Abstract
The present study was conducted to identify the novel QTLs controlling salinity and sodicity tolerance using indica MAGIC rice population. Phenotyping was carried out in salinity (EC ~ 10 dS/m) and sodicity (pH ~ 9.8) at the seedling stage. Among 391 lines, 43 and 98 lines were found tolerant and moderately tolerant to salinity. For sodicity condition, 2 and 45 lines were showed tolerance and moderately tolerance at seedling stage. MAGIC population was genotyped with the help of genotyping by sequencing (GBS) and filtered 27041SNPs were used for genome wide marker trait association studies. With respect to salinity tolerance, 25 SNPs were distributed on chromosomes 1, 5, 11 and 12, whereas 18 SNPs were mapped on chromosomes 6, 4 and 11 with LOD value of > 3.25 to sodicity tolerance in rice. The candidate gene analysis detected twelve causal genes including SKC1 gene at Saltol region for salinity and six associated genes for sodic stress tolerance. The significant haplotypes responsible for core histone protein coding gene (LOC_Os12g25120) and three uncharacterized protein coding genes (LOC_Os01g20710, LOC_Os01g20870 and LOC_Os12g22020) were identified under saline stress. Likewise, five significant haplotypes coding for ribose 5-phosphate isomerise (LOC_Os04g24140), aspartyl protease (LOC_Os06g15760), aluminum-activated malate transporter (LOC_Os06g15779), OsFBX421-Fbox domain containing protein (LOC_Os11g32940) and one uncharacterized protein (LOC_Os11g32930) were detected for sodic stress tolerance. The identified novel SNPs could be the potential candidates for functional characterization. These candidate genes aid to further understanding of genetic mechanism on salinity and sodicity stress tolerance in rice. The tolerant line could be used in future breeding programme to enhance the salinity and sodicity tolerance in rice.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-022-01174-8.
Keywords: GWAS, MAGIC, GBS, Salinity, Sodicity, Rice
Introduction
Rice is one of the important staple food crops, about 50 per cent of the global population consumes it as main source of energy. The global population is increasing day by day and it is expected to reach around 9.5 billion in 2050. Therefore, world food production needs to be increased by 70% (FAO 2013). Hence, there is a need to increase the productivity of rice by combating its low productivity caused by various biotic and abiotic stresses. Among the abiotic stresses, salinity and sodicity are the most important environmental factors hampering the crop productivity by ionic disequilibrium and disrupting the metabolic activities of the plants (Munns and Tester 2008). Salinity and sodicity stresses are highly influenced by environmental fluctuations like rise in temperature and relative humidity (Tack et al. 2015). Salt stress affects all the stages of plants but early seedling and reproductive stage are most sensitive in rice (Singh et al. 2010; Singh and Flowers 2010). Salinity stress reduces root length and shoot length at seedling stage, while tiller number, number of spikes, panicle length and spikelet fertility at reproductive stage (Ali et al. 2013; Krishnamurthy et al. 2016a). Sodicity also affects crop productivity. Many researchers have reported the effect of sodic soils on growth and development in rice (Singh et al. 2013; Krishnamurthy et al. 2017). However, less information is available on molecular basis for sodicity tolerance. Thus, it is necessary to study the molecular mechanisms underlying sodic stress.
It is very important to know the effect of salt stress on several traits in rice to breed the well adoptable salt tolerant varieties (Arshadullah et al. 2011). Various studies have been done on molecular breeding for abiotic stresses in rice. Several QTLs have been identified for salinity tolerance in rice (Lang et al. 2000; Tuan et al. 2000; Warraich et al. 2020; Mazumder et al. 2020). Saltol QTL was mapped on chromosome 1 using RIL population of Pokkali /IR29 by AFLP markers (Gregorio 1997). Saltol QTLs facilitates marker-assisted backcrossing breeding (MABB) to introgress it to popular rice varieties (Thomson et al. 2010) and has been introgressed into mega rice varieties (Singh et al. 2016; Babu et al. 2017; Geetha et al. 2017; Bhandari et al. 2019; Jaiswal et al. 2019; Krishnamurthy et al. 2021; Yadav et al. 2020). Majority of the salt tolerant rice varieties were developed based on the single crosses between two diverse parents. However, attempts were made to develop varieties employing multiple crosses involving three parents (Jansen et al. 2003). Due to technical complications like large population size and precision these multiple crosses were not frequently used by breeders. Only few reports were available on development of varieties by multi-parent advanced generation inter cross (MAGIC) population and also on identification of the QTLs for salt tolerance in rice. GWAS utilizes a wide range of diversity based on the groups of non-related individuals and take the advantage of the past crossing over and recombination events that have accrued over the generations (Korte and Farlow, 2013). Complex agronomic traits such as flowering time and grain related traits in rice have been detected by this approach (Huang et al. 2011). Linkage disequilibrium (Visscher et al. 2012) and non-identification of phenotypes caused by rare alleles (Kover and Mott, 2012) limits the application of GWAS by breeders. Earlier investigators have proposed the development of advanced intercross lines (AILs) to increase the number of recombination (Darvasi and Soller, 1995). But epistasis and the effect of genetic background were not considered in these populations. Recently, this AIL methodology has been extended into multiple parent populations. Multi-parent advanced generation intercross (MAGIC) populations has been used in model plant species Arabidopsis for the first time to make accessible of distended recombination and high mapping resolution (Cavanagh et al. 2008). MAGIC has a better control over population structure along with kinship and highly effective in identifying major genes through GWAS strategy (Bandillo et al. 2013; Ponce et al. 2020). This MAGIC population was employed in different cereal crops by many workers. With this background, the purpose of the present study was to (i) know the effect of salinity and sodicity on indica MAGIC population (ii) identify the novel QTLs for salinity and sodicity tolerance at seedling stage in rice.
Materials and methods
Plant materials
The indica MAGIC population was developed using eight originator lines that include both elite and modern varieties. These founder lines known to exhibit tolerance to both biotic and abiotic stresses along with good grain quality (Table 1).
Table 1.
Agronomic relevance of the 8 founder lines used in developing the indica MAGIC population
| Germplasm/variety | Varietal type | Origin | Agronomic relevance |
|---|---|---|---|
| Fedearroz 50 | Indica | Colombia | Popular variety in several countries, with stay green/delayed senescence & quality traits, disease tolerance, progenitor of many breeding lines |
| Shan-Huang Zhan-2 (SHZ-2) | Indica | China | Blast resistant, high yielding; in the pedigrees of many varieties in south China |
| IR64633-87–2-2–3-3 (PSBRc82) | Indica | IRRI | High yielding and most popular variety of the Philippines |
| IR77186-122–2-2–3 (PSBRc 158) | Indica / tropical japonica background | IRRI | High yielding variety in New Plant Type II background |
| IR77298-14–1-2–10 | Indica | IRRI | Drought tolerant in lowlands with IR64 background and tungro resistance |
| IR4630-22–2-5–1-3 | Indica | IRRI | Good plant type, salt tolerant at seedling and reproductive stages |
| IR45427-2B-2-2B-1–1 | Indica | IRRI | Fe toxicity tolerant |
| Sambha Mahsuri + Sub1 | Indica | IRRI | Mega variety with wide compatibility, good grain quality and submergence tolerance |
Phenotyping of MAGIC population in saline stress
A set of 391 indica MAGIC lines procured from IRRI, Philippines were evaluated for salinity tolerance along with 8 parents, FL478 (tolerant check) and IR29 (sensitive check) using Yoshida nutrient solution (Yoshida et al. 1976). The experiment was piloted in a controlled glasshouse at Central Soil Salinity Research Institute (CSSRI), Karnal under two different stress conditions, namely salinity stress (EC ~ 10 dS/m) and normal (EC ~ 1.2 dS/m) by hydroponic technique at seedling stage (IRRI, 1996). The nutrient solution used in hydroponics was salinized by adding NaCl to acquire the desired levels of salinity. Saline stress of EC ~ 10 dS/m was induced at 14 days after sowing (DAS) and the anticipated level of salinity was maintained for the next 14 days. The revised standard evaluation system (SES) was used in screening the visual symptoms of salt injury (1 = highly tolerant, 3 = tolerant, 5 = moderately tolerant, 7 = susceptible and 9 = highly susceptible) and genotypes were scored after 14 days of salinization for salinity tolerance (IRRI, 1996). In this experiment, the data was recorded on salt injuries score (1–9), root length (cm) and shoot length (cm).
Phenotyping of MAGIC population in alkaline stress
The same set of 391 indica MAGIC lines was evaluated for sodic stress in a controlled condition at CSSRI, Karnal. The experiment was carried out using a sodic soil in the tray. To obtain the desired level of sodicity (pH ~ 9.7 to 9.8), required amount of sodium bicarbonate (NaHCO3) and sodium carbonate (Na2CO3) were added to the soil that simulating the field conditions. Visual indications of the salt injury (1 = highly tolerant, 3 = tolerant, 5 = moderately tolerant, 7 = susceptible and 9 = highly susceptible) was recorded by the revised standard evaluation system (SES) (and scoring of genotypes was done after 14 days of stress (IRRI, 1996). The observation was taken on salt injuries score (1–9) and shoot length (cm) for this investigation.
