termites collected in the Japan archipelago.
Termites.
Six colonies of the termite R. speratus (Kolbe, order Isoptera, family Rhinotermitidae) were collected from forests in Japan (Table 1). One colony was collected from each sampling point except for the Tokyo area, where three colonies were sampled.
TABLE 1.
Grouping of SSU rRNA clones amplified and isolated from R. speratus hindgut contents
| Area | Colony | No. of clones in symbiotic type
|
|||||
|---|---|---|---|---|---|---|---|
| 1A | 1B | 1C | 1D | 2 | 3 | ||
| Tokyo | RS1 | 4 | 4 | 0 | 0 | 1 | 1 |
| RS2 | 5 | 5 | 0 | 0 | 0 | 0 | |
| RS4 | 1 | 6 | 1 | 0 | 0 | 2 | |
| M4a | 1 | ||||||
| Kobe | RS5 | 8 | 2 | 0 | 0 | 0 | 0 |
| Yamaguchi | RS3 | 7 | 0 | 0 | 3 | 0 | 0 |
| Okinawa | RS6 | 6 | 4 | 0 | 0 | 0 | 0 |
| Total | 31 | 21 | 1 | 3 | 1 | 3 | |
Sequence of the clone M4 amplified from hindgut DNA of R. speratus was reported by Ohkuma et al. (9). M4 was grouped in type 1B.
DNA extraction.
Ten individual workers from each colony were selected for the DNA extraction. After surface sterilization with 70% ethanol, the digestive tracts of the termites were pulled out with a pair of fine-tipped forceps. The gut tissues and their contents were crushed with a Teflon homogenizer in 1 ml of extraction buffer consisting of 100 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 0.1% sodium dodecyl sulfate. The mixture was frozen in liquid nitrogen and thawed at 57°C five times and then treated with 0.5 mg of proteinase K per ml at 57°C for 16 h. The solution was extracted with phenol and chloroform, and the DNA was precipitated with ethanol (11).
PCR amplification and sequence analysis.
PCR primers were designed to amplify a part of the archaeal small-subunit (SSU) rRNA gene. The primers used were ME855F (5′-TTAA AGGAATTGGCGGGGGA-3′) and ME1354R (5′-TGACGGGCGGTGTGTGCAAG-3′). The PCR reaction was performed with a thermal program which comprised 40 cycles at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 90 sec. The amplified DNA fragments were ligated with a ddT-tailed vector as described by Holton and Graham (5) and cloned. The nucleotide sequences were determined by the dideoxynucleotide chain termination method (12) on an A. L. F. model II DNA sequencer (Pharmacia Biotech).
Phylogenetic analysis.
Phylogenetic trees were constructed by neighbor-joining distance matrix methods (10) with the programs in the software package PHYLIP, version 3.572 (J. Felsenstein and the University of Washington). The aligned sequences were also analyzed by maximum-parsimony and maximum-likelihood methods to check the tree topology.
Phylogeny of symbiotic methanogens of R. speratus.
Ten clones were isolated for each termite colony, and the sequences of 60 clones were determined in total. Clones which had evolutionary distances within 0.02 substitutions (sequence identity, >98%) were grouped together. As a result, the clones were classified into six types: 1A, 1B, 1C, 1D, 2, and 3 (Table 1).
