Abstract
Melatonin exerts a wide range of effects among various tissues and organs. However, there is currently no study to investigate the genetic determinants of melatonin secretion. Here, we conducted a genome-wide association study (GWAS) for melatonin secretion using morning urine 6-hydroxymelatonin sulfate-to-creatinine ratio (UMCR). We initially enrolled 5000 participants from Taiwan Biobank in this study. After excluding individuals that did not have their urine collected in the morning, those who had history of neurological or psychiatric disorder, and those who failed to pass quality control, association of single nucleotide polymorphisms with log-transformed UMCR adjusted for age, sex and principal components of ancestry were analyzed. A second model additionally adjusted for estimated glomerular filtration rate (eGFR). A total of 2373 participants underwent the genome-wide analysis. Five candidate loci associated with log UMCR (P value ranging from 6.83 × 10−7 to 3.44 × 10−6) encompassing ZFHX3, GALNT15, GALNT13, LDLRAD3 and intergenic between SEPP1 and FLJ32255 were identified. Similar results were yielded with further adjustment for eGFR. Interestingly, the identified genes are associated with circadian behavior, neuronal differentiation, motor disorders, anxiety, and neurodegenerative diseases. We conducted the first GWAS for melatonin secretion and identified five candidate genetic loci associated with melatonin level. Replication and functional studies are needed in the future.
Subject terms: Genetics, Endocrinology
Introduction
Melatonin is a pleiotropic hormone primarily synthesized and secreted from the pineal gland. Many other tissues can also produce it, including leukocytes, bone marrow, gastrointestinal tract, neuronal cells, and gonads1–3. Melatonin regulates various physiological processes, including circadian and seasonal rhythms, energy and glucose metabolism, antioxidant effects, anti-inflammatory actions, and immune function1,3–6. There are many studies showing associations between melatonin and many disorders, including certain types of mental illness, cancer, cardiovascular disease, metabolic syndrome, type 2 diabetes, and obesity6–11. Melatonin is secreted into the circulation following a circadian rhythm with peak levels at night12. Aging was once thought to be directly associated with decreased melatonin secretion. However, there was no significant difference between circadian amplitude of the plasma melatonin between healthy elderlies and young adults13. Instead of aging, the degree of pineal calcification was associated with melatonin excretion amount14.
Substantial evidence suggests genetic factors also play a significant role in melatonin secretion15,16. Genome-wide association study (GWAS) has been introduced as a powerful tool to identify common genetic variants of complex diseases or quantitative traits17. Currently, there is no published GWAS regarding melatonin levels. Here, we conducted the first GWAS of urine melatonin metabolite, 6-hydroxymelatonin sulfate (aMT6s), which surrogate the circulating melatonin level18.
Materials and methods
Study population
Five thousand individuals aged 30–70 years old and without cancer history were enrolled from Taiwan Biobank. Biological specimens, personal and clinical information as delinked data were used in this study. Individuals with a record of neurological disorders or psychiatric illnesses were excluded from this study as these conditions may affect melatonin secretion19,20. This study was approved by the Institutional Review Board of Chang Gung Medical Foundation and the Institutional Review Board of National Taiwan University Hospital. All subjects have provided written informed consent, and all methods were carried out in accordance with relevant guidelines and regulations.
Urine aMT6s and creatinine measurement
It is infeasible to draw blood samples from volunteers in the middle of the night for serum melatonin levels. Urinary aMT6s is the major metabolite of melatonin excreted from the kidneys21. Thus, measuring morning urine aMT6s level is a practical alternative for serum melatonin level at night18. For better correlation, spot urine aMT6s level should be creatinine-corrected to adjust the effect of variable urinary dilution22. Urine aMT6s-to-creatinine ratio (UMCR) was calculated from urinary aMT6s divided by urine creatinine level. The concentration of aMT6s was measured in the urine of Taiwan Biobank subjects by an enzyme-linked immunosorbent assay (ELISA) kit using the manufacturer's protocol (Human Melatonin Sulfate ELISA kit, Elab science). No significant cross-reactivity or interference between melatonin sulfate and analogs was observed. All standards via serial dilution were assayed in duplicates. The urine creatinine level was measured using a chemistry analyzer (AU5800, Beckman Coulter) with compensated Jaffe method.