Genotyping by sequencing (GBS) for genome-wide SNP analysis in indica MAGIC population
Genotyping by sequencing (GBS) is a novel technique for genotyping single nucleotide polymorphism (SNP) which has been utilized in implementation of GWAS (genome-wide association study) in different populations for crop improvement (Elshire et al. 2011; Huang et al. 2010). The raw GBS data of 391 indica MAGIC population was processed and 109,610 SNP markers were obtained. The processed data contains a large proportion of missing calls along with many heterozygous SNPs in the population. Further, filtering of data was done with missing rates > 30% and minor allele frequencies < 5%. Finally, we obtained 27,041 SNP sites and that was used for Marker Trait Association studies (MTA) (Raghavan et al. 2017).
Statistical analysis
The filtered GBS data provided a total of 27,041 SNP marker sites across all the 12 chromosomes and that were used to create kinship matrix with the help of Trait Analysis by Association, Evolution and Linkage (TASSELV5.2) program (Bradbury et al. 2007). Genotypic, kinship matrix and phenotypic data of SES score for salinity and sodicity were combined for Marker Trait Association (MTA) analysis by mixed linear model (MLM) in 391 MAGIC lines. The model statistics gives the p-value along with R2 value for the marker and we have used a cut-off P value < 0.001 for detection of trait association (Zhang et al. 2010). The Manhattan and Q-Q plots were visualized through the qqman package in R programme (Turner, 2014). The significant SNPs (LOD of > 3.25) associated with traits were taken into consideration for candidate gene analysis by using the Rice Annotation Project (RAP) database genome browser (http://rapdb.dna.affrc.go.jp/viewer/gbrowse/irgsp1). The SNP positions of genes associated to trait of interest were recognized as candidate genes. The haplotype analysis for detected candidate genes was conducted by CandiHap V2. (https://github.com/xukaili/CandiHap).
Results
In order to identify QTLs for salinity and sodic stress, we screened the 391 indica MAGIC populations in glasshouse condition and then 27,041 SNP sites were used for association studies. The results of Marker Trait Association (MTA) are presented below.
Phenotyping of parents and MAGIC lines for non-stress and salinity stress at seedling stage
Based on the results of vigor score (SES-score), shoot and root length among eight indica parents, IR4630-22-2-5-1-3 and IR77186-122-2-2-3 (PSBRc 158) genotypes exhibited moderately tolerant response (SES-5); Fedearroz 50 and IR45427-2B-2-2B-1–1 genotypes were found susceptible (SES-7); remaining parents like Shan–Huang Zhan-2 (SHZ-2), IR64633-87-2-2-3-3 (PSBRc82), IR77298-14-1-2-10 and Sambha Mahsuri + Sub1 showed highly susceptible reaction (SES-9) with respect to salinity stress (supplementary table 1).The results of mean, range and per cent reduction of different traits for 391 MAGIC lines are represented in supplementary table 2. In general, all the MAGIC lines were showing reduction in their root and shoot length and these reductions varied among the lines. The frequency distribution of 391 MAGIC rice lines were estimated in all the stresses and furnished in supplementary Fig. 1.The findings indicate that tolerant and sensitive groups of MAGIC lines differed significantly in all the parameters, including reduction in shoot length, root length and vigor score (SES). Per cent root length reduction was 23% and shoot length reduction accounts for 30.56% by salinity stress in all the MAGIC lines. The root length (cm) ranged from 2.33 (IR 93,347:21-B-7-7-5-1RGA-2RGA-1-B-B) to 21.3 (IR 93,337:28-B-9-3-20-1RGA-2RGA-1-B-B) in non-stress and 2.27 (IR 93,327:27-B-8-19-6-1RGA-2RGA-1-B-B) to 14.50 (IR 93,353:15-B-3-22-7-1RGA-2RGA-1-B-B) in saline stress condition. The range of shoot length (cm) was 19.20 (IR 93,336:31-B-16–23-6-1RGA-2RGA-1-B-B) to 61.80 (IR 93,350:45-B-23–16-15-1RGA-2RGA-1-B-B) in non-stress and 14.40 (IR 93,343:14-B-12–5-20-1RGA-2RGA-1-B-B) to 35.73 (IR 93,347:2-B-10–14-4-1RGA-2RGA-1-B-B) in salt stress environment (supplementary table 2). With respect to SES, maximum number of lines scored 7 (sensitive) under salinity stress at seedling stage. Out of 391 lines, 43 genotypes were salinity tolerant and 98 genotypes were moderately tolerant (supplementary table 3). The lines expressing the tolerance to salinity are listed in supplementary table 4.
Phenotyping of MAGIC lines for sodicity stress at seedling stage
Shoot length (cm) was ranged from 6.44 (IR 93,346:35-B-23–2-7-1RGA-2RGA-1-B-B) to 48.34 (IR 93,341:59-B-3–13-10-1RGA-2RGA-1-B-B) in sodic conditions (supplementary table 2). Maximum number of genotypes scored 9 (highly sensitive) under sodicity. Among the 391 lines, two genotypes were tolerant and 45 genotypes were found moderately tolerant to sodicity at seedling stage (supplementary table 3). The genotypes, IR 93,326:18-B-12–7-15-1RGA-2RGA-1-B-B and IR 93,328:13-B-8–6-23-1RGA-2RGA-1-B-B were found to be tolerant in terms of sodicity stress (supplementary table 5).
Association mapping and candidate genes haplotype analysis for salinity tolerance at seedling stage
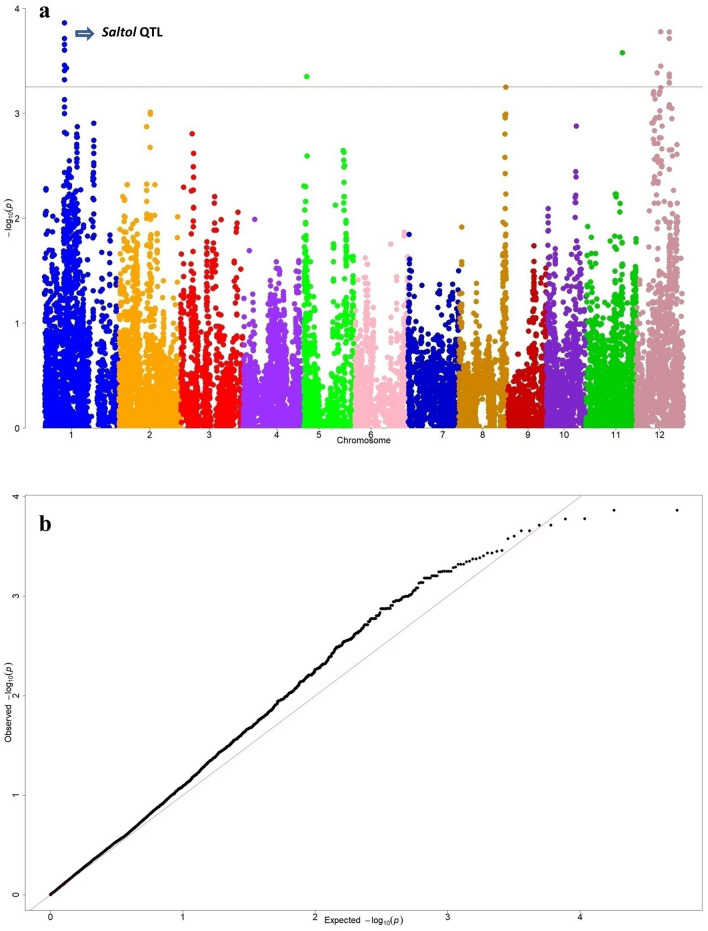
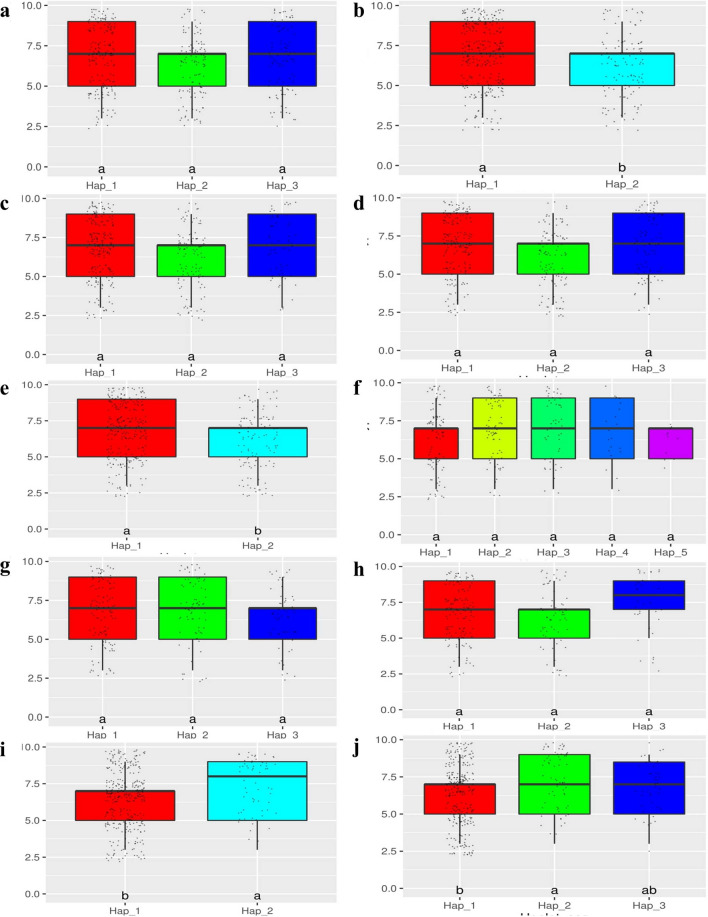