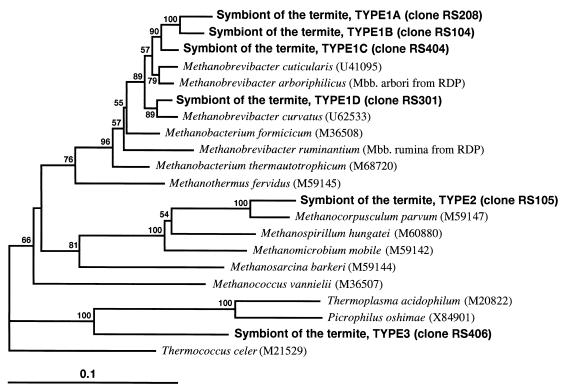
The phylogenetic tree shown in Fig. 1 was constructed by the neighbor-joining method. Types 1, 2, and 3 were separated from each other by more than 0.1 evolutionary distances. The clones belonging to types 1A to 1D were located in the order Methanobacteriales. Ohkuma et al. (9) have reported on the amplification and cloning of SSU ribosomal DNA sequences of methanogens from R. speratus. The sequence of the clone M4 reported by Ohkuma et al. was found to be type 1B. Methanobrevibacter-related methanogens were detected from all colonies used in this study. Methanobrevibacter-related methanogens seem to be most abundant in R. speratus. Leadbetter and Breznak (6) have recently isolated Methanobrevibacter cuticularis and Methanobrevibacter curvatus from R. flavipes. These previously isolated methanogens are also in the genus Methanobrevibacter. Methanobrevibacter-related methanogens may be the major phylum of symbiotic methanogens in termites. However, it should be kept in mind that PCR may have some biases in the amplification of sequences.
FIG. 1.
Phylogenetic positions of the PCR clones obtained from R. speratus hindgut contents within the members of Euryarchaeota. Type clones of respective groups of the clones were used to construct the tree and are indicated in parentheses. The tree was inferred from 499 unambiguously aligned nucleotide sequences of SSU rRNA by using the neighbor-joining method. Thermococcus celer was used as the out-group. Bar, 0.1 substitutions per nucleotide position. Numbers are bootstrap values (3). The bootstrap confidence interval was calculated from 1,000 repetitions of resampling.
The type 2 clone fell in the order Methanomicrobiales (Fig. 1). This clone was closely related to Methanocorpusculum parvum with 95.3% sequence identity. The clustering of the clone and M. parvum was strongly supported by a bootstrap value of 100%.
In the present study, three clones of type 3 were obtained from R. speratus hindgut microflora. The closest neighbors of these clones were Thermoplasma relatives (Picrophilus oshimae, with 82.5% identity), and the second closest one was Thermoplasma acidophilum, showing a similarity of 81.3%. Type 3 clones formed a monophyletic clade with species of Thermoplasmales, which was supported by a relatively high bootstrap value (90%). Many sequences which are related to those of Thermoplasma have been amplified from various environmental samples and reported (1, 2, 4). However, these sequences could not be used in the phylogenetic analysis because of the differences in the region determined. Judging from the sequence analysis, the type 3 symbiont seemed to be a Thermoplasma-related novel archaeon. Known Thermoplasma species grow in acidic and hot environments. The type 3 symbiont must be a neutrophilic and mesophilic archaeon, because the physical conditions of the Reticulitermes hindgut are at a nearly neutral pH (8) and of course at an ambient temperature.
In situ hybridization.
We also performed whole-cell hybridizations with fluorescent oligonucleotide probes. Oligonucleotide probes were designed to bind to the SSU rRNAs of the methanogens detected in this study (Fig. 2). Two archaeon-specific probes, ARC344 (5′-GCGCCTGCTGCGCCCGT-3′) and ARC915 (5′-GTGCTCCCCGCCAATTCCT-3′), de-scribed by Stahl et al. (14), were also used as positive controls.
FIG. 2.
Alignment of the oligonucleotide probe sequences and the corresponding SSU rRNA sequences of termite symbiont clones, methanogens, and Escherichia coli. Only nucleotides which are different from those of the type 1 sequence are shown.
A worker from colony RS1, sampled in the Tokyo area, was used in this analysis. Samples were prepared basically according to the work of Stahl et al. (14). The hindgut contents were collected by centrifugation at 16,000 rpm (15,000 × g) for 10 min and fixed in 1% paraformaldehyde in phosphate-buffered saline at 4°C for 30 min. The hindgut contents of the termite were hybridized with the archaeon-specific probes labeled with fluorescein and with those specific to cloned sequences that had been labeled with rhodamine. They were inspected with a laser scanning confocal imaging system (MRC-600; Bio-Rad) with a krypton-argon mixed-gas laser generator.