Genotyping, quality control and imputation
Genotyping with the Axiom-Taiwan Biobank Array Plate (TWB chip; Affymetrix Inc, Santa Clara, California) was performed at the National Center for Genome Medicine of Academia Sinica23. We use PLINK (version 1.9), an open-source whole-genome data analysis toolset, for quality control procedures24. For SNPs with batch effect, their genotypes were set as missing. SNPs were excluded if missing genotype rate was high (> 5%), minor allele frequency was low (< 1%) or deviated from Hardy–Weinberg equilibrium (P value < 10−5). Individuals with discordant sex (self-reported sex incongruent to genetic sex, where genetic male or female was defined by X chromosome homozygosity estimate above 0.8 or below 0.2), high missing genotyping rate (> 5%), extreme heterozygosity rate (more than 5 standard deviations away from the mean) or high identity-by-descent score (≥ 0.1875) implying close relatedness were excluded from subsequent analyses. We computed the principal components on a linkage disequilibrium (LD)-pruned (r2 < 0.2) set of autosomal variants obtained by removing high-LD regions via PLINK. Genotype imputation was carried out with SHAPEIT25 and IMPUTE226. We applied1000 Genomes Project Phase 3 East Asian Ancestry as the reference population. For gene annotation, Genome Reference Consortium Human Build 37 was used. Imputed SNPs with low-quality scores (info score27 lower than 0.8) were excluded. Indels were removed by using VCFtools28.
Statistical analyses
Age and estimated glomerular filtration rate (eGFR) were expressed as mean and standard deviation. Urine aMT6s and UMCR were expressed as median and interquartile range. Logarithmic transformation of UMCR was done to normalize the data. GWAS analysis was carried out via PLINK v1.9 with an additive genetic model. We applied linear regression for analyzing associations between SNPs and log UMCR. Covariate adjustment in Model 1 included age, sex, and the first ten principal components of ancestry. eGFR was additionally adjusted in Model 2. We used a genome-wide significance threshold of P < 5.0 × 10−829. Since this threshold is very conservative for small sample size, we set the level for suggestive significance at P < 5 × 10−630,31. The Manhattan plot and quantile–quantile plot were generated by the qqman R package32,33. Regional association plots were made via LocusZoom34. The proportion of phenotypic variance explained by SNP was calculated using the following items: effect size estimate of each minor allele on log UMCR, standard error of the effect size, sample size, and minor allele frequency for the SNP35. The statistical power of this study was calculated using methods for quantitative GWAS36.
Bioethics statement