The GWAS was conducted for salt injury score (1–9 vigour score) at salinity stress to identify causal genes. The genome wide Manhattan plot for salinity vigor score and Q-Q plot are shown in Fig. 1. In the present study, we detected a total of 25 SNPs significantly associated with (LOD value of > 3.25) the trait vigour score under salt stress at seedling stage. The associated markers were distributed across the chromosomes, among them 13 SNPs found on chromosome 1, 10 SNPs on chromosome 12 and single peaks were observed on chromosome 5 and 11. The chromosome 1 and 12 consists of QTL regions for salinity tolerance at seedling stage. The origin of GWAS peaks pertaining to salinity tolerance at chromosome 1 was tracked between the 128 kb region from 11.523 to 11.647 Mb and located eleven SNP markers. These markers situated near to Saltol region, a known and highly considered region for salinity tolerance at seedling stage in rice. Similarly, seven peak SNP markers on chromosome 12 were positioned in between 36 kb region from 19.369 to 19.405 Mb. At the chromosome 1, we found a peak at 11.523233 which is 60 kb apart from SKC1, the well known gene responsible for salinity tolerance at seedling stage. The identified SNPs responsible for salt tolerance were used for candidate gene analysis and detected eleven putative candidate genes lying in trait associated regions are presented in Table 2. Out of eleven genes, five genes LOC_Os01g20710, LOC_Os01g20870, LOC_Os12g22020, LOC_Os12g32150 and LOC_Os12g32160 codes for uncharacterized hypothetical proteins. Remaining six genes were annotated for stress related proteins like C-NBS-LRR domain (LOC_Os01g20720), heavy metal and copper transport associated protein (LOC_Os01g20830), retrotransposon protien containing pectin lyase fold domain (LOC_Os01g22590), WRKY-109 comprising of DNA binding domain (LOC_Os05g03900) and core histone related (H2A/H2B/H3/H4) protein (LOC_Os12g25120) for salt tolerance at seedling stage. Candidate gene haplotype analysis was carried out for these genes along with SKC1 (LOC_Os01g20160) which confers seedling stage salinity tolerance in rice is presented in Table 3. We observed a range of two to eight haplotypes for each putative locus. The significant haplotypes were found for LOC_Os01g20710, LOC_Os01g20870, LOC_Os12g22020 and LOC_Os12g25120 (Fig. 2 and supplementary Fig. 2) genes. The haplotype analysis indicates three haplotypes for SKC1 (LOC_Os01g20160) gene which was present on chromosome 1 between 11.458 Mb to 11.466 Mb (Fig. 2a). The probable candidate genes haplotype contributing parents and haplotype specific MAGIC lines for salinity tolerance are presented in supplementary table 6.
Fig. 1.
a Genome wide distribution of SNPs presented in Manhattan plot for salt injury score (vigour score) for salinity stress through MLM model, b The expected and observed -log10 probability distribution of SNPs presented in Q-Q plot for salt injury score (vigour score) for salinity stress through MLM model. Saltol region, a known QTL responsible for salinity tolerance at seedling stage on chromosome 1 was highlighted
Table 2.
Associated QTLs with SNP position and probable candidate genes for salinity tolerance in MAGIC population
| Sl.No | Chr | Peak SNP | P value | LOD | MarkerR2 | FDR | Locus ID | Gene annotation |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 11,523,233 | 4.81E-04 | 3.32 | 0.031 | 0.416 | LOC_Os01g20710 | Uncharecterized expressed protein |
| 2 | 1 | 11,529,325 | 4.81E-04 | 3.32 | 0.031 | 0.416 | LOC_Os01g20720 | CC-NBS-LRR,Putative expressed and response to stress |
| 3 | 1 | 11,529,368 | 4.81E-04 | 3.32 | 0.031 | 0.416 | ||
| 4 | 1 | 11,587,558 | 3.49E-04 | 3.46 | 0.032 | 0.416 | ||
| 5 | 1 | 11,596,445 | 3.94E-04 | 3.40 | 0.032 | 0.416 | ||
| 6 | 1 | 11,601,232 | 2.21E-04 | 3.66 | 0.035 | 0.416 | ||
| 7 | 1 | 11,611,121 | 2.21E-04 | 3.66 | 0.035 | 0.416 | LOC_Os01g20830 | Heavy metal associated protein |
| 8 | 1 | 11,623,953 | 1.38E-04 | 3.86 | 0.037 | 0.416 | ||
| 9 | 1 | 11,624,067 | 1.38E-04 | 3.86 | 0.037 | 0.416 | ||
| 10 | 1 | 11,624,734 | 1.94E-04 | 3.71 | 0.035 | 0.416 | ||
| 11 | 1 | 11,647,832 | 2.51E-04 | 3.60 | 0.034 | 0.416 | LOC_Os01g20870 | Expressed hpothetical protein |
| 12 | 1 | 12,702,701 | 3.71E-04 | 3.43 | 0.032 | 0.416 | LOC_Os01g22590 | Retrotransposon pectin lyase fold domain containing protein |
| 13 | 1 | 12,702,721 | 3.71E-04 | 3.43 | 0.032 | 0.416 | ||
| 14 | 5 | 1,743,516 | 4.47E-04 | 3.35 | 0.031 | 0.416 | LOC_Os05g03900 | WRKY 109- DNA-binding domain containing protein |
| 15 | 11 | 21,055,879 | 2.66E-04 | 3.58 | 0.034 | 0.416 | LOC_Os11g35870 | RING finger family RWD domain containing protein |
| 16 | 12 | 12,383,067 | 4.14E-04 | 3.38 | 0.032 | 0.416 | LOC_Os12g22020 | Expressed protein |
| 17 | 12 | 14,424,756 | 1.67E-04 | 3.78 | 0.036 | 0.416 | LOC_Os12g25120 | Core histone H2A/H2B/H3/H4, |
| 18 | 12 | 14,430,101 | 3.56E-04 | 3.45 | 0.032 | 0.416 | ||
| 19 | 12 | 19,369,099 | 4.23E-04 | 3.37 | 0.031 | 0.416 | ||
| 20 | 12 | 19,369,102 | 4.23E-04 | 3.37 | 0.031 | 0.416 | ||
| 21 | 12 | 19,392,199 | 1.94E-04 | 3.71 | 0.035 | 0.416 | LOC_Os12g32150 | Expressed conserved hypothetical protein |
| 22 | 12 | 19,392,202 | 1.68E-04 | 3.77 | 0.036 | 0.416 | ||
| 23 | 12 | 19,392,459 | 5.18E-04 | 3.29 | 0.031 | 0.416 | ||
| 24 | 12 | 19,400,490 | 5.11E-04 | 3.29 | 0.031 | 0.416 | ||
| 25 | 12 | 19,405,473 | 4.52E-04 | 3.34 | 0.031 | 0.416 | LOC_Os12g32160 | Expressed hpothetical protein |
Table 3.
