Abstract
Fruit morphology and dehiscence-related genes were analyzed in dehiscent N4P and dehiscent P12 Bixa orellana accessions. Fruit architecture (exocarp and pericarp cells, trichomes, vascular bundles, vesicles, and bixin cells) documented by Scanning electron microscopy (SEM) morphology, blue toluidine stain, and phloroglucinol and hydrochloric acid (PHCL) stain was similar in both accessions. Although, the dehiscent zone (DZ) was higher in the indehiscent P12 B. orellana accession, lignification values, obtained by phloroglucinol and hydrochloric acid stain, within the DZ remain was similar in both variants being lower at 34 days after floral anthesis in the dehiscent N4P B. orellana accession. Dehiscence-related genes APETALA (AP2), SHATTERPROOF (SHP), and SPATULA (SPT) were identified on the reported B. orellana transcriptome (SRX1117606). Real-time quantitative polymerase chain reaction primers build by using these genes allow observing a differential expression during six fruit development stages. In both B. orellana accessions, the AP2 transcripts have a reduced expression, whereas the SHP transcripts were significantly higher during the first two days and ten days of development. SPT transcripts show an expression differential between both accessions being significantly higher in the dehiscent N4P, peaking with 9.66% at 42 days after floral anthesis (DAFA) of development. SPT transcription profile suggested that this gene has an important role during the fruit opening in the dehiscent N4P B. orellana accession.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-022-01180-w.
Keywords: Bixa orellana; Dehiscence; SPATULA gene, APETALA gene, SHATTERPROOF gene
Introduction
Bixa orellana L. plants are the source of the commercial pigment bixin, an important food colorant that accumulates around seeds (Rodríguez-Ávila et al. 2011). The fruits are formed by dehiscent or indehiscent valves according to the B. orellana accession (Trujillo-Hdz et al., 2016; Valdez-Ojeda et al. 2010). Bixin pigment is sensitive to environmental factors such as light, air, and temperature (Rivera-Madrid et al. 2016), and therefore, dehiscent phenomena might affect bixin yield. It has been demonstrated that the accessions with dehiscent fruits provide the highest amount of bixin compared to the indehiscent ones (Trujillo-Hdz et al., 2016).
Dehiscent phenomena have been well characterized in Arabidopsis thaliana, where different fruit tissues, including ovarium tissue, form valves that are separated by a thin cell layer called replum. In the margin of the valves, a dehiscent zone (DZ) is formed, which consists of a cell separation layer and a lignified cell layer (Ferrándiz 2002). At the end of the ripening process, an enzymatic and mechanical process in the DZ leads to the valve being detached and the seed being released (Ferrándiz 2002; Roeder and Yanofsky 2006). Gene participation during fruit development and dehiscence phenomena in A. thaliana have been identified (Roeder and Yanofsky 2006). In the current study, attention was given to the basic helix-loop-helix transcription factor SPATULA (SPT), which is expressed in developing carpel margins, leaves, and petals, as well as the dehiscence zone of fruits and anthers (Groszmann et al. 2010). This gene functions downstream of two MADS-box genes, FRUITFULL and SHATTERPROOF (SHP) (Zumajo-Cardona et al. 2017). The SHP gene, which belongs to the MADS-box family (Callens et al. 2018), establishes valve and valve margin identity, thus delineating the DZ (Ferrándiz et al. 2000; Liljegren et al. 2000; Zumajo-Cardona et al. 2017). Mutations in SHP genes lead to the loss of valve margin lignification and separation layer formation (Ferrándiz et al. 2000). Therefore, the SHP gene was also selected in the current study. In addition, the floral homeotic gene AP2, which is a member of the Arabidopsis family of transcription factors (Jofuku et al. 2005), was selected. AP2 has also been demonstrated to have a critical regulatory role in seed (Ohto et al. 2009) and fruit development since it acts as a brake on valve margin and replum growth (Ripoll et al. 2011). Gene expression analysis was conducted in parallel to morphological fruit characterization during the ripening stages. These characterizations will improve our understanding of the phenomena of dehiscence in B. orellana.
Materials and methods
Plant material
B. orellana L. accessions P12 and N4P were selected in this study. These accessions were previously characterized using a combined approach that considered some morphological characteristics and their bixin content (Carballo-Uicab et al. 2019). P12 exhibits white flowers, fruit with green indehiscent valves and green trichomes and a bixin content of 8.84 mg/g dry weight, whereas N4P exhibits pink flowers, fruits with green dehiscent valves and red trichomes, and a bixin content of 16.04 mg/g dry weight (Trujillo-Hdz et al. 2016). Currently, both accessions are preserved in the Yucatan Center for Scientific Research collections in Yucatán, México.
Fruit dehiscence characterization
Six fruit development stages were considered to conduct this analysis based on the number of days after floral anthesis (DAFA), Days 2, 10, 18, 26, 34, and 42 (Fig. 1). Three fruits were collected in each stage to determine their morphological characteristics and evaluate them microscopically. First, the morphological characterization was documented by using a Leica MZFLIII stereomicroscope, with special emphasis on the dehiscence zone. Objective lenses 2X, 1.5X, and 1X were used in the 2 and 10, 18 and 26, and 34 and 42 DAFA stages, respectively. Then, cross-sections of the fruit at different stages were made. For this, tissue sections including the dehiscent zone were washed for 10 min in phosphate-buffered saline (PBS), cut into squares (2 cm), fixed in formaldehyde at 4% for 24 h at 4 °C, and subjected to sucrose gradient infiltration to cryoprotect the tissue. The gradient consisted of 10%, 20%, and 30% sucrose in 0.16 M phosphate buffer (Na2 HPO4 and NaH2PO4) plus 0, 3, and 6 drops of tissue freezing medium (Leica cat # 14,020,108,926; Buffalo Grove, IL, USA), respectively. The infiltration steps were performed for 1 h each at room temperature. Embedded fruit tissues (in tissue freezing medium) were frozen at − 27 °C in a CM1950 S Cryostat (Leica, Buffalo Grove, IL, USA). The frozen block was cut into 20 μm thick sections from the region of interest. Microscopy documentation of these tissue cuts was conducted by using a Microscopy Axioplan (Zeiss Germany), emphasizing the dehiscence zone and characterization of specific tissues, such as valves, pericarp, exocarp, endocarp, vesicles, trichomes, and bixin specialized cells.