This study was approved by the Institutional Review Board of Chang Gung Medical Foundation and the Institutional Review Board of National Taiwan University Hospital. All subjects have provided written informed consent and all methods were carried out in accordance with relevant guidelines and regulations.
Results
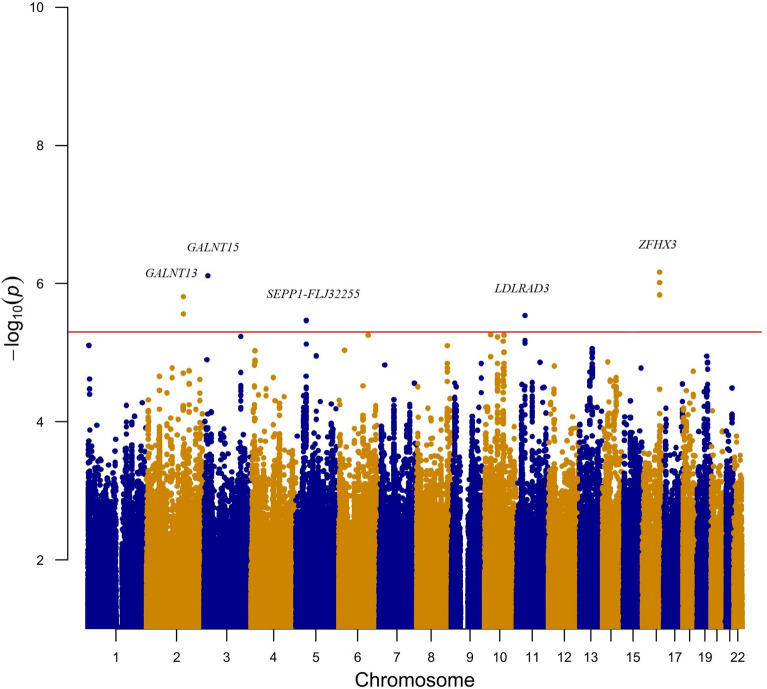
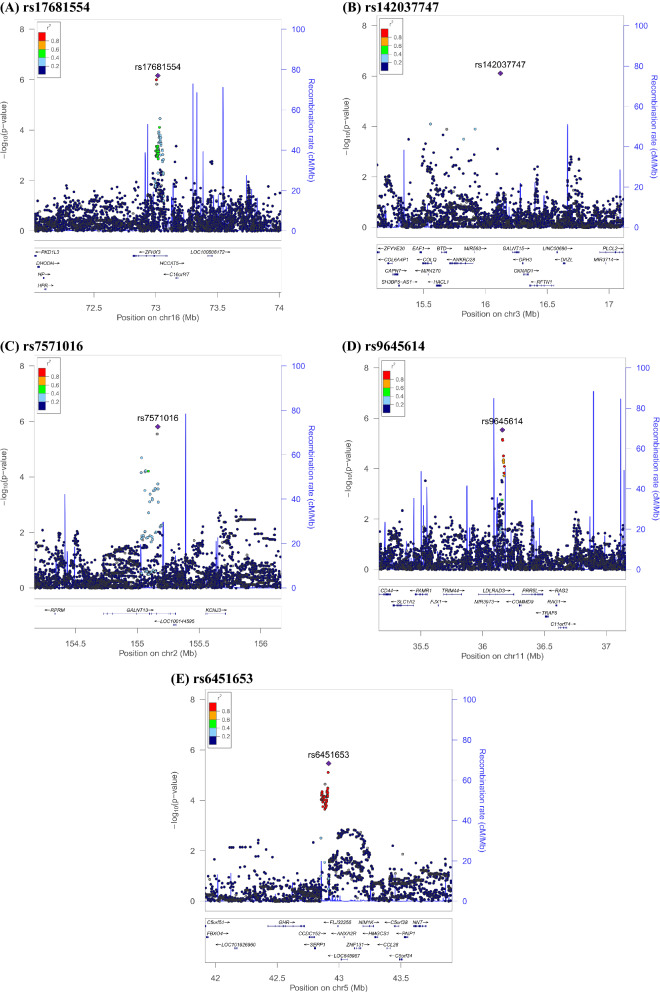
Five thousand subjects were enrolled from Taiwan Biobank initially. One withdrew from the study. 2361 did not have their urine collected in the morning and were excluded. 128 people had documented neurological or psychiatric illness. 137 individuals did not pass quality control procedures. After imputation and quality control, 7,897,704 autosomal SNPs remained. After log transformation of UMCR, data is still not normalized, but the shape of the histogram is better. We performed a GWAS analysis for log UMCR in the remaining 2373 subjects. The characteristics of our study population are listed in Table 1. Age is not significantly associated with log UMCR (Pearson’s r = 0.031; P = 0.137). There is no significant gender difference of log UMCR shown by T test (males 1.208, females 1.205; P = 0.857). eGFR is also not significantly associated with log UMCR (Pearson’s r = 0.001; P = 0.973). Variants with the strongest association in each region regarding to log UMCR are shown in Table 2. Melatonin production is known to be decreased with advanced chronic kidney disease37; thus, we adjusted eGFR additionally in Model 2. There was no evidence of liver disorders in these 2373 individuals, and thus no additional model was generated. Figures 1 and 2 are the Manhattan plot and quantile–quantile plot. The genomic inflation factor, defined as the median of the observed chi-squared test statistics divided by the expected median of the corresponding chi-squared distribution, was 1.006. Regional association plots of the top SNPs are shown in Fig. 3. Five loci showed suggestive significance, with one within the ZFHX3 gene on chromosome 16 (rs17681554; P = 6.83 × 10−7, Fig. 3a), another near the GALNT15 gene on chromosome 3 (rs142037747; P = 7.82 × 10−7, Fig. 3b), a third within the GALNT13 gene on chromosome 2 (rs7571016; P = 1.53 × 10−6, Fig. 3c), a fourth within the LDLRAD3 gene on chromosome 11 (rs9645614; P = 2.90 × 10−6, Fig. 3d), and the other between SEPP1 and FLJ32255 on chromosome 5 (rs6451653; P = 3.44 × 10−6, Fig. 3e).
Table 1.
Descriptive characteristics of study subjects.
| Characteristics | |
|---|---|
| Total participants, N | 2373 |
| Age, year | 50.75 ± 10.83 |
| Males, N (%) | 890 (37.51) |
| eGFR, ml/min/1.73 m2 | 108.20 ± 27.83 |
| Urine aMT6s, ng/ml | 20.41 (11.88–30.19) |
| UMCR, ng/mg | 16.98 (10.32–27.33) |
Data are mean ± SD, median (IQR) or number (%), as appropriate.
Age is at specimen collection.
eGFR estimated glomerular filtration rate (by modification of diet in renal disease equation), UMCR urine aMT6s/creatinine ratio.
Table 2.