Alternative haplotypes for candidate genes for salinity tolerance identified by GWAS in MAGIC population
| Locus ID | Haplo group | Number of lines | SNP positions | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LOC_Os01g20160 | 11,460,324 | 11,460,344 | ||||||||
| Hap_1 | 180 | GG | GG | |||||||
| Hap_2 | 115 | GG | TT | |||||||
| Hap_3 | 96 | CC | GG | |||||||
| LOC_Os01g20710 | 11,523,233 | |||||||||
| Hap_1 | 276 | CC | ||||||||
| Hap_2 | 115 | TT | ||||||||
| LOC_Os01g20720 | 11,529,325 | 11,529,363 | 11,529,368 | |||||||
| Hap_1 | 214 | GG | GG | GG | ||||||
| Hap_2 | 115 | TT | AA | AA | ||||||
| Hap_3 | 62 | GG | AA | GG | ||||||
| LOC_Os01g20830 | 11,611,121 | 11,611,147 | ||||||||
| Hap_1 | 181 | GG | TT | |||||||
| Hap_2 | 116 | TT | CC | |||||||
| Hap_3 | 94 | GG | CC | |||||||
| LOC_Os01g20870 | 11,647,832 | |||||||||
| Hap_1 | 274 | GG | ||||||||
| Hap_2 | 117 | AA | ||||||||
| LOC_Os01g22590 | 12,701,866 | 12,702,701 | 12,702,721 | 12,704,071 | 12,704,146 | 12,704,338 | ||||
| Hap_1 | 96 | TT | TT | TT | CC | GG | GG | |||
| Hap_2 | 73 | TT | AA | CC | GG | GG | AA | |||
| Hap_3 | 48 | CC | AA | CC | CC | GG | AA | |||
| Hap_4 | 20 | TT | TT | TT | CC | CC | GG | |||
| Hap_5 | 11 | CC | AA | CC | CC | GG | GG | |||
| Hap_6 | 2 | TT | AA | CC | CC | GG | GG | |||
| LOC_Os05g03900 | 1,743,516 | 1,743,571 | ||||||||
| Hap_1 | 116 | AA | TT | |||||||
| Hap_2 | 77 | AA | CC | |||||||
| Hap_3 | 57 | TT | TT | |||||||
| LOC_Os11g35870 | 21,055,876 | 21,055,879 | 21,055,916 | 21,055,974 | ||||||
| Hap_1 | 142 | GG | GG | GG | TT | |||||
| Hap_2 | 70 | TT | GG | AA | CC | |||||
| Hap_3 | 38 | TT | AA | GG | TT | |||||
| LOC_Os12g22020 | 12,383,067 | |||||||||
| Hap_1 | 329 | GG | ||||||||
| Hap_2 | 62 | CC | ||||||||
| LOC_Os12g25120 | 14,424,738 | 14,424,756 | ||||||||
| Hap_1 | 281 | CC | CC | |||||||
| Hap_2 | 68 | CC | TT | |||||||
| Hap_3 | 42 | TT | TT | |||||||
| LOC_Os12g32150 | 19,392,188 | 19,392,199 | 19,392,202 | 19,392,459 | 19,392,469 | 19,392,470 | 19,392,479 | 19,392,522 | ||
| Hap_1 | 157 | CC | AA | AA | AA | CC | AA | CC | AA | |
| Hap_2 | 132 | TT | GG | GG | GG | AA | CC | GG | GG | |
| Hap_3 | 53 | CC | GG | GG | GG | AA | CC | CC | GG | |
| Hap_4 | 42 | CC | GG | GG | GG | CC | AA | CC | AA | |
| Hap_5 | 3 | TT | GG | GG | AA | CC | AA | CC | AA | |
| Hap_6 | 2 | CC | GG | AA | AA | CC | AA | CC | AA | |
| Hap_7 | 1 | CC | AA | AA | GG | AA | CC | CC | GG | |
| Hap_8 | 1 | TT | GG | GG | GG | CC | AA | CC | GG | |
| LOC_Os12g32160 | 19,405,449 | 19,405,473 | 19,405,682 | 19,408,151 | 19,408,157 | |||||
| Hap_1 | 89 | GG | AA | GG | CC | GG | ||||
| Hap_2 | 79 | AA | GG | GG | TT | TT | ||||
| Hap_3 | 47 | GG | GG | GG | CC | GG | ||||
| Hap_4 | 34 | GG | GG | AA | CC | GG | ||||
| Hap_5 | 1 | GG | AA | GG | TT | TT | ||||
Fig. 2.
Boxplots indicating phenotypic responses for salinity vigor score among indica MAGIC rice lines with alternative haplotypes for candidate genes determined from significantly associated SNPs by GWAS analysis (X-axis = Haplotypes; Y-axis = Vigor score). a LOC_Os01g20160; b LOC_Os01g20710; c LOC_Os01g20720; d LOC_Os01g20830; e LOC_Os01g20870; f LOC_Os01g22590; g LOC_Os05g03900; h LOC_Os11g35870; i LOC_Os12g22020; j LOC_Os12g25120
Association mapping and candidate genes haplotype analysis for sodicity tolerance at seedling stage
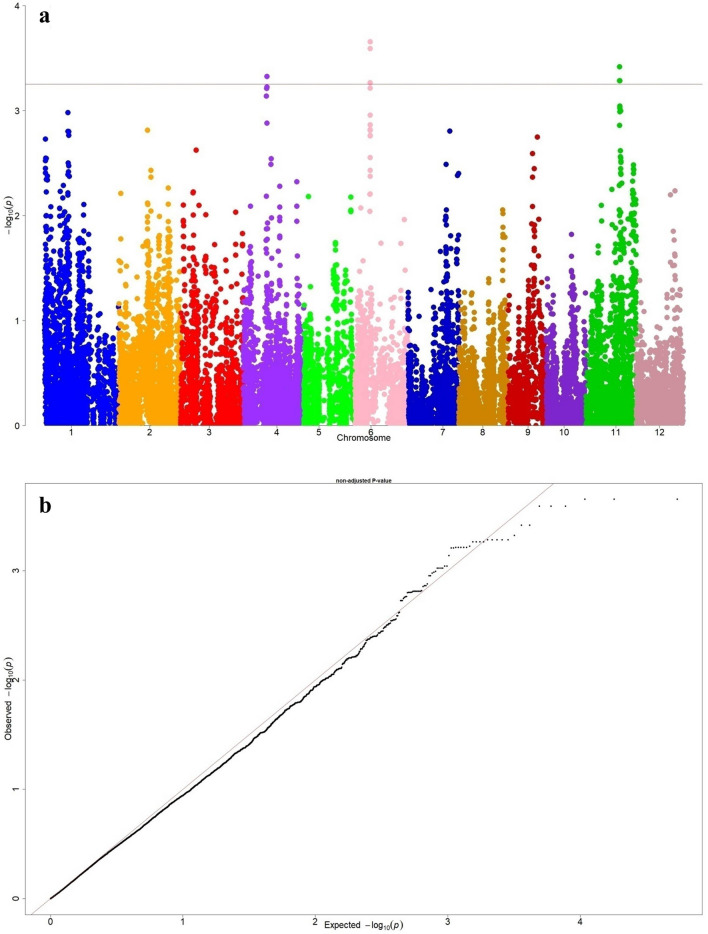
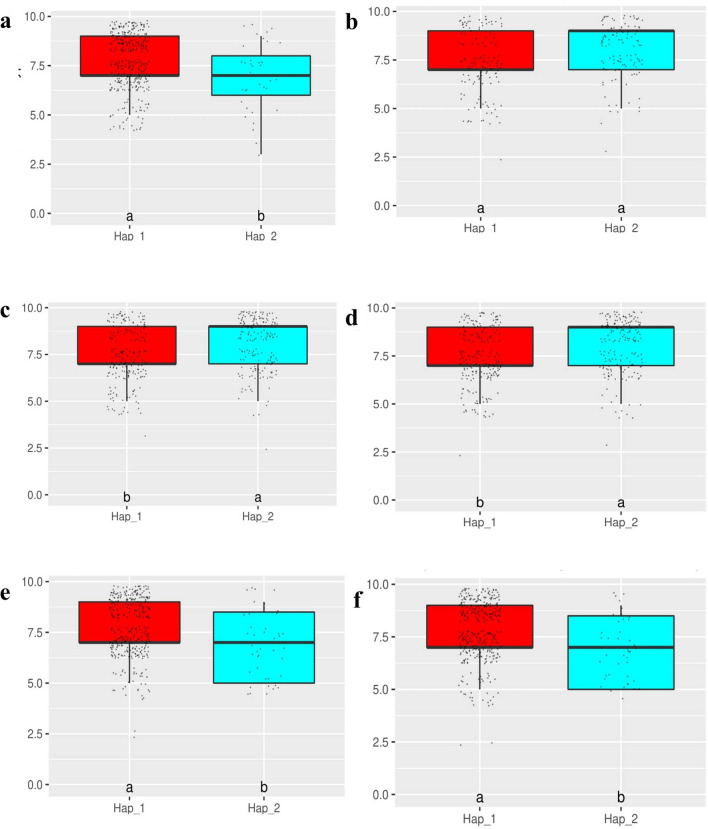
As similar to salinity, GBS data of 391 indica MAGIC rice lines was used for association analysis with respect to vigor score (SES) under sodic stress at seedling stage.The genome wide Manhattan plot for sodicity vigor score and Q-Q plot are presented in Fig. 3. GWAS peaks revealed that, QTLs mapped on chromosome 6, 4 and 11 plays an important role in controlling the sodicity tolerance at seedling stage. Significant GWAS loci peaks related to sodicity at chromosome 6 was formed between the coordinates of 8.91 and 8.96 Mb in 45 kb region, whereas at chromosome 11 it was traced between 19.4 to 19.5 Mb in 80 kb region. For sodicity tolerance, around 18 SNPs found distributed on three chromosomes i.e., one on chromosome 4, ten on chromosome 6 and seven on chromosome 11 with LOD value of > 3.25. The associated 18 SNPs were subjected to candidate gene analysis and identified six putative causal genes prevailing in genomic regions are presented in Table 4. Among six genes, LOC_Os11g32930 gene on chromosome 11 is uncharacterized and responsible for expression of hypothetical protein. There is a peak on chromosome 4 that mapped a candidate gene LOC_Os04g24140 (13.8–13.805 Mb) which is known for ribose‐5‐phosphate isomerase. The three candidate genes were detected on chromosome 6 between 8.91 and 8.96 Mb of 45 kb region. The gene LOC_Os06g15730 regulates NB-ARC domain containing protein and it is similar to disease resistance protein RGA3 (XM_026025943.1). LOC_Os06g15760 locus encodes aspartyl protease and peptidase domain containing protein, while the locus LOC_Os06g15779 plays an crucial role for aluminium-activated malate transporter (ALMT1) which is one of the key genes that performs the multiple functions like osmotic adjustment, maintenance of electro neutrality and influencing tolerance at sodicity (pH ~ 9.8). We found a candidate gene LOC_Os11g32940 on chromosome 11 related to OsFBX421 an F-box domain containing protein. Candidate gene haplotype analysis was performed for the six candidate genes that were involved in the sodic stress tolerance in rice and are presented in Table 5. We noticed two significant haplotypes for the genes LOC_Os04g24140, LOC_Os06g15760, LOC_Os06g15779, LOC_Os11g32930, LOC_Os11g32940 and four non-significant haplotypes for the candidate gene LOC_Os06g15730 which are shown in Fig. 4. The probable candidate genes haplotype contributing parents and haplotype specific MAGIC lines for sodicity tolerance are provided in supplementary table 6.