Fig. 1.
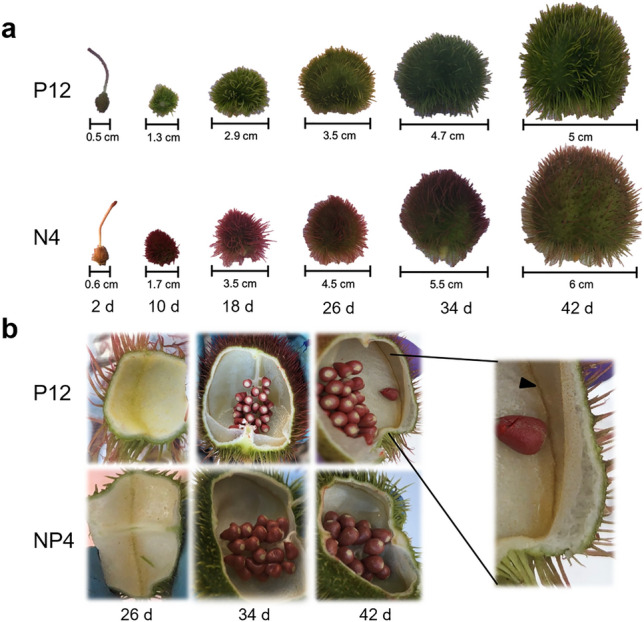
Fruit development stages after floral anthesis in the B. orellana P12 (indehiscent) and N4P (dehiscent) accessions. a Fruit sizes at 2, 10, 18, 26, 34, and 42 d after floral anthesis. b Fruits were cut to expose the seeds and the dehiscence zone (head arrow)
Histochemical analysis
Blue toluidine staining was used to support the microscopic characterization. For this, fruit tissue Sects. (20 μm) were stained with a blue toluidine solution at 1% for 5 min and 0.5% for 1 min (Pradhan Mitra and Loqué, 2014). Stained fruit sections were mounted on microscope slides and visualized using a fluorescence microscope (Axioplan Zeiss, Germany) at a magnification of 10X. Histochemical staining of lignin was also conducted on fruit tissue Sects. (20 μm), which were dipped in a phloroglucinol and hydrochloric acid (PHCL) solution of two volumes of phloroglucinol (3% [vol/vol] in 95% ethanol) and one volume of 18% HCl solution for 10 min (Pradhan Mitra and Loqué, 2014). Stained fruit sections were mounted on microscope slides with DPX mounting media. The edges were sealed with nail polish and visualized immediately using a microscope (Axioplan Zeiss, Germany). Lignin accumulation was analyzed using ImageJ software (Schneider et al. 2012). The ZD area was established on each DAFA in each accession by using the stained images. The straight-line function was used to produce a line of known distance on the rule, then the “Analyze” menu was opened, “Set Scale” was selected, and the “known distance” was entered as well as the “Unit of length” to confirm a length of 0.5 mm in pixels. Red color in the PHC stained indicates the staining of lignin (Nakano and Meshitsuka 1992; Zheng et al. 2017). Therefore, the lignin area was quantified using the color threshold function (RGB Threshold = 0, 100, 120) within the ZD area with the Shanbhag method, white color, and a dark background blocked in ImageJ software.
Scanning electron microscopy
Scanning electron microscopy (SEM) images were also obtained to support the microscopic characterization. The tissues were fixed in formaldehyde in 4% for 24 h at 4 °C and dehydrated in increasing ethanol concentrations. They were subsequently embedded (in tissue freezing medium), frozen and cut using a Leica cryostat. Next, tissues were critical point dried using liquid carbon dioxide and a critical point dryer (Samdri-795, MD, USA). Then, the fruit tissue sections were coated with a gold layer using a vacuum sputter-coater (Denton Vacuum, NJ, USA) to improve the sample conductivity and viewed using a JSM 6360LV scanning electron microscope (Jeol, Tokyo, Japan). The microscope was operated at 10 kV, and images were acquired at different magnifications.
AP2, SPT, and SHP gene identification
A BLASTX analysis using the A. thaliana genes (AP; AAM28448.1, SHP; AAM28448.1, and SPT; AAG33640.1) of the B. orellana transcriptome (SRX1117606; Cárdenas-Conejo et al. 2015) was conducted. Sequences with the highest identity for these genes were selected for further analysis (Table 1). Nucleotide sequences were used with the BLASTX program to identify the proteins or other related predictable proteins. Gene identity was supported for the MAD-box gene to include the closest relatives of the PLE and euAG lineages. For all the sequences selected, the serial Cloner 2.5 program was used to confirm the predictable ORF. AP2, SHP, and SPT proteins of the species A. thaliana, Solanum lycopersicum, and Theobroma cacao were selected to better support the identification. Tomato plants were selected for their high pigment accumulation, a phenotypic trait related to B. orellana, and cacao plants were selected as they belong to the same order (Malvales) as B. orellana. Identity was supported by conducting a phylogenetic tree analysis using the maximum likelihood (ML) method with the JTT matrix model with a bootstrap of 500.
Table 1.