Association of genetic loci with log UMCR in a Taiwan Han Chinese population.
| SNP | Chr | Position | Nearest gene | UMCR increasing allele | Other allele | UMCR increasing allele frequency | Model 1 | Model 2 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| P value | Beta (SEM) | P value | Beta (SEM) | |||||||
| rs17681554 | 16 | 73,016,768 | ZFHX3 | A | C | 0.804 | 6.86 × 10−7 | 0.068 (0.014) | 6.83 × 10−7 | 0.068 (0.014) |
| rs142037747 | 3 | 16,121,712 | GALNT15 | G | A | 0.989 | 7.82 × 10−7 | 0.256 (0.052) | 7.73 × 10−7 | 0.256 (0.052) |
| rs7571016 | 2 | 155,166,873 | GALNT13 | A | G | 0.617 | 1.53 × 10−6 | 0.056 (0.011) | 1.54 × 10−6 | 0.056 (0.012) |
| rs9645614 | 11 | 36,159,947 | LDLRAD3 | A | G | 0.953 | 2.90 × 10−6 | 0.124 (0.026) | 2.91 × 10−6 | 0.124 (0.026) |
| rs6451653 | 5 | 42,915,584 | SEPP1-FLJ32255 | G | A | 0.869 | 3.44 × 10−6 | 0.080 (0.017) | 3.42 × 10−6 | 0.080 (0.017) |
Model 1 was adjusted for age, sex and the first ten principal components of ancestry.
Model 2 was additionally adjusted for eGFR.
SNP single nucleotide polymorphism, Chr chromosome.
SNPs are imputed with high info score (0.831, 0.988 and 0.946 for rs142037747, rs9645614 and rs6451653, respectively).
Figure 1.
Manhattan plot of the GWAS results for log UMCR. SNPs are plotted on the x axis according to their chromosome position against association with log UMCR on the y axis. The red horizontal line represents the suggestive association threshold of P = 5.0 × 10−6.
Figure 2.
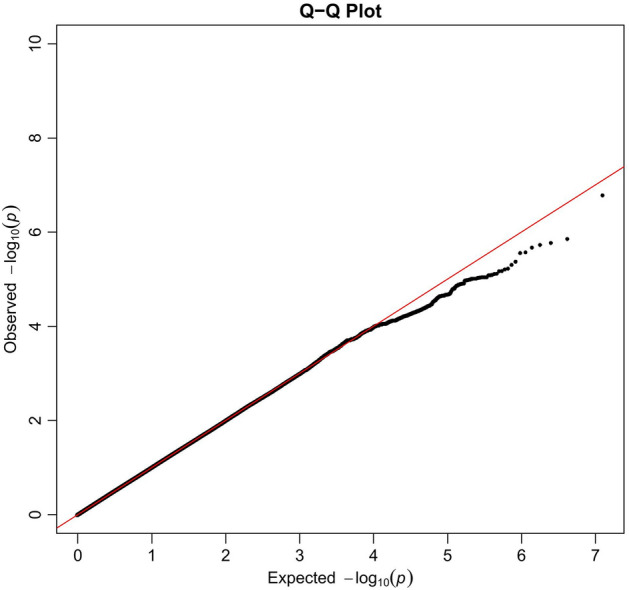
Quantile–quantile plots of log UMCR.
Figure 3.
Regional association plots of log UMCR. (A) rs17681554, (B) rs142037747, (C) rs7571016, (D) rs9645614, (E) rs6451653.
Proportion of variance explained by the individual SNPs are 0.98%, 1.01%, 1.08%, 0.95% and 0.92% for rs17681554, rs142037747, rs7571016, rs9645614 and rs6451653 respectively. The calculated power of this GWAS was 56%.
Discussion
In this first GWAS on melatonin secretion, we identified five suggestive loci associated with variation in log UMCR. rs17681554 is located within ZFHX3 (Zinc Finger Homeobox 3). ZFHX3 is a transcriptional regulator which contains four homeodomains and seventeen zinc fingers38. During neuronal differentiation, there is a preferential expression pattern of ZFHX3 isoforms39. In addition, circadian behavior alteration is shown in inducible conditional Zfhx3 knockout adult mice40. Further studies are needed to elucidate a direct linkage between ZFHX3 and melatonin.
rs142037747 and rs7571016 are located near GALNT15 (polypeptide N-acetylgalactosaminyltransferase 15) and within GALNT13 (polypeptide N-acetylgalactosaminyltransferase 13), respectively. These two polypeptide N-acetylgalactosaminyltransferases of the same family catalyze initiation of mucin-type O-linked glycosylation by adding N-acetylgalactosamine to serine or threonine residues of the polypeptide chain41,42. Glycosylation is associated with cell adhesion, signal transduction, molecular trafficking, and differentiation in central nervous system development43. Whether and how GALNT15 or GALNT13 significantly affects melatonin levels remains determined.