Fig. 3.
a Genome wide distribution of SNPs presented in Manhattan plot for sodic injury score (vigour score) under sodic stress through MLM model b The expected and observed -log10 probability distribution of SNPs presented in Q-Q plot for sodic injury score (vigour score) under sodic stress through MLM model
Table 4.
Associated QTLs with SNP position and probable candidate genes for sodicity tolerance in MAGIC population
| Sl.No | Chr | SNP position | P value | LOD | MarkerR2 | FDR | Locus ID | Gene annotation |
|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 13,803,235 | 4.75E-04 | 3.32 | 0.032 | 0.641 | LOC_Os04g24140 | Ribose 5-phosphate isomerase family protein |
| 2 | 6 | 8,917,185 | 5.44E-04 | 3.26 | 0.031 | 0.641 | LOC_Os06g15730 | NB-ARC domain containing protein part of the gene is similer to disease resistance protein RGA3 (XM_026025943.1) |
| 3 | 6 | 8,917,933 | 5.44E-04 | 3.26 | 0.031 | 0.641 | ||
| 4 | 6 | 8,917,936 | 5.44E-04 | 3.26 | 0.031 | 0.641 | ||
| 5 | 6 | 8,917,962 | 5.44E-04 | 3.26 | 0.031 | 0.641 | ||
| 6 | 6 | 8,921,200 | 2.57E-04 | 3.59 | 0.035 | 0.641 | ||
| 7 | 6 | 8,921,201 | 2.57E-04 | 3.59 | 0.035 | 0.641 | ||
| 8 | 6 | 8,932,488 | 2.57E-04 | 3.59 | 0.035 | 0.641 | ||
| 9 | 6 | 8,956,202 | 2.22E-04 | 3.65 | 0.036 | 0.641 | LOC_Os06g15760 | Aspartyl protease/Peptidase A1 domain containing protein |
| 10 | 6 | 8,962,742 | 2.22E-04 | 3.65 | 0.036 | 0.641 | LOC_Os06g15779 | Aluminum-activated malate transporter protein |
| 11 | 6 | 8,962,796 | 2.22E-04 | 3.65 | 0.036 | 0.641 | ||
| 12 | 11 | 19,448,111 | 5.19E-04 | 3.28 | 0.031 | 0.641 | LOC_Os11g32930 | Expressed protein |
| 13 | 11 | 19,448,156 | 5.19E-04 | 3.28 | 0.031 | 0.641 | ||
| 14 | 11 | 19,452,795 | 5.19E-04 | 3.28 | 0.031 | 0.641 | LOC_Os11g32940 | OsFBX421—F-box domain containing protein |
| 15 | 11 | 19,468,613 | 3.83E-04 | 3.42 | 0.033 | 0.641 | ||
| 16 | 11 | 19,468,625 | 3.83E-04 | 3.42 | 0.033 | 0.641 | ||
| 17 | 11 | 19,528,488 | 5.19E-04 | 3.28 | 0.031 | 0.641 | ||
| 18 | 11 | 19,528,524 | 5.19E-04 | 3.28 | 0.031 | 0.641 |
Table 5.
Alternative haplotypes for candidate genes for sodicity tolerance identified by GWAS in MAGIC population
| Locus ID | Haplo group | Number of lines | SNP positions | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LOC_Os04g24140 | 13,803,235 | |||||||||
| Hap_1 | 356 | CC | ||||||||
| Hap_2 | 35 | GG | ||||||||
| LOC_Os06g15730 | 8,914,643 | 8,914,650 | 8,914,651 | 8,914,671 | 8,914,769 | 8,917,185 | 8,917,933 | 8,917,936 | ||
| Hap_1 | 136 | CC | GG | CC | TT | TT | CC | TT | GG | |
| Hap_2 | 112 | TT | AA | TT | CC | CC | TT | CC | CC | |
| Hap_3 | 1 | TT | AA | TT | CC | CC | CC | TT | GG | |
| Hap_4 | 1 | CC | GG | CC | TT | TT | TT | CC | CC | |
| LOC_Os06g15760 | 8,956,202 | |||||||||
| Hap_1 | 210 | CC | ||||||||
| Hap_2 | 181 | GG | ||||||||
| LOC_Os06g15779 | 8,962,742 | 8,962,796 | ||||||||
| Hap_1 | 210 | TT | TT | |||||||
| Hap_2 | 181 | CC | CC | |||||||
| LOC_Os11g32930 | 19,448,111 | 19,448,156 | ||||||||
| Hap_1 | 349 | CC | GG | |||||||
| Hap_2 | 42 | TT | CC | |||||||
| LOC_Os11g32940 | 19,452,795 | |||||||||
| Hap_1 | 349 | CC | ||||||||
| Hap_2 | 42 | TT | ||||||||
Fig. 4.
Boxplots indicating phenotypic responses for sodic vigor score among indica MAGIC rice lines with alternative haplotypes for candidate genes determined from significantly associated SNPs by GWAS analysis (X-axis = Haplotypes; Y-axis = Vigor score). a LOC_Os04g24140; b LOC_Os06g15730; c LOC_Os06g15760; d LOC_Os06g15779; e LOC_Os11g32930; f LOC_Os11g32940
Discussion
Agricultural production is adversely influenced by the presence of excess salts in the soil. According to statistical data, nearly four million hectare of rice crop is affected badly by salinity in India. Salt tolerance is governed by many genes with different genetic interactions and it is regulated by multigenic traits in rice. Identification of QTLs which contribute to the salt tolerance helps in improving this trait through marker assisted back cross breeding and producing new rice cultivars with desirable traits DNA markers linked to the salt tolerant trait can considerably shorten breeding time to develop varieties for salt tolerance.
Effect of salinity and sodicity on phenotypic traits
Significant variation was observed in salt injury score (vigor) and other growth parameters in MAGIC lines. This was reflected due to strong interaction in the lines and could be from salt tolerant donor parent IR4630-22–2-5–1-3 or IR77186-122–2-2–3. Among the 391 genotypes, 43 were tolerant to salinity and 2 were tolerant to sodicity. Reduction in seedling height was the major trait that clearly showed variation between the tolerant and sensitive lines under salt stress. Decline in survival and growth of seedlings were the major causes of reduction in plant height, biomass, root length and crop stand failure which leads to yield loss in salt-affected areas (Singh et al. 2014; Heenan et al. 1988; Zeng and Shannon, 2000a, b). Root length and shoot length reduction was noticed in all the lines for salinity stress. Earlier researchers found that the degree of decrease in shoot height was due to intensity and duration of saline stress (Singh et al. 2013; Munns and Tester, 2008; Zeng and Shannon, 2000b; Pundir et al. 2016).