Primers designed for the AP2, SHP and SPT genes
| Gene | Code | Sequence (5´–3´) | Size (pb) | Product size (pb) |
|---|---|---|---|---|
| AP2 | AP2 Fwd | CGTTGTGATCGAGGACGGAT | 20 | 196 |
| AP2 Rev | ACTTCCACTTCCGCCCAAAG | 20 | ||
| SHP | SHP Fwd | AAGAGGGAAATCGAGCTGCAA | 21 | 212 |
| SHP Rev | TCGTGGCTGGAGTAATGGTT | 20 | ||
| SPT | SPT Fwd | ATGTGCTTACCCACTGGACT | 20 | 100 |
| SPT Rev | GTGCCTGTTCCTGCATTAGG | 20 |
RNA isolation, cDNA synthesis, and qPCR experiments
Total RNA was isolated from fruit in the six developmental stages in triplicate from three different N4P and P12 individuals. Tissues (50 mg) were ground in liquid nitrogen using a mortar and pestle. Total RNA was extracted with the PureLink RNA Mini Kit (Cat. No. 12183018A; Ambion, Carlsbad, CA, USA) following the manufacturer’s instructions. Genomic DNA traces were eliminated by a 15 min DNase I treatment (DNase I amplification grade, Cat. No 18068–015; Invitrogen, Carlsbad, CA, USA). The extracted RNA was stored at − 80 °C until analysis (Rodríguez-Ávila et al. 2009). RNA quality was verified using 1.2% agarose gels that were stained with ethidium bromide. cDNA was synthesized from 100 ng of total RNA by using SuperScript™ III Reverse Transcriptase (Cat. No. 18080–093; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and stored at − 20 °C.
Nucleotide sequences belonging to the open reading frame (ORF) selected for each B. orellana gene were used to design real-time PCR primers using the Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Then, in silico PCR (http://insilico.ehu.es/PCR/) was performed on the contig sequences to support probable PCR amplification. Table 1 shows the expected contigs, primers, and PCR product sizes. The quantitative RT–PCR (qRT–PCR) assay was carried out using 200 ng of cDNA in an iCycler IQ real-time PCR detection system (Bio–Rad, Hercules, CA, USA) with SYBR Green qPCR SuperMix-UDG (Cat. No. 11730046; Invitrogen, Carlsbad, CA, USA). The qRT–PCR program included an initial 2- and 1-min periods at 50 and 95 °C, respectively, to activate the polymerase. Optimization of the annealing temperature was assayed at various temperatures (65.0, 64.6, 63.9, 62.5, 60.8, 59.4, 58.5 and 58.0 °C), and the resulting melting curves were compared with the 18S gene primer amplification. The real-time amplification program included 32 cycles of denaturation at 95 °C for 30 s, annealing at 60, 62.5 or 60.8 °C for 30 s for the SHP, AP2 and SPT genes, respectively (see primers in Table 1), and a final extension step at 72 °C for 30 s. Three independent replicates per sample were analyzed. First, the relative transcript levels were determined using the Formula 2-ΔΔCt (Livak and Schmittgen 2001). The 18S gene (GenBank: AF206868) with the primers FW 5′ CGGCTACCACATCCAAGGAA3′ and RW 5′ GCTGGAATTACCGCGGCT3´ was used to normalize transcript abundance in each sample to allow for standardizing the comparisons among gene expression of fruit development stages. Significant differences (p < 0.05) among the mean values were determined by Tukey’s test using one-way ANOVA and StatSoft Statistica software, v. 8.0 (https://www.statistica.com/en/).
Results
Fruit characterization
Gradual fruit development was observed in both B. orellana accessions. The dehiscent N4P accession was more significant than its indehiscent counterpart, P12, with a ratio of 6 cm and maximum fruit circumference of 19 cm (42 DAFA), considering the trichomes, with respect to the ratio of 5 cm and 16.5 cm circumference obtained for P12 at the same development stage (Fig. 1a). For both B. orellana accessions, there were 10–50 seeds inside the fruits, the endocarp was observed as a white membrane, the two parietal placentas were 3–4 mm long at 42 DAFA, and the dehiscence zone was observed (Fig. 1b).
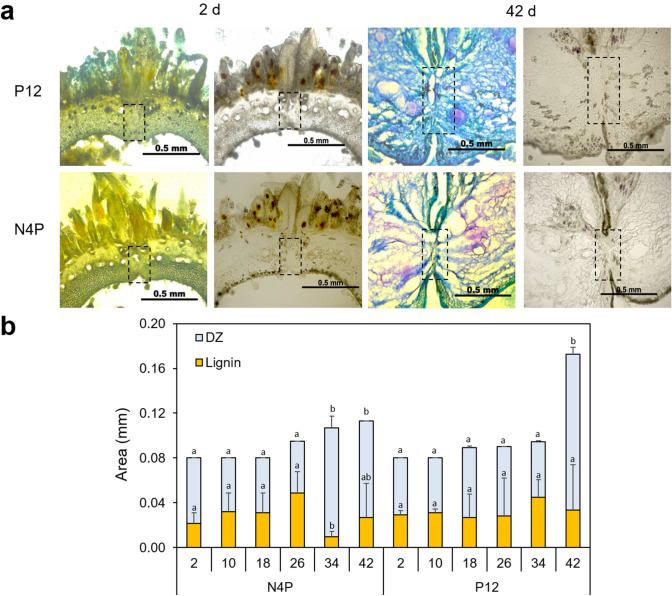
The fruit size peaked at 18.38 and 14.66 cm for the dehiscent N4P and indehiscent P12 accessions, respectively (Fig. 2). Blue toluidine and PHCL staining allowed for the identification of the DZ during fruit development, which exhibited a similar length in both accessions until 42 DAFA, when the DZ increased in the indehiscent P12 B. orellana accession (Fig. 2a). The lignification values within the DZ determined by PHCL staining remained similar, 0.03–0.04 mm2 in both accessions; the main differences were found at 34 DAFA in dehiscent N4P plants, with a reduction to 0.01 mm2. The indehiscent P12 accession showed a substantial reduction in the lignified area compared to the DZ area at 42 DAFA as a consequence of the fruit size increase (Fig. 2b). In general, lignin accumulation was reduced within the DZ and in other fruit sections in both B. orellana accessions, with some exceptions, such as the trichomes, vascular bundles, and pericarp and endocarp cells, where PHCL staining was clearly observed (Figs. 2a and 3c). Blue toluidine staining also supports the identification of pericarp and endocarp cells, where a high amount of lignin is observed (Mori and Bellani 2009), specifically in the margin of the valve (Figs. 2a and 3b). The trichomes in both accessions were large and surrounded the fruit exocarp (Fig. 3a).