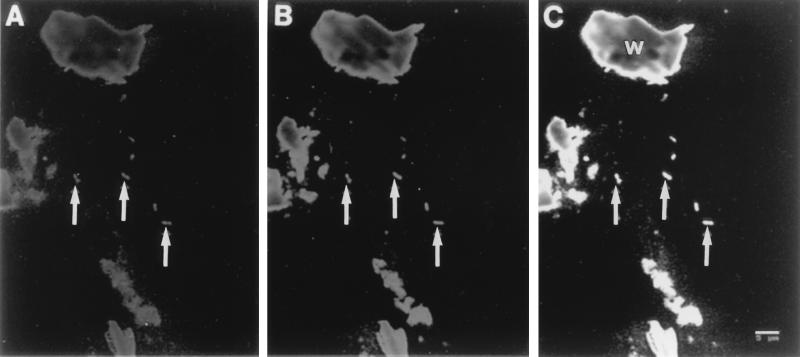
Though wood particles present in the sample emitted fluorescence with a wide spectrum, prokaryotic cells could be morphologically distinguished from the particles (Fig. 3). The cells that hybridized with the archaeon-specific probes were all straight rods, and they frequently existed as two cells attached to each other. The morphological features of the cells are consistent with those of some observed on the epithelial surface of the hindgut. The cells that hybridized with the archaeon-specific probes also hybridized with type 1-specific probes (Fig. 3).
FIG. 3.
Whole-cell in situ hybridization of hindgut contents from R. speratus. (A) In this image taken with fluorescein-labeled archaea-specific probes, ARC344 and ARC915, hybridized cells are indicated by arrows. (B) The same field as in panel A, showing the rhodamine fluorescence of type 1-specific probes 1109TY1R and 1174TY1R. (C) Merged field of panels A and B. The cells hybridized with both sets of probes appear yellow on the real merged field. Wood particles (W) which emitted fluorescence in scans for both fluorescein and rhodamine were also yellow on the real merged field.
In summary, we report that the methanogen community in the hindgut of R. speratus consists mainly of Methanobrevibacter-related methanogens, which had some divergence. The type 1 symbiotic methanogens were detected by in situ hybridization analysis.
Acknowledgments
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan (07265209 and 08228221).
REFERENCES
- 1.Delong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delong E F, Wu K Y, Prezelin B B, Jovine R V M. High abundance of archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 4.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holton T A, Graham M W. A simple and efficient method for direct cloning of PCR product using ddT-tailed vectors. Nucleic Acids Res. 1990;19:1156. doi: 10.1093/nar/19.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leadbetter J R, Breznak J A. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl Environ Microbiol. 1996;62:3620–3631. doi: 10.1128/aem.62.10.3620-3631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messer A C, Lee M J. Effect of chemical treatments on methane emission by the hindgut microbiota in the termite Zootermopsis angusticollis. Microb Ecol. 1989;18:275–284. doi: 10.1007/BF02075814. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien R W, Slaytor M. Role of microorganisms in the metabolism of termites. Aust J Biol Sci. 1982;35:239–262. [Google Scholar]
- 9.Ohkuma M, Noda S, Horikoshi K, Kudo T. Phylogeny of symbiotic methanogens in the gut of the termite Reticulitermes speratus. FEMS Microbiol Lett. 1995;134:45–50. doi: 10.1111/j.1574-6968.1995.tb07912.x. [DOI] [PubMed] [Google Scholar]
- 10.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 12.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinzato N, Yoshino H, Yara K. Methane production by microbial symbionts in the lower and higher termites of the Ryukyu Archipelago. In: Sato S, Ishida M, Ishikawa H, editors. Endocytobiology V. Tübingen, Germany: Tübingen University Press; 1993. pp. 161–166. [Google Scholar]
- 14.Stahl D A, Amann R I, Poulsen L K, Raskin L, Capman W C. Use of fluorescent probes for determinative microscopy of methanogenic archaea. In: Sowers K R, Schreier H J, editors. Archaea: a laboratory manual. Methanogens. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 111–121. [Google Scholar]