Association studies for salinity and sodicity tolerance
GWAS in indica MAGIC population was conducted by using their corresponding genotype datasets. QTLs for salt stress have been mapped repeatedly on chromosome- 1, 4, 6 and 7, whereas very few on chromosome 2, 3, 5, 8, 9, 10, 11 and 12 (Tiwari et al. 2016; Negrao et al. 2011; Pandit et al. 2010). Salinity and sodicity tolerance are highly influenced by environment (Krishnamurthy et al. 2016b) as it is controlled by multiple complex genes. These environmental factors affect the early vigor and grain yield. The success of marker assisted back cross breeding (MABB) encouraged the rice breeders in further exploration of the novel QTLs that covers the complex traits. Thus, mapping of QTLs for salinity tolerance is very helpful for rice breeders to enhance the salt tolerance. In this experiment, we carried out GWAS in MAGIC population and it provides adequate power for association analysis because of its larger population size and wider genetic diversity. The combination of expression pattern and QTL mapping for salt tolerance offers significant information on expression of genes associated with a particular QTL region that are affected by a salt stress condition.
Earlier workers have reported the presence of salinity tolerance genes using bi-parental population on chromosome 1 (Ganie et al. 2014; Thomson et al. 2010). Previous studies focused on the Saltol QTL regions marks the presence of SKC1, SalT and pectin esterase genes as the key factors for the salinity tolerance (Bonilla et al. 2002; Claes et al. 1990; Ren et al. 2005). The SKC1 gene was found to be overexpressed under salinity stress in Arabidopsis (Ren et al. 2005). This gene could be one of the potential targets that are being affected during salinity stress. Our findings showed the presence of a peak (60 kb) near to SKC1 gene at chromosome 1 of rice. But, non-significant haplotype results was observed for SKC1 (LOC_Os01g20160) locus, this may be due to its association with K+/Na+ phenotypic value under salt stress and not because of SES-vigor score. Similarly, the stress responsive CC-NBS-LRR-encoding gene (LOC_Os01g20720) belongs to a class of resistance gene which helps in the recognition of pathogen-derived avirulence protein. It provides a durable blast resistance in rice through the activation of signal transduction pathway by interacting with WRKY45 transcription factor (Inoue et al. 2013). It was also involved in the drought tolerance mechanism by the overexpression of ADR1 gene encoding a CC-NBS-LRR in A. thaliana (Chini et al. 2004). Leon et al.(2016) recorded the QTL region between 11.53 to 11.58 Mb on chromosome 1 which confers high shoot K+ concentration (qK1.11) in the vicinity of LOC_Os01g20720 from pokkali derived RIL population. Another candidate gene LOC_Os01g20830 responsible for heavy metal transport/detoxification superfamily protein was overexpressed during biotic stress (Magnaporthe oryzae) in rice was identified in this study (Zhang et al. 2010). Retrotransposon protein containing pectin lyase fold domain was encoded by the LOC_Os01g22590 gene. The pectin lyase is an extracellular enzyme and induced by pectin. Divergent functions of the pectin lyase members for stress response was reported in Arabidopsis (Cao 2012). LOC_Os05g03900 gene regulates WRKY 109 DNA-binding domain containing protein. Usually, WRKY TF family plays a key role in controlling the tolerance to biotic and abiotic stresses response (Li et al. 2020). Likewise, LOC_Os11g35870 controlling Really Interesting Novel Gene (RING) finger family protein was involved in the ubiquitination pathway. The stress-induced RING E3 ligase related to RING family genes inferred to play a role in regulation of different physiological responses like protein stabilization, maintenance of cell membrane integrity, stomatal opening, heavy metal levels and reactive oxygen species through ubiquitination in plants (Chapagain et al. 2017). However, haplotype analysis of these candidate genes showed non-significant results. It may be due to the use of broad ranged phenotype (vigor score) for identification of QTLs. Even though vigor score is an important trait for salt injury, which may be unable to capture the minute phenotypic differences caused from each of the candidate genes. The significant haplotypes were observed for the genes LOC_Os01g20710, LOC_Os01g20870 and LOC_Os12g22020 related to uncharacterized proteins and the gene LOC_Os12g25120 regulating core histone H2A/H2B/H3/H4 protien. Histones play an important role in chromosomal stability, transcription regulation, DNA repair and replication. The modification of histone protein like acetylation associated with up-regulation of the cell wall related genes under salt stress condition (Li et al. 2014).
Sodicity is believed to be one of the major factors of abiotic stresses which limit the crop productivity. The effect of sodicity was investigated in the different crops like leaf and roots of a xerophilous grass (Leymus chinensis) (Jin et al. 2006), tomato (Biatczyk et al. 1994) and wheat (Millar et al. 2007) reveals that the plants have developed various tolerant mechanisms to adapt themselves under sodic stress conditions during the course of evolution. No such studies have been reported in rice. Our investigation targeted the genes related to sodicity in rice using MAGIC population. Out of six candidate genes, all the genes showed significant haplotypes except the locus LOC_Os06g15730 which codes for NB-ARC domain containing disease resistance protein. These genes recognizes the pathogen-associated molecular patterns (PAMPs) during the pathogen attack, while plants also sense the abiotic stress through the surface-localized pattern recognition receptors (PRRs) that were present in the NB-ARC domain containing protiens (Głowacki et al. 2011; Fujita et al. 2006; Kim et al. 2014). The gene, LOC_Os06g15760 encoding Aspartyl protease domain containing protein regulates biotic and abiotic stressors. Overexpression of grapevine aspartic protease (AP17) gene in transgenic Arabidopsis plants exhibited salt- and drought-tolerance (Guo et al. 2015). An aluminium activated malate transporter (which is an ortholog for ALMT1 gene in Arabidopsis) confers sodicity stress on chromosome 6. The AtALMT1 gene was found to be one of the several genes critical for aluminium toxicity tolerance in Arabidopsis (Hoekenga et al. 2006). Recently, an aluminium resistant gene ral1 has been isolated and characterized on chromosome 6 in rice (Liu et al. 2016), Barley (Delhaize et al. 2004) and Wheat (Sasaki et al. 2004). Sodicity tolerance seems to be controlled by the LOC_Os06g15779 gene. Aluminium is considered as toxic to plants at high pH, which considerably reduces the growth and development of plants more than that caused by sodicity (high pH) alone. The impact of both sodicity and aluminium became evident at pH ~ 9.0 and waning at pH > 9.2. Hence, it is important to know the pH where aluminium becomes dominant, that probably can cause phytotoxicity (Brautigan et al. 2012). Therefore, correlation of markers associated with aluminium toxicity and sodicity needs to be further studied. LOC_Os04g24140 (Ribose-5-phosphate isomerase) is one of the key genes influencing tolerance at sodicity (pH ~ 9.8). Deficiency in a cytosolic ribose-5-phosphate isomerase causes chloroplast dysfunction, late flowering and premature cell death in Arabidopsis (Xiong et al. 2009). Thus, this gene also plays an important role in expressing tolerance at sodic stress in rice. OsFBX421 F-box domain containing protein was controlled by the LOC_Os11g32940 gene. F-box proteins constitute large family encoding genes for different abiotic stress conditions, floral transition, panicle and seed development (Jain et al. 2007). The function of LOC_Os11g32940 gene in cold tolerance at germination stage in chromosome segment substitution lines (CSSL) developed from cold tolerant wild rice donor Y11 (Oryza rufipogon Griff.) was reported by Pan et al. (2020). Identified salinity and sodicity tolerant SNPs and respective candidate genes responsible for abiotic stress can be further reinvestigated to confirm their role in salt tolerance in rice. After testing in multiplication trail identified tolerant lines could be released as salt tolerant commercial variety and they can also be used as salt tolerant donor for future breeding programme.
Conclusions
The results revealed that, salinity and sodicity plays a significant role in the seedling stage of rice growth and development. In this study, we identified 25 SNPs associated with salinity tolerance along with twelve candidate genes including SKC1 gene on chromosome 1, 5, 11 and 12. The GWAS analysis detected a Saltol region, a major QTL with 11 SNPs related to salinity on chromosome 1. Trait linked 18 SNPs and six candidate genes were noticed on chromosome 4, 6 and 11 for sodicity tolerance. An aluminium activated malate gene transporter conferring sodic stress tolerance was located on chromosome 6. About 10 candidate genes showing significant haplotypes were recorded for salinity and sodicity tolerance in rice. The genotypes tolerant to salinity and sodicity stress could be used in the development of salt tolerant variety. This population could be further used in the QTL mapping at the reproductive stage.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Indian Council of Agricultural Research (ICAR), India and International Rice Research Institute (IRRI), Philippines for funding and sparing breeding materials, the GWAS, MAGIC team at IRRI for sparing GBS data and advice in data analysis and Central Soil Salinity Research Institute Karnal (PME Cell reference no Research Article/95/2019).