Fig. 2.
Increase in the dehiscence zone (DZ) during fruit development in the accessions P12 (indehiscent) and N4P (dehiscent) of B. orellana. a Contrasting fruit development stages (2 d and 42 d) after staining with blue toluidine (left) and PHCL (right), showing the DZ (dotted rectangles). b Increased DZ (blue bars) and PHCL staining areas (orange bars) indicate the lignification process. Numbers on the X-axis indicate the days (d) after floral anthesis (DAFA), and Numbers on the Y-axis indicate the area in mm. a and b indicate the minimum significant difference for each group (significance of 0.05 with the Tukey test)
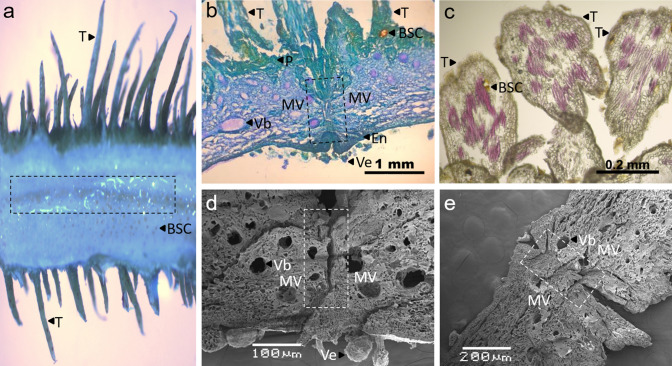
Fig. 3.
Representative morphological traits of fruits DZ of N4P accession (dehiscent). a Longitudinal cut showing the DZ (dotted rectangle) at 42 d. b Longitudinal cut showing the DZ (dotted rectangle) staining with blue toluidine at 42 d. c PHCL staining on trichomes at 10 d, showing bixin specialized cells (BSC). d SEM image at 18 d. e SEM image at 42 d. Valve mesophyll (MV), pericarp (P), endocarp (En), vesicles (Ve), vascular bundles (Vb), and trichomes (T)
The locations of the valves, pericarp, exocarp, vesicles, vascular bundles, trichomes and specialized bixin cells were similar in both varieties. Figure 3 shows these structures in the fruits of dehiscent accession N4P. This figure shows the bixin cells dispersed in the fruit mesophyll (Fig. 3a) and the trichomes surrounding the fruit exocarp (Fig. 3c). In addition, the vesicle number and size changed during all fruit development stages, and were more abundant at the beginning and more prominent at the end (Fig. 3b and d). Finally, SEM images generally support the structural architecture observed by the fluorescence microscopy analysis, showing a clear DZ, vesicles, and vascular bundles (Fig. 3d and e). The DZ could also be observed by the naked eye in longitudinal fruit cuts in both B. orellana accessions since 26 DAFA (Supplementary Fig. S1).
Gene expression analysis
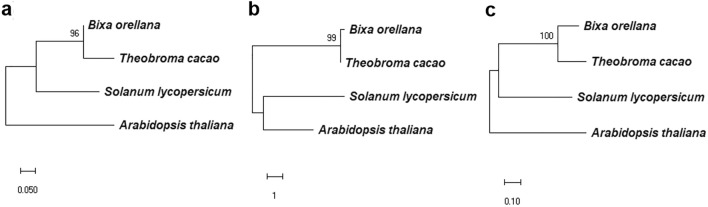
It was possible to obtain transcriptomic sequences with high BLAST identity (> 99%) for A. thaliana genes (AP2; AAM28448.1, SHP; AAM28448.1, and SPT; AAG33640.1). ORF localization and six-frame translation allow for the detection of protein sequences of 332, 232, and 172 amino acids for the AP2, SHP, and SPT proteins, respectively (accession numbers OME647688, 0M674689 and OM674690). An initial BLASTX analysis using these sequences identified diverse plant gene orthologs with high identity (> 99%) for each gene, supporting the clear identification of the AP2 and SPT genes (Fig. S2 and S3). However, the SHP gene shows high identity with different members of the MAD-box family (data not shown). A second analysis was conducted on this MAD-box sequence. This analysis included accessions of the PLE, euAG and AGL11 lineages (Callens et al. 2018) of the species A. thaliana, Petunia x hybrida, Malus domestica, Cucumis sativus, Nicotiana tabacum, and Solanum lycopersicum and allowed for us to identify this sequence as an SHP-encoding gene (Fig. S4). Then, the predicted protein of each B. orellana gene was aligned with the AP2, SPT, and SHP predicted proteins of Arabidopsis, tomato, and cacao to support better identification (Fig. 4). In all cases, the ML analysis of the B. orellana sequences shows a clear grouping with the T. cacao accessions because both plant genera belong to the same order (Malvales).
Fig. 4.