rs9645614 is located within LDLRAD3 (low density lipoprotein receptor class A domain containing 3). LDLRAD3 alters the proteolysis of amyloid precursor protein and increases the production of amyloid beta-peptide (Aβ)44. The primary pathogenesis of Alzheimer's disease (AD) has been attributed to the extracellular aggregation of Aβ45. Melatonin levels are altered in AD patients, possibly due to decreased suprachiasmatic nucleus cell number and functional pineal gland volume46. Patients with neurodegenerative disorder such as Alzheimer's disease exhibit reduced serum and cerebrospinal fluid melatonin levels comparing to age-matched controls47,48. Our present unbiased genetic study, revealing the LDLRAD3 variant associated with melatonin secretion from pineal gland, provides additional evidence for potential mechanistic explanation in AD patients with altered melatonin levels.
rs6451653 is located between SEPP1 (selenoprotein P, or SELENOP) and pseudogene FLJ32255. SEPP1 serves as a phospholipid hydroperoxide glutathione peroxidase and thus protect the plasma membrane from oxidative damage and is expressed in all brain tissues49. SEPP1 is secreted from astrocytes to neurons for prevention of oxidative damage50. Several studies demonstrated that Sepp1 knockout mice displayed cerebellar ataxia, anxiety, impaired spatial memory, and widespread neurodegeneration in various studies51–55. Also, deletion of SEPP1 in dogs resulted in central nervous system atrophy and cerebellar ataxia56. It is convincing that the SEPP1 variant is associated with melatonin levels.
This study also showed borderline significance regarding the positive correlation between age and log UMCR. Since aging causes sarcopenia, the subsequently decreased creatinine excretion from urine increases the substance-to-creatinine ratio. Our results support the current concept that aging itself will not cause a decrease in melatonin secretion or excretion.
There was a concern that aMT6s excretion may be altered when renal function declines. A previous study enrolling 20 elderlies demonstrated that 24-h urine aMT6s was a reliable surrogate for plasma melatonin level, at least among individuals with GFR 24.6 ml/min or above57. Our study confirmed that morning UMCR is not significantly correlated with eGFR, and adjusting eGFR in GWAS analysis essentially did not influence the results.
There are limitations to our study. First, it lacks replication of the result in another cohort. We searched in the UK Biobank, but melatonin as phenotype does not exist in the database. Moreover, the sample size is relatively small; thus, for the time being, these SNPs can only be seen as suggestive signals. The statistical power of this GWAS is only 56%, and therefore there are true loci that remain to be identified and validated. Also, there might be individuals receiving medications that can affect melatonin levels but not documented in the Taiwan Biobank data due to the imprecise nature of the questionnaire survey. However, production or selling of melatonin pills is illegal in Taiwan, and thus this important confounding factor may not be significant in our study.
In summary, we have performed the first GWAS regarding melatonin secretion to date. This GWAS identified five highly suggestive genetic loci encompassing genes that demonstrated potential functional connectivity between the genes-associated melatonin level and circadian behavior, neuronal differentiation, cerebellar ataxia, neurodegeneration and Alzheimer's disease. Replication and functional studies of these genetic variations are warranted to understand better the regulation of melatonin secretion and related clinical disorders.
Acknowledgements
This work is supported by Grants from the Ministry of Science and Technology in Taiwan (MOST 105-2314-B-182-062, MOST 106-2314-B-182-043), the Translational Medical Research Program of Academia Sinica (ASTM-108-01-04), National Taiwan University Hospital, Yunlin Branch Intramural Grant (NTUHYL106.X003, NTUHYL.107S004, NTUHYL 111.X019, NTUHYL110.X017) and Chang Gung University, Taoyuan, Taiwan (NMRPD1F154, NMRPD1G0711, and BMRPD08). We would like to thank Taiwan Biobank for providing the biological specimens and information for our research, and the staff of the Core Lab, Department of Medical Research, National Taiwan University Hospital for technical support during the study. We would also like to thank Professor Lee-Ming Chuang at the Department of Internal Medicine, National Taiwan University Medical College for the critical review and valuable discussion about this manuscript. Lastly, we thank the Data Science Statistical Cooperation Center of Academia Sinica (AS-CFII-108-117) for statistical support.
Author contributions
P.-H.L., G.-T.C. and Y.-C.C. contributed to the experimental design. Y.-S.W., H.-C.K., Y.-C.L., Y.-S.C., Y.-Y.H., C.-H.L., W.-Y.L., J.-W.L., Chih-Neng H., J.-J.H, M.-L.H., H.-L.L. and Y.-C.C. contributed to the funding, sample procurement and data generation. G.-T.C. was responsible for data analysis; P.-H.L., Chia-Ni H., and C.-Y.S. provided statistical consultation. K.C.-W.L. wrote the revised discussion. Manuscript writing was done by G.-T.C., P.-H.L. and Y.-C.C.