Author contributions
SLK, PCS, RKS, HL design the experiment, edit the manuscript, SLK, ASW conducted experiments, draft the manuscript and analyze the data, DD, SR NMV, ASW, BML performed the analysis of data, wrote and revised the manuscript.
Declaration
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
S. L. Krishnamurthy, Email: krishnagene@gmail.com
P. C. Sharma, Email: pcsharma.knl@gmail.com
R. K. Singh, Email: rksinghirri@gmail.com
References
- Alam R, Sazzadur Rahman M, Seraj Z, Thomson M, Ismail A, Tumimbang-Raiz E, Gregorio G. Investigation of seedling-stage salinity tolerance QTLs using backcross lines derived from Oryzasativa L. Pokkali. Plant Breed. 2011;130(4):430–437. doi: 10.1111/j.1439-0523.2010.01837.x. [DOI] [Google Scholar]
- Ali S, Gautam R, Mahajan R, Krishnamurthy S, Sharma S, Singh R, et al. Stress indices and selectable traits in SALTOL QTL introgressed rice genotypes for reproductive stage tolerance to sodicity and salinity stresses. Field Crop Res. 2013;154:65–73. doi: 10.1016/j.fcr.2013.06.011. [DOI] [Google Scholar]
- Arshadullah M., Rasheed, M. and Zaidi.S.A.R. (2011) Salt tolerance of different rice cultivars for their salt tolerance under salt-affected soils.International Research Journal of Agricultural Science and Soil Science 1:183–184.
- Babu N, Krishnan S, Vinod K, Krishnamurthy S, Singh V, Singh M et al. (2017) Marker Aided Incorporation of Saltol, a Major QTL Associated with Seedling Stage Salt Tolerance, into Oryzasativa ‘Pusa Basmati 1121’. Frontiers in Plant Science 8. [DOI] [PMC free article] [PubMed]
- Bandillo N, Raghavan C, Muyco P, Sevilla M, Lobina I, Dilla-Ermita C, et al. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice. 2013;6:11. doi: 10.1186/1939-8433-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari A, Jayaswal P, Yadav N, Singh R, Singh Y, Singh B et al. (2019) Genomics-assisted backcross breeding for infusing climate resilience in high-yielding green revolution varieties of rice. Indian Journal of Genetics and Plant Breeding (The) 79.
- Biatczyk J, Lechowski Z, Libik A. Growth of tomato seedlings under different HCO-3concentration in the medium. J Plant Nutr. 1994;17:801–816. doi: 10.1080/01904169409364768. [DOI] [Google Scholar]
- Bonilla P, Mackell D, Deal K, Gregorio G. RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryzasativa L.) using recombinant inbred lines. Philippine Agricultural Scientist. 2002;65(1):68–76. [Google Scholar]
- Bradbury P, Zhang Z, Kroon D, Casstevens T, Ramdoss Y, Buckler E. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Brautigan D, Rengasamy P, Chittleborough D. Aluminium speciation and phytotoxicity in alkaline soils. Plant Soil. 2012;360:187–196. doi: 10.1007/s11104-012-1232-5. [DOI] [Google Scholar]
- Cao J. The pectin Lyases in arabidopsis thaliana: evolution, selection and expression profiles. PLoS ONE. 2012;7(10):e46944. doi: 10.1371/journal.pone.0046944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh C, Morell M, Mackay I, Powell W. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Opin Plant Biol. 2008;11:215–221. doi: 10.1016/j.pbi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Chapagain S, Park YC, Jang CS. Functional diversity of RING E3 ligases of major cereal crops in response to abiotic stresses. J Crop Sci Biotech. 2017;20(5):351–357. doi: 10.1007/s12892-017-0104-0. [DOI] [Google Scholar]
- Chini A, Grant J, Seki M, et al. Drought tolerance established by enhanced expression of CC-NBS-LRR gene ADR1 requires salicylic acid, EDS1, and ABI1. Plant J. 2004;38:810–822. doi: 10.1111/j.1365-313X.2004.02086.x. [DOI] [PubMed] [Google Scholar]
- Claes B, Dekeyser R, Villarroel R, den Bulcke M, Bauw G, Montagu M, et al. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell. 1990;2:19. doi: 10.1105/tpc.2.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H. Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA. 2004;101:15249–15254. doi: 10.1073/pnas.0406258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit S, Swamy B, Vikram P, Ahmed H, Sta Cruz M, Amante M, et al. Fine mapping of QTLs for rice grain yield under drought reveals sub-QTLs conferring a response to variable drought severities. Theor Appl Genet. 2012;125:155–169. doi: 10.1007/s00122-012-1823-9. [DOI] [PubMed] [Google Scholar]
- Elshire R, Glaubitz J, Sun Q, Poland J, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2013) Global cereals forecast to increase by 7 percent in 2013. http://wwwfao.org/asiapacific/rap/home/news/detail/en/?newsuid=180032.
- Fujita M, Fujita Y, Noutoshi Y, et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. CurrOpin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Ganie SA, Karmakar J, Roychowdhury R, Mondal TK, Dey N. Assessment of genetic diversity in salt-tolerant rice and its wild relatives for ten SSR loci and one allele mining primer of salT gene located on 1st chromosome. Plant Syst Evol. 2014;300:1741–1747. doi: 10.1007/s00606-014-0999-7. [DOI] [Google Scholar]
- Geetha S, Vasuki A, Selvam P, Saraswathi R, Krishnamurthy S, Palanichamy M, et al. Development of sodicity tolerant rice varieties through marker assisted backcross breeding. Electron J Plant Breed. 2017;8:1013. doi: 10.5958/0975-928X.2017.00151.X. [DOI] [Google Scholar]
- Głowacki S, Macioszek VK, Kononowicz AK. R proteins as fundamentals of plant innate immunity. Cell MolBiolLett. 2011;16:1–24. doi: 10.2478/s11658-010-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio GB (1997) Tagging salinity tolerance genes in rice using amplified fragment length polymorphism (AFLP). Ph.D Thesis, University of the Philippines Los Banõs, Laguna
- Guo R, Zhao J, Wang XX, et al. Constitutive expression of a grape aspartic protease gene in transgenic arabidopsis confers osmotic stress tolerance. Plant Cell Tissue Organ Cult. 2015;121:275–287. doi: 10.1007/s11240-014-0699-6. [DOI] [Google Scholar]
- Heenan D, Lewin L, McCaffery D. Salinity tolerance in rice varieties at different growth stages. Animal Product Science. 1988;28:343–349. doi: 10.1071/EA9880343. [DOI] [Google Scholar]
- Hoekenga O, Maron L, Pineros M, Cancado G, Shaff J, Kobayashi Y, et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminium tolerance in Arabidopsis. Proc Natl Acad Sci. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2011;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- Inoue H, Hayashi N, Matsushita A, et al. Blast resistance of CC-NBS-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. ProcNatlAcadSci USA. 2013;110:9577–9582. doi: 10.1073/pnas.1222155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Rita-Arora R, et al. F-Box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007;143(4):1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Gautam R, Singh R, Krishnamurthy S, Ali S, Sakthivel K, et al. Harmonizing technological advances in phenomics and genomics for enhanced salt tolerance in rice from a practical Perspective. Rice. 2019;12:89. doi: 10.1186/s12284-019-0347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Jannink J, Beavis W. Mapping quantitative trait loci in plant breeding populations. Crop Sci. 2003;43:829–834. [Google Scholar]
- Jin H, Plaha P, Park J, Hong C, Lee I, Yang Z, et al. Comparative EST profiles of leaf and root of Leymuschinensis, a xerophilous grass adapted to high pH sodic soil. Plant Sci. 2006;170:1081–1086. doi: 10.1016/j.plantsci.2006.01.002. [DOI] [Google Scholar]
- Kim Y, Tsuda K, Igarashi D, et al. Signaling mechanisms underlying the robustness and tunability of the plant immune network. Cell Host Microbe. 2014;15:84–94. doi: 10.1016/j.chom.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 2013;9:1. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, Sharma S, Sharma D, Singh Y, Mishra V, et al. Analysis of stability and G × E interaction of rice genotypes across saline and alkaline environments in India. Cereal Res Commun. 2016;44:349–360. doi: 10.1556/0806.43.2015.055. [DOI] [Google Scholar]
- Krishnamurthy S, Gautam R, Sharma P, Sharma D. Effect of different salt stresses on agro-morphological traits and utilisation of salt stress indices for reproductive stage salt tolerance in rice. Field Crop Res. 2016;190:26–33. doi: 10.1016/j.fcr.2016.02.018. [DOI] [Google Scholar]