Simplified distance trees based on the ML method for the AP2 (a), SHP (b) and SPT (c) genes, considering the respective gene orthologs of Arabidopsis, tomato and cacao. Protein and nucleotide (within parenthesis) accession numbers in (a) AAC13770.1 (U12546.1) APETALA2 A. thaliana, NP_001233886.1 (NM_001246957.1) APETALA2 S. lycopersicum, and XP_007047337.2 (XM_007047275) APETALA2 T. cacao. (b) AGB51141.1 (JQ973091) SHATTERPROOF A. thaliana, NP_001300859.1 (NM_001313930.1) TAGL1 S. lycopersicum and XP_007048826.1 (XM_007048764.2) Agamous AGL1 T. cacao. (c) XP_007017187.2 (XM_007017125.2) SPATULA X2 T. cacao, NP_001361318.1 (NM_001374389.1) SPATULA S. lycopersicum, and AAG33640.1 (AF319540.1) SPATULA A. thaliana
AP2, SHP, and SPT gene sequences from B. orellana were used as a template to design PCR primers (Table 1). The primers were first validated in silico and then on the original contigs to observe the expected PCR products (data not shown). Real-time PCR conditions, particularly the annealing temperatures, were also established for each gene. Gene expression analysis was conducted during fruit development. The AP2 transcript accumulation exhibited differential expression during fruit development in both B. orellana accessions. In general, this gene showed a reduced expression level compared with other genes in the study, with less than 1% of the transcripts, with the exception of the 42 DAFA indehiscent P12 accession, where values were slightly higher than 1% and significantly different with respect to dehiscent N4P accession (Fig. 5a). The SHP transcript accumulation showed a higher relative expression during fruit development in both B. orellana accessions, which was significantly higher on the first days (2 and 10 DAFA) of fruit development, 9%. Then, SHP transcript expression was reduced in both B. orellana accessions and dropped to 3.9% at 26 DAFA for the indehiscent P12 accession (Fig. 5b). The SPT transcript profile generally exhibited larger differences in expression for both B. orellana accessions, as the indehiscent P12 accession was higher in all fruit development stages than the dehiscent (N4P) accession. The largest differences were observed at the end of fruit ripening (42 DAFA), 9.66 and 0.77% for the P12 and N4P accessions, respectively (Fig. 5c). In general, the indehiscent P12 accession plant shows approximately half of the level of transcripts than their dehiscent counterpart, suggesting the importance of the SPT gene in dehiscent phenomena.
Fig. 5.
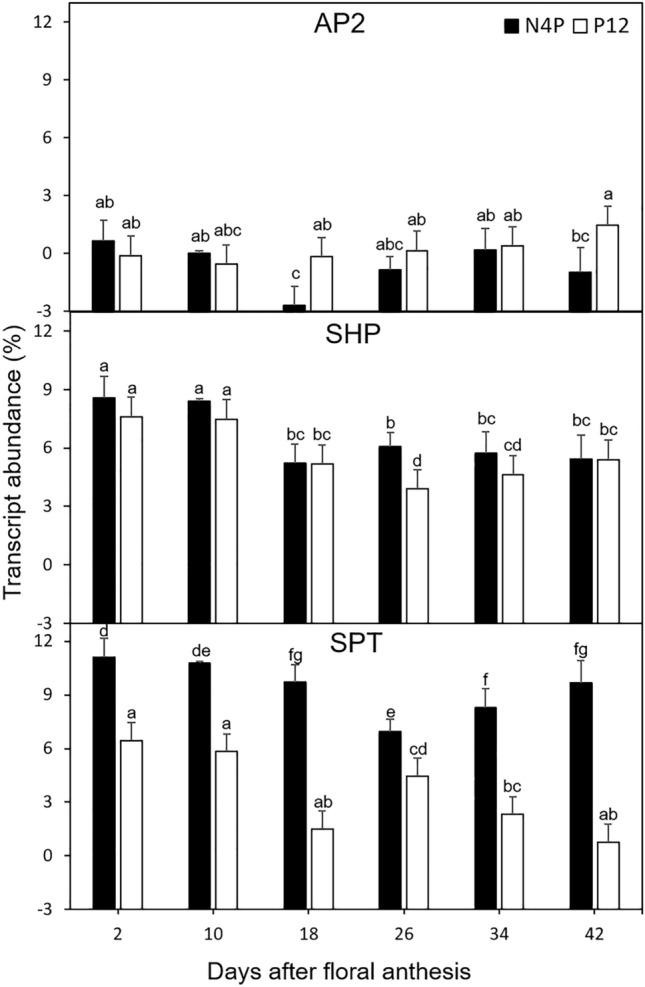
Global expression of dehiscent-related genes in dehiscent N4P (black) and indehiscent P12 accessions (white) at 2, 10, 26, 34, and 42 d after floral anthesis. a, b, c, d, e, f, and g indicate the minimum significant difference for each group (significance of 0.05 with the Tukey test)
Discussion
The current study characterized fruit during six fruit stages after floral anthesis (DAFA) in dehiscent and indehiscent B. orellana accessions. Both accessions maintained a dissimilar seed number and fruit architecture. Previous morphological and pigment-productive characterization of these B. orellana accessions grouped them according to their dehiscent phenotype and showed that the plants with dehiscent fruits have higher bixin content in their immature seeds (Trujillo-Hdz et al. 2016). However, dehiscence phenomena might have an essential effect on the bixin content in mature fruits where seeds are exposed to sunlight and other environmental factors (Rivera-Madrid et al. 2016). Therefore, in the current study, special attention was given to morphological and genetic traits related to dehiscent phenomena. The SEM images and blue toluidine and PHCL staining support the identification of diverse structures in the fruits, including trichomes, valves, vesicles, pericarp cells, exocarp cells, endocarp cells, and bixin specialized cells. The blue toluidine staining supports the identification of exocarp and pericarp cells, which are stained greenish-blue, indicating a high amount of lignin (Wongkaew et al. 2021). Vascular bundles in B. orellana are also stained by blue toluidine but with a pinkish purple color, as previously described (Mori and Bellani 2009). PHC staining also supports lignin accumulation in the trichomes, vascular bundles, pericarp and endocarp cells. Lignin is predominantly deposited in xylem and phloem fibers, and it is crucial for stability and structural integrity (Liu et al. 2018). In addition, stereoscopic analysis shows bixin cells dispersed in the fruits (mesophyll cells and trichomes). It has been documented in B. orellana that bixin deposits occur mainly in the seed aril (Carvalho et al. 2010; Rodríguez-Ávila et al. 2011). These bixin cells have been considered carotenoid storage cells in seeds (Louro and Santiago 2016). However, more recently, bixin cells have been described as pigment glands that may be dispersed throughout the plant body, including the fruit tissues (Almeida et al. 2021), as in the current study. Significant differences between both B. orellana accessions (Trujillo-Hdz et al. 2016) were also found in the DZ, and were more considerable in the indehiscent P12 accession. Lignin values were reduced within the DZ in both B. orellana accessions, only showing differences at 36 DAFA in the dehiscent N4P plant, with a slight reduction to 0.01 mm2. Lignification in the DZ, particularly in the valve margin cells, is essential for dehiscent phenomena and has been described in various genetic studies with several dehiscent-related genes (Liljegren et al. 2004; Mitsuda and Ohme-Takagi 2008).