Data availability
Individual researchers may request to use the data for specific projects on a collaborative basis. Our data has been submitted to the NHGRI-EBI GWAS Catalog (accession ID: GCST90101875).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pi-Hua Liu and Gwo-Tsann Chuang.
References
- 1.Galano A, Tan DX, Reiter RJ. Melatonin as a natural ally against oxidative stress: A physicochemical examination. J. Pineal Res. 2011;51:1–16. doi: 10.1111/j.1600-079X.2011.00916.x. [DOI] [PubMed] [Google Scholar]
- 2.Hardeland R, Cardinali DP, Srinivasan V, Spence DW, Brown GM, Pandi-Perumal SR. Melatonin—A pleiotropic, orchestrating regulator molecule. Prog. Neurobiol. 2011;93:350–384. doi: 10.1016/j.pneurobio.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Favero G, Rodella LF, Reiter RJ, Rezzani R. Melatonin and its atheroprotective effects: A review. Mol. Cell. Endocrinol. 2014;382:926–937. doi: 10.1016/j.mce.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Reiter RJ. Melatonin: Clinical relevance. Best Pract. Res. Clin. Endocrinol. Metab. 2003;17:273–285. doi: 10.1016/S1521-690X(03)00016-2. [DOI] [PubMed] [Google Scholar]
- 5.Claustrat B, Brun J, Chazot G. The basic physiology and pathophysiology of melatonin. Sleep Med. Rev. 2005;9:11–24. doi: 10.1016/j.smrv.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Nduhirabandi F, Lochner A. Melatonin and the metabolic syndrome. In: Srinivasan V, Brzezinski A, Oter S, Shillcutt SD, editors. Melatonin and Melatonergic Drugs in Clinical Practice, Chapter 6. 1. Springer; 2014. pp. 71–95. [Google Scholar]
- 7.Peschke E. Melatonin, endocrine pancreas and diabetes. J. Pineal Res. 2008;44:26–40. doi: 10.1111/j.1600-079X.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 8.Tengattini S, Reiter RJ, Tan DX, Terron MP, Rodella LF, Rezzani R. Cardiovascular diseases: Protective effects of melatonin. J. Pineal Res. 2008;44:16–25. doi: 10.1111/j.1600-079X.2007.00518.x. [DOI] [PubMed] [Google Scholar]
- 9.Espino J, Pariente JA, Rodríguez AB. Role of melatonin on diabetes-related metabolic disorders. World J. Diabetes. 2011;2:82–91. doi: 10.4239/wjd.v2.i6.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardeland R, Madrid JA, Tan DX, Reiter RJ. Melatonin, the circadian multioscillator system and health: The need for detailed analyses of peripheral melatonin signaling. J. Pineal Res. 2012;52:139–166. doi: 10.1111/j.1600-079X.2011.00934.x. [DOI] [PubMed] [Google Scholar]
- 11.Sigurdardottir LG, Markt SC, Rider JR, Haneuse S, Fall K, Schernhammer ES, Tamimi RM, Flynn-Evans E, Batista JL, Launer L, et al. Urinary melatonin levels, sleep disruption, and risk of prostate cancer in elderly men. Eur. Urol. 2015;67:191–194. doi: 10.1016/j.eururo.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arendt J. Melatonin: Characteristics, concerns, and prospects. J. Biol. Rhythms. 2005;20:291–303. doi: 10.1177/0748730405277492. [DOI] [PubMed] [Google Scholar]
- 13.Zeitzer JM, Daniels JE, Duffy JF, Klerman EB, Shanahan TL, Dijk DJ, Czeisler CA. Do plasma melatonin concentrations decline with age? Am. J. Med. 1999;107:432–436. doi: 10.1016/S0002-9343(99)00266-1. [DOI] [PubMed] [Google Scholar]
- 14.Kunz D, Schmitz S, Mahlberg R, Mohr A, Stöter C, Wolf KJ, Herrmann WM. A new concept for melatonin deficit: On pineal calcification and melatonin excretion. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 1999;21:765–772. doi: 10.1016/S0893-133X(99)00069-X. [DOI] [PubMed] [Google Scholar]
- 15.Wetterberg L, Iselius L, Lindsten J. Genetic regulation of melatonin excretion in urine. A preliminary report. Clin. Genet. 1983;24:399–402. doi: 10.1111/j.1399-0004.1983.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 16.Hallam KT, Olver JS, Chambers V, Begg DP, McGrath C, Norman TR. The heritability of melatonin secretion and sensitivity to bright nocturnal light in twins. Psychoneuroendocrinology. 2006;31:867–875. doi: 10.1016/j.psyneuen.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 17.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 18.Graham C, Cook MR, Kavet R, Sastre A, Smith DK. Prediction of nocturnal plasma melatonin from morning urinary measures [published correction appears in Journal of Pineal Research 1999 26 128] J. Pineal Res. 1998;24:230–238. doi: 10.1111/j.1600-079X.1998.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 19.Meng H, Liu T, Borjigin J, Wang MM. Ischemic stroke destabilizes circadian rhythms. J. Circadian Rhythms. 2008;6:9. doi: 10.1186/1740-3391-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pacchierotti C, Iapichino S, Bossini L, Pieraccini F, Castrogiovanni P. Melatonin in psychiatric disorders: A review on the melatonin involvement in psychiatry. Front. Neuroendocrinol. 2001;22:18–32. doi: 10.1006/frne.2000.0202. [DOI] [PubMed] [Google Scholar]