- Krishnamurthy S, Pundir P, Warriach A, Rathor S, Lokeshkumar B, Singh N, et al. IntrogressedSaltol QTL lines improve the salinity tolerance in rice at seedling stage. Front Plant Sci. 2021;11:833. doi: 10.3389/fpls.2020.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, Sharma P, Sharma D, Ravikiran K, Singh Y, Mishra V, et al (2017) Identification of mega-environments and rice genotypes for general and specific adaptation to saline and alkaline stresses in India. Sci Rep 7 [DOI] [PMC free article] [PubMed]
- Kumar V, Singh A, Mithra S, Krishnamurthy S, Parida S, Jain S, et al. Genome-wide association mapping of salinity tolerance in rice (Oryzasativa) DNA Res. 2015;22:133–145. doi: 10.1093/dnares/dsu046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang N, Yanagihara S, Buu BC. Quantitative trait loci for salt tolerance in rice via molecular markers. Omonrice. 2000;8:37–48. [Google Scholar]
- De Leon TB, Steven Linscombe S, Subudhi PK. Molecular dissection of seedling salinity tolerance in rice (Oryzasativa L) using a high-density GBS-Based SNP linkage map. Rice. 2016;9(1):52. doi: 10.1186/s12284-016-0125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yan S, Zhao L, et al. Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 2014;14:105. doi: 10.1186/1471-2229-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Pang S, Lu Z, et al. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants (basel) 2020;9(11):1515. doi: 10.3390/plants9111515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Gao H, Wu X, Fang Q, Chen L, Zhao F, et al. Isolation and characterization of an aluminium-resistant mutant in rice. Rice. 2016;9(1):60. doi: 10.1186/s12284-016-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I, Powell W. Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 2007;12(5):7–63. doi: 10.1016/j.tplants.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Mazumder A, Rohilla M, Bisht D, Krishnamurthy S, Barman M, Sarma R, et al. Identification and mapping of quantitative trait loci (QTL) and epistatic QTL for salinity tolerance at seedling stage in traditional aromatic short grain rice landrace Kolajoha (Oryzasativa L.) of Assam, India. Euphytica. 2020;216:75. doi: 10.1007/s10681-020-02602-0. [DOI] [Google Scholar]
- McWilliam JR. The national and international importance of drought and salinity effects on agricultural production. Funct Plant Biol. 1986;13:1–13. doi: 10.1071/PP9860001. [DOI] [Google Scholar]
- Millar A, Rathjen A, Cooper D. Genetic variation for subsoil toxicities in high pH soils. In: Buck HT, Nisi JE, Salomön N, editors. Wheat production in stressed environments. Springer; 2007. pp. 395–401. [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Negrao S, Courtois B, Ahmadi N, Abreu I, SaiboNand OMM. Recent updates on salinity stress in rice: from physiological to molecular responses. Crit Rev Plant Sci. 2011;30:329–377. doi: 10.1080/07352689.2011.587725. [DOI] [Google Scholar]
- Pan Y, Chen L, Yang X, et al (2020) Mapping quantitative trait loci for cold tolerance in rice under germination stage by whole genome resequencing and analysis of candidate genes. Guangxi Academy AgriSci https://orcid.org/0000-0003-0782-6158
- Pandit A, Rai V, Bal S, Sinha S, Kumar V, Chauhan M, et al. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryzasativa L.) Mol Genet Genomics. 2010;284:121–136. doi: 10.1007/s00438-010-0551-6. [DOI] [PubMed] [Google Scholar]
- Ponce K, Zhang Y, Guo L, et al. Genome-wide association study of grain size traits in indica rice multiparent advanced generation Intercross (MAGIC) population. Front Plant Sci. 2020 doi: 10.3389/fpls.2020.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pundir P, Sharma P, Krishnamurthy S, Devi A, Warraich A, Sharma A. Utilization of salt stress indices and genetic variability in F2 population (PS5×CSR10) of rice for salinity tolerance at reproductive stage. J Soil Salin Water Quality. 2016;8:14–24. [Google Scholar]
- Raghavan C, Mauleon R, Lacorte V, Jubay M, Zaw H, Bonifacio J, Singh RK, Huang BE, Leung H. Approaches in characterizing genetic structure and mapping in a rice multiparental population. G3: Genes Genomes Genetics. 2017;7(6):1721–1730. doi: 10.1534/g3.117.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Gao J, Li L, Cai X, Huang W, Chao D, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Matsumoto H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004;37:645–653. doi: 10.1111/j.1365-313X.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- Singh R, Flowers T. The physiology and molecular biology of the effects of salinity on rice. In: Pessarakli M, editor. Handbook of plant and crop stress. Handbook of Plant and Crop Stress. 3. Florida: Taylor and Francis; 2010. pp. 901–942. [Google Scholar]
- Singh R, Redoña E, Refuerzo L. Varietal improvement for abiotic stress tolerance in crop plants, special reference to salinity in rice. In: Pareek A, Sopory SK, Bohnert HJ, Govindjee, editors. Abiotic stress adaptation in plants, physiological, molecular and genomic foundation. New York: Springer; 2010. pp. 387–415. [Google Scholar]
- Singh Y, Singh D, Sharma S, Krishnamurthy S. Evaluation of rice genotypes for yield, physiological and biological traits in sodic soil. J Soil Salin Water Quality. 2013;5:40–49. [Google Scholar]
- Singh Y, Singh D, Krishnamurthy S. Grouping of advanced rice breeding lines based on grain yield and Na: K ratio under alkaline conditions. J Soil Salin Water Quality. 2014;6:21–27. [Google Scholar]
- Singh R, Singh Y, Xalaxo S, Verulkar S, Yadav N, Singh S, et al. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 2016;242:278–287. doi: 10.1016/j.plantsci.2015.08.008. [DOI] [PubMed] [Google Scholar]
- Storey J, Tibshirani R. Statistical significance for genome wide studies. Proc Natl Acad Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack J, Singh R, Nalley L, Viraktamath B, Krishnamurthy S, Lyman N, et al. High vapor pressure deficit drives salt-stress-induced rice yield losses in India. Glob Change Biol. 2015;21:1668–1678. doi: 10.1111/gcb.12803. [DOI] [PubMed] [Google Scholar]
- Thomson M, De Ocampo M, Egdane J, Rahman M, Sajise A, Adorada D, et al. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010;3:148–160. doi: 10.1007/s12284-010-9053-8. [DOI] [Google Scholar]
- Tiwari S, Krishnamurthy S, Kumar V, Singh B, Rao A, Mithra S, et al. Mapping QTLs for salt tolerance in rice (Oryzasativa L.) by bulked segregant analysis of recombinant inbred lines using 50K SNP Chip. PLoS ONE. 2016;11:e0153610. doi: 10.1371/journal.pone.0153610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan V, Fukuta Y, Mand Ban T. Mapping quantitative trait loci for salinity tolerance in rice. Omonrice. 2000;8:27–35. [Google Scholar]
- Turner S. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014 doi: 10.1101/005165. [DOI] [Google Scholar]
- Visscher P, Brown M, McCarthy M, Yang J. Five years of GWAS discovery. Am J Human Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warraich A, Krishnamurthy S, Sooch B, Vinaykumar N, Dushyanthkumar B, Bose J, et al. Rice GWAS reveals key genomic regions essential for salinity tolerance at reproductive stage. Acta Physiol Plant. 2020;42:134. doi: 10.1007/s11738-020-03123-y. [DOI] [Google Scholar]
- Xiong Y, DeFraia C, Williams D, Zhang X, Mou Z. Deficiency in a cytosolic ribose-5-phosphate isomerase causes chloroplast dysfunction, late flowering and premature cell death in Arabidopsis. Physiol Plant. 2009;137:249–263. doi: 10.1111/j.1399-3054.2009.01276.x. [DOI] [PubMed] [Google Scholar]
- Yadav A, Kumar A, Grover N, Ellur R, Krishnan S, Bollinedi H, et al. Marker aided introgression of 'Saltol', a major QTL for seedling stage salinity tolerance into an elite Basmati rice variety 'Pusa Basmati 1509'. Sci Rep. 2020;10(1):13877. doi: 10.1038/s41598-020-70664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno D, Cock J, Gomez K. Laboratory manual for physiological studies of rice. Las Banos: IRRI; 1976. [Google Scholar]
- Zeng L, Shannon MC. Salinity effects on seedling growth and yield components of rice. Crop Sci. 2000;40:996–1003. doi: 10.2135/cropsci2000.404996x. [DOI] [Google Scholar]
- Zeng L, Shannon MC. Effects of salinity on grain yield and yield components of rice at different seeding densities. Agron J. 2000;92:418–423. doi: 10.2134/agronj2000.923418x. [DOI] [Google Scholar]
- Zhang Z, Ersoz E, Lai C, Todhunter R, Tiwari H, Gore M, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao J, Li Y, et al. Transcriptome analysis highlights defense and signaling pathways mediated by rice pi21 gene with partial resistance to magnaportheoryzae. Front Plant Sci. 2010;7:1834. doi: 10.3389/fpls.2016.01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.