The expression of specific genes related to pigments and mevalonate pathways has been previously analyzed during fruit development in different accessions of B. orellana (Narváez et al. 2001; Rodríguez-Ávila et al. 2011). However, to our knowledge, no study has focused on dehiscence-related genes in this plant. The transcription factors SPT, SHP2, and AP2, which are involved in floral, fruit, and seed development, were selected (Groszmann et al. 2010; Huang et al. 2017; Ripoll et al. 2011; Zumajo-Cardona et al. 2017). It was possible to identify these genes in the reported B. orellana transcriptome (Cárdenas-Conejo et al. 2015) by using Arabidopsis gene orthologs. In general, B. orellana genes were most closely related to the cacao gene and then the tomato gene orthologs. AP2 transcript accumulation was evaluated during fruit development in both B. orellana accessions to consider specific gene expression. This gene acts as a regulator of diverse genes related to floral and seed development (Huang et al. 2017; Ohto et al. 2009). Specifically, it acts as a brake on valve margin and replum growth (Ripoll et al. 2011) and in floral and seed development (Huang et al. 2017; Ohto et al. 2009). AP2 also negatively regulates valve margin formation since it causes the ectopic activity of the SHP and INDEHICENT (IND) genes during Arabidopsis fruit development (Ripoll et al. 2011). Consistent with their known functions, the AP2 transcript profile exhibited a reduced expression level during fruit development in both B. orellana accessions.
SHP transcript accumulation was also recorded. SHP genes are MADS-box transcription factors that act redundantly in Arabidopsis to control the dehiscence of pods in the DZ during late fruit development (Colombo et al. 2010; Liljegren et al. 2000). The SHP gene establishes the valve and valve margin identity, thus delineating the DZ (Ferrándiz et al. 2000; Liljegren et al. 2000; Zumajo-Cardona et al. 2017). SHP transcripts in B. orellana were significantly higher at 2 and 10 DAFA. However, at 26 DAFA, these values were reduced overall in the indehiscent P12 accession. In Arabidopsis, SHP transcripts act downstream of the b-HLH transcription factors IND and ALCATRAZ, which are important in dehiscent phenomena (Liljegren et al. 2000). Therefore, the significant decrease in SHP transcripts after 26 DAFA in the P12 accession might be related to their indehiscent trait. Another topic that equally notable about the SHP gene is its relationship with pigment accumulation. In tomato, a plant with high carotenoid accumulation, the transcription factor TAGL1, a homolog of the Arabidopsis SHP, has evolved novel functions to those in the Arabidopsis plant (Vrebalov et al. 2009). This is probably due to the wide fruit development differences between both plants since Arabidopsis produces silique and tomato produces fleshy and nondehiscent berry fruits (Garceau et al. 2017). In tomato, TAGL1 has important roles in fleshy fruit ripening, including in ethylene production, fruit stiffness, and carotenoid metabolism (Garceau et al. 2017; Gimenez et al. 2016; Vrebalov et al. 2009). Through RNA interference repression, transgenic plants with reduced TAGL1 expression produced fruits with defects in their ripening and showed reduced carotenoid production with low lycopene levels and downregulation of the phytoene synthase and lycopene-β-cyclase genes (Vrebalov et al. 2009). Notably, B. orellana fruits are capsules that greatly differ from the siliques of Arabidopsis and fleshy fruits of tomato. Fruit adaptations may generate different adaptations in homeotic genes involved in fruit development (Garceau et al. 2017). In B. orellana, the seeds remain attached to the fruits even in dehiscent fruits (Rodríguez-Ávila et al. 2011). In the current study, the lignification pattern was similar in both B. orellana accessions. In Arabidopsis, the mutant (alc) shows disrupted dehiscence phenomena; the SEM morphology and lining-specific PHCL staining show a similar morphological and lignification pattern between this mutant and the wild type (Rajani and Sundaresan 2001). These results indicate the importance of the valve aperture during dehiscence in B. orellana. Valve aperture is mediated by environmental factors and physical forces in the tissue, which affects the properties of the lignified and nonlignified cell walls around the valve suture (Dong and Wang 2015). In Arabidopsis, the valves (seed pod walls) and the central replum tissues are separated by narrow files of highly specialized tissue called valve margins. These tissues are formed by lignification and a layer of small cells, which secrete cell wall-degrading enzymes to promote cell separation and allow for fruit opening and seed dispersal when the fruit reaches maturity (Girin et al. 2011). The INDEHISCENT (IND) gene regulates lignin-degrading cells (Liljegren et al. 2004), which interact genetically and via protein–protein contact with the SPT gene to mediate fruit opening in Arabidopsis (Girin et al. 2011). SPT genes are basic helix-loop-helix transcription factors expressed in developing carpel margins, leaves and petals, and the dehiscence zone of fruits and anthers (Groszmann et al. 2010). This gene functions downstream of two MADS-box genes, FRUITFULL and SHP (Zumajo-Cardona et al. 2017). In the current study, SPT transcripts showed strong expression during fruit development, which was higher in the indehiscent P12 accession than in the dehiscent plant N4P counterpart during all fruit development stages. The highest differences were observed at the end of fruit development, suggesting the important role of the SPT gene during dehiscent phenomena, consistent with its known function in fruit opening in Arabidopsis (Girin et al. 2011).