- 21.Kopin IJ, Pare CM, Axelrod J, Weissbach H. The fate of melatonin in animals. J. Biol. Chem. 1961;236:3072–3075. doi: 10.1016/S0021-9258(19)76431-X. [DOI] [PubMed] [Google Scholar]
- 22.Klante G, Brinschwitz T, Secci K, Wollnik F, Steinlechner S. Creatinine is an appropriate reference for urinary sulphatoxymelatonin of laboratory animals and humans. J. Pineal Res. 1997;23:191–197. doi: 10.1111/j.1600-079X.1997.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 23.Chen CH, Yang JH, Chiang C, Hsiung CN, Wu PE, Chang LC, Chu HW, Chang J, Song IW, Yang SL, et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum. Mol. Genet. 2016;25:5321–5331. doi: 10.1093/hmg/ddw346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 26.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 28.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics (Oxford, England) 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 30.Fallin MD, Kao WH. Is “X”-WAS the future for all of epidemiology? [Published correction appears in Epidemiology 2011 22 881] Epidemiology. 2011;22:457–468. doi: 10.1097/EDE.0b013e31821d3a9f. [DOI] [PubMed] [Google Scholar]
- 31.Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genom. 2008;9:516. doi: 10.1186/1471-2164-9-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner S. qqman: An R package for visualizing GWAS results using Q–Q and Manhattan plots. J. Open Source Softw. 2018;3:731. doi: 10.21105/joss.00731. [DOI] [Google Scholar]
- 33.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2018. [Google Scholar]
- 34.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics (Oxford, England) 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shim H, Chasman DI, Smith JD, Mora S, Ridker PM, Nickerson DA, Krauss RM, Stephens M. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS ONE. 2015;10:e0120758. doi: 10.1371/journal.pone.0120758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delongchamp R, Faramawi MF, Feingold E, Chung D, Abouelenein S. The association between SNPs and a quantitative trait: Power calculation. Eur. J. Environ. Public Health. 2018;2:10. doi: 10.20897/ejeph/3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch BC, van der Putten K, Van Someren EJ, Wielders JP, Ter Wee PM, Nagtegaal JE, Gaillard CA. Impairment of endogenous melatonin rhythm is related to the degree of chronic kidney disease (CREAM study) Nephrol. Dial. Transplant. Off. Publ. Eur. Dial. Transplant Assoc. Eur. Ren. Assoc. 2010;25:513–519. doi: 10.1093/ndt/gfp493. [DOI] [PubMed] [Google Scholar]
- 38.Morinaga T, Yasuda H, Hashimoto T, Higashio K, Tamaoki T. A human alpha-fetoprotein enhancer-binding protein, ATBF1, contains four homeodomains and seventeen zinc fingers. Mol. Cell. Biol. 1991;11:6041–6049. doi: 10.1128/mcb.11.12.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura Y, Tam T, Ido A, Morinaga T, Miki T, Hashimoto T, Tamaoki T. Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J. Biol. Chem. 1995;270:26840–26848. doi: 10.1074/jbc.270.45.26840. [DOI] [PubMed] [Google Scholar]
- 40.Wilcox AG, Vizor L, Parsons MJ, Banks G, Nolan PM. Inducible knockout of mouse Zfhx3 emphasizes its key role in setting the pace and amplitude of the adult circadian clock. J. Biol. Rhythms. 2017;32:433–443. doi: 10.1177/0748730417722631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng L, Tachibana K, Iwasaki H, Kameyama A, Zhang Y, Kubota T, Hiruma T, Tachibana K, Kudo T, Guo JM, Narimatsu H. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T15. FEBS Lett. 2004;566:17–24. doi: 10.1016/j.febslet.2004.03.108. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Iwasaki H, Wang H, Kudo T, Kalka TB, Hennet T, Kubota T, Cheng L, Inaba N, Gotoh M, Togayachi A, Guo J, Hisatomi H, Nakajima K, Nishihara S, Nakamura M, Marth JD, Narimatsu H. Cloning and characterization of a new human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, designated pp-GalNAc-T13, that is specifically expressed in neurons and synthesizes GalNAc alpha-serine/threonine antigen. J. Biol. Chem. 2003;278:573–584. doi: 10.1074/jbc.M203094200. [DOI] [PubMed] [Google Scholar]