Conclusion
The fruit morphology assessed by SEM, blue toluidine, and PHCL staining showed a similar architecture in both B. orellana accessions. Some morphological features were identified, such as exocarp and pericarp cells, trichomes, vascular bundles, vesicles, and bixin storage cells. The DZ was more significant in the indehiscent P12 accession. However, the lignification values within the DZ were similar in both accessions, and were lower at 34 DAFA in the dehiscent N4P accession. The AP2, SHP, and SPT genes identified in the transcriptome were differentially expressed during the six fruit development stages. Specifically, AP2 transcripts showed reduced expression, whereas SHP transcripts were slightly more highly expressed at the beginning of fruit development in both accessions. SPT transcripts show high expression during all fruit development stages in the dehiscent N4P accession. Major differences between the B. orellana accessions were found at the end of fruit development, where SPT transcript levels were 9.66 and 0.77% for the dehiscent N4P and the indehiscent P12 accessions, respectively. Therefore, SPT gene expression might be related to valve detachment in the dehiscent N4P B. orellana accession.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Consejo Nacional de Ciencia y Tecnología (CONACYT) (CB-2008-01, proyecto No. 220259; Fronteras de la Ciencia No. 2016-01-1716). M.D. Rocio Tamayo-García was financially supported by M.D. and Ph.D scholarship (No. 706229; 8006).
Author contributions
T-GR and N-ZJA: are co-first authors; these authors contributed equally to this investigation: Conceptualization, Investigation, Data curation, Reviewing, Editing, and Writing. All authors contributed to the Material preparation, data collection and analysis. K-GA, A-EM and G-PLC: Methodology, Investigation, and Data curation. R-MRConceptualization, Resources, Funding acquisition, Reviewing and Writing. R-MR and N-ZJA: Supervising T-GR: work.
Funding
The Consejo Nacional de Ciencia y Tecnología provided the grant for this study.
Declarations
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Consent for publication
Authors hereby give our consent for the publication of this study to be published in Physiology and Molecular Biology of Plants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
R. Tamayo-García and J. A. Narváez-Zapata these authors have contributed equally to this work and share first authorship.
Contributor Information
R. Tamayo-García, Email: bio.rociotamayo@gmail.com
J. A. Narváez-Zapata, Email: jnarvaez@ipn.mx
A. Ku-González, Email: angela@cicy.mx
M. Aguilar-Espinosa, Email: mgf@cicy.mx
L. C. Gutiérrez-Pacheco, Email: lcgp@cicy.mx
R. Rivera-Madrid, Email: renata@cicy.mx
References
- Almeida AL, Freitas PF, Ferreira CP, Ventrella MC. Syncytial development of annatto (Bixa orellana L.) pigment gland: A curious type of anastomosed articulated laticifer. Flora. 2021;274:151727. doi: 10.1016/J.FLORA.2020.151727. [DOI] [Google Scholar]
- Callens C, Tucker MR, Zhang D, Wilson ZA. Dissecting the role of MADS-box genes in monocot floral development and diversity. J Exp Bot. 2018;69:2435–2459. doi: 10.1093/JXB/ERY086. [DOI] [PubMed] [Google Scholar]
- Cárdenas-Conejo Y, Carballo-Uicab V, Lieberman M, Aguilar-Espinosa M, Comai L, Rivera-Madrid R. De novo transcriptome sequencing in Bixa orellana to identify genes involved in methylerythritol phosphate, carotenoid and bixin biosynthesis. BMC Genomics. 2015;16:1–19. doi: 10.1186/S12864-015-2065-4/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo-Uicab VM, Cardenas-Conejo Y, Vallejo-Cardona AA, Aguilar-Espinosa M, Rodriguez-Campos J, Serrano-Posada H, Narváez-Zapata JA, Vázquez-Flota F, Rivera-Madrid R. Isolation and functional characterization of two dioxygenases putatively involved in bixin biosynthesis in annatto ( Bixa orellana L.) PeerJ. 2019;7:e7064. doi: 10.7717/peerj.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho PRN, da Silva MG, Fabri EG, Tavares PE, Martins ALM, Spatti LR. Concentração de bixina e lipídios em sementes de urucum da coleção do instituto agronômico (IAC) Bragantia. 2010;69:519–524. doi: 10.1590/S0006-87052010000300002. [DOI] [Google Scholar]
- Colombo M, Brambilla V, Marcheselli R, Caporali E, Kater MM, Colombo L. A new role for the SHATTERPROOF genes during Arabidopsis gynoecium development. Dev Biol. 2010;337:294–302. doi: 10.1016/J.YDBIO.2009.10.043. [DOI] [PubMed] [Google Scholar]
- Dong Y, Wang YZ. Seed shattering: from models to crops. Front Plant Sci. 2015;6:1–13. doi: 10.3389/FPLS.2015.00476/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C. Regulation of fruit dehiscence in Arabidopsis. J Exp Bot. 2002;53:2031–2038. doi: 10.1093/JXB/ERF082. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289:436–438. doi: 10.1126/SCIENCE.289.5478.436. [DOI] [PubMed] [Google Scholar]
- Garceau DC, Batson MK, Pan IL. Variations on a theme in fruit development: the PLE lineage of MADS-box genes in tomato (TAGL1) and other species. Planta. 2017;246:313–321. doi: 10.1007/S00425-017-2725-5/FIGURES/3. [DOI] [PubMed] [Google Scholar]
- Gimenez E, Castañeda L, Pineda B, Pan IL, Moreno V, Angosto T, Lozano R. TOMATO AGAMOUS1 and ARLEQUIN/TOMATO AGAMOUS-LIKE1 MADS-box genes have redundant and divergent functions required for tomato reproductive development. Plant Mol Biol. 2016;91:513–531. doi: 10.1007/S11103-016-0485-4/FIGURES/9. [DOI] [PubMed] [Google Scholar]