- 43.Iqbal S, Ghanimi Fard M, Everest-Dass A, Packer NH, Parker LM. Understanding cellular glycan surfaces in the central nervous system. Biochem. Soc. Trans. 2019;47:89–100. doi: 10.1042/BST20180330. [DOI] [PubMed] [Google Scholar]
- 44.Ranganathan S, Noyes NC, Migliorini M, Winkles JA, Battey FD, Hyman BT, Smith E, Yepes M, Mikhailenko I, Strickland DK. LRAD3, a novel low-density lipoprotein receptor family member that modulates amyloid precursor protein trafficking. J. Neurosci. 2011;31:10836–10846. doi: 10.1523/JNEUROSCI.5065-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics [published correction appears in Science 2002 297 2209] Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 46.Nous A, Engelborghs S, Smolders I. Melatonin levels in the Alzheimer's disease continuum: A systematic review. Alzheimer’s Res. Ther. 2021;13:52. doi: 10.1186/s13195-021-00788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu YH, Swaab DF. The human pineal gland and melatonin in aging and Alzheimer's disease. J. Pineal Res. 2005;38:145–152. doi: 10.1111/j.1600-079X.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 48.Baskett JJ, Cockrem JF, Antunovich TA. Sulphatoxymelatonin excretion in older people: Relationship to plasma melatonin and renal function. J. Pineal Res. 1998;24:58–61. doi: 10.1111/j.1600-079X.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 49.Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein p. J. Biol. Chem. 1999;274:2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- 50.Steinbrenner H, Sies H. Selenium homeostasis and antioxidant selenoproteins in brain: Implications for disorders in the central nervous system. Arch. Biochem. Biophys. 2013;536:152–157. doi: 10.1016/j.abb.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 51.Peters MM, Hill KE, Burk RF, Weeber EJ. Altered hippocampus synaptic function in selenoprotein P deficient mice. Mol. Neurodegener. 2006;1:12. doi: 10.1186/1750-1326-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caito SW, Milatovic D, Hill KE, Aschner M, Burk RF, Valentine WM. Progression of neurodegeneration and morphologic changes in the brains of juvenile mice with selenoprotein P deleted. Brain Res. 2011;1398:1–12. doi: 10.1016/j.brainres.2011.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raman AV, Pitts MW, Seyedali A, Hashimoto AC, Seale LA, Bellinger FP, Berry MJ. Absence of selenoprotein P but not selenocysteine lyase results in severe neurological dysfunction. Genes Brain Behav. 2012;11:601–613. doi: 10.1111/j.1601-183X.2012.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Byrns CN, Pitts MW, Gilman CA, Hashimoto AC, Berry MJ. Mice lacking selenoprotein P and selenocysteine lyase exhibit severe neurological dysfunction, neurodegeneration, and audiogenic seizures. J. Biol. Chem. 2014;289:9662–9674. doi: 10.1074/jbc.M113.540682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burk RF, Hill KE, Motley AK, Winfrey VP, Kurokawa S, Mitchell SL, Zhang W. Selenoprotein P and apolipoprotein E receptor-2 interact at the blood–brain barrier and also within the brain to maintain an essential selenium pool that protects against neurodegeneration. FASEB J. 2014;28:3579–3588. doi: 10.1096/fj.14-252874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christen M, Högler S, Kleiter M, Leschnik M, Weber C, Thaller D, Jagannathan V, Leeb T. Deletion of the SELENOP gene leads to CNS atrophy with cerebellar ataxia in dogs. PLoS Genet. 2021;17:e1009716. doi: 10.1371/journal.pgen.1009716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou JN, Liu RY, Kamphorst W, Hofman MA, Swaab DF. Early neuropathological Alzheimer's changes in aged individuals are accompanied by decreased cerebrospinal fluid melatonin levels. J. Pineal Res. 2003;35:125–130. doi: 10.1034/j.1600-079X.2003.00065.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Individual researchers may request to use the data for specific projects on a collaborative basis. Our data has been submitted to the NHGRI-EBI GWAS Catalog (accession ID: GCST90101875).