- Girin T, Paicu T, Stephenson P, Fuentes S, Körner E, O’Brien M, Sorefan K, Wood TA, Balanzá V, Ferrándiz C, Smyth DR, Østergaard L. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell. 2011;23:3641–3653. doi: 10.1105/tpc.111.090944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Bylstra Y, Lampugnani ER, Smyth DR. Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J Exp Bot. 2010;61:1495–1508. doi: 10.1093/JXB/ERQ015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Shi T, Zheng B, Yumul RE, Liu X, You C, Gao Z, Xiao L, Chen X. APETALA2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in Arabidopsis thaliana. New Phytol. 2017;215:1197–1209. doi: 10.1111/NPH.14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, Omidyar PK, Gee Z, Okamuro JK. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc Natl Acad Sci U S A. 2005;102:3117. doi: 10.1073/PNAS.0409893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowmant JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AHK, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116:843–853. doi: 10.1016/S0092-8674(04)00217-X. [DOI] [PubMed] [Google Scholar]
- Liu Q, Luo L, Zheng L. Lignins: biosynthesis and biological functions in plants. Int J Mol Sci. 2018;19:335. doi: 10.3390/IJMS19020335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/METH.2001.1262. [DOI] [PubMed] [Google Scholar]
- Louro RP, Santiago LJM. Development of carotenoid storage cells in Bixa orellana L. seed arils. Protoplasma. 2016;253:77–86. doi: 10.1007/S00709-015-0789-2/FIGURES/6. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 2008;56:768–778. doi: 10.1111/J.1365-313X.2008.03633.X. [DOI] [PubMed] [Google Scholar]
- Mori B, Bellani LM. Differential staining for cellulosic and modified plant cell walls. Biotech Histochem. 2009;71:71–72. doi: 10.3109/10520299609117136. [DOI] [PubMed] [Google Scholar]
- Nakano J, Meshitsuka G. The detection of lignin. In: Lin SY, Dence CW, editors. Methods in lignin chemistry. Berlin, Heidelberg: Springer; 1992. pp. 23–32. [Google Scholar]
- Narváez JA, Canto-Canché BB, Pérez PF, Rivera-Madrid R. Differential expression of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) during flower and fruit development of Bixa orellana. J Plant Physiol. 2001;158:1471–1477. doi: 10.1078/0176-1617-00614. [DOI] [Google Scholar]
- Ohto MA, Floyd SK, Fischer RL, Goldberg RB, Harada JJ. Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod. 2009;22:277–289. doi: 10.1007/S00497-009-0116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan Mitra P, Loqué D. Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J vis Exp. 2014;87:51381. doi: 10.3791/51381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani S, Sundaresan V. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol. 2001;11:1914–1922. doi: 10.1016/S0960-9822(01)00593-0. [DOI] [PubMed] [Google Scholar]
- Ripoll JJ, Roeder AHK, Ditta GS, Yanofsky MF. A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development. 2011;138:5167–5176. doi: 10.1242/DEV.073031. [DOI] [PubMed] [Google Scholar]
- Rivera-Madrid R, Aguilar-Espinosa M, Cárdenas-Conejo Y, Garza-Caligaris LE. Carotenoid derivates in achiote (Bixa orellana) seeds: synthesis and health promoting properties. Front Plant Sci. 2016;7:1406. doi: 10.3389/FPLS.2016.01406/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ávila NL, Narváez-Zapata JA, Aguilar-Espinosa ML, Rivera-Madrid R. Full-length gene enrichment by using an optimized RNA isolation protocol in Bixa orellana recalcitrant tissues. Mol Biotechnol. 2009;42:84–90. doi: 10.1007/s12033-008-9138-4. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ávila NL, Narváez-Zapata JA, Ramírez-Benítez JE, Aguilar-Espinosa ML, Rivera-Madrid R. Identification and expression pattern of a new carotenoid cleavage dioxygenase gene member from Bixa orellana. J Exp Bot. 2011;62:5385–5395. doi: 10.1093/jxb/err201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AHK, Yanofsky MF. Fruit development in Arabidopsis. Arabidopsis Book. 2006 doi: 10.1199/TAB.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/NMETH.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo-Hernandez JA, Cárdenas-Conejo Y, Turriza PE, Aguilar-Espinosa M, Carballo-Uicab V, Garza-Caligaris LE, Comai L, Rivera-Madrid R. Functional polymorphism in lycopene beta-cyclase gene as a molecular marker to predict bixin production in Bixa orellana L. (achiote) Mol Breed. 2016;36:1–15. doi: 10.1007/S11032-016-0555-Y. [DOI] [Google Scholar]
- Valdez-Ojeda R, Quiros CF, Aguilar-Espinosa M, Rivera-Madrid R. Outcrossing rates in annatto determined by sequence-related amplified polymorphism. Agron J. 2010;102:1340–1345. doi: 10.2134/AGRONJ2009.0510. [DOI] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, Irish VF. Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009;21:3041–3062. doi: 10.1105/TPC.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongkaew M, Kittiwachana S, Phuangsaijai N, Tinpovong B, Tiyayon C, Pusadee T, Chuttong B, Sringarm K, Bhat FM, Sommano SR, Cheewangkoon R. Fruit characteristics, peel nutritional compositions, and their relationships with mango peel pectin quality. Plants. 2021;10:1148. doi: 10.3390/PLANTS10061148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Chen J, Shi Y, Li Y, Yin Y, Yang D, Luo Y, Pang D, Xu X, Li W, Ni J, Wang Y, Wang Z, Li Y. Manipulation of lignin metabolism by plant densities and its relationship with lodging resistance in wheat. Sci Rep. 2017;7:41805. doi: 10.1038/SREP41805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumajo-Cardona C, Ambrose BA, Pabón-Mora N. Evolution of the SPATULA/ALCATRAZ gene lineage and expression analyses in the basal eudicot, Bocconia frutescens L. (Papaveraceae) Evodevo. 2017;8:5. doi: 10.1186/S13227-017-0068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.